Abstract
Developing B and T lymphocytes generate programmed DNA Double Strand Breaks (DSBs) during the V(D)J recombination process that assembles exons that encode the antigen-binding variable regions of antibodies. In addition, mature B lymphocytes generate programmed DSBs during the Immunoglobulin Heavy chain (IgH) Class Switch Recombination (CSR) process that allows expression of different antibody heavy chain constant regions that provide different effector functions. During both V(D)J recombination and CSR, DSB intermediates are sensed by the ATM-dependent DSB response (DSBR) pathway, which also contributes to their joining via Classical Non-Homologous End-Joining (C-NHEJ). The precise nature of the interplay between the DSBR and C-NHEJ pathways in the context of DSB repair via C-NHEJ remains under investigation. Recent studies have shown that the XLF C-NHEJ factor has functional redundancy with several members of the ATM-dependent DSBR pathway in C-NHEJ, highlighting unappreciated major roles for both XLF as well as the DSBR in V(D)J recombination, CSR and C-NHEJ in general. In this review, we discuss current knowledge of the mechanisms that contribute to the repair of DSBs generated during B lymphocyte development and activation with a focus on potential functionally redundant roles of XLF and ATM-dependent DSBR factors.
Keywords: Cernunnos, ATM, DNA-PKcs, NHEJ, V(D)J recombination, 53BP1
1. Repair of Programmed DSBs during Lymphocyte Development and Activation
Classical Non-Homologous End-Joining (C-NHEJ) is a major mammalian double-strand break (DSB) repair pathway that is active throughout the cell cycle but which has particular importance in the G1 cell cycle phase. There are two programmed DNA recombination processes in lymphocytes that employ C-NHEJ: V(D)J recombination in developing B and T lymphocytes, and IgH Class Switch Recombination (CSR) in activated mature B lymphocytes [1–4]. In addition to C-NHEJ, homologous recombination (HR) is the other known major DSB repair pathway in mammalian cells and predominates in the S/G2 phases of the cell cycle. Moreover, one or more Alternative End-Joining (A-EJ) DSB repair mechanisms have been observed to fuse at least certain classes of DSBs in the absence of C-NHEJ; however, the overall functional significance of A-EJ in the presence of C-NHEJ is yet to be determined [1, 2].
In developing B and T cell lymphocytes, variable (V), diversity (D) and joining (J) gene segments are assembled to form V(D)J exons that encode the N-terminal “variable” portion of immunoglobulin (Ig) or T cell receptor (TCR) proteins that provide antigen binding specificity. Together with downstream exons that encode C-terminal “constant” region portions of Ig or TCR proteins, they form complete Ig or TCR genes [5]. V(D)J recombination is initiated by the Recombination activating gene 1 and 2 proteins, which together form an endonuclease (RAG) that recognizes short recombination signal (RS) sequences that flank V, D, or J coding sequences [6] and then introduces DSBs between an appropriate set of RSs and V, D, or J coding gene segments [7, 8]. RAG generates DSBs at these sequences in the form of blunt 5′-phosphorylated signal RS ends (“SEs”) and hairpin-sealed coding ends (“CEs”) [9]. The RAG complex holds cleaved V(D)J CEs and SEs in a post-cleavage synaptic complex and, in a yet to be determined manner, directs joining of two appropriate CEs to each other to form V(D)J coding joins (“CJs”) and joining of the two SEs to each other to form signal joins (“SJs”), by the C-NHEJ pathway [10] (Figure 1).
Figure 1. V(D)J recombination in developing lymphocytes requires the RAG endonuclease and C-NHEJ factors.
This Figure depicts an example of a V(D)J recombination event occurring between a V gene segment and a rearranged DJ gene segment to form a complete V-D-J exon.
(A) The RAG1/2 endonuclease (green ovals) is recruited to recombination signal (RS) sequences flanking a “V” gene segment and a “DJ” gene segment. The white triangle represents a 23-RSS, and the black triangle represents a 12-RSS.
(B) After DSB induction, RAG holds hairpin-sealed coding ends and blunt signal ends in a post-cleavage synaptic complex.
(C) The Ku70/Ku80 heterodimer (denoted by the light blue semicircle and dark blue semicircle, respectively) recognizes and binds DNA ends. Hairpin-sealed coding ends require DNA-PKcs (light blue oval) and the nuclease Artemis (red circle) to open and process the hairpins before ligation.
(D) DNA ligase 4 (Lig4, yellow oval) and XRCC4 (orange oval) are recruited to the DNA repair complex in a Ku-dependent manner. XLF (orange circle) is also recruited to the DNA repair complex by Ku.
(E) The XRCC4/Lig4 complex ligates signal ends and coding ends, resulting in signal joins (left) and coding joins (right), respectively.
V(D)J recombination end-joining occurs exclusively by C-NHEJ and does not occur at all in the absence of core C-NHEJ factors [11]. In this regard, RAG2, through unknown mechanisms, functions to exclude potential A-EJ repair pathways [12]. Consistent with V(D)J joining occurring only by C-NHEJ, the entire V(D)J recombination reaction occurs only in the G1 cell cycle phase, a restriction solidified by the degradation of RAG2 at the G1/S transition [13, 14]. Overall, the “V(D)J recombinase” consists of the lymphocyte-specific RAG cleavage component and lymphocyte-specific TdT diversification component (see below), along with more generally expressed C-NHEJ proteins that comprise the joining component. As discussed below, recent studies also implicate ATM-dependent DSBR factors as potential joining components of the V(D)J recombinase. It is notable that during V(D)J recombination, RAG must alter the normal process of C-NHEJ from simply rejoining the two ends of a given DSB to direct joins of CEs to CEs and SEs to SEs from two separate DSBs, leading to an inversional or deletional outcome depending on the relative chromosomal orientation of the initiating RSs to each other [13, 15, 16].
Mature B cells are generated through the productive assembly of an IgH V(D)J exon and an Ig light chain (IgL) VJ exon; which, respectively, lead to production of IgH and IgL proteins that together form the B cell antigen receptor (“BCR”). In the peripheral immune system (e.g. spleen and lymph nodes), mature B cells may encounter antigens that bind to their BCR, causing them to become activated and to undergo additional genomic alterations including IgH Class Switch Recombination (CSR). Through CSR, the set of exons encoding the initially expressed IgH constant region (Cμ) are replaced with one of several sets of downstream CH exons (Cγ, Cα, Cε), leading to IgH class switching from IgM to IgG, IgA, or IgE. CSR results in secretion of specific antibodies that are endowed with optimal effector functions for elimination of particular pathogens. CSR is initiated by the Activation-induced cytidine deaminase (AID), which deaminates C’s to U’s within long (i.e. up to 10kb), highly repetitive switch (“S”) regions that lie just upstream of each set of CH exons, thereby triggering downstream mechanisms that generate S region DSBs that are requisite intermediates for CSR [1, 2]. To complete CSR, DSBs in a donor S region upstream of Cμ(S μ) and a downstream acceptor S region (e.g. Sγ, Sα, Sε) are fused by C-NHEJ. However, in the complete absence of C-NHEJ, CSR, unlike V(D)J recombination, can still be completed, albeit at somewhat reduced efficiency, by A-EJ [2, 17–19].
2. C-NHEJ in the repair of DSB during V(D)J recombination and other processes
The Ku70, Ku80, XRCC4 and DNA Ligase 4 (Lig4) factors are often described as the evolutionarily conserved “core” C-NHEJ factors [1, 2, 20, 21]. Ku70 and Ku80 form a dimer (“Ku”) that recognizes and binds to DSBs [22]. XRCC4 and Lig4 form a complex that is requisite for the ligation phase of C-NHEJ [1, 20, 21]; these proteins apparently are recruited to DSB ends by Ku [23, 24]. Due to their recognition and joining functions, respectively, Ku and XRCC4/Lig4 are required for all known forms of C-NHEJ; for example, during V(D)J recombination they are required for joining of blunt SEs and for joining of hairpin-sealed CEs [1, 2]. Accordingly, deficiency for any of the core C-NHEJ factors (Ku70, Ku80, XRCC4 or Lig4) in mice leads to inability to join CEs or SEs during V(D)J recombination, resulting in a complete block in B and T cell development and a Severe Combined Immunodeficiency (SCID), that is essentially as severe as that of RAG-deficient mice where V(D)J recombination cannot be initiated [2, 25, 26].
