Abstract
Single stranded DNA binding (SSB) proteins play central roles in genome maintenance in all organisms. Plasmodium falciparum, the causative agent of malaria, encodes an SSB protein that localizes to the apicoplast and likely functions in the replication and maintenance of its genome. Pf-SSB shares a high degree of sequence homology with bacterial SSB proteins, but differs in the composition of its C-terminus, which in E. coli SSB interacts with more than a dozen other proteins. Using sedimentation methods we show that Pf-SSB forms a stable homo-tetramer alone and when bound to single stranded DNA. We also present a crystal structure at 2.1 Å resolution of the Pf-SSB tetramer bound to two (dT)35 molecules. The Pf-SSB tetramer is structurally similar to the E. coli SSB tetramer and ssDNA wraps completely around the tetramer with a “baseball seam” topology that is similar to E. coli SSB in its “65 binding mode”. However, the polarity of the ssDNA wrapping around Pf-SSB is opposite to that observed for E. coli SSB. The interactions between the bases in the DNA and the amino acids side chains also differ from those observed in the E. coli SSB-DNA structure suggesting that other differences may exist in the DNA binding properties of these structurally similar proteins.
Keywords: DNA repair, recombination, replication, structure, Plasmodium, malaria
Introduction
Plasmodium falciparum is a eukaryotic parasite and the causative agent for over 250 million cases of malaria that result in five million deaths annually1. It contains a unique non-photosynthetic plastid-like organelle called the apicoplast, which is involved in a variety of biosynthetic pathways of the parasite. A single apicoplast is present in each cell and functions in isoprenoid, fatty acid and heme synthesis/metabolism and is critical to parasite survival and pathogenesis making it a logical target for anti-malarial drugs. The ~35 kb apicoplast genome contains 68 open reading frames which encode a variety of ribosomal proteins, tRNAs, RNA polymerase, chaperones and other proteins of unknown function2. However, proteins involved in DNA metabolism are encoded by the nuclear DNA and targeted for transport to the apicoplast by an apicoplast localization signal (ALS) which is cleaved upon delivery to the apicoplast3. The single stranded (ss) DNA binding (SSB) protein from P. falciparum (Pf-SSB) is encoded in the nucleus and transported to the apicoplast where it likely functions in the replication and maintenance of the apicoplast genome4.
SSB proteins are present in nearly all organisms and bind to ssDNA intermediates produced transiently during replication, repair and recombination. E. coli SSB (Ec-SSB) is a well characterized prototype of bacterial SSB proteins5 and shares high sequence homology with Pf-SSB (39 % identity and 66 % homology)4. Ec-SSB functions as a homo-tetramer with each subunit consisting of two domains, an N-terminal OB-fold containing the ssDNA binding site and an unstructured C-terminal tail. Ec-SSB also interacts with more than a dozen other proteins involved in DNA metabolism8. These interactions are primarily mediated through a conserved stretch of 8–10 amino acids located at the end of its unstructured C-termini8.
In the case of Ec-SSB, it has been shown that at moderate to high salt concentrations a ssDNA ~65 nucleotides long can fully wrap around the tetrameric DNA binding core to form the so-called (SSB)65 binding mode11. However, due to its four potential DNA binding sites, Ec-SSB can also bind to long ssDNA in a number of different binding modes that differ by the number of subunits (OB-folds) within the tetramer that contact the DNA. In the (SSB)65 mode ssDNA interacts with all four subunits and displays little tendency to form cooperative clusters along ssDNA. The low cooperative, fully wrapped (SSB)65 binding mode has been proposed to facilitate RecA mediated DNA strand exchange during homologous recombination. In fact, Ec-SSB, while bound in its (SSB)65 mode is able to diffuse along ssDNA and transiently melt DNA hairpins, thus facilitating RecA filament formation along ssDNA18. Here we present a structural study of the Pf-SSB protein and its complexes with ssDNA including a crystal structure of a Pf-SSB tetramer in complex with ssDNA in a fully wrapped binding mode allowing a detailed comparison of its structure with Ec-SSB.
Results
Pf-SSB forms a stable homo-tetramer in solution
SSB proteins can exist in a variety of oligomeric states including monomers (e.g., T4 phage gp32)19, dimers (e.g., D. radiodurans SSB)20, trimers (e.g., eukaryotic RPA)21, tetramers (most bacterial SSBs)8 and pentamers (e.g., D. radiodurans DdrB)22. Based on dynamic light scattering and sucrose density gradient analysis, a histidine tagged version of recombinant Pf-SSB appears to behave as a homo-tetramer in solution4. Here, we examined the assembly state of an untagged version of Pf-SSB using analytical sedimentation methods. In sedimentation velocity experiments of Pf-SSB in the absence of reducing agents (3 µM Pf-SSB, buffer H0.2 at 25°C), we observe three or more distinct species with average sedimentation coefficients of 5.3 ± 0.2, 8.1 ± 0.4 and 11.2 ± 0.7 S with predicted molecular weights corresponding to a tetramer (92.7 kDa), octamer (176 kDa) and dodecamer (280 kDa; Figure S1A). Since Pf-SSB has a native cysteine at position 93, we tested whether disulfide bond formation influences the oligomerization by repeating the experiments in the presence of reducing agents (either 5 mM 2–mercaptoethanol (2-ME) or 1 mM tris(2-carboxyethyl)phosphine(TCEP)). In the presence of reducing agents in buffer H0.2, Pf-SSB displays a single symmetrical peak in a continuous sedimentation [c(s)] analysis consistent with a single species (Figure S1b), with a weight average sedimentation coefficient of 5.28 ± 0.16 S, corresponding to a predicted molecular weight of 92.5 kDa and a frictional coefficient ratio, f/f0 = 1.39 ± 0.03. This is close to the expected molecular weight for a Pf-SSB homo-tetramer (98,296 Da) as calculated from its amino acid composition.
We also used sedimentation equilibrium to obtain a rigorous molecular weight estimate of Pf-SSB. The results of experiments performed at three Pf-SSB concentrations (1.04, 3.13 and 6.03 µM (tetramer)) and four speeds (9.5, 11.5, 14 and 17 k rpm) are shown in Figure 1B. Global non-linear least squares (NLLS) analysis of these data gave results consistent with a single ideal species (eq 1, Methods section) with an average molecular mass of Mr= 98824 ± 221 Da. This value agrees well with the predicted molecular weight of 98296 Da for a Pf-SSB homo-tetramer. Hence Pf-SSB is a stable homo-tetramer over a concentration range from 0.5–6 µM (tetramer).
Figure 1. Pf-SSB is a stable homotetramer in solution.
(A) Domain architecture of Pf- and Ec-SSB proteins. (B) Sedimentation equilibrium experiments indicate that Pf-SSB is a stable homotetramer in solution. Experiments were performed at three different protein concentrations as indicated in the plot, and at four rotor speeds (9.5 k – green, 11.5 k – blue, 14 k - red and 17 k – black). The smooth black lines depict the fits to a single-species model and the appropriate residuals are also shown.
