Introduction
A new and exciting phase of muscle disease research has recently been entered. The application of next generation sequencing technology has spurred an unprecedented era of gene discovery for both myopathies and muscular dystrophies. Gene-based therapies for Duchenne muscular dystrophy have entered clinical trial, and several pathway-based therapies are doing so as well for a handful of muscle diseases. While many factors have aided the extraordinary developments in gene discovery and therapy development, the zebrafish model system has emerged as a vital tool in these advancements. In this review, we will highlight how the zebrafish has greatly aided in the identification of new muscle disease genes and in the recognition of novel therapeutic strategies. We will start with a general introduction to the zebrafish as a model, discuss the ways in which muscle disease can be modeled and analyzed in the fish, and conclude with observations from recent studies that highlight the power of the fish as research tool for muscle disease.
Why zebrafish?
There are numerous factors that make zebrafish a particularly appealing model organism. Such advantages have been extensively enumerated in several outstanding recent reviews, and include especially: (1) the ability to quickly generate a large number of offspring (100-300 embryos per cross), (2) the rapid ex utero development of the zebrafish, (3) the optical transparency of zebrafish embryos and larvae, (4) the ease of genetic manipulations, and (5) the ready ability of the zebrafish embryo to absorb drugs [1, 2]. In addition, zebrafish offer several specific qualities as a model for human muscle disorders. They have reproducible, quantitative and easily measured motor behaviors that are present from one day of life onward. Their skeletal muscle shares many molecular and histological features with mammalian muscle (Figure 1A-C), and appears nearly identical to human muscle at the ultrastructural level. This includes preservation of the components of the dystrophin-associated glycoprotein complex, the excitation-contraction coupling machinery, and of the contractile apparatus, three fundamental muscle structures that are key to studying and understanding human muscle disease pathogenesis [3-7]. In addition, skeletal muscle is the largest and most prominent organ system of the developing zebrafish and, due to the optical transparency of the zebrafish, can be easily visualized in live animals. Finally, the phenotypes of several zebrafish models more closely approximate the severity of the human clinical presentation than corresponding mouse models [8-10]. For example, dystrophin-deficient zebrafish exhibit a severe motor phenotype by 4 days of life and die by 2 weeks of life, while dystrophin-deficient mdx mice have a very mild phenotype that only moderately impacts muscle function and survival [11, 12].
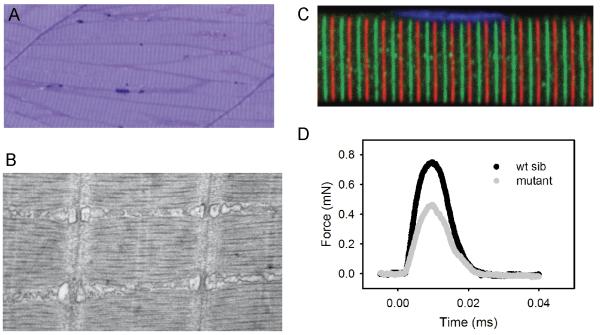
Figure 1. Properties of zebrafish skeletal muscle.
(A) Toluidine blue stained semi thin longitudinal section from a 3 day old zebrafish. Note the obvious striations corresponding to the Z lines as well as the rare, eccentric nuclei. The thick vertical lines connecting the myofibers are the myotendinous junctions. (20×) (B) Electron microscopy of 7 day old zebrafish skeletal muscle. All of the salient features of mammalian muscle (Z bands, sarcomeres, thin and thick filaments, triad) are observed in fish muscle at this age. (10,500×) (C) Immunostaining of an isolated myofiber from a 3 day old zebrafish. Green is tropomodulin and red is a actinin. This co-stain highlights the ability to detect and visualize common markers of skeletal muscle structure. (120×) (D) Maximal force generation curve for wild type (black) and candyfloss (gray) embryos. These curves recapitulate force curves measured in mouse muscle and human myofibers.
How are zebrafish models of muscle disease made?
There are 3 primary techniques for generating zebrafish models of muscle disease, with the basic overall strategy centered on manipulating a gene in a manner analogous to what is observed in human patients. Morpholino-based gene knockdown is a rapid but transient strategy, forward genetic screens identify germline mutations primarily based on muscle-related phenotypes, and genome editing techniques create targeted mutations in specific genes of interest. Despite some limitations, particularly related to recapitulating dominant mutations and specific point mutations, these techniques have been used with great success to model numerous muscle diseases (Table I).
TABLE I.