The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and Artemis, which is a DNA-PKcs-activated endonuclease, are C-NHEJ factors that have specialized functions in C-NHEJ that are required for joining DNA ends that must be processed before joining. For example, joining of hairpin-sealed CEs is nearly abrogated in the absence of DNA-PKcs or Artemis due to their inability to be opened in the absence of DNA-PKcs-activated Artemis endonuclease activity [27]. In accord with the requirement for DNA-PKcs and Artemis to generate V(D)J recombination CJs, complete deficiency for DNA-PKcs or Artemis in mice also results in a block in B and T cell development and a SCID due to inability to join V(D)J CEs and thereby form functional Ig and TCR genes required for development beyond progenitor stages [2, 25]. Processing of DSBs for C-NHEJ also may involve various DNA polymerases, such as DNA polymerase μ and DNA polymerase λ [20]. In addition, the developing lymphocyte specific terminal deoxynucleotidyl transferase enzyme (TdT), the first recognized component of the V(D)J recombinase [28], adds non-templated nucleotides to V(D)J junctions prior to their ligation, thereby greatly increasing junctional diversity and vastly expanding the diversity of antibody and TCR repertoires [28–30]. Once DSB ends are processed, if necessary, the XRCC4/Lig4 complex ligates them to complete the reaction.
DNA-PKcs clearly has functions in C-NHEJ beyond activating Artemis. In this regard, ligation of blunt SEs occurs normally in the absence of Artemis; but SE joining is variably impaired, depending on cell type, both with respect to frequency and fidelity in the absence of DNA-PKcs. Thus, SE joining occurs normally in DNA-PKcs-deficient embryonic stem (ES) cells but shows variable impairment in developing lymphocytes and somatic cell lines [31–38]. Among other possibilities, such variability in SE joining among different DNA-PKcs-deficient cell types might be explained by differential expression in different cell types of factors that are functionally redundant with DNA-PKcs C-NHEJ functions beyond those associated with Artemis activation. As discussed below, two known factors, XLF and ATM, can, at least partially, compensate for DNA-PKcs in SE joining. Non-Artemis-related C-NHEJ functions of DNA-PKcs revealed by defective SE joining during V(D)J recombination in DNA-PKcs-deficient cells have been suggested to include DNA end-bridging functions based on biochemical studies [39–41]. While DNA-PKcs may also play such end-bridging functions in C-NHEJ more generally, such a role is difficult to study in the context of V(D)J recombination CE joining due to requisite function of DNA-PKcs in the opening of hairpin-sealed CEs prior to joining.
C-NHEJ is involved in repair of DSBs more generally. Deficiency for core C-NHEJ factors in lymphoid or non-lymphoid cells leads to IR sensitivity and increased genomic instability, associated defects in repair of DSBs ranging from DSBs introduced by the I-SceI enzyme in reporter constructs [42, 43] to DSBs introduced during CSR in activated B cells [17–19, 42] and to DSBs resulting from unknown factors during neuronal development. In the latter context, deficiency of XRCC4 or Lig4 results in severe and widespread p53-mediated apoptotic death of newly generated neurons in response to DSBs [44–48]. This severe neuronal death is associated with late embryonic lethality of XRCC4- and Lig4-deficient mice, which can be rescued by p53 deficiency [45, 46]. Ku deficiency in mice also leads to increased p53-mediated death of newly generated neurons [49], but not as severely as that seen in XRCC4 or Lig4 deficiency and, correspondingly, Ku deficiency does not lead to embryonic lethality [50, 51]. Why Ku deficiency is less severe in this context is unknown but one hypothesis, based on the finding that Ku-deficiency can rescue the embryonic lethality of Lig4 deficiency [52, 53], is that Ku binding may block access of ends to alternative DSB repair pathways in XRCC4- or Lig4-deficient cells [1]. Deficiency for non-core C-NHEJ factors has a more variable and less severe effect for C-NHEJ events beyond the joining of CEs duringV(D)J recombination. DNA-PKcs deficiency also leads to increased IR-sensitivity, genomic instability and reduced CSR but to a much lesser degree than observed for core C-NHEJ factor deficiencies [31, 32, 54–56]. Artemis deficiency also may variably impact these processes but to an even lesser extent than DNA-PKcs deficiency [32, 54, 57, 58]. Correspondingly, neither DNA-PKcs- nor Artemis-deficient mice have growth defects or neuronal defects [31, 50, 58].
C-NHEJ-deficient mice are not highly disposed to development of lymphoid or other cancers even though their developing and activated lymphocyte Ig and TCR loci translocate due to mis-joining of persistent RAG-initiated DSBs. Lack of tumor development in C-NHEJ-deficient mice is thought to be due to elimination of cells with persistent DSBs or oncogenic translocations via the p53-dependent G1/S checkpoint. Correspondingly, all tested C-NHEJ/p53 double-deficient mice, except XLF-deficient mice (see below), rapidly develop RAG-dependent pro-B lymphomas with IgH locus translocations that lead to c-myc or, in the case of Artemis deficiency, N-myc oncogene amplification [45, 46, 59–62]. Core C-NHEJ-deficient mice that are also p53-deficient consistently develop medulloblastoma brain tumors, consistent with an important, but unknown, role of C-NHEJ in development of the nervous system [44, 48, 63, 64].
The XRCC4-like factor (XLF) [65, 66] has also been implicated in joining of DSBs, although its requirement for C-NHEJ appears variable and, in that regard, it is not required for robust developmental V(D)J recombination in mice [67, 68], due to a functional redundancy between XLF and various DSBR factors in C-NHEJ ([69–71]; discussed below) and a functional redundancy with DNA-PKcs in SE joining [32]. Correspondingly, germline deficiency for XLF in mice does not lead to any major impacts on survival or development, including that of lymphocytes. In the latter context, while there are modest effects on B and T cell development, these largely may be due to impacts on repair of DSBs other than those involved in V(D)J recombination [67, 68]. Also, consistent with functionally redundant factors that could compensate for XLF in end-joining, there is no obvious impact of XLF deficiency on neuronal development in mice. Due to the compensatory functions of XLF and the ATM-dependent DSBR, we will discuss XLF in more detail later in the review.
3. ATM-dependent DNA double-strand break response Proteins
The Ataxia telangiectasia (AT) mutated (ATM) protein kinase is a key upstream member of the ATM-dependent DSBR pathway [72]. ATM belongs to the phosphoinositide 3-kinase related protein kinase (PIKK) family that includes DNA-PKcs and Ataxia telangiectasia and Rad3-related protein (ATR) [73]. After DSB generation in G1, ATM activates several downstream factors including p53. Activation of p53 mediates the p53-dependent G1/S checkpoint to arrest cells with unrepaired DSBs to facilitate proper DSB repair or to cause apoptosis of cells with persistent DSBs [74–76]. The DSBR also participates directly in repair of DSBs, including those involved in V(D)J recombination [77, 78] and those involved in CSR [79]. Following activation via DSBs, ATM phosphorylates a set of proteins that includes histone H2AX, mediator of DNA damage checkpoint 1 (MDC1) and the p53-binding protein 1 (53BP1), which generate large foci in chromatin flanking DSBs [73]. In this regard, phosphorylated histone H2AX (“γ-H2AX”) promotes recruitment of MDC1 [80], which contributes to the generation of a positive feedback loop that promotes spreading of phosphorylated H2AX over hundreds of kilobases (kb) within chromatin on either side of the DSB [81–84]. MDC1 also recruits ubiquitin ligases RNF8 and RNF168, the latter of which modifies H2A family histones (H2A and H2AX) to promote stable 53BP1 association within these foci [85–88]. Beyond potential roles in checkpoint signaling, formation of these ATM-dependent foci have been proposed to tether DSB ends for re-joining via C-NHEJ [89]. DSBR factors downstream of ATM also have been implicated in directing repair into C-NHEJ versus HR or A-EJ, for example by preventing end resection [90–94].