Pf-SSB binds tightly to ssDNA
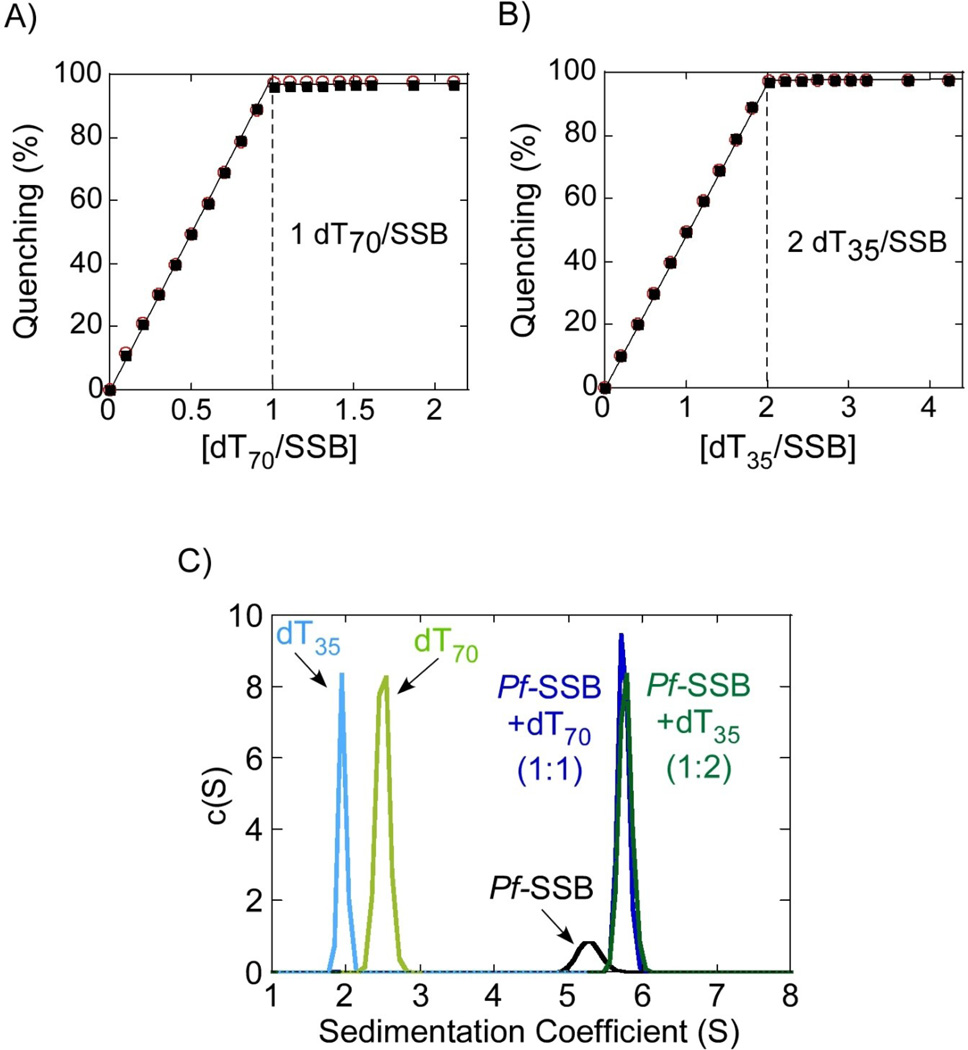
Ec-SSB contains 4 Trp residues per monomer (3 in the DNA binding core) and ssDNA binding can be monitored by the quenching of its intrinsic Trp fluorescence (~90% at saturation). The Ec-SSB tetramer binds to DNA with very high affinity and 65 nucleotides of poly (dT) are required to fully wrap around the tetramer10. As such, an Ec-SSB tetramer can bind either one molecule of (dT)70 or two molecules of (dT)3526. Pf-SSB contains 3 Trp residues per monomer in the same conserved positions within the DNA binding core and its Trp fluorescence is also quenched upon binding ssDNA (see below). For Pf-SSB, we have measured an occluded site size of 62 ± 2 nucleotides per Pf-SSB tetramer on poly (dT) at high [NaCl] (> 0.2 M) (Antony et. al., accompanying paper). In preparation for attempts at crystallizing Pf-SSB with ssDNA, we examined the binding of Pf-SSB to both (dT)70 and (dT)35 by monitoring the accompanying change in Pf-SSB tryptophan fluorescence (buffer H0.2 at 25°C) as shown in Figure 2A and B. These titrations show that under these conditions, Pf-SSB binds very tightly to both (dT)70 and (dT)35 such that we can only estimate a binding stoichiometry, but not an affinity. The Pf-SSB tetramer can bind either one molecule of (dT)70 (Figure 2A) or two molecules of (dT)35 (Figure 2B) per tetramer and in both cases almost complete quenching (96 – 98 %) of the Trp fluorescence is observed. By comparison with the Trp fluorescence quenching observed for (dT)70 and (dT)35 binding of Ec-SSB, these results suggest that one molecule of (dT)70 or two molecules of (dT)35 can bind to Pf-SSB, both resulting in complete wrapping of DNA around the Pf-SSB tetramer. One interesting difference between Pf-SSB and Ec-SSB is the lack of apparent negative cooperativity in the binding of the second molecule of (dT)35 to the Pf-SSB tetramer (see Antony et al., accompanying paper), whereas Ec-SSB tetramer shows clear negative cooperativity.
Figure 2. Pf-SSB binds stoichiometrically to DNA.
Fluorescence experiments show the quenching of tryptophan fluorescence upon binding to (dT)70 (A) or (dT)35 (B) DNA oligonucleotides. The dotted lines show the stoichiometric binding of either one (dT)70 or two (dT)35 molecules per Pf-SSB tetramer. (○) and (■) represent experiments done at either 0.1 µM or 0.3 µM Pf-SSB in the reaction respectively. (C) Continuous sedimentation coefficient distribution c(s) analysis of Pf-SSB in the presence or absence of ssDNA. Pf-SSB sediments as a single tetramer in the absence of DNA (green) and is capable of stoichiometrically binding two (dT)35 molecules (when mixed in a 1:2 ratio, dark green trace) or one (dT)70 molecule (mixed in a 1:1 ratio, dark blue). Sedimentation profiles of (dT)35 (light green) or (dT)70 (light blue) in the absence of protein are also depicted.
We also examined Pf-SSB and its ssDNA complexes using sedimentation velocity. Pf-SSB tetramers, when bound to either two molecules of (dT)35 or one molecule of (dT)70, displayed c(s) profiles23 with single symmetrical peaks with s̅20,w = 5.73 ± 0.07 S and 5.76 ± 0.09 S, respectively (Figure 2C), corresponding to predicted molecular weights of 133.6 and 133.58 kD, respectively. The f/f0 values calculated from these experiments are 1.39 ± 0.02, 1.63 ± 0.02 and 1.63 ± 0.03 for Pf-SSB alone and the Pf-SSB-(dT)35 and Pf-SSB-(dT)70 complexes, respectively. Therefore, Pf-SSB bound to either one (dT)70 or two (dT)35 molecules displays similar hydrodynamic properties.
Crystal Structure of Pf-SSB is similar to E. coli SSB
We have solved a crystal structure at 2.1 Å resolution of Pf-SSB in complex with two molecules of (dT)35. The Pf-SSB used in the crystallization contained amino acid residues 77–284. Residues 1–76 are part of the apicoplast localization signal (Figure 1A) and are cleaved upon arrival at the apicoplast, hence we did not include these in the recombinant protein that we expressed and purified. We observed two crystal forms with either a monomer or a tetramer in the asymmetric unit (Figure S2). In both cases, we only observe electron density for the amino acids that form the DNA binding core (residues 77–194). Upon analysis of the crystals using SDS-PAGE, the protein component that had crystallized migrated faster than the full length starting protein suggesting that the C-terminal part of the protein was at least partially cleaved (Figure S2 C). Since we observe excellent density for residues 77–194, the cleavage must occur after residue 194; however, we have not determined the precise site of cleavage. Similar proteolytic cleavage has been observed during crystallization of the Ec-SSB protein. Since the structural details of both Pf-SSB-(dT)35 crystal forms are similar, we discuss only the structure derived from the crystal form showing the tetramer in the asymmetric unit.
