Abstract
Concatenation of engineered biocatalysts into multistep pathways dramatically increases their utility, but development of generalizable assembly methods remains a significant challenge. Herein we evaluate ‘bioretrosynthesis’, which is an application of the retrograde evolution hypothesis, for biosynthetic pathway construction. To test bioretrosynthesis, we engineered a pathway for synthesis of the antiretroviral nucleoside analog didanosine (2,3-dideoxyinosine). Applying both directed evolution and structure-based approaches, we began pathway construction with a retro-extension from an engineered purine nucleoside phosphorylase and evolved 1,5-phosphopentomutase to accept the substrate 2,3-dideoxyribose 5-phosphate with a 700-fold change in substrate selectivity and 3-fold increased turnover in cell lysate. A subsequent retrograde pathway extension, via ribokinase engineering, resulted in a didanosine pathway with a 9,500-fold change in nucleoside production selectivity and 50-fold increase in didanosine production. Unexpectedly, the result of this bioretrosynthetic step was not a retro-extension from phosphopentomutase, but rather the discovery of a fortuitous pathway-shortening bypass via the engineered ribokinase.
INTRODUCTION
Many societally important molecules are currently generated using biocatalytic processes. In most cases, individual enzymes are optimized for incorporation into multi-step synthetic routes, such as in the syntheses of the blockbuster drugs sitagliptin1, montelukast2, and simvastatin3, among others4. With less frequency, secondary metabolites, their intermediates, and/or analogs are synthesized via recapitulating and improving existing biosynthetic pathways (for example, artimesinic acid5, taxadiene6) or by modifying native pathways (such as, pactamycin7, macbecin8). Given the apparent multiplicative benefits of combining biotransformations into pathways, it is notable that de novo multistep engineered biosynthetic pathways for the synthesis of unnatural compounds are quite uncommon (1,2,4-butanetriol9, 1,4-butanediol10). The small subset of such pathways that employ cascades of more than one engineered/evolved enzyme, as in the production of the atorvastatin side chain11, are exceptional, as these can be the most difficult to design and implement. Improved pathway evolution paradigms would enable broader production of valuable compounds via engineered biocatalysts.
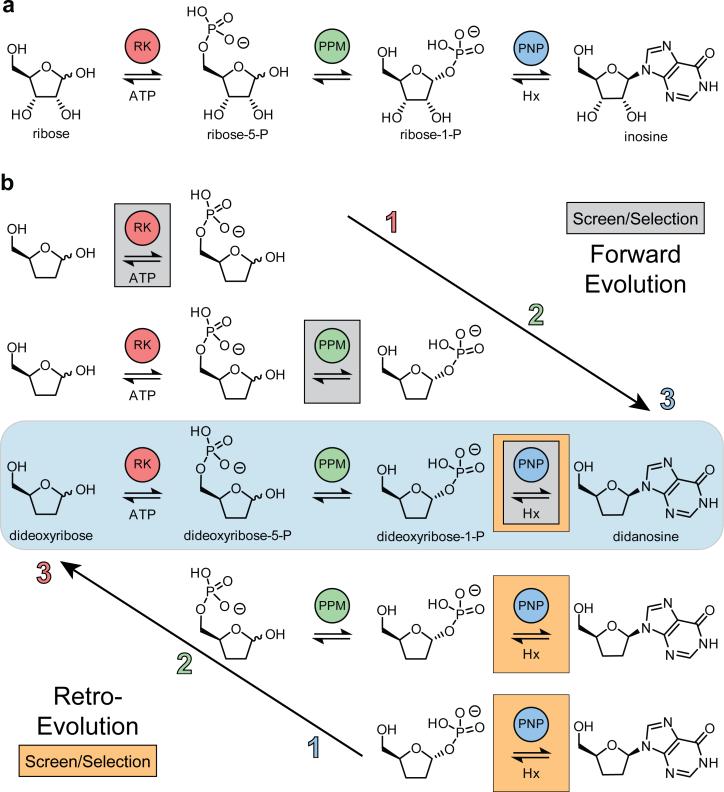
Herein we describe an in vitro pathway for the nucleoside analog didanosine (2,3-dideoxyinosine, ddI), an off-patent inhibitor of HIV-1 reverse transcriptase. The selection of this target was based on the following criteria: (1) didanosine represents a widely prescribed class of antiviral and anticancer drugs with new members currently in clinical trials, (2) manufacturing costs contribute over 75% of the therapy costs12, and (3) metabolic progenitor enzymes can be identified for each step. We developed our pathway using parallels to sugar and nucleoside metabolism, with inosine as a model natural nucleoside (Fig. 1a). In our proposed pathway (Fig. 1b, blue box), 2,3-dideoxyribose 5-phosphate is generated by phosphorylation of 2,3-dideoxyribose by ribokinase (RK) and is converted to 2,3-dideoxyribose 1-phosphate using 1,5-phosphopentomutase (PPM). Finally, didanosine is generated by addition of hypoxanthine to 2,3-dideoxyribose 1-phosphate by purine nucleoside phosphorylase (PNP).