The human AT syndrome includes progressive ataxia, immunodeficiency, radio-sensitivity, genomic instability, increased Ig and TCR locus translocations in normal lymphocytes, and B and T cell lymphomas [95, 96]. The phenotype of ATM-deficient mice overlaps with that of AT patients and includes general cellular radio-sensitivity and genomic instability (as determined cytogenetically), modest immunodeficiency, IgH CSR defects (30–50% of normal), and susceptibility to T cell lymphomas that all carry recurrent chromosomal translocations involving the TCRδ locus [97, 98]. Cytogenetic studies showed that most chromosomal aberrations in ATM-deficient cells, similar to those of C-NHEJ deficient cells [79, 99–101], occur in the form of chromosomal breaks and translocations, supporting the notion that ATM plays a most critical role during DSB repair in pre-replicative (e.g. G1) cells. While chromosomal translocations have not been well-characterized in immature ATM-deficient human T cell acute lymphocyte leukemias (“T-ALLS”), it seems likely that, as has recently been reported for human T-ALLs more generally, TCRδ locus translocations will also be a major feature of immature ATM-deficient human T-ALLs [102]. The CSR defects in ATM-deficient mice are associated with substantially impaired end-joining during CSR, which leads to unrepaired AID-initiated S region DSBs in activated B cells. These breaks progress at high frequency to chromosomal breaks and translocations observed by metaphase fluorescence in situ hybridization (FISH), supporting the notion that the DSBR contributes to tethering S region DSBs for end-joining during CSR[79].
While there is clearly a substantial level of normal V(D)J recombination in AT patients and ATM-deficient mice, the frequent Ig or TCR locus translocations observed in normal T and B cells of AT patients and ATM-deficient mice were noted to be consistent with a V(D)J recombination joining defect [95]. With respect to roles of ATM in V(D)J recombination, studies that employed ATM-deficient mouse pro-B cell lines demonstrated that ATM deficiency or inhibition of ATM kinase activity leads to release of some CEs from RAG-held V(D)J recombination post-cleavage complexes, leading to unrepaired RAG-generated DSBs that can progress to chromosomal breaks and translocations [78]. Such “free” CEs are also joined at increased frequency to cleaved SEs to generate hybrid or, potentially, open and shut joins instead of normal CJs [78]. Thus, ATM contributes to promoting proper C-NHEJ during V(D)J recombination by stabilizing RAG-mediated DSB post-cleavage complexes during V(D)J recombination [78]. Furthermore, ATM also has a role in SE joining that is revealed in the absence of DNA-PKcs [33, 103], that could reflect overlapping functions in phosphorylating common DNA repair substrates and/or in more direct roles in DNA end-tethering. With respect to downstream substrates, deficiencies for H2AX, MDC1 and 53BP1 DSBR factors all also have clear-cut impacts on general C-NHEJ as best illustrated by effects on CSR; however, deficiencies for these factors have quite modest impacts on V(D)J recombination and lymphocyte development [81, 104, 105].
H2AX- and MDC1-deficient mice appear relatively normal, although their cells have increased IR-sensitivity and cytogenetic genomic instability [79, 81, 104]. Correspondingly, activated H2AX-deficient or MDC1-deficient B cells have a modest reduction in CSR associated with substantial levels of IgH locus chromosomal breaks, indicating roles for both in the end-joining phase of CSR. Cytogenetic studies revealed that H2AX and MDC1 likely function in DSB repair both during pre-replicative and post-replicative cell cycle phases [79, 81, 101], as evidenced by an increase in both chromosomal and chromatid breaks in H2AX-deficient or MDC1-deficient cells [79, 104, 106]. Neither H2AX deficiency nor MDC1 deficiency had any readily detectable impact on V(D)J recombination in vivo or on lymphocyte development. Yet, in a p53-deficient background, H2AX deficiency, or haplo-insufficiency, predisposed to thymic lymphomas and to pro-B and B cell lymphomas [77]. Notably, H2AX/p53 double-deficient proB lymphomas harbored oncogenic IgH locus translocations with junctions that involve V(D)J recombination-associated DSBs, a finding that led to the proposal that H2AX may function in suppressing occasional generation of unrepaired DSBs during V(D)J recombination [77, 107]. In this regard, H2AX was subsequently shown to protect persistent RAG-initiated DSBs in C-NHEJ-deficient (e.g. Artemis- or Lig4-deficient) pro-B lines from undergoing aberrant resection [90] (also see below).
53BP1-deficient mice also appear relatively normal but again their cells have increased IR sensitivity [105, 108]. Yet, other than CSR-activated B cells, 53BP1-deficient cells have only modest, if any, cytogenetic instability. However, cytogenetic analyses of those cells that do show genomic instability indicate that 53BP1 deficiency mainly leads to chromosomal breaks consistent with a major 53BP1 role in genome stability maintenance occurring in pre-replicative cells as observed for ATM [70, 79, 99, 108, 109]. 53BP1-deficient mice have modestly reduced lymphocyte numbers and their differentiating T lymphocytes have very modest V(D)J recombination defects [105, 110, 111]. Strikingly, though, CSR is nearly abrogated in the absence of 53BP1 [105, 110]. In this context, CSR-activated 53BP1-deficient B cells have much greater levels of cytogenetic instability than other 53BP1-deficient cell types but nearly all of it is associated with AID-dependent IgH locus breaks and translocations.
The more dramatic CSR deficiency and associated IgH locus instability identified in 53BP1-deficient B cells compared to other DSB-deficient B cells indicates a more specialized role for 53BP1 in CSR, beyond that of the ATM-dependent DSBR[79]. Roles of 53BP1 that might contribute to CSR defects include protection of DSB ends from resection and potentially promoting their joining in the context of the DSBR [2, 79, 91–93, 105, 110, 111], although the overall mechanisms by which 53BP1 plays an especially crucial role in CSR compared to other DSBR factors remains to be elucidated. Finally, while 53BP1-deficient mice are not cancer prone, 53BP1-deficient mice that are also p53-deficient develop thymic or B cell lymphomas [109, 112], and 53BP1-deficient mice that have deregulated AID expression may also develop B cell lymphomas [113].
4. XLF is a C-NHEJ Factor but is not required for Normal V(D)J Recombination in mice
The XRCC4-like factor (XLF, also known as Cernunnos, or Nhej1) was identified as a potential C-NHEJ factor through both cDNA complementation of cells derived from an IR-sensitive human immunodeficiency patient and through a yeast two-hybrid screen for XRCC4-interacting proteins [65, 66]. In humans, inactivating mutations of XLF result in an autosomal recessive disorder characterized by immunodeficiency that may become progressively more severe; but which, in general, is not as severe as the complete SCID phenotypes associated with human Artemis or DNA-PKcs deficiency [66, 114–116]. Similar to patients with hypomorphic mutations in Lig4, human XLF deficiency also is associated with microcephaly and radio-sensitivity [66, 116]. Moreover, XLF-deficient human fibroblasts that ectopically expressed RAG are substantially impaired for ability to undergo V(D)J recombination in the context of episomal and intrachromosomal V(D)J recombination substrates, consistent with a role for XLF in C-NHEJ [66, 117].
Two independent XLF-deficient mouse lines were both relatively normal overall; but XLF-deficient MEFs and ES cells, similar to cells from XLF-deficient patients, had significant V(D)J recombination defects [67, 68]. Yet, XLF-deficient mice have only very mildly reduced peripheral T and B cell numbers. Moreover, distributions of developing B and T cells in the bone marrow and thymuses of XLF-deficient mice are comparable to those of WT counterparts [67, 68], in striking contrast to the block in lymphocyte differentiation at the progenitor stage of core C-NHEJ-deficient mice and DNA-PKcs- or Artemis-deficient mice [2, 25]. Abelson Murine Leukemia virus transformed pro-B cells (Abl pro-B cells) treated with the Abl kinase inhibitor Gleevec (STI571) undergo G1 cell cycle arrest, induce RAG expression, and carry out V(D)J recombination at the endogenous Igκ light chain locus, as well as in the context of transient or chromosomally-integrated V(D)J recombination substrates [78]. XLF-deficient Abl pro-B lines performed V(D)J recombination on such substrates similarly to WT Abl pro-B lines, despite the fact that these lines were mildly IR-sensitive, suggesting a potential differential effect of XLF deficiency on DSB repair during V(D)J recombination versus more general DSB repair in these cells [32, 67, 70, 71].