Each subunit contains an oligosaccharide/oligonucleotide-fold (OB-fold) found in many SSB proteins34 (Figure 3A), and is composed of five beta strands (β1– β5) and one alpha helix (α) connected by short linkers (L1–1’, L1’–2, L2–3, L3-α, Lα-4, L4–4’, L4–5, and L5–5’; Figure 3B). All four subunits in the tetramer have similar conformations with RMSD values ranging from 0.12 to 0.26 Å for 90–95 Cα atoms. The overall structure resembles the Ec-SSB tetramer and 77 Cα atoms of Pf-SSB and Ec-SSB monomers align with RMSD of 0.8 Å (Figure 3C). Alignment was performed using Py Mol with the default cutoff level of 2 Å. Noticeable differences in the Pf-SSB structure are the more ordered 4–5 and 2–3 β-sheets and the disordered tips of the 4–5 loops (Figure 3C). Pf-SSB and Ec-SSB tetramers align with RMSD of 8.6 Å for 306 Cα atoms reflecting minor differences in the mutual orientation of individual subunits (Figure S3). Both proteins show a high degree of structural similarity in the overall architecture of the DNA binding domains and the organization of the homotetramer.
Figure 3. Crystal structure of Pf-SSB.
(A) Sequence alignment of Pf-SSB and Ec-SSB and a schematic representation of their secondary structure. Strictly conserved residues are marked by an asterisk (*). (B) Architecture of a single monomer in Pf-SSB. (C) Overlay of a single monomer from Pf-SSB (cyan) and Ec-SSB (orange) highlight the similarity in protein structure between the two proteins.
ssDNA wraps around the Pf-SSB tetramer interacting with all four subunits
Since Pf-SSB binds tightly to either one molecule of (dT)70 or two molecules of (dT)35 (Figure 2), we formed and obtained crystals of each of these complexes. Crystals of Pf-SSB with two bound (dT)35 molecules diffracted to 2.1Å, although interpretable density is observed for only 25 nts per (dT)35 molecule (Figure 4A), and a majority of the modeled DNA is well ordered (Figure S4). We have numbered the nucleotides T1 – T14 and T18 – T28 (Figure 4B) (since the DNA is an oligo-(dT), T-1 may not be the actual 5′ end of the DNA molecule). We assumed that the gap between the two ordered ssDNA segments was missing three nucleotides based on the distance between the ends of the ssDNA segments and the presence of weak electron density potentially corresponding to the missing nucleotides. Although we observe some density consistent with two or three bases corresponding to residues T15–T17, the density cannot be fit to a single conformation, suggesting that this region is disordered. We also obtained small crystals that diffracted to 3.8 Å for Pf-SSB bound to a molecule of (dT)70, but were unable to obtain a structure of this complex due to the poor quality of the crystals. These crystals were of the same space group as the Pf-SSB-(dT)35 complex suggesting that (dT)70 is bound in a similar conformation to that of the two (dT)35 molecules.
Figure 4. A homotetramer of Pf-SSB binds to ssDNA.
(A) The subunits of the Pf-SSB tetramer are colored cyan, violet, green and red with the two bound (dT)35 molecules shown as sticks (black). (B) Schematic of DNA residues 1 – 13 that are observed in the monomer bound to the purple subunit and residues in Pf-SSB that interact with the DNA are shown. The residues colored in red and blue are from the red and blue subunits respectively and all other residues are from the violet subunit. The bold-green arrows denote stacking interactions between the bases and the aromatic amino acid side chains. Details of the interactions between DNA bound to the violet subunit in Pf-SSB and the amino acid side chains are depicted with respect to nucleotides T1–T3 (C), T4 – T5 (D), T6 and T8 (E), T9 and T13: front view (F), back view (G), (F). The orange spheres denote density for either water or probable ion molecules in the structure that mediate specific interactions between the protein and the DNA.
Protein-DNA contacts
In the Pf-SSB-(dT)35 structure, the residues that contact the DNA are identical in all four subunits (Figure 4). This differs from the asymmetric contacts observed in the Ec-SSB-(dC)35 structure11. A schematic of the specific contacts between amino acids in one monomer and the DNA is shown in Figure 4B. Residues from three different subunits contact each half of the (dT)35 molecule. We also observe density for several water and/or ion molecules; since it is difficult to distinguish between electron density for water molecules versus ions at 2.1 Å resolution, all solvent molecules in the structure are modeled and discussed as waters. The first nucleotide in the DNA for which we observe density, denoted T-1, contacts both the 2–3 loop of one subunit and the 1–2 loop of the adjacent subunit. R164 and Y137 contact T-1 through a water molecule and H162 and W117 (subunit I) form stacking interactions with T-1. D130 contacts both T-1 and H162 through water molecules. S184 contacts both the backbone phosphate and the sugar moiety (Figure 4 C). T-2 makes a contact with K98 through another water molecule and T-3 contacts N101 (Figure 4 D). R164 also contacts both the T-4 and T-5 base and their respective sugars along with its aforementioned contact with T-1. R133 contacts T-4 and forms a network with E180, R164 and the DNA. Two threonines (T163 and T179) from subunit I coordinate the T-5 and T-6 bases through two waters and the backbone of T-6 makes a contact with S110 (Figure 4E). W166 forms a stacking interaction with T-5 and this conserved residue assists in bending of the DNA around the individual monomer. W131 is homologous to the functionally critical W54 in Ec-SSB38, and makes a base-stacking interaction with T-8. W131 also makes a polar contact with T-6 at which point the DNA bends around the 1–2 loop and funnels towards the other half of the subunit (Figure 4E).
S110 contacts T-8, N114 and T129 contacts T-9 and along with E79 from subunit I interacts with the bend in the DNA formed by residues T-8 through T-10. T-12 and T-13, the last two residues for which we observe electron density interact with the N-terminus of subunit I (E79 and K80) and the C-terminus of the subunit IV (R153, D189 and F192). Together with R154 they form a series of well networked connections that may control the entry of the DNA into the next subunit (Figures 4F and G).
Topology of DNA wrapping around the Pf-SSB tetramer
Although we were unable to solve a structure of the Pf-SSB tetramer bound to (dT)70, we were able to use the structure of the complex with two (dT)35 molecules bound to identify the most likely path of the ssDNA in a fully wrapped complex (Figure 5A). There are four DNA fragments in our structure that show clear electron density for 13–14 nts bound per subunit (Figure 5B). We observe weak density for 2–4 nts that lie between the DNA fragments bound to subunits I and II and between subunits III and IV (Figure 5B) and the spacing in these gaps is small (< 18 Å). Hence we assume that these short gaps reflect disordered regions of the two (dT)35 molecules (Figure 5B). We next need to decide the path that a (dT)70 molecule would follow in a fully wrapped structure. The 3′ end of the DNA bound to subunit I and the 5′ end of the DNA bound to subunit III would likely be connected if part of a (dT)70 molecule since the ends are unobstructed in the structure and the distance between them is ~ 30 Å (Figures 5A and 5B). Based on this, a model for how DNA might wrap completely around the Pf-SSB tetramer is shown in Figure 5C. This proposed path of the ssDNA follows a topology similar to the seams on a baseball as previously found for the Ec-SSB-(dC)35 structure11 (Figure 5D), although the backbone polarity is opposite to that observed in the Ec-SSB structure (see below for discussion of polarity).