| Human gene |
Zebrafish gene |
Zebrafish model | Reference | Human condition |
|---|---|---|---|---|
| DMD | dmd | morpholino mutant (sapje) mutant (sapje-like) |
Guyon 2003 Bassett 2003; Berger2011 Guyon 2009 |
Duchenne muscular dystrophy |
| LAMA2 | Iama2 | mutant (caf) mutant (cl501) |
Hall 2007 Gupta 2012 |
Congenital muscular dystrophy (MDC1A) |
| ITGA7 | itga7 | morpholino | Postel 2008 | Congenital muscular dystrophy |
| LMNA | Imna | morpholino | Vogel et al 2009; Koshimizu 2011 | Multiple forms of dystrophy |
| COL6A1 | col6a1 | morpholino | Telfer 2010 | Ullrich congenital muscular dystrophy; Bethlem myopathy |
| COL6A3 | col6a3 | morpholino | Telfer 2010 | Ullrich congenital muscular dystrophy; Bethlem myopathy |
| DUX4 | overexpression | Wallace 2011; Mitsuhashi 2013 | Facioscapulohumeral muscular dystrophy | |
| DNAJB6 | dnajb6b | morpholino | Sarparanta 2012 | Limb-girdle muscular dystrophy |
| DAG1 | dag1 | morpholino mutant |
Parsons 2002 Gupta 2011 |
Limb-girdle muscular dystropy (MDDGC9) |
| SGCD | sgcd | morpholino | Guyon 2005; Cheng 2006; Vogel 2009 | Limb-girdle muscular dystrophy |
| TCAP | tcap | morpholino | Vogel 2009; Zhang 2009 | Limb-gidle muscular dystrophy |
| DUX4 | overexpression | Mitsuhashi 2013 | Facioscapulohumeral muscular dystrophy | |
| DYSF | dysf | morpholino | Kawahara 2011; Roostalu 2012 | Myoshi myopathy; limb-girdle muscular dystrophy |
| POMT1 | pomtl | morpholino | Avsar-Ban 2010 | Multiple forms of dystroglycanopathy |
| POMT2 | pomt2 | morpholino | Avsar-Ban 2010 | Multiple forms of dystroglycanopathy |
| FKRP | fkrp | morpholino | Thornhill 2008; Karahara 2010; Lin 2011; Wood 2011 |
Multiple forms of dystroglycanopathy |
| FKTN | fktn | morpholino | Lin 2011; Wood 2011 | Multiple forms of dystroglycanopathy |
| ISPD | ispd | morpholino | Roscioli 2012 | Multiple forms of dystroglycanopathy |
| B3GNT1 | b3gnt1 | morpholino | Buysse 2013 | Walker-Warburg syndrome |
| GTDC2 | gtdc2 | morpholino | Manzini 2012 | Walker-Warburg syndrome |
| MTM1 | mtml | morpholino | Dowling 2009 | Myotubular myopathy |
| DNM2 | overexpression | Gibbs 2013 | Centronuclear myopathy | |
| CCDC78 | ccdc78 | morpholino | Majczenko 2012 | Congenital myopathy with internal nuclei and cores |
| NEB | neb | morpholino | Telfer 2012 | Nemaline myopathy |
| RYR1 | ryrlb | mutant (ryr) | Hirata 2007 | Multiple forms of congenital myopathy |
| SEPN1 | sepnl | morpholino | Deniziak 2007; Jurynec 2008 | Multiple forms of congenital myopathy |
| DES |
desma
desmb |
morpholino morpholino |
Vogel 2009; Li 2013 Li 2013 |
Myofibrillar myopathy |
| CAV3 | cav3 | morpholino | Nixon 2005 | Caveolinopathies |
Morpholino-mediated knockdown has emerged as an exceptionally useful technology, particularly for examining gene loss-of-function in zebrafish embryos (although some dominant muscle diseases have been modeled as well with morpholinos). This is largely because of the rapidity by which “morphant” zebrafish can be created and analyzed. Morpholinos are synthetic oligonucleotides similar to RNA, with an uncharged morpholino ring in place of a ribose. When targeted against the start codon or splice sites of a gene of interest, morpholinos can transiently reduce or eliminate protein expression by inhibiting translation or pre-mRNA processing. Morpholinos provide a means to knockdown gene expression quickly and with relatively low cost. Most notably, as discussed below, morpholino-mediated gene manipulation has emerged as a vital approach for validating new gene mutations discovered by next generation sequencing [4, 5, 13].
Because of their widespread use, it is important to point out some of the caveats associated with morpholinos in zebrafish. Many genes are duplicated in the teleost genome, making it critical to carefully look for multiple putative orthologs to a human gene of interest when designing a knockdown experiment. Morpholino knockdown is transient, and the efficacy of morpholinos typically fades by 3 days post fertilization. Gene knockdown may only be partial in many cases, making it important to confirm loss of expression using western blot or antibody staining. Additionally, morpholinos may have off-target effects. To ensure that a phenotype is gene-specific, careful validation of morpholino knockdown is necessary. Controls for morpholino experiments include injecting a mismatched control, using multiple morpholinos targeted to the same gene, and rescuing a morpholino phenotype with co-injection of capped mRNA.
Forward genetic screens have proven to be an excellent source of zebrafish mutants that model human disease. The first muscle mutants emerged from the original ENU mutagenesis-based screen by Nüsslein-Vollhard and colleagues, and include the first identified zebrafish mutant strains for dystrophin (sapje), titin (runzel), and laminin α2 (candyfloss) [14, 15]. More recently, ENU and transposon-based screens have been performed by Kunkel and Beggs (using birefringence as a screen) and by Kuwada and colleagues (using impaired motility as the output), and have resulted in the identification of several strains with mutations in muscular dystrophy and congenital myopathy genes [16-19]. Finally, the Wellcome Trust Sanger Institute has created an emerging source of genetic mutants based on a saturating ENU mutagenesis screen (http://www.sanger.ac.uk/Projects/D_rerio/zmp/). Their mutation resource contains a large bank of strains with mutations that are identified based on a gene sequencing strategy called TILLING. By using TILLING, they have identified mutations in several genes associated with human muscle disease, though only a few resulting mutant strains have been characterized (including nebhu2849 and dag1hu3072) and the majority of the strains have yet to be analyzed [20, 21].
Until recently, there has not been a method for the targeted generation of stable zebrafish knockouts. However, new advances in genome editing technologies have made it possible to generate gene-specific lesions in the zebrafish genome. Two of these technologies, zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), utilize the non-specific cleavage domain of the FokI endonuclease to introduce a double-stranded break in genomic DNA. In the case of ZFNs, these cleavage domains are targeted to a precise genomic location by a pair of DNA-binding zinc finger domains assemblies [22]. TALENs are a similar form of genomic editing that utilize DNA-binding domains from transcription activator-like (TAL) effectors, and Ekker and colleagues have developed a publicly-available TALEN toolkit for targeting the zebrafish genome [23, 24]. Both ZFNs and TALENs have been successfully used to generate somatic and germline mutations in zebrafish embryos; however, efficiently engineering a target-specific domain remains a bottleneck with both technologies. A third technology utilizing bacterial CRISPR (clustered regularly interspaced short palindromic repeats) offers a promising alternative approach for inducing targeted lesions in genomes [25, 26]. CRISPR-based targeting is RNA-guided, bypassing some of the technical challenges involved in assembling appropriately-targeted ZFNs or TALENs. In zebrafish embryos, CRISPR-based has been shown to induce targeted gene lesions in somatic cells with high efficiency [27, 28]. These new genome editing technologies open up exciting new avenues for zebrafish research, and while muscle disease mutants have yet to be published, they promise ultimately to be a transformative technology in the generation of zebrafish models.