Despite the lack of a V(D)J recombination defect in developing lymphocytes, other types of XLF-deficient cells show manifestations of C-NHEJ deficiency. Both XLF-deficient MEFs and XLF-deficient ES cells were IR-sensitive and impaired for joining V(D)J CEs and SEs within transient V(D)J recombination substrates [67, 68, 118], but in neither case was the impact as severe as is observed for XRCC4-deficient cells. Cytogenetic genomic instability is present in XLF-deficient MEFs and is primarily in the form of chromosomal breaks, implicating a role for XLF in repair of pre-replicative DSBs [32, 67, 70, 118]. Also consistent with an effect of XLF deficiency on general C-NHEJ, XLF-deficient mature B cells have reduced CSR coupled with increased unrepaired IgH chromosome breaks [67]. XLF/p53 double-deficient mice, unlike other C-NHEJ/p53 double-deficient mice, rarely die of pro-B cell lymphomas; but rather die of T cell lymphomas characteristic of p53 deficiency [67]. As C-NHEJ/p53 double-deficient pro-B cell lymphomas routinely contain oncogenic translocations involving RAG-generated IgH locus DSBs, the rarity of this type of tumor in XLF/p53 double-deficient mice is consistent with normal V(D)J recombination in developing XLF-deficient lymphocytes. Notably, however, many XLF/p53 double-deficient mice also develop medulloblastomas (MBs), as seen in other C-NHEJ/p53 double-deficient mice [17, 119, 120]. The occurrence of recurrent MBs in XLF/p53 double-deficient mice is consistent with a general C-NHEJ defect in MB progenitors and warrants deeper investigation into underlying mechanisms. As XLF clearly functions as a C-NHEJ factor in a broad variety of cell types, the relatively normal V(D)J recombination in XLF-deficient progenitor lymphocytes and Abl pro-B cell lines coupled with the normal lymphocyte development in XLF-deficient mice [67, 68], suggested the existence of factors that compensate for XLF function in C-NHEJ, specifically in the context of V(D)J recombination in developing lymphocytes [67].
Most current models for XLF function derive from biochemical and structural assays, which suggest that XLF largely works in concert with the structurally similar XRCC4 factor. Both XLF and XRCC4 contain a globular head domain, an alpha-helical stalk domain, and an unstructured C-terminal domain [65, 121]. In this regard, XLF forms heterodimers with XRCC4 through interactions of their respective globular head domains [65, 121]. Known functions of XRCC4 include stabilization of Lig4 through direct interaction [122] and stimulation of Lig4 activity [123]. XLF/XRCC4 heterodimers form protein filaments [121, 124–127] that tether DNA ends in vitro and which have been speculated toen hance XRCC4-dependent recruitment of Lig4 to DSBs [124, 126]. In biochemical assays, XLF also enhances efficiency of the XRCC4/Lig4 complex to ligatelinearized plasmids in vitro [128]. Notably, non-lymphoid cells that express mutant forms of XRCC4 that are unable to interact with XLF carry out joining of SEs, but not CEs, in the context of extra-chromosomal V(D)J recombination substrates [129], suggesting potential functional compensation for XLF-XRCC4 end-bridging by RAG or other factors in the post-cleavage synaptic complex for joining of SEs [121, 129, 130].
The findings that XLF is not required for V(D)J recombination in normal developing lymphocytes but is required for ectopic V(D)J recombination in non-lymphoid cells led to the hypothesis that other factors may functionally compensate for XLF to promote V(D)J recombination in developing lymphocytes [67]. As mentioned above, one such factor might be RAG, which holds CEs and SEs in a post-cleavage synaptic complex and directs C-NHEJ-mediated joining of two CEs and two SEs, respectively, to each other [131, 132]. In this context, SEs are held more tightly than CEs in the RAG post-cleavage synaptic complex [133]; which in the context of proposed XLF synapsis functions could explain the lack of impact on SE joining versus CE joining when XRCC4-XLF interaction is disrupted [129]. If RAG does function redundantly with XLF during V(D)J recombination in developing lymphocytes, but not in non-lymphoid cells, it is conceivable that normal physiological V(D)J recombination may have evolved to optimize ability of RAG, for example via post-translational modifications, to hold CEs and/or SEs in post-cleavage synaptic complexes for proper end-joining and/or to contribute to C-NHEJ factor recruitment [12, 131, 132, 134]. However, additional studies, described below, demonstrated that factors other than RAG provide functional redundancy with XLF in V(D)J recombination and C-NHEJ more generally.
5. XLF has functional redundancy with ATM
The ATM-dependent DSBR is activated in response to endogenous RAG-initiated DSBs at antigen receptor loci [135, 136], and deficiencies for ATM or several ATM downstream factors (H2AX and 53BP1) impair end-joining during V(D)J recombination, with impacts ranging from moderate for ATM [78] to very modest for H2AX [79, 104, 106]. Such studies led to the proposal that the ATM-dependent DSBR, beyond activating checkpoints, may contribute to C-NHEJ through end-tethering during V(D)J recombination, CSR, and more generally [1, 2, 79, 89, 137]. Indeed, ATM was found to stabilize RAG-mediated V(D)J breaks in the context of the post-cleavage complex [78]. Together, these findings led to the evaluation of ATM and downstream DSBR factors as candidates for having functional redundancy with XLF, potentially through roles in end-tethering. Correspondingly, XLF/ATM double-deficient mice were observed to have a severe block in B and T cell development at the progenitor stage when V(D)J recombination occurs, very reminiscent of the SCID phenotype of C-NHEJ-deficient mice [71]. In the XLF/ATM double-deficient background, the B cell developmental block was substantially rescued by introduction of germline alleles containing pre-assembled IgH and IgL variable region exons (“HL alleles”), consistent with impairment resulting largely from a V(D)J recombination defect [71]. Correspondingly, XLF/ATM double-deficient Abl pro-B cells arrested in G1 to activate V(D)J recombination accumulated unrepaired breaks in the endogenous Igκ locus [71], and were severely impaired in ability to join CEs and SEs of RAG-initiated DSBs within chromosomally integrated V(D)J recombination substrates. Thus, combined deficiency for ATM and XLF essentially abrogates chromosomal V(D)J recombination in developing lymphocytes (Figure 2).
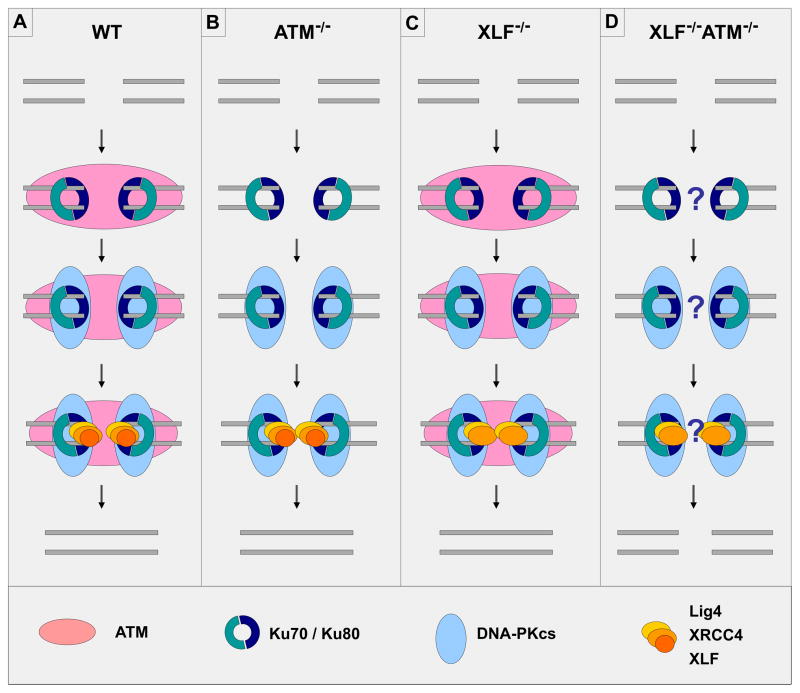
Figure 2. XLF has functional redundancy with ATM and ATM substrates (H2AX, 53BP1) during C-NHEJ.