Figure 5. Models for DNA wrapping around Pf-SSB.
(A) Shows a spaced filled version of the Pf-SSB structure with the bound DNA colored according to its B-factor. The predicted path of a (dT)70 DNA molecule is shown by dotted lines traversing across the tetramer and the path of the missing DNA residues in each (dT)35 molecule is also denoted by dotted lines. (B) Is a schematic depicting the polarity of each (dT)35 molecule bound to the four subunits in the Pf-SSB tetramer. The black dotted lines represents regions of the DNA for which we observe weak density and the grey dotted lines, connecting the two (dT)35 molecules, is a predicted path for wrapping of a (dT)70 molecule. (C) Is a cartoon representation showing the front and back views of DNA wrapping around the Pf-SSB tetramer. (D) Depicts the path of DNA wrapping around the Ec-SSB tetramer11.
Based on this crystal structure, we cannot completely exclude the alternate pathway for DNA wrapping around the tetramer, where the 5′ end of the DNA bound to subunit II connects with the 3′ end of the DNA bound to subunit III (Figures 5A and 5B). If this were to be the case, then a ‘baseball seam’ like topology for wrapping would not hold true. However, to accommodate this alternate path, the 5′ end of the DNA bound to subunit II, which is buried within the protein due to the closure of the L2–3 and L1–2 loops must become available, and would require significant movement of both these loops away from each other to facilitate this alternate pathway for wrapping.
We observe ~26 nucleotides bound to each half of the tetramer and the ~ 30 Å gap between the 5′ and 3′ ends of the DNA can be filled with ~ 5 nucleotides (Figure 5B). The shorter ~ 18 Å gap would accommodate ~ 3 nucleotides (Figure 5B), hence ~61– 64 nts of ssDNA would be required to completely wrap around the entire tetramer. Indeed this estimate is consistent with the occluded site size of 62 ± 2 nt for the Pf-SSB tetramer on poly (dT) DNA measured under high salt conditions (buffer H0.2M) (Antony et. al., accompanying paper).
We observe excellent density for the DNA in our structure and the 2.1 Å resolution is sufficient to determine the backbone polarity of the DNA bound to Pf-SSB. Interestingly, the backbone polarity of the (dT)35 molecules within the Pf-SSB complex (Figure 6A) is opposite to that observed in the Ec-SSB-(dC)35 structure (Figure 6B). In the Pf-SSB structure, the 5′ end of the DNA binds to the L1–2 loop and is extended through contacts with the L4–5 loop towards loop L2–3 (Figure 6A). In the DNA bound structures of Mycobacteria smegmatis SSB (Ms-SSB) (PDB code: 3A5U) and Helicobacter pylori SSB (Hp-SSB)39, the polarity of the bound DNA is the same as observed in the Pf-SSB structure.
Figure 6. ssDNA wraps around Pf-SSB with polarity opposite to Ec-SSB.
Polarity of the ssDNA bound across a monomer in (A) Pf-SSB and (B) Ec-SSB. The DNA is bound with a 5′-3′ polarity from top to bottom in the Pf-SSB structure and with opposite polatiry in the Ec-SSB structure.
Discussion
We describe a crystal structure of the Plasmodium falciparum SSB tetramer bound to ssDNA at 2.1 Å resolution. All four subunits interact with the ssDNA and the topology of the DNA path resembles the “seams of a baseball” as observed for E. coli SSB in its fully wrapped (SSB)65 DNA binding mode (Figure S5)11. Although crystal structures of SSB proteins from multiple organisms have been reported in their apo-form, only three SSB-DNA complex structure shave been reported. Of these, only the crystal structure of the Ec-SSB DNA complex was crystallized with the entire tetramer in the asymmetric unit and density for 26–28 of the 35 nucleotides in each of the two bound (dC)35 DNA molecules was observed thereby providing sufficient information to determine the topology of the wrapped ssDNA11. In the Pf-SSB-(dT)35 structure reported here, we also observe electron density for the entire tetramer and for 25–26 nts of each of the two molecules of (dT)35 bound to the protein (Figure S4). We find that ssDNA wraps around the Pf-SSB tetramer with a topology similar to Ec-SSB, but with opposite polarity. There are three notable differences between the Pf-SSB and Ec-SSB structures: a) protein-DNA contacts, b) the symmetry of the DNA contacts in the four subunits, and c) the protein-protein contacts between adjacent tetramers. If and how these factors contribute to the observed difference in polarity of DNA wrapping, DNA binding activity or DNA binding modes remains to be determined (see Antony et. al., accompanying paper).
In the Pf-SSB structure, an extensive network of interactions between the amino acid side chains and the bases of the DNA is apparent (Figure 4). These interactions can be divided into stacking interactions between the aromatic protein side chains and the nucleotides, ionic and polar contacts, and contacts between the protein and the phosphate backbone of the DNA. In both the Ec-SSB and Pf-SSB structures, significant interactions occur through stacking interactions between the bases in the DNA and three tryptophan residues per subunit (W117, W131 and W166 in Pf-SSB). The aromatic side chains of these tryptophans stack against the pyrimidine bases T1, T8 and T5, respectively (Figure 4). These residues are also conserved in Ec-SSB (W40, W54 and W88) but only two of the three Trp residues (W54 and W88) base stack with similar orientations in all four subunits. W40 is located on the 2–3 loop and adopts multiple conformations in the four subunits of Ec-SSB due to the flexible nature of the loop. The homologous W117 residue in Pf-SSB is not positioned on the 2–3 loop, but is located on the structured β-sheet 2 and adopts the same conformation in all four subunits. In Pf-SSB we observe 98% quenching of the Trp fluorescence upon saturation with two molecules of (dT)35 or one molecule of (dT)70. This is consistent with the observation that all three of the Trp residues in Pf-SSB form stacking interactions with the DNA bases and thus the fluorescence of each is essentially fully quenched. Pf-SSB also uses a unique set of charged residues to mediate electrostatic interactions with the DNA phosphates. R90, K128, R153 and R154 form electrostatic interactions with the DNA, but these residues are not conserved in Ec-SSB (Figure 3). In Ec-SSB, Histidine 55 contributes to the stability of the tetramer in that a H55Y mutation (ssb-1) destabilizes the tetramer in favor of monomers. This ssb-1 mutation results in a temperature sensitive phenotype in vivo. The homologous residue, H132in Pf-SSB, is also located at the same position and makes inter-subunit contacts with N83 and L160 (N6 and L83 are the homologous residues in Ec-SSB).
Another difference between the Ec-SSB and Pf-SSB structures is that the protein-DNA contacts are more symmetric within the Pf-SSB complex. In the Pf-SSB-(dT)35 structure, the DNA contacts are the same within each subunit (Figures 3 and 4), whereas in the Ec-SSB-(dC)35 structure, a subset of the contacts differ among the subunits11. A part from the conformational differences observed for W40 in the Ec-SSB structure, it has been hypothesized that the (SSB)35 binding mode is mediated by the asymmetry in the stacking interaction between the DNA and W54 situated on β3 extension which is observed in only three of the four subunits11. In Pf-SSB, the homologous W166 residue shows stacking interactions within all four subunits of the tetramer. An E. coli mutant with either a W54S or W88T mutation shows increased sensitivity to UV, however this is not the case for a W40T substitution51. However, biochemical studies suggest interactions between W40 and the ssDNA52. A W54S mutation also results in a relative stabilization of the (SSB)35 DNA binding mode in Ec-SSB36 and in both the Ec-SSB and Pf-SSB structures this residue forms a stacking interaction with the nucleotide. This suggests that W54 is important for promoting the fully wrapped (SSB)65 DNA binding mode. The E. coli ssb-3 mutation (G15 to D) shows extreme sensitivity to UV53 and is positioned close to the ssDNA in the crystal structure11. This residue is also conserved in Pf-SSB (G92) and is also positioned close to the DNA suggesting a conservation of key amino acid residues between the two proteins.