How are zebrafish models of muscle disease analyzed?
As the number of zebrafish models for muscle disease expands, a useful repertoire of techniques have been in parallel been developed for the analysis of muscle structure and function in zebrafish. First and foremost, developing zebrafish embryos exhibit highly stereotyped motor behaviors that can be easily measured without the need for special equipment [29]. Zebrafish embryos begin coiling in a steady rhythmic pattern independent of stimulation within the first day of life, providing a simple measure of early slow muscle dependent muscle function (Video 1) [7, 9, 20]. By two days post fertilization, zebrafish embryos have developed a robust escape response, rapidly swimming away from mechanical stimuli (Video 2). Beginning at approximately 3 days post fertilization, larvae begin swimming spontaneously and swim behaviors such as velocity and turn frequency can be precisely quantified, especially through the use of proprietary imaging systems like the Noldus DanioVision or the Viewpoint Zebrabox. Together, these sequential motor behaviors provide simple and non-invasive measures of muscle function, and can thus serve as a platform for assessing pharmacologic and genetic interventions.
A number of zebrafish muscle studies have utilized electrophysiological and biomechanical measurements adapted from other model organisms. Several studies have used in vivo patch clamp recording of larval skeletal muscle to establish whether a motor defect is caused by defects in the central nervous system, the neuromuscular junction or in the muscle fiber itself. To characterize the relatively relaxed and accordian behavioral mutants, Kuwada and colleagues analyzed the fictive swimming response from both mutants and demonstrated normal rhythmic input to the muscle in response to tactile stimulation, showing that the primary functional defect must be downstream of myofiber excitation from the nervous system and NMJ [5, 30]. Similarly, injecting current into zebrafish muscle can evoke contractions, and this technique has been used to measure muscle fatigue and relaxation in models of muscle disease [7, 30, 31]. To directly measure muscle force generation, Brooks and colleagues developed a novel system adapted from studies of murine muscle (Figure 1D). Using this system, they showed that a zebrafish model of nemaline myopathy exhibits defects in contractile properties nearly identical to those observed in nebulin knockout mice and patient myofibers [20].
In addition to these functional measures of muscle output, the striking similarities between fish and mammalian muscle has allowed important insights to be gained from the analysis of zebrafish muscle using both standard light microscopic analysis and transmission electron microscopy. Skeletal muscle can be readily visualized in whole zebrafish using immunohistochemistry or by using fluorescent markers driven by muscle-specific promoters. Furthermore, the transparency of zebrafish embryos permits elegant optical assays of muscle structure and function that would not be possible in mammalian systems. Roostalu and colleagues recently used high-resolution imaging to visualize real-time membrane resealing in myofibers from whole embryos, elucidating a sequential repair program that occurs following sarcolemmal damage [32]. In vivo calcium imaging is another optical assay that has provided insight into the nature of muscle defects. Calcium flux in zebrafish myofibers is monitored in vivo by injecting embryos with chemical (such as dextran green) or genetically-encoded calcium indicators (such as Gcamp3), which can reveal defects in calcium release or uptake [5, 30].
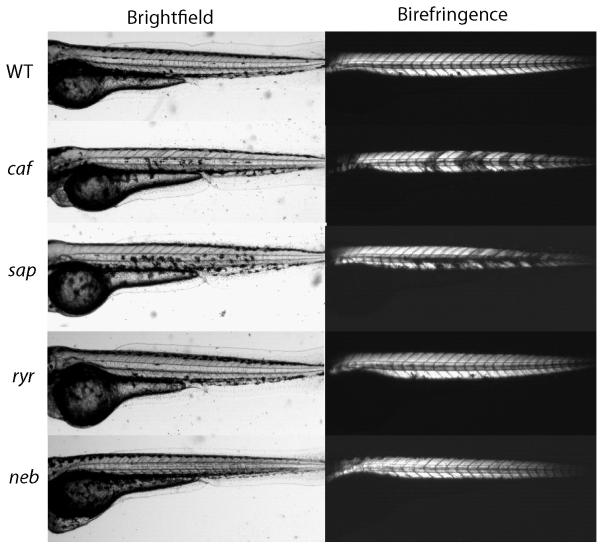
One notable observation in developing zebrafish muscle is that myofiber integrity can be easily viewed using an optical phenomenon called birefringence. When viewed under polarized light, detached myofibers appear as dark patches in the skeletal muscle of zebafish embryos (Figure 2). This phenomenon can be directly quantified to measure loss of muscle integrity in dystrophic larval muscle, and it has been particularly useful in identifying novel muscle mutants in forward screens and for testing therapies in known dystrophy models [14, 33, 34]. Figure 2 depicts birefringence patterns for several well-characterized mutants. This simple visual assay can be combined with other measures of muscle integrity to provide substantial insight into the dynamics of myofiber damage, retraction and death in dystrophic mutants. Evans blue dye, a small molecule that only enters damaged myofibers, can be injected into zebrafish larvae to assay for sarcolemmal damage. Currie and colleagues have visualized myofiber detachment in real time in candyfloss and sapje dystrophic mutants by anesthetizing embryos for the first three days of life, which preserved muscle integrity by eliminating any muscle contraction during development [8, 13]. When anesthetic was washed out, fish embryos embedded in agarose or methylcellulose contract against surrounding media, inducing myofiber detachment.
Figure 2. Birefringence in representative zebrafish models of human muscle disease.
(Left) Brightfield images of the trunk and tail regions of the following zebrafish models: wild type (WT), candyfloss (lama2 mutant; caf), sapje (dystrophin mutant, sap), relatively relaxed (RYR1 mutant, ryr), and neb (nebulin mutant). (Right) birefringence images as obtained by viewing zebrafish with a plane polarizing filter. Note the patchy birefringence of caf and sap, indicative of muscle detachment in these modes, and the reduced birefringence in neb, indicative of the generalized and global sarcomeric disorganization.