(A) Factors of the DSBR pathway (ATM, γH2AX, 53BP1; pink oval) accumulate around a DSB. Ku70/Ku80 (light blue and dark blue semicircles) bind DNA ends. DNA-PKcs (light blue oval), XRCC4/Lig4 (orange and yellow ovals) and XLF (orange circle) are recruited to the C-NHEJ DNA repair complex by Ku.
(B) and (C) In the absence of ATM (or H2AX, or 53BP1), C-NHEJ is functional. In the absence of XLF, C-NHEJ is also largely functional.
(D) In cells with combined deficiency for XLF and ATM, C-NHEJ is nearly abrogated. In cells with combined deficiency for XLF and H2AX, or XLF and 53BP1, C-NHEJ is dramatically reduced. It remains unclear which stage of C-NHEJ is defective in the combined absence of XLF and ATM or its substrates (as denoted by the question marks).
The studies of developing lymphocytes or pro-B cell lines clearly demonstrate that XLF-deficient B lineage cells require ATM and ATM-deficient B cells require XLF to perform C-NHEJ during V(D)J recombination [71]. XLF/ATM double-deficient mouse fibroblasts have substantially increased cytogenetic instability as compared to fibroblasts deficient for either factor alone (Oksenych and Alt, unpublished), which among other possibilities, suggests that ATM and XLF may have functional redundancy in C-NHEJ more generally. Indeed, studies of CSR in the XLF/ATM double-deficient background strongly support this notion. Thus, CSR to IgG1 in mature XLF/ATM double-deficient B cells generated in a background containing HL alleles was reduced to the residual levels observed in core C-NHEJ-deficient cells (Figure 2). As residual CSR in C-NHEJ-deficient B cells is known to occur via A-EJ, these findings also suggested that the observed functional redundancy for ATM and XLF primarily occurs in the context of C-NHEJ and not for A-EJ [71].
Assays for V(D)J recombination within extra-chromosomal substrates introduced into XLF/ATM double-deficient Abl pro-B cells revealed that both CE and SE joining occurred at levels similar to those of WT Abl pro-B cells, in striking contrast to the essentially complete block in chromosomal V(D)J recombination in XLF/ATM double-deficient lines [71]. This finding suggested that XLF and ATM functional redundancy may be more specific to chromosomal V(D)J recombination in the context of chromatin [71]. In this regard, inhibition of ATM kinase activity in XLF-deficient Abl pro-B cells also led to a block in joining of RAG-cleaved CEs and SEs within chromosomally integrated V(D)J recombination substrates, suggesting that XLF-redundant functions of ATM in chromosomal V(D)J recombination joining may be mediated, at least in part, by downstream ATM substrates in the context of chromatin [71]. These findings led to additional studies that implicated H2AX and 53BP1 as factors that have functional redundancy with XLF in C-NHEJ during V(D)J recombination.
6. XLF has redundant functions with ATM substrates H2AX and 53BP1
Combined deficiency for XLF and histone H2AX results in early embryonic lethality. The cause of such embryonic lethality remains to be determined. One proposed model is that embryonic lethality may be caused by XLF and H2AX functional redundancy in DNA repair processes beyond those shared with ATM alone. Such functions, for example, might be related to potential ATM-independent post-replicative DNA repair roles for H2AX as suggested by the increased levels of chromatid breaks in H2AX-deficient versus ATM-deficient cells [71, 79, 81, 101]. In this context, H2AX can be activated independently of ATM by ATR, DNA-PKcs [71, 138–141] and, conceivably, by other kinases. If this model is correct, one might also expect MDC1 deficiency, which also leads to defects in replicative or post-replicative DSB repair defects, to lead to embryonic lethality when combined with XLF deficiency. Another, non-mutually exclusive model, in analogy to p53 rescue of embryonic lethality of Lig4- or XRCC4-deficient mice [46], would be that checkpoint defects associated with ATM deficiency, but not H2AX deficiency, rescue potential embryonic lethal DNA repair defects of XLF/ATM deficient mice [71]. If correct, p53 deficiency might rescue the embryonic lethality of XLF/H2AX deficiency. However, it is also notable that XLF/H2AX embryonic lethality occurs much earlier than the late embryonic lethality of Lig4 or XRCC4 deficiency, both of which are associated with dramatic neuronal apoptosis [45–47, 71].
It was not possible to assess potential defects in lymphocyte development or DNA repair in the context of XLF/H2AX double-deficient mice due to early embryonic lethality. However, conditional inactivation of H2AX in XLF-deficient Abl pro-B lines had no impact on cellular viability. In these lines, combined H2AX and XLF deficiency also results in severely impaired joining of RAG-generated CEs and SEs; albeit not quite to the same extent as seen with dual ATM- and XLF-deficiency [71] (Figure 2). However, while in XLF/ATM double-deficient Abl pro-B lines, un-joined CEs and SEs appeared largely intact, in XLF/H2AX double-deficient Abl pro-B lines most un-joined CEs and SEs were highly resected, consistent with findings that H2AX protects such ends from resection in C-NHEJ-deficient backgrounds [90]. At first glance, this differential impact of ATM and H2AX deficiency on the fate of un-joined CEs and SEs in an XLF-deficient background might not seem consistent with such H2AX functions lying downstream of ATM. However, ATM deficiency has a dual impact on resection of un-joined CEs and SEs during V(D)J recombination, both protecting these ends from resection via generation of phophorylated γH2AX and also promoting their resection by activating CtIP [90]. Thus, the defect in CE and SE joining per se in the combined absence of H2AX and XLF may reflect H2AX functioning downstream of ATM activation at a DSB, both to promote joining, for example via an end-tethering function, and also to specifically prevent end resection by ATM-activated CtIP. Correspondingly, inhibition of ATM kinase activity in XLF/H2AX double-deficient Abl pro-B cell lines substantially restored accumulation of un-joined CEs and SEs that were not markedly resected.
XLF and 53BP1 double-deficient mice are live born, but compared to XLF-deficient or 53BP1-deficient mice, are growth retarded, their fibroblasts have increased cytogenetic genomic instability manifested primarily as chromosome breaks, and they are more prone to thymic lymphoma [69, 70]. Moreover, like XLF/ATM double-deficient mice, XLF/53BP1 double-deficient mice have a SCID phenotype with lymphocyte development essentially blocked at the progenitor B and T cell stages, consistent with impaired V(D)J recombination [69, 70]. Indeed, analyses of XLF/53BP1 double-deficient Abl pro-B cell lines revealed a combined impact on ability to join RAG-generated CEs and SEs during V(D)J recombination that was strikingly similar to the combined impact of dual XLF/H2AX deficiency (Figure 2), including severely impaired joining of both CEs and SEs and dramatically increased resection of the un-joined CEs and SEs that was rescued by treatment of cells with an ATM inhibitor [69, 70]. The end protection role of 53BP1 is consistent with such roles in other contexts including CSR [91–94]. While the V(D)J recombination defects of XLF/53BP1 double-deficient Abl pro-B lines was substantial, as seen with dual XLF/H2AX deficiency, some residual joining remained [69, 70]. In this context, the essentially complete SCID phenotype of XLF/53BP1 double-deficient mice may reflect both greatly impaired V(D)J recombination, as well as an impact on developing or more mature lymphocytes due to effects on general C-NHEJ as suggested by the increased genomic instability of XLF/53BP1 double-deficient fibroblasts.