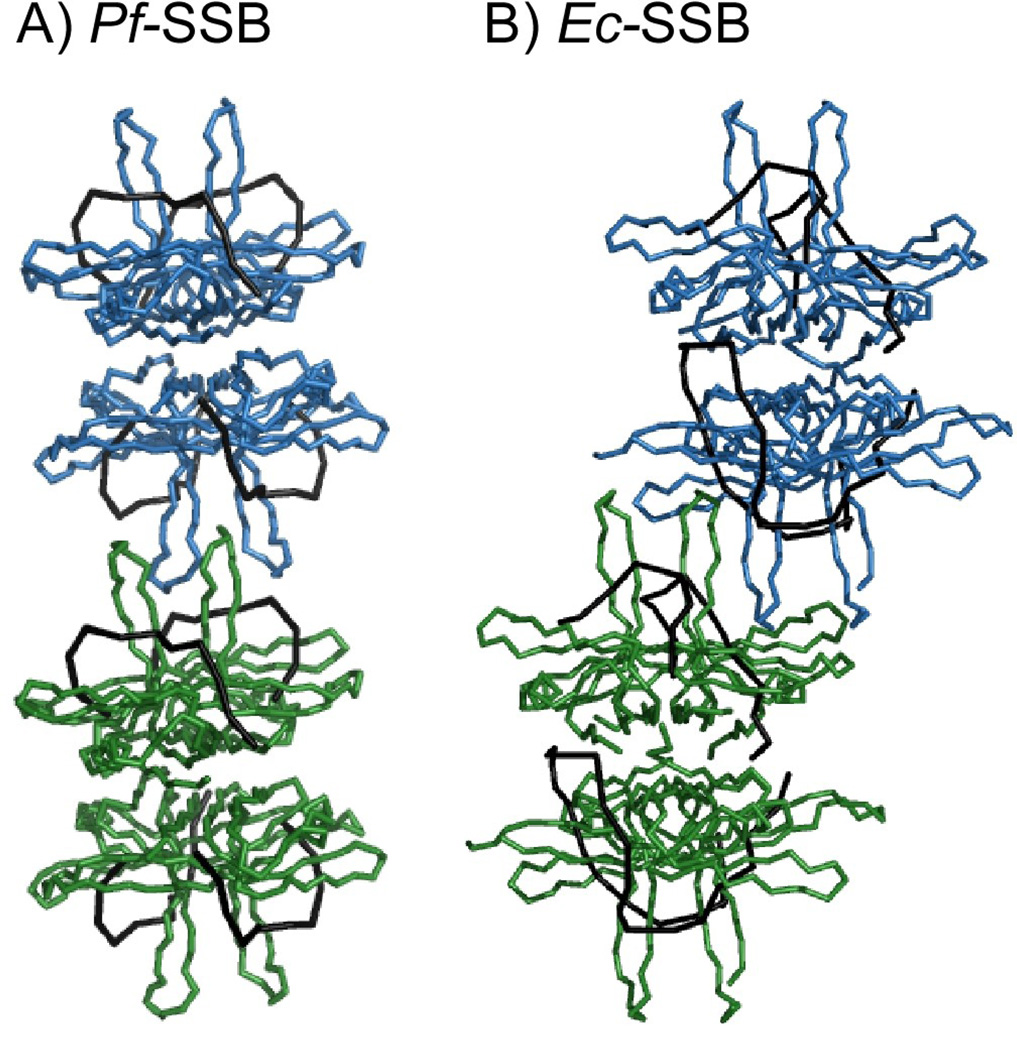
The third major difference between the two structures lies in the tetramer-tetramer interface between symmetry related molecules (Figure 7). In all Ec-SSB structures solved to date, the L4–5 loops from neighboring tetramers pack against each other (Figure 7B). This led to the hypothesis that this tetramer-tetramer interface might be involved in the cooperative (SSB)35 DNA binding mode in Ec-SSB11. We do not observe such an interface in the Pf-SSB crystals (Figure 7A). As shown in the accompanying paper (Antony et. al.), Pf-SSB also does not appear able to forma stable (SSB)35 DNA binding mode. The structure and the length of the L4–5 loops in the various apo- or DNA bound crystal structures of homologous bacterial SSB proteins also appears to be similar. In the Ec-SSB structures, only the basal half of this loop contacts the DNA. Moreover, the top parts of the L4–5 loops are disordered in all structures except Ec-SSB, where they form inter-tetrameric contacts that may be important for cooperative binding in the (SSB)35 mode. The conserved size of the L4–5 loop suggests that it may be important for tetramer-tetramer interactions in other SSB homologs as well.
Figure 7. Protein-protein contacts between SSB tetramers.
Contacts between tetramers in neighboring unit cells in Pf-SSB (A) and Ec-SSB (B) are shown. The bound DNA in each structure is represented by the black ribbons.
It is possible that some of these differences may result from the different solution conditions under which the two structures were crystallized. The Pf-SSB-(dT)35 crystals only grew in either 0.2 M Na [Br, Cl or SO4] whereas the Ec-SSB-(dC)35 crystals were obtained in the absence of any added salt11. Multiple DNA binding modes have been observed with Ec-SSB that are dependent upon the salt concentration. The (SSB)35 DNA binding mode is observed at low NaCl concentrations (< 10 mM), whereas the fully wrapped (SSB)65 binding mode is observed at higher NaCl concentrations (> 200 mM)13. Our attempts to obtain crystals of Pf-SSB-(dT)35 complexes under the low salt conditions used to obtain the Ec-SSB-dC35 crystals were not successful.
A final striking difference between the Pf- and Ec-SSB proteins is the sequence divergence of the unstructured C-terminus and the composition of the C-terminal amino acid end (Pf: MNVQEFEE versus Ec: DFDDDIPF). However, it is not likely that this region plays a role in determining the polarity of the bound ssDNA. In Plasmodium, compounds inhibiting the activity of apicoplast proteins have been used as successful anti-malarial drugs54. Mutations in the Ec-SSB C-terminus render E. coli severely impaired for DNA repair and replication or result in lethality8. Small molecule inhibitors that inhibit the interaction of the E. coli SSB C-terminal tails with an array of other proteins have emerged as a new class of potential antibiotics55. It remains to be determined whether the sequence of the Pf-SSB C-terminus is important for any Pf-SSB interactions with any proteins important for its function in the apicoplast, although this possibility seems likely.
Materials and Methods
Buffers
Buffer H0.08 is 10 mM Hepes, pH 8.1, 1 mM EDTA, 0.08 M NaCl and 1 mM tris(2-carboxyethyl)phosphine (TCEP). Buffer H0.2 is 10 mM Hepes, pH 8.1, 0.1 mM Na3EDTA, 200 mM NaCl and 5 mM 2-ME. Lysis buffer is 50 mM Tris-Cl, pH 8.3, 1 mM EDTA, 200 mM NaCl, 10% sucrose, and 15 mM spermidine. Buffer Tx is 50 mM Tris-Cl, pH 8.3, 1 mM EDTA, 5 % (v/v) glycerol, where “x” denotes the molar concentration of NaCl. Storage buffer is 20 mM Tris-Cl, pH 8.3, 1 mM Na3EDTA, 500 mM NaCl, 5 mM 2-mercaptoethanol, 50% (v/v) glycerol.