For experiments that cannot easily be performed in an intact embryo, myofibers from the developing zebrafish can be isolated and maintained ex vivo (Horstick et al., in press). Such preparations contain a mixture of myofibers and other cell types, with myofibers representing the predominant cellular subtype. A range of experimentation is feasible on isolated myofibers - including immunocytochemistry, calcium imaging, and electrophysiological recording - and these preparations can allow more precise measurements of myofiber size and protein subcellular localization.
Models of muscle disorders
As summarized in Table 1, zebrafish have provided useful models for many different muscle disorders. Recent reviews, in particular by Peter Currie, have extensively summarized several of these models [10, 35]. While we present a brief listing of the major available models in Table I, this review will instead focus on a few notable studies that reflect the power of the zebrafish to provide insight into muscle disease
The zebrafish and new gene discovery: the perfect partner for next generation sequencing
It is estimated that the genetic basis of disease is not known for 25-50% of all genetic muscle disorders. This knowledge gap presents a significant barrier to clinical practice and to research into disease pathogenesis and therapy development. Next generation sequencing, including especially whole exome capture followed by massively parallel sequencing, is rapidly filling in the missing genetic pieces. A particular challenge with next generation sequencing, however, is that it identifies numerous variants of unclear significance that are not present in the existing variant databases. Given that sequencing efforts are often focused on isolated probands with a similar disease or else small families with very rare myopathies, identifying the true pathogenic variants is often difficult in the absence of supporting experimentation. The zebrafish has turned out to be the ideal system for validating novel variants, as it provides a rapid way of genetically modeling and testing a newly-identified putative mutation.
There are now multiple examples, both in the muscle literature and for other monogenetic disorders, where zebrafish modeling has proven critical for the validation of a new gene mutation. We present a recent example from our laboratory for a novel congenital myopathy, and an example from Hans van Bokhoven and colleagues describing a new gene associated with a severe form of congenital muscular dystrophy.
CCDC78 mutation as the cause of a unique congenital myopathy
We identified 5 individuals in a 3 generation dominant pedigree with moderate distal weakness, excessive fatigue, and a muscle biopsy with the unusual combination of excessive central nuclei, core-like lesions, and desmin and actin accumulations [36]. We used the combination of SNP linkage on the pedigree and whole exome sequencing of two affected individuals to identify a splice site sequence variant in the previously uncharacterized gene CCDC78. Based on RT-PCR data from patient lymphoblastoid cells, we predicted that this variant resulted in disease by allowing inclusion of an in-frame intron of CCDC78. To validate the pathogenicity of this variant, we developed a morpholino to zebrafish ccdc78 targeted the exon-intron boundary affected by the human sequence variant. Injection of this morpholino into the developing zebrafish results in aberrant processing of ccdc78 RNA, with inclusion of the in-frame intron and production of a higher molecular protein. The consequences associated with this morpholino included impaired motor function and abnormal larval morphology. Most notably, the morpholino-induced histopathological changes in the muscle, including myofibrillar disruption, triad abnormalities, and desmin and RYR1 accumulations, mirror exactly the findings observed in the patient biopsies. The similarities in both RNA processing and histopathology in the zebrafish morphants enabled us to conclude that the sequence variant in CCDC78 identified in the patients was the cause of disease.
Mutations in ISPD cause Walker-Warburg syndrome
Mutant and morphant zebrafish have provided exceptionally useful models for a clinically diverse group of muscle disorders known as dystroglycanopathies, recessive disorders characterized by reduced O-mannose glycosylation of alpha-dystroglycan (DAG1). The phenotypes of dystroglycanopathies range from relatively mild limb-girdle muscular dystrophy to Walker-Warburg syndrome, a severe form of congenital muscular dystrophy associated with eye and brain defects. DAG1 plays an essential role in mammalian development, limiting the usefulness of mouse models in studying dystroglycanopathies, and complete knockout of dystroglycan or certain dystroglycan processing genes leads to embryonic lethality in mice [37-39]. While the dag1 glycosylation pathway appears to be largely conserved in zebrafish, it is not essential for early development, and knockdown of most known dystroglycanopathy genes have been shown recapitulate many aspects of human disorders [21, 40-45].
Recently, van Bokhoven and colleagues identified mutations in the ISPD gene in individuals from 9 families with Walker-Warburg-like features [46]. To elucidate the role of ISPD in this disorder, they injected zebrafish with morpholinos to ispd. Consistent with histopathology in patient muscle and other models of dystroglycanopathy, ispd morphants exhibit hypoglycosylation of dag1. The ispd morphants also have a dystrophic muscle phenotype, and sarcolemma integrity is disrupted. In addition to muscle abnormalities, zebrafish recapitulate a severe hydrocephalus phenotype seen in patients. These findings demonstrate the value of zebrafish in validating and characterizing rare variants associated with muscle disease. Of note, morpholino knockdown has also been used to functionally validate mutations in GTDC2 and B3GNT1, two additional genes in which mutations were identified in patients with Walker-Warburg and related dystroglycanopathies. Both gtdc2 and b3gnt1 morphants exhibited severe muscle, eye and brain defects, as well as reduced glycosylation of alpha-dystroglycan [44, 45].
The zebrafish as a tool for identifying novel pathogenic mechanisms
Much of what is known concerning disease pathogenesis in most skeletal muscle disorders has been derived from the combination of cell culture studies and experimentation in murine models. The zebrafish has recently entered the arena of pathogenesis studies, and has helped reveal novel aspects of disease that were not previously detected in mice or cultured cells. Here we describe findings in two such studies and highlight the contributions they have made to the understanding of pathomechanisms in human muscle disease.