Overall, the impact of H2AX or 53BP1 deficiency on V(D)J recombination and C-NHEJ in an XLF-deficient background is consistent with both DSBR proteins having critical functions in both end-joining per se (e.g. by end-tethering roles) and end protection from aberrant resection in the context of the ATM-dependent DSBR. As 53BP1 may also have functions at DSBs independent of ATM, as it likely does during CSR, a functional redundancy with XLF in this context cannot be ruled out. XLF, on its own, does not appear to have a major role in protecting un-joined RAG-initiated CEs and SEs from resection. Thus, these ends are highly resected in Lig4/H2AX double-deficient or Artemis/H2AX double-deficient Abl pro-B cells [90]. Likewise, in Artemis-, DNA-PKcs-, or XRCC4-deficient Abl pro-B cells that are also XLF-deficient, persistent, unrepaired CEs generated during attempted V(D)J recombination do not undergo increased resection [32].
7. XLF and DNA-PKcs have Functional Overlaps in V(D)J recombination and C-NHEJ
DNA-PKcs and ATM are related PIKKs with shared substrates [140], are functionally redundant with respect to SE joining during V(D)J recombination [33, 103], and have both been suggested to potentially play a role in tethering DSB ends during C-NHEJ [39–41, 78]. Correspondingly, DNA-PKcs also appears to have functional overlap with XLF in C-NHEJ. Thus, XLF/DNA-PKcs double-deficient fibroblasts have increased levels of chromosomal breaks compared to that of DNA-PKcs- or XLF-deficient fibroblasts [32]. Moreover, combined deficiency for XLF and DNA-PKcs abrogates joining of RAG-initiated SEs within chromosomally integrated V(D)J recombination substrates in Abl pro-B cells [32]. Given that XLF/Artemis double-deficient Abl pro-B cells did not display such an impact on SE joining, the dramatically impaired SE joining in XLF/DNA-PKcs double-deficient Abl pro-B cells involves Artemis-independent DNA-PKcs functions. In this context, XLF/DNA-PKcs double-deficient mice are live born but die shortly after birth for unknown reasons; whereas XLF/Artemis double-deficient mice do not exhibit early post-natal lethality, consistent with XLF/DNA-PKcs functional redundancy separate from DNA-PKcs function in Artemis activation [32]. Finally, the apparent DNA-PKcs redundant function with XLF is mediated by its kinase activity, as DNA-PKcs kinase inhibition in XLF-deficient Abl pro-B cells or mature B cells abrogates SE joining and reduces CSR levels, respectively [32]. The latter finding also is consistent with a broader functional redundancy between XLF and DNA-PKcs in C-NHEJ in general.
8. Perspectives
The finding that the ATM-dependent DNA damage response results in the formation of large foci in chromatin surrounding DSBs [142, 143] raised the possibility of a direct role for the DSBR in DSB repair, and, in particular, in C-NHEJ. Likewise, the discovery that XLF is required for protection from ionizing radiation and also for joining of RAG-initiated DSBs in patient fibroblasts implicated this XRCC4-related factor as a potential C-NHEJ component [66, 144]. A confounding issue, however, in considering potential roles of ATM-dependent DSBR factors and the XLF protein as C-NHEJ factors was the relative dispensability of these factors for C-NHEJ during V(D)J recombination. However, the finding that XLF shares redundant functions with ATM and several of its downstream DSBR factors with respect to C-NHEJ during V(D)J recombination in developing lymphocytes greatly clarified the critical roles of XLF and the DSBR in this process. Thus, in the absence of XLF, developing B and T lymphocytes are totally reliant on ATM and ATM downstream factors to carry out normal chromosomal V(D)J recombination; likewise, in the absence of ATM or downstream factors, developing B and T cells require XLF for C-NHEJ during V(D)J recombination. Indeed, while XLF deficiency has little impact on lymphocyte development, and ATM deficiency has only a modest impact, combined ATM and XLF deficiency results in a SCID phenotype reminiscent of that observed in the context of C-NHEJ deficiency [71]. A further surprising recent finding is the additional functional redundancy between XLF and the DNA-PKcs in C-NHEJ [32].
The ongoing challenge is to elucidate the nature of the redundant functions of XLF and DSBR factors (and DNA-PKcs) and whether they have overlapping roles in the same general function (e.g. end-tethering) (Figure 3) or whether they represent roles in different processes that provide different functions (e.g. end-tethering versus C-NHEJ factor recruitment) that are compensatory for each other in C-NHEJ [71]. A potential example of compensatory but different functions might include DNA end-tethering and C-NHEJ factor recruitment (or activation) (Figure 4). Thus, efficient tethering of DSBs may keep them together long enough for ligation even at reduced repair efficiency due to impaired C-NHEJ factor recruitment; conversely, efficient C-NHEJ recruitment might rapidly repair ends before they separate due to reduced tethering activity (Figure 4). With respect to such potential roles in C-NHEJ, DNA-PKcs and XLF have been directly implicated in DNA end-tethering [39–41, 121, 124–127], while ATM and its downstream DSBR factors have been suggested to play such roles either directly or indirectly [78, 79, 89, 105, 111]. With respect to C-NHEJ factor recruitment or activation, XLF was originally described as a part of the XLF/XRCC4/Lig4 complex with potential roles in the ligation phase of the reaction [65, 128, 145], and XLF has been found to stimulate Lig4 activity [128, 145].
Figure 3. XLF and ATM (and DSBR factors) have redundant functions during C-NHEJ.
(A) Both ATM (pink oval) and XLF (orange circle) may stimulate Lig4 (yellow oval) activity, and/or stabilize the XRCC4/Lig4 (orange and yellow ovals) complex at a DSB.
(B) ATM and its downstream substrates, combined with XLF/XRCC4 filaments, may tether DNA ends in close proximity before ligation.
Figure 4. XLF and ATM (and DSBR factors) have complementary functions during C-NHEJ.
(A) ATM (pink oval) may function to hold DNA ends, while XLF (orange circle) stimulates XRCC4/Lig4 (orange and yellow ovals) activity.
(B) ATM may stimulate XRCC4/Lig4, while XLF/XRCC4 filaments hold DNA ends.
It is notable that single deficiencies for XLF or individual DSBR factors generally have a less pronounced effect on V(D)J recombination than on other forms of C-NHEJ-mediated DSB repair including CSR. In theory, the lower V(D)J recombination impact might reflect a contribution by RAG in holding CE and SE in synaptic complexes and/or promoting their joining by C-NHEJ [12, 131, 132]. In this context, ATM or DNA-PKcs phosphorylation of RAG is not required for its function in normal cells [146]; but whether such an activity could contribute to the impact of ATM or DNA-PKcs deficiency on V(D)J recombination in the absence of XLF remains to be tested. Finally, functional redundancies between XLF and DSBR factors or RAG might also contribute to the phenotypic diversity of deficiencies for these factors in human patients or between humans and mice. For example, the marked variation in degree of lymphopenia observed in XLF-deficient patients [66, 116] might reflect variations in the expression of XLF compensatory proteins. Conversely, the impact of ATM deficiency in humans can vary among patients [95, 147, 148], and ATM-deficient mice do not exhibit the overt neurological defects observed in humans [95, 98]. Potentially, such variations could also reflect, at least in part, varying degrees of XLF compensation for ATM function in these different settings.
Bullet points.
XLF is a DNA repair protein that functions in C-NHEJ.
XLF has functional redundancy with multiple DSB response factors including ATM.
XLF and the DSB response have redundant roles in B and T cell development.
XLF and several DSB response factors have redundant roles in general DSB repair.
XLF and certain DSB response factors have redundant roles in mouse development.