Expression and Purification of Pf-SSB
The Pf-SSB gene was amplified from genomic DNA (3D7 isolate, a kind gift from Dr. Daniel Goldberg, Washington University) using the following primers: Forward: 5′-AATTCATATGAATGAGAAATCATTAAAT-3’ and Reverse: 5′-AATTGGATCCTCATTCTTCAAATTCTTGG -3′, and cloned into the pET21a vector (Novagen Inc.) using NdeI and BamHI restriction sites. The DNA encoding for the N-terminal amino acids 1–76 was omitted since it encodes the apicoplast localization signal (ALS)4. Furthermore, constructs containing the ALS signal sequence did not overexpress in E. coli. We refer to this version of Pf-SSB (residues 77–284) that lacks the ALS signal sequence as the wild type protein.
Pf-SSB was overexpressed in BL21(DE3) cells and purified using a procedure similar to that described for E. coli SSB. All steps were carried out at 4°C. 30 g of cell paste was resuspended in lysis buffer (150 mL) and lysed using an Avestin cell disrupter (Avestin Inc., Canada) and Pf-SSB and DNA in the clarified lysate were precipitated by adding polyethyleneimine (PEI) to 0.2% (final). The protein was resuspended from the PEI pellet using 200 ml of buffer T0.4. Pf-SSB from the PEI-resuspension was precipitated by adding solid ammonium sulfate (144 g/L) (25% saturation) and the pellet containing >90 % pure Pf-SSB was resuspended in buffer T0.3 (200 mL). The resuspended protein was loaded onto a ssDNA cellulose column (50 mL resin with ~3 mg/mL binding capacity) and eluted using a linear NaCl gradient (0.3 – 2 M) in buffer T. Fractions containing Pf-SSB were pooled and precipitated with 30.8% ammonium sulfate (170 g/L). The resulting precipitate was resuspended in 10 mL of storage buffer, dialyzed and stored as 0.5 mL aliquots at −20°C. The concentration of Pf-SSB was determined spectrophotometrically using an extinction coefficient of ε280 = 9.58 × 104 M−1 (tetramer) cm−1. Using this procedure the typical yield of Pf-SSB is around 15 mg per gram of cell paste. The extinction coefficient in buffer H0.2 and T0.2 was determined by comparing the absorbance of Pf-SSB in buffer H0.2 and T0.2 with its absorbance in 6 M Guanidium-HCl, 10 mM Tris-Cl, pH 8.1, 0.25 mM Na3EDTA, and 1 mM 2-mercaptoethanol at 25°C. The extinction coefficient of the denatured Pf-SSB in 6 M Guanidinium HCl was calculated as the sum of the extinction coefficients of the 3 Trp, 4 Tyr and 3 Phe in 6 M Guanidinium HCl57. Pf-SSB was dialyzed extensively at 4°C versus the buffers used in each experiment using a 10,000 Da molecular weight cut-off dialysis membrane (Spectrum Inc., Houston, TX). Pf-SSB has a single exposed cysteine and in the absence of reducing agent (5 mM 2-mercaptoethanol or 1 mM TCEP (tris(2-carboxyethyl)phosphine)) it forms higher order oligomers in solution (Figure S1). For this reason, all experiments were performed in the presence of 1 mM TCEP.
DNA
The oligodeoxynucleotides, (dT)35 and (dT)70, were synthesized and purified as described58. All ssDNA concentrations were determined spectrophotometrically using the extinction coefficient ε260 = 8.1 × 103 M−1 (nucleotide) cm−1 for oligo (dT) in buffer H0.08.
Analytical Ultracentrifugation
Sedimentation experiments were performed using an Optima XL-A analytical ultracentrifuge equipped with an An50Ti rotor (Beckman Coulter, Fullerton, CA) at 25°C. For sedimentation equilibrium experiments, 120 µL of protein solution was loaded into each of the three channels of an Epon charcoal-filled six-channel centerpiece with 130 µl of buffer in each reference channels. Protein concentration was monitored by absorbance at 280 nm at three different protein concentrations (1.04 µM, 3.13 µM and 6 µM Pf-SSB in buffer H0.1M). Data were collected with a spacing of 0.001 cm with an average of ten scans per step at three rotor speeds: 9500, 11500, and 14000 rpm. At each speed sedimentation equilibrium was determined when successive scans measured over a 2 hour time window were super imposable. Data sets were edited and extracted using SEDFIT followed by analysis by nonlinear least squares (NLLS) using the program SEDPHAT59. Apparent molecular weights were obtained by fitting the data to eq 1:
| (1) |
where AT is the total absorbance at radial position r, A0,i is the absorbance of component i at the reference radial position (rref), b is the baseline offset, σi= [Mi(1−∂̅iρ)ω2]/RT, Mi and ∂i are the molecular mass and partial specific volume of component i, respectively (calculated using SEDENTREP60). For Pf-SSB the ∂i value (0.7191 mL g−1 at 25°C) was calculated based on its amino acid composition (residues 77–284). The solution density ρ for buffer H0.1M was 1.0026 (calculated using SEDENTREP). ω is the angular velocity, R is the ideal gas constant and T is the absolute temperature. A global NLLS fit to eq 1 of the nine absorbance files was used to calculate the molecular weight.
Sedimentation velocity experiments (Figure 3B), were performed using 3 µM Pf-SSB alone or in complex with 3 µM (dT)70 (1:1 molar ratio) or 6 µM (dT)35 (1:2 molar ratio). Experiments were also performed on both (dT)70 and (dT)35 DNA molecules alone. 380 µL of sample and 392 µL of buffer were loaded into the appropriate sectors of an Epon charcoal-filled two-sector centerpiece and centrifuged at 42000 rpm (25°C) and the absorbance was monitored at 280 nm. The continuous sedimentation coefficient distribution, c(s), was calculated using the program SEDFIT.
DNA Binding
Pf-SSB binding to the oligodeoxynucleotides, (dT)70 or (dT)35, was monitored by the quenching of the intrinsic Trp fluorescence of Pf-SSB using a PTI QM-2000 fluorometer (Photon Technologies, Inc., Lawrenceville, NJ) [λεx = 296 nm, 2 nm excitation band-pass, and λem = 345 nm, 2–5 nm emission band-pass] with corrections applied as described previously57. The experiments were performed in Buffer H0.2 at 25°C using either 0.1 or 0.3 µM Pf-SSB tetramer. Under these conditions, the binding affinities of Pf-SSB for (dT)70 and (dT)35 are too large to measure (i.e., the titrations were stoichiometric) and the intersection of the linear parts of the titration curves was used to determine the stoichiometry of DNA binding per Pf-SSB tetramer.