Vitamin-based rescue in a zebrafish model of muscular dystrophy
Using a dag1 morphant zebrafish model, Henry and colleagues demonstrated a remarkable rescue of dystrophic muscle by supplementation with coenzyme NAD+ [47]. In untreated dag1 morphants, muscle degeneration began at 3 days post fertilization. Morphants supplemented with NAD+ exhibited reduced fiber detachment, improved organization of the surrounding basement membrane, and increased swim speed in touch-evoked escape responses. Intriguingly, the group achieved a similar rescue by the simple addition of a vitamin supplement containing niacin, a precursor to NAD+.
To identify the mechanism underlying the protective effect of NAD+ supplementation, they used morpholinos against components of the dystrophin-glycoprotein complex or the α7β1 integrin complex, two of the major adhesion complexes in muscle. Although they demonstrated that full NAD+-mediated rescue is dependent on expression of either dag1 or integrin alpha-7, they find that an additional receptor (integrin alpha-6) is also required for rescue. These findings suggest that integrin alpha-6 may be part of a previously unrecognized integrin adhesion complex in skeletal muscle. Therefore, along with identifying a novel therapeutic strategy, this study highlights the utility of the zebrafish model in teasing out the molecular underpinnings of muscle disorders.
Oxidative stress in a zebrafish model of RYR1-related myopathies
Ryanodine receptors are calcium release channels that play a critical role in muscle excitation and contraction. Mutations in ryanodine receptor 1 (RYR1) cause a diverse range of dominant and recessive muscle disorders, including especially central core disease and minicore myopathy [48-53]. The spontaneous relatively relaxed mutant (ryr) zebrafish mutant, identified by a slow swimming phenotype, carries an insertional mutation in ryr1b, the fast-twitch fiber zebrafish ortholog to RYR1 [5]. Using a differential RNA microarray-based approach to identify expression, our laboratory found that ryr zebrafish exhibit upregulation of numerous genes associated with cellular stress, and follow up biochemical studies uncovered increased intracellular oxidant activity [54]. Based on this result, ryr zebrafish were treated with the anti-oxidant N-acetylcysteine (NAC), and treated larvae exhibited significant increases in swimming behavior and substantial improvements in muscle histopathology. These results were corroborated by Ana Ferreiro and colleagues by the observation of increased oxidative stress in myotubes derived from patients with RYR1-related myopathies, suggesting that this is a clinically relavant pathomechanism, and NAC is now considered an excellent therapeutic candidate for patients with RYR1 mutations.
Zebrafish as an ideal system for novel drug discovery
Large-scale drugs screens are not feasible in mammals due to cost and time required but are well suited to invertebrate models or to cultured cells. However, such assays in invertebrates or cells are often far removed from the human disease, and often lack functional outcome measures that provide relevance for the identification of drugs that will ultimate translate to the clinical setting. In principal, the zebrafish offers the ideal combination of invertebrate advantages (small size, rapid development, large numbers) and vertebrate biological relevancy. In practice, only one large-scale drug screen (described below) has been published to date using a zebrafish model of muscular dystrophy. However, the success of zebrafish screens in other disease spheres - particular melanoma, where a novel drug identified in a large-scale screen has successfully translated to the clinic - has engendered optimism that screens in models of muscle disease will yield similar results [55].
Kunkel and colleagues performed a large-scale drug screen in dystrophin-null zebrafish (sapje and sapje-like), using birefringence (which is a marker of muscle integrity and is markedly abnormal in sapje) as the outcome measure [56]. This straightforward read-out permitted the rapid screening of 1120 chemicals for compounds that prevented the loss of muscle organization that typically occurs in sapje during the first few days of development. In the initial screen, they tested pools of 8 chemicals on embryos from heterozygous sapje parents, with the expectation that approximately 25% of embryos would exhibit abnormal birefringence when treated with a pool of non-effective compounds. Chemicals pools that had reduced numbers of embryos with abnormal birefringence (≤7.5%) were then separated and individually screened to determine which compounds were associated with the decrease. Using this two-step screening strategy, they identified 7 compounds that preserved muscle integrity in sapje mutants. They further demonstrated that long-term treatment with one compound, a phosphodiesterase (PDE) inhibitor, was able to both extend survival and restore muscle integrity in older affected sapje zebrafish. Given that PDE inhibitors have been shown to modulate disease in mouse models of DMD and in DMD patient myotubes, this result provides important proof-of-principle of the suitability of zebrafish as a tool for drug development [57, 58]. In addition, the study highlights the tremendous potential of the zebrafish in large-scale drug screens, and provides a path forward for drug discovery in zebrafish models of muscle disease.
Conclusions and Prospective
In this review, we have outlined the utility of the zebrafish as a model for human muscle diseases and have highlighted several recent publications that illuminate the power of the model for gene discovery and therapy development. At present, the most important area of impact for the fish model has been as a tool for validating new mutations discovered by next generation sequencing, and we predict that such experimentation will become increasingly important as whole exome and genome sequencing are applied more extensively to isolated probands and individual families. An ongoing challenge related to the application of zebrafish to mutation validation is the proper use of the system. We believe the strongest and most support outcomes to measure in the fish are those that reflect on specific features of the human disease, such as dystrophic changes (like patchy birefringence) or specific histopathologic features (like rods or actinin accumulations in models of nemaline myopathy), and not on non specific outcomes like motor function and overall fish morphology. In addition, while speed is often desired for new mutation validation, the need for well considered and controlled zebrafish experiments is paramount to reduce the chance that non-pathogenic variants are falsely considered disease causing.
Moving forward, perhaps the most exciting area for fish research relates to therapy development, as the ability to perform large-scale chemical and/or genetic screens in the zebrafish has amazing potential. To date, research has only scratched the surface of this potential, and it is exciting to speculate that such experimentation, because it is based on whole organism screening, will yield unexpected disease insights and novel therapeutic targets. The true translatability of the zebrafish, however, is still an unknown, and will not truly be understood until targets first identified in the fish are brought to the clinical trial arena.