Acknowledgments
V.K. is supported by an NIH T32 training grant GM007226-38. F.W.A. is supported by NIH grant AI076210. F.W.A. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Advances in immunology. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Gostissa M, Hildebrand DG, Becker MS, Boboila C, Chiarle R, Lewis S, Alt FW. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Advances in immunology. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annual review of immunology. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 5.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 6.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 7.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 8.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 9.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 10.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annual review of immunology. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 11.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 12.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, Meek K, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 13.Bednarski JJ, Sleckman BP. Lymphocyte development: integration of DNA damage response signaling. Advances in immunology. 2012;116:175–204. doi: 10.1016/B978-0-12-394300-2.00006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desiderio S. Temporal and spatial regulatory functions of the V(D)J recombinase. Seminars in immunology. 2010;22:362–369. doi: 10.1016/j.smim.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annual review of genetics. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 16.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 17.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 18.Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, Alt FW. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, Nussenzweig A, Nussenzweig M, Alt FW. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. The Journal of experimental medicine. 2010;207:417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 22.Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutation research. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 23.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Molecular and cellular biology. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Advances in immunology. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 26.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 28.Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 30.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver DT, Alt FW. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 32.Oksenych V, Kumar V, Liu X, Guo C, Schwer B, Zha S, Alt FW. Functional redundancy between the XLF and DNA-PKcs DNA repair factors in V(D)J recombination and nonhomologous DNA end joining. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2234–2239. doi: 10.1073/pnas.1222573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zha S, Jiang W, Fujiwara Y, Patel H, Goff PH, Brush JW, Dubois RL, Alt FW. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2028–2033. doi: 10.1073/pnas.1019293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Errami A, He DM, Friedl AA, Overkamp WJ, Morolli B, Hendrickson EA, Eckardt-Schupp F, Oshimura M, Lohman PH, Jackson SP, Zdzienicka MZ. XR-C1, a new CHO cell mutant which is defective in DNA-PKcs, is impaired in both V(D)J coding and signal joint formation. Nucleic acids research. 1998;26:3146–3153. doi: 10.1093/nar/26.13.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukumura R, Araki R, Fujimori A, Mori M, Saito T, Watanabe F, Sarashi M, Itsukaichi H, Eguchi-Kasai K, Sato K, Tatsumi K, Abe M. Murine cell line SX9 bearing a mutation in the dna-pkcs gene exhibits aberrant V(D)J recombination not only in the coding joint but also in the signal joint. The Journal of biological chemistry. 1998;273:13058–13064. doi: 10.1074/jbc.273.21.13058. [DOI] [PubMed] [Google Scholar]
- 36.Fukumura R, Araki R, Fujimori A, Tsutsumi Y, Kurimasa A, Li GC, Chen DJ, Tatsumi K, Abe M. Signal joint formation is also impaired in DNA-dependent protein kinase catalytic subunit knockout cells. J Immunol. 2000;165:3883–3889. doi: 10.4049/jimmunol.165.7.3883. [DOI] [PubMed] [Google Scholar]
- 37.Kurimasa A, Ouyang H, Dong LJ, Wang S, Li X, Cordon-Cardo C, Chen DJ, Li GC. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, Priestley A, Jackson SP, Marshak Rothstein A, Jeggo PA, Herrera VL. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 39.Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Advances in immunology. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 40.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Molecular cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 41.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. The EMBO journal. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boboila C, Oksenych V, Gostissa M, Wang JH, Zha S, Zhang Y, Chai H, Lee CS, Jankovic M, Saez LM, Nussenzweig MC, McKinnon PJ, Alt FW, Schwer B. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1) Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2473–2478. doi: 10.1073/pnas.1121470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Molecular cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Yan CT, Kaushal D, Murphy M, Zhang Y, Datta A, Chen C, Monroe B, Mostoslavsky G, Coakley K, Gao Y, Mills KD, Fazeli AP, Tepsuporn S, Hall G, Mulligan R, Fox E, Bronson R, De Girolami U, Lee C, Alt FW. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7378–7383. doi: 10.1073/pnas.0601938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 46.Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Molecular cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 47.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 48.Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME, Alt FW. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Liu N, Rajendran GK, Gernon TJ, Rockhill JK, Schwartz JL, Gu Y. A role for endogenous and radiation-induced DNA double-strand breaks in p53-dependent apoptosis during cortical neurogenesis. Radiation research. 2008;169:513–522. doi: 10.1667/RR1230.1. [DOI] [PubMed] [Google Scholar]
- 50.Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt FW. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, Stanhope-Baker P, Schlissel MS, Roth DB, Alt FW. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 52.Karanjawala ZE, Adachi N, Irvine RA, Oh EK, Shibata D, Schwarz K, Hsieh CL, Lieber MR. The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA repair. 2002;1:1017–1026. doi: 10.1016/s1568-7864(02)00151-9. [DOI] [PubMed] [Google Scholar]
- 53.Adachi N, Ishino T, Ishii Y, Takeda S, Koyama H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: Implications for DNA double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12109–12113. doi: 10.1073/pnas.201271098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franco S, Murphy MM, Li G, Borjeson T, Boboila C, Alt FW. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. The Journal of experimental medicine. 2008;205:557–564. doi: 10.1084/jem.20080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manis JP, Dudley D, Kaylor L, Alt FW. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 2002;16:607–617. doi: 10.1016/s1074-7613(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 56.Rooney S, Alt FW, Sekiguchi J, Manis JP. Artemis-independent functions of DNA-dependent protein kinase in Ig heavy chain class switch recombination and development. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2471–2475. doi: 10.1073/pnas.0409857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. The Journal of experimental medicine. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Molecular cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 59.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanasse GJ, Halbrook J, Thomas S, Burgess A, Hoekstra MF, Disteche CM, Willerford DM. Genetic pathway to recurrent chromosome translocations in murine lymphoma involves V(D)J recombinase. The Journal of clinical investigation. 1999;103:1669–1675. doi: 10.1172/JCI6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gladdy RA, Taylor MD, Williams CJ, Grandal I, Karaskova J, Squire JA, Rutka JT, Guidos CJ, Danska JS. The RAG-1/2 endonuclease causes genomic instability and controls CNS complications of lymphoblastic leukemia in p53/Prkdc-deficient mice. Cancer cell. 2003;3:37–50. doi: 10.1016/s1535-6108(02)00236-2. [DOI] [PubMed] [Google Scholar]
- 62.Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA, Hasty P. Analysis of ku80-mutant mice and cells with deficient levels of p53. Molecular and cellular biology. 2000;20:3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee Y, McKinnon PJ. DNA ligase IV suppresses medulloblastoma formation. Cancer research. 2002;62:6395–6399. [PubMed] [Google Scholar]
- 64.Holcomb VB, Vogel H, Marple T, Kornegay RW, Hasty P. Ku80 and p53 suppress medulloblastoma that arise independent of Rag-1-induced DSBs. Oncogene. 2006;25:7159–7165. doi: 10.1038/sj.onc.1209704. [DOI] [PubMed] [Google Scholar]
- 65.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 66.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 67.Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Molecular cell. 2008;31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vera G, Rivera-Munoz P, Abramowski V, Malivert L, Lim A, Bole-Feysot C, Martin C, Florkin B, Latour S, Revy P, de Villartay JP. Cernunnos deficiency reduces thymocyte life span and alters the T cell repertoire in mice and humans. Molecular and cellular biology. 2013;33:701–711. doi: 10.1128/MCB.01057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, Jiang W, Dubois RL, Yamamoto K, Wolner Z, Zha S. Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3903–3908. doi: 10.1073/pnas.1120160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oksenych V, Alt FW, Kumar V, Schwer B, Wesemann DR, Hansen E, Patel H, Su A, Guo C. Functional redundancy between repair factor XLF and damage response mediator 53BP1 in V(D)J recombination and DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2455–2460. doi: 10.1073/pnas.1121458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS letters. 2010;584:3675–3681. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 76.Rotman G, Shiloh Y. ATM: a mediator of multiple responses to genotoxic stress. Oncogene. 1999;18:6135–6144. doi: 10.1038/sj.onc.1203124. [DOI] [PubMed] [Google Scholar]