Crystallization and Structure Determination
Pf-SSB (6 mg/ml: 61 µM tetramer) was mixed with 122 µM (dT)35 (1:2 ratio) in buffer H0.2 (10 mM Hepes, pH 8.1, 0.1 mM Na3EDTA, 200 mM NaCl and 5 mM 2-mercaptoethanol) and dialyzed extensively vs. buffer H0.2 at 4°C. The concentration of protein after dialysis was ~ 3 mg/mL. Crystals were grown by vapor diffusion using the sitting drop method in 96-well plates using a crystallization robot (Phoenix – Art Robbins Instruments, Sunnyvale, CA). The first Pf-SSB-(dT)35 crystal form (bi-pyramidal) was observed in several commercial PEG based screens (PEG/Ion HT - Hampton Research, Aliso Viejo, CA, PEGs Suite – Qiagen, Valencia, CA, and PACT premier - Molecular Dimensions, Apopka, FL) at 20°C after 3–4 days. The second Pf-SSB-(dT)35 crystal form (long rods) was observed at 4°C after 4–5 weeks in 0.1 M Bis-Tris Propane, pH 8.5, 20 % PEG 3350 and either 0.2 M sodium bromide or 0.2 M sodium sulfate. Crystals of a Pf-SSB-(dT)35 complex, grown in 0.1 M Bis-Tris Propane, pH 8.5, 0.15 M sodium sulfate and 24 % PEG 3350 at 15°C, were harvested into cryo-protectant solution (20 % ethylene glycol, 5 % PEG 3350 and 60 % mother liquor) and flash frozen in liquid nitrogen. Diffraction data were collected using a 1.2 kW MM007 Rigaku generator with VHF optics and Raxis-IV++ image detector under cryo-stream with the X-stream cryo-cooling system. Data were processed with the HLK2000 program61. Initial phases were obtained by the molecular replacement method using Phaser within the CCP4i program suit using the structure of the E. coli SSB/DNA complex11 (PDB code 1EYG).
Initial bi-pyramidal crystals belonging to the tetragonal space group I422 with unit cell parameters a= b= 83.1, and c=136.7 Å contained one Pf-SSB monomer per asymmetric unit. A second crystal form was also obtained belonging to a monoclinic space group (Table 1) with one tetramer per asymmetric unit. Since the structure of the monomer in the tetragonal group was close to that of the monoclinic form, we describe the structure of the tetramer. The model building and refinement were completed using Arp/Warp66, Coot67 and Refmac68. Non-crystallographic averaging was not utilized during the initial model building and refinement steps. The following residues were disordered and not modeled due to poor electron density: chain A 169–172, chain B 121 and 171–173, chain C 170–171 and chain D 121–122 and 169–171. The model was refined to a resolution of 2.1 Å with R= 22.8 and Free R = 27.5 with excellent geometry (Table 1).
Table 1.
Data collection and refinement statistics.
| Space group | C2 |
|---|---|
| Unit cell (Å) | a=118.0; b=82.8; c=87.6; d=99.57 |
| Data collection resolution | 50-2.1 |
| R-merge (%)* | 6.8 (52.9) |
| Completeness* | 99.4 (97.3) |
| I/σ high resolution shell | 3 |
| Refinement resolution (Å) | 30-2.1 |
| # protein non-H atoms | 3638 |
| # DNA atoms | 1022 |
| # water molecules | 232 |
| # reflections | 45791 |
| # reflection test set (5%) | 2438 |
| R (%)* | 21.8 (28.3) |
| Free R (%)* | 27.6 (35.4) |
| Rmsd bonds (Å) | 0.01 |
| Angles | 1.5 |
| Overall B factor, protein (Å2) | 35 |
| Overall B factor, DNA (Å2) | 57 |
| Overall B factor, solvent (Å2) | 43 |
| Ramachandran plot** | |
| Most favored regions (%) | 90.0 |
| Allowed regions (%) | 9.0 |
| Generously allowed (%)*** | 1.0 |
| Disallowed (%) | 0.0 |
Values for highest resolution for data collection 2.10–2.14, and for refinement 2.10–2.15 Å are shown in parentheses.
Ramachandran plot parameters were calculated by program PROCHECK69
Residues in generously allowed conformation are in poorly structured loops.
Supplementary Material
Highlights.
DNA binding properties of Plasmodium falciparum SSB are different from E. coli SSB.
Unlike Ec-SSB, the Pf-SSB does not possess the (SSB)35 DNA binding mode.
Pf-SSB has unique DNA binding properties.
Pf-SSB DNA binding activity might be specific for biological function in Plasmodium.
Acknowledgements
The Plasmodium falciparum genomic DNA was a kind gift from Dr. Daniel Goldberg (Washington University School of Medicine). We thank Drs. Alex Kozlov and Binh Nguyen for significant technical advice. Thang Ho for synthesis and purification of the oligodeoxynucleotides. This work was supported in part by grants from the NIH to T.M.L. (GM30498, GM45948) and S.K. (GM073837).
Abbreviations
- SSB
Single strand DNA binding protein
- ssDNA
single stranded DNA
- dsDNA
double stranded DNA
- Pf-SSB
Plasmodium falciparum SSB
- Ec-SSB
Escherichia coli SSB
- SIPs
SSB interacting proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers: The coordinates of the Pf-SSB-(dT)35 complex have been deposited in the Protein Data Bank with PDB ID: 3ULP.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson RJ, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, Williamson DH. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- 3.Dahl EL, Rosenthal PJ. Apicoplast translation, transcription and genome replication: targets for antimalarial antibiotics. Trends Parasitol. 2008;24:279–284. doi: 10.1016/j.pt.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Prusty D, Dar A, Priya R, Sharma A, Dana S, Choudhury NR, Rao NS, Dhar SK. Single-stranded DNA binding protein from human malarial parasite Plasmodium falciparum is encoded in the nucleus and targeted to the apicoplast. Nucleic Acids Res. 2010;38:7037–7053. doi: 10.1093/nar/gkq565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 6.Lohman TM, Bujalowski W, Overman LB. E. coli single strand binding protein: a new look at helix-destabilizing proteins. Trends Biochem Sci. 1988;13:250–255. [PubMed] [Google Scholar]
- 7.Sancar A, Williams KR, Chase JW, Rupp WD. Sequences of the ssb gene and protein. Proc Natl Acad Sci U S A. 1981;78:4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc Natl Acad Sci U S A. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 11.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 12.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 13.Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, binding density effects. J Biol Chem. 1988;263:4629–4640. [PubMed] [Google Scholar]
- 14.Lohman TM, Overman LB, Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 15.Bujalowski W, Lohman TM. Limited co-operativity in protein-nucleic acid interactions. A thermodynamic model for the interactions of Escherichia coli single strand binding protein with single-stranded nucleic acids in the "beaded", (SSB)65 mode. J Mol Biol. 1987;195:897–907. doi: 10.1016/0022-2836(87)90493-1. [DOI] [PubMed] [Google Scholar]
- 16.Griffith JD, Harris LD, Register J., 3rd Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb Symp Quant Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 17.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein DA, Eggington JM, Killoran MP, Misic AM, Cox MM, Keck JL. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc Natl Acad Sci U S A. 2004;101:8575–8580. doi: 10.1073/pnas.0401331101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 22.Norais CA, Chitteni-Pattu S, Wood EA, Inman RB, Cox MM. DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J Biol Chem. 2009;284:21402–21411. doi: 10.1074/jbc.M109.010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dam J, Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- 24.Schuck P. Sedimentation analysis of noninteracting and self-associating solutes using numerical solutions to the Lamm equation. Biophys J. 1998;75:1503–1512. doi: 10.1016/S0006-3495(98)74069-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krauss G, Sindermann H, Schomburg U, Maass G. Escherichia coli single-strand deoxyribonucleic acid binding protein: stability, specificity, and kinetics of complexes with oligonucleotides and deoxyribonucleic acid. Biochemistry. 1981;20:5346–5352. doi: 10.1021/bi00521a040. [DOI] [PubMed] [Google Scholar]