In all, we anticipate widespread and increased use of the zebrafish to study muscle disease and to develop therapies, as more and more researchers discover the ease of the use of the system, the facility for genetic manipulation, and the power of the muscle related outcomes it provides.
Supplementary Material
Acknowledgements
This work was supported in part by the National Institutes of Health (K08AR054835 to JJD), the Muscular Dystrophy Association (MDA 186999 to JJD), and the Taubman Medical Institute at the University of Michigan.
Footnotes
SUPPORTING INFORMATION
Supplemental Video 1 (coiling video): Spontaneous coiling in a 1 day old embryo
Supplemental Video 2 (escape video): Touch evoked escape response in a 3 day old embryo
REFERENCES
- 1.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. doi: nrg2091 [pii] 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 2.Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. doi: 60434 [pii] 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons MJ, Campos I, Hirst EM, Stemple DL. Removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos. Development. 2002;129:3505–3512. doi: 10.1242/dev.129.14.3505. [DOI] [PubMed] [Google Scholar]
- 4.Guyon JR, Mosley AN, Zhou Y, O’Brien KF, Sheng X, Chiang K, Davidson AJ, Volinski JM, Zon LI, Kunkel LM. The dystrophin associated protein complex in zebrafish. Hum Mol Genet. 2003;12:601–615. [PubMed] [Google Scholar]
- 5.Hirata H, Watanabe T, Hatakeyama J, Sprague SM, Saint-Amant L, Nagashima A, Cui WW, Zhou W, Kuwada JY. Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development. 2007;134:2771–2781. doi: 10.1242/dev.004531. doi: dev.004531 [pii] 10.1242/dev.004531. [DOI] [PubMed] [Google Scholar]
- 6.Dou Y, Andersson-Lendahl M, Arner A. Structure and function of skeletal muscle in zebrafish early larvae. J Gen Physiol. 2008;131:445–453. doi: 10.1085/jgp.200809982. doi: jgp.200809982 [pii] 10.1085/jgp.200809982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowling JJ, Vreede AP, Low SE, Gibbs EM, Kuwada JY, Bonnemann CG, Feldman EL. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5:e1000372. doi: 10.1371/journal.pgen.1000372. doi: 10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger J, Berger S, Hall TE, Lieschke GJ, Currie PD. Dystrophin-deficient zebrafish feature aspects of the Duchenne muscular dystrophy pathology. Neuromuscul Disord. 2010;20:826–832. doi: 10.1016/j.nmd.2010.08.004. doi: S0960-8966(10)00591-2 [pii] 10.1016/j.nmd.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Telfer WR, Busta AS, Bonnemann CG, Feldman EL, Dowling JJ. Zebrafish models of collagen VI-related myopathies. Hum Mol Genet. 2010;19:2433–2444. doi: 10.1093/hmg/ddq126. doi: 10.1093/hmg/ddq126 ddq126 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger J, Currie PD. Zebrafish models flex their muscles to shed light on muscular dystrophies. Dis Model Mech. 2012;5:726–732. doi: 10.1242/dmm.010082. doi: 5/6/726 [pii] 10.1242/dmm.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassett DI, Bryson-Richardson RJ, Daggett DF, Gautier P, Keenan DG, Currie PD. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130:5851–5860. doi: 10.1242/dev.00799. doi: 10.1242/dev.00799 130/23/5851 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 13.Hall TE, Bryson-Richardson RJ, Berger S, Jacoby AS, Cole NJ, Hollway GE, Berger J, Currie PD. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc Natl Acad Sci U S A. 2007;104:7092–7097. doi: 10.1073/pnas.0700942104. doi: 0700942104 [pii] 10.1073/pnas.0700942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- 15.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Gupta V, Kawahara G, Gundry SR, Chen AT, Lencer WI, Zhou Y, Zon LI, Kunkel LM, Beggs AH. The zebrafish dag1 mutant: a novel genetic model for dystroglycanopathies. Hum Mol Genet. 2011;20:1712–1725. doi: 10.1093/hmg/ddr047. doi: ddr047 [pii] 10.1093/hmg/ddr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta VA, Kawahara G, Myers JA, Chen AT, Hall TE, Manzini MC, Currie PD, Zhou Y, Zon LI, Kunkel LM, et al. A splice site mutation in laminin-alpha2 results in a severe muscular dystrophy and growth abnormalities in zebrafish. PLoS One. 2012;7:e43794. doi: 10.1371/journal.pone.0043794. doi: 10.1371/journal.pone.0043794 PONE-D-12-12852 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirata H, Wen H, Kawakami Y, Naganawa Y, Ogino K, Yamada K, Saint-Amant L, Low SE, Cui WW, Zhou W, et al. Connexin 39.9 protein is necessary for coordinated activation of slow-twitch muscle and normal behavior in zebrafish. J Biol Chem. 2012;287:1080–1089. doi: 10.1074/jbc.M111.308205. doi: M111.308205 [pii] 10.1074/jbc.M111.308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saint-Amant L, Sprague SM, Hirata H, Li Q, Cui WW, Zhou W, Poudou O, Hume RI, Kuwada JY. The zebrafish ennui behavioral mutation disrupts acetylcholine receptor localization and motor axon stability. Dev Neurobiol. 2008;68:45–61. doi: 10.1002/dneu.20569. doi: 10.1002/dneu.20569. [DOI] [PubMed] [Google Scholar]