Figure 1. Model inosine biosynthetic pathway and proposed bioretrosynthesis of didanosine.
(a) The three enzyme metabolic pathway for inosine used as a model to construct a didanosine pathway. (b) Comparison of the forward and retro-evolution strategies of pathway construction. Enzymes evolved in the forward direction proceed in the order of biosynthesis (ribokinase, RK; phosphopentomutase, PPM; then purine nucleoside phosphorylase, PNP), requiring individual screening assays for each enzymatic step (gray boxes). Retro-evolution requires one screening assay for terminal enzyme activity (orange boxes) for evolution of each enzyme in the reverse order of biosynthesis (PNP, PPM then RK) in increasingly tandem assays.
Beyond the multifold challenges entailed in designing new biochemical pathways13 and engineering multiple enzymes for unnatural substrates14, a requirement for advancing unnatural pathway engineering is the development of generalizable methods for the assembly and optimization of unnatural biosynthetic pathways. Construction of an engineered multistep pathway can progress in one of two directions. In the forward direction, enzyme recruitment, evolution, and assembly are performed in the order of biosynthesis (Fig. 1b, top). Engineering a pathway in this fashion requires developing a unique selection/selection strategy for each step (Fig 1b, gray boxes). Assay design can be challenging and, in the event of an intransigent intermediary step, new pathway strategies may be required. An alternative approach may be inspired by the hypothesis of retrograde evolution15. This hypothesis asserts that biosynthetic pathways may be assembled in a stepwise fashion, but that the order of enzyme optimization is in the reverse direction of synthesis, beginning with the terminal step (Fig 1b, bottom). This approach can be experimentally applied to non-natural pathway construction and evolution of biosynthetic enzymes in a process we have termed ‘bioretrosynthesis’16, due to its similarity with the chemical and biochemical processes of retrosynthetic analysis17,18. In this approach, pathway product formation is the sole selection criterion for evolution at each retro-consecutive step (Fig 1b, orange boxes). As a result, screening is reduced to a single product selection modality. A key benefit is therefore in reducing the time consuming and difficult design of a unique assay for selection or screening each biosynthetic step.
In this work, we assembled and evolved a didanosine synthesis pathway in a retrograde fashion, according to a bioretrosynthetic process. This five enzyme biosynthetic pathway includes an ATP regeneration cycle. The goals of the enzyme engineering aspect of this work are both to increase turnover of the non-natural substrate, and to improve substrate selectivity of each enzyme for the non-natural substrate. Modification of both of these parameters to favor the non-natural substrate will facilitate eventual in vivo production of didanosine by organisms harboring engineered enzymes. Indeed, abrogation of activity toward natural substrates is critical since residual activity could have undesired effects on sugar and nucleoside metabolism. As described herein, these goals were attainable and nucleoside production via the resulting engineered pathway in vitro displays a 9,500-fold change in pathway substrate selectivity and a 50-fold increase in didanosine production in comparison to the progenitor pathway. An unanticipated result of the bioretrosynthetic strategy was the discovery of a pathway-shortening biochemical bypass and a previously unreported phosphorylation activity elicited by RK. This underlines the potential for bioretrosynthesis as a practical method for the engineering of unnatural biosynthetic pathways.