- 26.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J Mol Biol. 1989;207:269–288. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- 27.Lohman TM, Bujalowski W. Negative cooperativity within individual tetramers of Escherichia coli single strand binding protein is responsible for the transition between the (SSB)35 and (SSB)56 DNA binding modes. Biochemistry. 1988;27:2260–2265. doi: 10.1021/bi00407a002. [DOI] [PubMed] [Google Scholar]
- 28.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I. Evidence and a quantitative model. J Mol Biol. 1989;207:249–268. doi: 10.1016/0022-2836(89)90454-3. [DOI] [PubMed] [Google Scholar]
- 29.Gallagher JR, Matthews KA, Prigge ST. Plasmodium falciparum apicoplast transit peptides are unstructured in vitro and during apicoplast import. Traffic. 2011;12:1124–1138. doi: 10.1111/j.1600-0854.2011.01232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McFadden GI. Plastids and protein targeting. J Eukaryot Microbiol. 1999;46:339–346. doi: 10.1111/j.1550-7408.1999.tb04613.x. [DOI] [PubMed] [Google Scholar]
- 31.Ng JD, McPherson A. Preliminary crystallographic analysis of a proteolytically modified form of E. coli single stranded DNA binding protein. J Biomol Struct Dyn. 1989;6:1071–1076. doi: 10.1080/07391102.1989.10506537. [DOI] [PubMed] [Google Scholar]
- 32.Thorn JM, Carr PD, Chase JW, Dixon NE, Ollis DL. Crystallization and low temperature diffraction studies of the DNA binding domain of the single-stranded DNA binding protein from Escherichia coli. J Mol Biol. 1994;240:396–399. doi: 10.1006/jmbi.1994.1453. [DOI] [PubMed] [Google Scholar]
- 33.Ollis D, Brick P, Abdel-Meguid SS, Murthy K, Chase JW, Steitz TA. Crystals of Escherichia coli single-strand DNA-binding protein show that the tetramer has D2 symmetry. J Mol Biol. 1983;170:797–800. doi: 10.1016/s0022-2836(83)80134-x. [DOI] [PubMed] [Google Scholar]
- 34.Murzin AG. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curth U, Bayer I, Greipel J, Mayer F, Urbanke C, Maass G. Amino acid 55 plays a central role in tetramerization and function of Escherichia coli single-stranded DNA binding protein. Eur J Biochem. 1991;196:87–93. doi: 10.1111/j.1432-1033.1991.tb15789.x. [DOI] [PubMed] [Google Scholar]
- 36.Curth U, Greipel J, Urbanke C, Maass G. Multiple binding modes of the single-stranded DNA binding protein from Escherichia coli as detected by tryptophan fluorescence and site-directed mutagenesis. Biochemistry. 1993;32:2585–2591. doi: 10.1021/bi00061a016. [DOI] [PubMed] [Google Scholar]
- 37.Ferrari ME, Fang J, Lohman TM. A mutation in E. coli SSB protein (W54S) alters intra-tetramer negative cooperativity and inter-tetramer positive cooperativity for single-stranded DNA binding. Biophys Chem. 1997;64:235–251. doi: 10.1016/s0301-4622(96)02223-5. [DOI] [PubMed] [Google Scholar]
- 38.Carlini LE, Porter RD. Analysis of ssb mutations in vivo implicates SSB protein in two distinct pathways of SOS induction and in recombinational DNA repair. Mol Microbiol. 1997;24:129–139. doi: 10.1046/j.1365-2958.1997.3431694.x. [DOI] [PubMed] [Google Scholar]
- 39.Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ. Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. J Mol Biol. 2009;388:508–519. doi: 10.1016/j.jmb.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 40.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc Natl Acad Sci U S A. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fedorov R, Witte G, Urbanke C, Manstein DJ, Curth U. 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins. Nucleic Acids Res. 2006;34:6708–6717. doi: 10.1093/nar/gkl1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaushal PS, Singh P, Sharma A, Muniyappa K, Vijayan M. X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures. Acta Crystallogr D Biol Crystallogr. 2010;66:1048–1058. doi: 10.1107/S0907444910032208. [DOI] [PubMed] [Google Scholar]
- 43.Saikrishnan K, Jeyakanthan J, Venkatesh J, Acharya N, Sekar K, Varshney U, Vijayan M. Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications. J Mol Biol. 2003;331:385–393. doi: 10.1016/s0022-2836(03)00729-0. [DOI] [PubMed] [Google Scholar]
- 44.Yang C, Curth U, Urbanke C, Kang C. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat Struct Biol. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 45.Yadav T, Carrasco B, Myers AR, George NP, Keck JL, Alonso JC. Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bujalowski W, Lohman TM. Monomers of the Escherichia coli SSB-1 mutant protein bind single-stranded DNA. J Mol Biol. 1991;217:63–74. doi: 10.1016/0022-2836(91)90611-9. [DOI] [PubMed] [Google Scholar]
- 47.Williams KR, Murphy JB, Chase JW. Characterization of the structural and functional defect in the Escherichia coli single-stranded DNA binding protein encoded by the ssb-1 mutant gene. Expression of the ssb-1 gene under lambda pL regulation. J Biol Chem. 1984;259:11804–11811. [PubMed] [Google Scholar]
- 48.Bujalowski W, Lohman TM. Monomer-tetramer equilibrium of the Escherichia coli ssb-1 mutant single strand binding protein. J Biol Chem. 1991;266:1616–1626. [PubMed] [Google Scholar]
- 49.Meyer RR, Glassberg J, Scott JV, Kornberg A. A temperature-sensitive single-stranded DNA-binding protein from Escherichia coli. J Biol Chem. 1980;255:2897–2901. [PubMed] [Google Scholar]
- 50.Meyer RR, Glassberg J, Kornberg A. An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc Natl Acad Sci U S A. 1979;76:1702–1705. doi: 10.1073/pnas.76.4.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlini LE, Porter RD, Curth U, Urbanke C. Viability and preliminary in vivo characterization of site-directed mutants of Escherichia coli single-stranded DNA-binding protein. Mol Microbiol. 1993;10:1067–1075. doi: 10.1111/j.1365-2958.1993.tb00977.x. [DOI] [PubMed] [Google Scholar]
- 52.Khamis MI, Casas-Finet JR, Maki AH, Murphy JB, Chase JW. Investigation of the role of individual tryptophan residues in the binding of Escherichia coli single-stranded DNA binding protein to single-stranded polynucleotides. A study by optical detection of magnetic resonance and site-selected mutagenesis. J Biol Chem. 1987;262:10938–10945. [PubMed] [Google Scholar]
- 53.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McFadden GI, Roos DS. Apicomplexan plastids as drug targets. Trends Microbiol. 1999;7:328–333. doi: 10.1016/s0966-842x(99)01547-4. [DOI] [PubMed] [Google Scholar]
- 55.Lu D, Bernstein DA, Satyshur KA, Keck JL. Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance. Proc Natl Acad Sci U S A. 2010;107:633–638. doi: 10.1073/pnas.0909191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 57.Lohman TM, Mascotti DP. Nonspecific ligand-DNA equilibrium binding parameters determined by fluorescence methods. Methods Enzymol. 1992;212:424–458. doi: 10.1016/0076-6879(92)12027-n. [DOI] [PubMed] [Google Scholar]
- 58.Ferrari ME, Bujalowski W, Lohman TM. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J Mol Biol. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 59.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 60.Laue TM, Shah BD, RIdgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. Royal Society of Cambridge, Cambridge, U.K. 1992;1 [Google Scholar]
- 61.Otwinowski ZaMW. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276 doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 62.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 65.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 69.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.