- 20.Telfer WR, Nelson DD, Waugh T, Brooks SV, Dowling JJ. Neb: a zebrafish model of nemaline myopathy due to nebulin mutation. Dis Model Mech. 2012;5:389–396. doi: 10.1242/dmm.008631. doi: dmm.008631 [pii] 10.1242/dmm.008631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YY, White RJ, Torelli S, Cirak S, Muntoni F, Stemple DL. Zebrafish Fukutin family proteins link the unfolded protein response with dystroglycanopathies. Hum Mol Genet. 2011;20:1763–1775. doi: 10.1093/hmg/ddr059. doi: 10.1093/hmg/ddr059 ddr059 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekker SC. Zinc finger-based knockout punches for zebrafish genes. Zebrafish. 2008;5:121–123. doi: 10.1089/zeb.2008.9988. doi: 10.1089/zeb.2008.9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. doi: nbt.1934 [pii] 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. doi: nature11537 [pii] 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. doi: science.1231143 [pii] 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. doi: science.1232033 [pii] 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. doi: nbt.2501 [pii] 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. doi: cr201345 [pii] 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saint-Amant L, Drapeau P. Time course of the development of motor behaviors in the zebrafish embryo. J Neurobiol. 1998;37:622–632. doi: 10.1002/(sici)1097-4695(199812)37:4<622::aid-neu10>3.0.co;2-s. doi: 10.1002/(SICI)1097-4695(199812)37:4<622::AID-NEU10>3.0.CO;2-S [pii] [DOI] [PubMed] [Google Scholar]
- 30.Hirata H, Saint-Amant L, Waterbury J, Cui W, Zhou W, Li Q, Goldman D, Granato M, Kuwada JY. accordion, a zebrafish behavioral mutant, has a muscle relaxation defect due to a mutation in the ATPase Ca2+ pump SERCA1. Development. 2004;131:5457–5468. doi: 10.1242/dev.01410. doi: dev.01410 [pii] 10.1242/dev.01410. [DOI] [PubMed] [Google Scholar]
- 31.Buss RR, Drapeau P. Physiological properties of zebrafish embryonic red and white muscle fibers during early development. J Neurophysiol. 2000;84:1545–1557. doi: 10.1152/jn.2000.84.3.1545. [DOI] [PubMed] [Google Scholar]
- 32.Roostalu U, Strahle U. In vivo imaging of molecular interactions at damaged sarcolemma. Dev Cell. 2012;22:515–529. doi: 10.1016/j.devcel.2011.12.008. doi: S1534-5807(11)00574-0 [pii] 10.1016/j.devcel.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Berger J, Sztal T, Currie PD. Quantification of birefringence readily measures the level of muscle damage in zebrafish. Biochem Biophys Res Commun. 2012;423:785–788. doi: 10.1016/j.bbrc.2012.06.040. doi: S0006-291X(12)01128-X [pii] 10.1016/j.bbrc.2012.06.040. [DOI] [PubMed] [Google Scholar]
- 34.Guyon JR, Goswami J, Jun SJ, Thorne M, Howell M, Pusack T, Kawahara G, Steffen LS, Galdzicki M, Kunkel LM. Genetic isolation and characterization of a splicing mutant of zebrafish dystrophin. Hum Mol Genet. 2009;18:202–211. doi: 10.1093/hmg/ddn337. doi: ddn337 [pii] 10.1093/hmg/ddn337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin YY. Muscle diseases in the zebrafish. Neuromuscul Disord. 2012;22:673–684. doi: 10.1016/j.nmd.2012.04.007. doi: S0960-8966(12)00124-1 [pii] 10.1016/j.nmd.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Majczenko K, Davidson AE, Camelo-Piragua S, Agrawal PB, Manfready RA, Li X, Joshi S, Xu J, Peng W, Beggs AH, et al. Dominant mutation of CCDC78 in a unique congenital myopathy with prominent internal nuclei and atypical cores. Am J Hum Genet. 2012;91:365–371. doi: 10.1016/j.ajhg.2012.06.012. doi: S0002-9297(12)00321-7 [pii] 10.1016/j.ajhg.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson RA, Henry MD, Daniels KJ, Hrstka RF, Lee JC, Sunada Y, Ibraghimov-Beskrovnaya O, Campbell KP. Dystroglycan is essential for early embryonic development: disruption of Reichert’s membrane in Dag1-null mice. Hum Mol Genet. 1997;6:831–841. doi: 10.1093/hmg/6.6.831. doi: dda114 [pii] [DOI] [PubMed] [Google Scholar]
- 38.Willer T, Prados B, Falcon-Perez JM, Renner-Muller I, Przemeck GK, Lommel M, Coloma A, Valero MC, de Angelis MH, Tanner W, et al. Targeted disruption of the Walker-Warburg syndrome gene Pomt1 in mouse results in embryonic lethality. Proc Natl Acad Sci U S A. 2004;101:14126–14131. doi: 10.1073/pnas.0405899101. doi: 10.1073/pnas.0405899101 0405899101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurahashi H, Taniguchi M, Meno C, Taniguchi Y, Takeda S, Horie M, Otani H, Toda T. Basement membrane fragility underlies embryonic lethality in fukutin-null mice. Neurobiol Dis. 2005;19:208–217. doi: 10.1016/j.nbd.2004.12.018. doi: S0969-9961(05)00002-1 [pii] 10.1016/j.nbd.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Thornhill P, Bassett D, Lochmuller H, Bushby K, Straub V. Developmental defects in a zebrafish model for muscular dystrophies associated with the loss of fukutin-related protein (FKRP) Brain. 2008;131:1551–1561. doi: 10.1093/brain/awn078. doi: awn078 [pii] 10.1093/brain/awn078. [DOI] [PubMed] [Google Scholar]
- 41.Kawahara G, Guyon JR, Nakamura Y, Kunkel LM. Zebrafish models for human FKRP muscular dystrophies. Hum Mol Genet. 2010;19:623–633. doi: 10.1093/hmg/ddp528. doi: ddp528 [pii] 10.1093/hmg/ddp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore CJ, Goh HT, Hewitt JE. Genes required for functional glycosylation of dystroglycan are conserved in zebrafish. Genomics. 2008;92:159–167. doi: 10.1016/j.ygeno.2008.05.008. doi: S0888-7543(08)00118-3 [pii] 10.1016/j.ygeno.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Avsar-Ban E, Ishikawa H, Manya H, Watanabe M, Akiyama S, Miyake H, Endo T, Tamaru Y. Protein O-mannosylation is necessary for normal embryonic development in zebrafish. Glycobiology. 2010;20:1089–1102. doi: 10.1093/glycob/cwq069. doi: cwq069 [pii] 10.1093/glycob/cwq069. [DOI] [PubMed] [Google Scholar]