- 77.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 78.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 79.Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, Lou Z, Bassing CH, Manis JP, Chen J, Carpenter PB, Alt FW. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Molecular cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 80.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 81.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Molecular cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 82.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 83.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO reports. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. The Journal of cell biology. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA repair. 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 86.Stewart GS. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 2009;8:1532–1538. doi: 10.4161/cc.8.10.8351. [DOI] [PubMed] [Google Scholar]
- 87.Noon AT, Goodarzi AA. 53BP1-mediated DNA double strand break repair: insert bad pun here. DNA repair. 2011;10:1071–1076. doi: 10.1016/j.dnarep.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 88.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-Leblanc J, Noordermeer SM, Sicheri F, Durocher D. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013 doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- 90.Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, McKinnon PJ, Zhang J, Bassing CH, Sleckman BP. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011;469:245–249. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, Wesemann DR, Lee JE, Tubbs A, Sleckman BP, Daniel JA, Ge K, Alt FW, Fernandez-Capetillo O, Nussenzweig MC, Nussenzweig A. 53BP1 Mediates Productive and Mutagenic DNA Repair through Distinct Phosphoprotein Interactions. Cell. 2013 doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. The Journal of experimental medicine. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Molecular cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 95.McKinnon PJ. ATM and the molecular pathogenesis of ataxia telangiectasia. Annual review of pathology. 2012;7:303–321. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 96.Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annual review of genetics. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- 97.Zha S, Bassing CH, Sanda T, Brush JW, Patel H, Goff PH, Murphy MM, Tepsuporn S, Gatti RA, Look AT, Alt FW. ATM-deficient thymic lymphoma is associated with aberrant tcrd rearrangement and gene amplification. The Journal of experimental medicine. 2010;207:1369–1380. doi: 10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lavin MF. The appropriateness of the mouse model for ataxia-telangiectasia: neurological defects but no neurodegeneration. DNA repair. 2013;12:612–619. doi: 10.1016/j.dnarep.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 99.Elson A, Wang Y, Daugherty CJ, Morton CC, Zhou F, Campos-Torres J, Leder P. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sekiguchi J, Ferguson DO, Chen HT, Yang EM, Earle J, Frank K, Whitlow S, Gu Y, Xu Y, Nussenzweig A, Alt FW. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3243–3248. doi: 10.1073/pnas.051632098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9302–9306. doi: 10.1073/pnas.0803520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Larmonie NS, Dik WA, Meijerink JP, Homminga I, van Dongen JJ, Langerak AW. Breakpoint sites disclose the role of the V(D)J recombination machinery in the formation of T-cell receptor (TCR) and non-TCR associated aberrations in T-cell acute lymphoblastic leukemia. Haematologica. 2013;98:1173–1184. doi: 10.3324/haematol.2012.082156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gapud EJ, Dorsett Y, Yin B, Callen E, Bredemeyer A, Mahowald GK, Omi KQ, Walker LM, Bednarski JJ, McKinnon PJ, Bassing CH, Nussenzweig A, Sleckman BP. Ataxia telangiectasia mutated (Atm) and DNA-PKcs kinases have overlapping activities during chromosomal signal joint formation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2022–2027. doi: 10.1073/pnas.1013295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nature immunology. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 106.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Molecular and cellular biology. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morales JC, Franco S, Murphy MM, Bassing CH, Mills KD, Adams MM, Walsh NC, Manis JP, Rassidakis GZ, Alt FW, Carpenter PB. 53BP1 and p53 synergize to suppress genomic instability and lymphomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3310–3315. doi: 10.1073/pnas.0511259103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, Nussenzweig MC, Chen J. 53BP1 is required for class switch recombination. The Journal of cell biology. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ward IM, Difilippantonio S, Minn K, Mueller MD, Molina JR, Yu X, Frisk CS, Ried T, Nussenzweig A, Chen J. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Molecular and cellular biology. 2005;25:10079–10086. doi: 10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jankovic M, Feldhahn N, Oliveira TY, Silva IT, Kieffer-Kwon KR, Yamane A, Resch W, Klein I, Robbiani DF, Casellas R, Nussenzweig MC. 53BP1 alters the landscape of DNA rearrangements and suppresses AID-induced B cell lymphoma. Molecular cell. 2013;49:623–631. doi: 10.1016/j.molcel.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moshous D, Callebaut I, Chasseval R de, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 115.van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, Mari PO, Tezcan I, Chen DJ, Zdzienicka MZ, van Dongen JJ, van Gent DC. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. The Journal of clinical investigation. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Du L, Peng R, Bjorkman A, Filipe de Miranda N, Rosner C, Kotnis A, Berglund M, Liu C, Rosenquist R, Enblad G, Sundstrom C, Hojjat-Farsangi M, Rabbani H, Teixeira MR, Revy P, Durandy A, Zeng Y, Gennery AR, de Villartay JP, Pan-Hammarstrom Q. Cernunnos influences human immunoglobulin class switch recombination and may be associated with B cell lymphomagenesis. The Journal of experimental medicine. 2012;209:291–305. doi: 10.1084/jem.20110325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malivert L, Ropars V, Nunez M, Drevet P, Miron S, Faure G, Guerois R, Mornon JP, Revy P, Charbonnier JB, Callebaut I, de Villartay JP. Delineation of the Xrcc4-interacting region in the globular head domain of cernunnos/XLF. The Journal of biological chemistry. 2010;285:26475–26483. doi: 10.1074/jbc.M110.138156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zha S, Alt FW, Cheng HL, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 120.Lee Y, McKinnon PJ. Responding to DNA double strand breaks in the nervous system. Neuroscience. 2007;145:1365–1374. doi: 10.1016/j.neuroscience.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 121.Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2013;91:31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutation research. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 123.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 124.Andres SN, Vergnes A, Ristic D, Wyman C, Modesti M, Junop M. A human XRCC4-XLF complex bridges DNA. Nucleic acids research. 2012;40:1868–1878. doi: 10.1093/nar/gks022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, Schriemer DC, Lees-Miller SP, Tainer JA. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. The Journal of biological chemistry. 2011;286:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ropars V, Drevet P, Legrand P, Baconnais S, Amram J, Faure G, Marquez JA, Pietrement O, Guerois R, Callebaut I, Le Cam E, Revy P, de Villartay JP, Charbonnier JB. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12663–12668. doi: 10.1073/pnas.1100758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu Q, Ochi T, Matak-Vinkovic D, Robinson CV, Chirgadze DY, Blundell TL. Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions. Biochemical Society transactions. 2011;39:1387–1392. doi: 10.1042/BST0391387. suppl 1382 p following 1392. [DOI] [PubMed] [Google Scholar]
- 128.Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. The Journal of biological chemistry. 2007;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- 129.Roy S, Andres SN, Vergnes A, Neal JA, Xu Y, Yu Y, Lees-Miller SP, Junop M, Modesti M, Meek K. XRCC4’s interaction with XLF is required for coding (but not signal) end joining. Nucleic acids research. 2012;40:1684–1694. doi: 10.1093/nar/gkr1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Deriano L, Roth DB. Modernizing the Nonhomologous End-Joining Repertoire: Alternative and Classical NHEJ Share the Stage. Annual review of genetics. 2013 doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 131.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 132.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 133.Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Raval P, Kriatchko AN, Kumar S, Swanson PC. Evidence for Ku70/Ku80 association with full-length RAG1. Nucleic acids research. 2008;36:2060–2072. doi: 10.1093/nar/gkn049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, Nussenzweig A. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Molecular cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bassing CH, Alt FW. The cellular response to general and programmed DNA double strand breaks. DNA repair. 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 138.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Current biology: CB. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 139.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. The Journal of biological chemistry. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 140.Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, Nussenzweig M. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Molecular cell. 2009;34:285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 142.Maser RS, Monsen KJ, Nelms BE, Petrini JH. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Molecular and cellular biology. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. The Journal of cell biology. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Revy P, Malivert L, de Villartay JP. Cernunnos-XLF, a recently identified non-homologous end-joining factor required for the development of the immune system. Current opinion in allergy and clinical immunology. 2006;6:416–420. doi: 10.1097/01.all.0000246623.72365.43. [DOI] [PubMed] [Google Scholar]
- 145.Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic acids research. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gapud EJ, Lee BS, Mahowald GK, Bassing CH, Sleckman BP. Repair of chromosomal RAG-mediated DNA breaks by mutant RAG proteins lacking phosphatidylinositol 3-like kinase consensus phosphorylation sites. J Immunol. 2011;187:1826–1834. doi: 10.4049/jimmunol.1101388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nature reviews Molecular cell biology. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 148.Ambrose M, Gatti RA. Pathogenesis of ataxia-telangiectasia: the next generation of ATM functions. Blood. 2013;121:4036–4045. doi: 10.1182/blood-2012-09-456897. [DOI] [PMC free article] [PubMed] [Google Scholar]