- 44.Manzini MC, Tambunan DE, Hill RS, Yu TW, Maynard TM, Heinzen EL, Shianna KV, Stevens CR, Partlow JN, Barry BJ, et al. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am J Hum Genet. 2012;91:541–547. doi: 10.1016/j.ajhg.2012.07.009. doi: S0002-9297(12)00366-7 [pii] 10.1016/j.ajhg.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buysse K, Riemersma M, Powell G, van Reeuwijk J, Chitayat D, Roscioli T, Kamsteeg EJ, van den Elzen C, van Beusekom E, Blaser S, et al. Missense mutations in beta-1,3-N-acetylglucosaminyltransferase 1 (B3GNT1) cause Walker-Warburg syndrome. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt021. doi: ddt021 [pii] 10.1093/hmg/ddt021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roscioli T, Kamsteeg EJ, Buysse K, Maystadt I, van Reeuwijk J, van den Elzen C, van Beusekom E, Riemersma M, Pfundt R, Vissers LE, et al. Mutations in ISPD cause Walker-Warburg syndrome and defective glycosylation of alpha-dystroglycan. Nat Genet. 2012;44:581–585. doi: 10.1038/ng.2253. doi: ng.2253 [pii] 10.1038/ng.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goody MF, Kelly MW, Reynolds CJ, Khalil A, Crawford BD, Henry CA. NAD+ biosynthesis ameliorates a zebrafish model of muscular dystrophy. PLoS Biol. 2012;10:e1001409. doi: 10.1371/journal.pbio.1001409. doi: 10.1371/journal.pbio.1001409 PBIOLOGY-D-12-01746 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Chen HS, Khanna VK, De Leon S, Phillips MS, Schappert K, Britt BA, Browell AK, MacLennan DH. A mutation in the human ryanodine receptor gene associated with central core disease. Nat Genet. 1993;5:46–50. doi: 10.1038/ng0993-46. doi: 10.1038/ng0993-46. [DOI] [PubMed] [Google Scholar]
- 49.Jungbluth H, Zhou H, Sewry CA, Robb S, Treves S, Bitoun M, Guicheney P, BujBello A, Bonnemann C, Muntoni F. Centronuclear myopathy due to a de novo dominant mutation in the skeletal muscle ryanodine receptor (RYR1) gene. Neuromuscul Disord. 2007;17:338–345. doi: 10.1016/j.nmd.2007.01.016. doi: S0960-8966(07)00021-1 [pii] 10.1016/j.nmd.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 50.Wilmshurst JM, Lillis S, Zhou H, Pillay K, Henderson H, Kress W, Muller CR, Ndondo A, Cloke V, Cullup T, et al. RYR1 mutations are a common cause of congenital myopathies with central nuclei. Ann Neurol. 2010;68:717–726. doi: 10.1002/ana.22119. doi: 10.1002/ana.22119. [DOI] [PubMed] [Google Scholar]
- 51.Bevilacqua JA, Monnier N, Bitoun M, Eymard B, Ferreiro A, Monges S, Lubieniecki F, Taratuto AL, Laquerriere A, Claeys KG, et al. Recessive RYR1 mutations cause unusual congenital myopathy with prominent nuclear internalization and large areas of myofibrillar disorganization. Neuropathol Appl Neurobiol. 2011;37:271–284. doi: 10.1111/j.1365-2990.2010.01149.x. doi: 10.1111/j.1365-2990.2010.01149.x. [DOI] [PubMed] [Google Scholar]
- 52.Dowling JJ, Lillis S, Amburgey K, Zhou H, Al-Sarraj S, Buk SJ, Wraige E, Chow G, Abbs S, Leber S, et al. King-Denborough syndrome with and without mutations in the skeletal muscle ryanodine receptor (RYR1) gene. Neuromuscul Disord. 2011;21:420–427. doi: 10.1016/j.nmd.2011.03.006. doi: S0960-8966(11)00074-5 [pii] 10.1016/j.nmd.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 53.Monnier N, Ferreiro A, Marty I, Labarre-Vila A, Mezin P, Lunardi J. A homozygous splicing mutation causing a depletion of skeletal muscle RYR1 is associated with multi-minicore disease congenital myopathy with ophthalmoplegia. Hum Mol Genet. 2003;12:1171–1178. doi: 10.1093/hmg/ddg121. [DOI] [PubMed] [Google Scholar]
- 54.Dowling JJ, Arbogast S, Hur J, Nelson DD, McEvoy A, Waugh T, Marty I, Lunardi J, Brooks SV, Kuwada JY, et al. Oxidative stress and successful antioxidant treatment in models of RYR1-related myopathy. Brain. 2012;135:1115–1127. doi: 10.1093/brain/aws036. doi: 10.1093/brain/aws036 aws036 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. doi: nature09882 [pii] 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawahara G, Karpf JA, Myers JA, Alexander MS, Guyon JR, l LM. Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2011;108:5331–5336. doi: 10.1073/pnas.1102116108. doi: 1102116108 [pii] 10.1073/pnas.1102116108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adamo CM, Dai DF, Percival JM, Minami E, Willis MS, Patrucco E, Froehner SC, Beavo JA. Sildenafil reverses cardiac dysfunction in the mdx mouse model of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2010;107:19079–19083. doi: 10.1073/pnas.1013077107. doi: 1013077107 [pii] 10.1073/pnas.1013077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Percival JM, Whitehead NP, Adams ME, Adamo CM, Beavo JA, Froehner SC. Sildenafil reduces respiratory muscle weakness and fibrosis in the mdx mouse model of Duchenne muscular dystrophy. J Pathol. 2012;228:77–87. doi: 10.1002/path.4054. doi: 10.1002/path.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.