RESULTS
To test the bioretrosynthetic strategy with the proposed didanosine pathway, we combined structure-guided site-saturation and whole-gene random mutagenesis to improve substrate selectivity and overall activity, respectively. For each enzyme in the pathway (PNP, PPM, RK), we screened for increases in activity toward the non-natural substrate by monitoring terminal product-formation with increasingly tandem assays. Improvements in catalytic parameters and pathway selectivity for each retrograde step were monitored by detection of final product or consumption of terminal cosubstrate. Validation and kinetic characterization of enzymes was performed using HPLC/MS analysis with comparison to authentic synthetic compounds and internal standards (Online Methods).
Bioretrosynthetic Step 1: PNP
In vitro production of didanosine by PNP
We previously improved the terminal reaction of the proposed pathway by evolving human PNP for the phosphorolysis of didanosine19, but had not evaluated improvements in the synthesis direction. We therefore began by assaying this enzyme for synthesis of inosine and didanosine in the wild-type and PNP-46D6 variants from the respective synthetic sugar 1-phosphates via HPLC/MS. Wild-type PNP exhibited a 660-fold higher turnover of ribose 1-phosphate than dideoxyribose 1-phosphate. This selectivity was reduced to 4.7-fold preference for ribose 1-phosphate in the PNP-46D6 variant and included a 16-fold increase in the rate of didanosine production and an 8.7-fold decrease in inosine formation (vide infra). While determining kinetic improvements for didanosine production is complicated by the instability of dideoxyribose 1-phosphate, the 16-fold increase in didanosine formation by the PNP-46D6 variant compares to the 22-fold increase in catalytic efficiency observed in the phosphorolysis direction19. These measurements facilitated the adaptation and validation of a colorimetric PNP assay to measure synthesis of didanosine, a critical requirement for using product formation as the single selection step for engineered the planned multi-step pathway. With a turnover rate of 0.37 μM/min/μM enzyme, the PNP-46D6 variant enzyme was considered to possess sufficient didanosine synthesis activity to allow product detection in the in vitro tandem assays with PPM and PPM-RK, a requirement for implementing a bioretrosynthetic strategy for didanosine synthesis.
Bioretrosynthetic Step 2: PPM
Structure Guided Mutagenesis of PPM
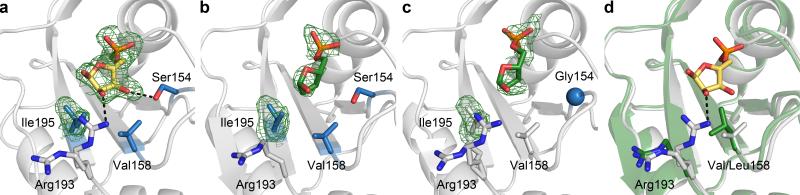
In the bioretrosynthetic strategy, the required substrate for PNP-46D6 is dideoxyribose 1-phosphate, and we searched for an enzyme that could catalyze its formation. Based on reports of a Bacillus enzyme that could measurably convert dideoxyribose 5-phosphate to our desired substrate dideoxyribose 1-phosphate (reportedly 12% in comparison to ribose 5-phosphate activity)20, we used the bioretrosynthetic strategy to assay B. cereus PPM for this activity by monitoring didanosine formation in a coupled assay with PNP-46D6. This confirmed detectable turnover of the non-natural substrate by wild-type PPM. The first strategy to improve PPM activity for the non-natural substrate was to target substrate-interacting residues for mutagenesis. We therefore determined the costructure of PPM with the target non-natural substrate dideoxyribose 5-phosphate (Supplementary Note) at 2.1 Å resolution (Supplementary Results, Supplementary Table 5). The electron density for the non-natural substrate was of lower quality as compared to the natural substrate ribose 5-phosphate (PDB entry 3M8Z21, Fig. 2a). The quality of this electron density suggests both lower occupancy of the non-natural substrate and rotational disorder of the dideoxy sugar ring, with multiple values of the glycosidic torsion angle. As a result, the non-natural substrate is not optimally aligned with key catalytic residues (Fig. 2b). From these structures, Ser154, Val158, and Ile195 were identified as candidates to target for improving selectivity in favor of the non-natural substrate (Fig. 2a-b). We analyzed the effects of Ser154 mutation on substrate orientation by determining costructures of the Ser154Ala mutant with either ribose 5-phosphate or dideoxyribose 5-phosphate (Supplementary Tables 4 and 5 and Supplementary Fig. 2c,d). We had anticipated that loss of a hydrogen-bond between the Ser154 ribose 5-phosphate in the Ser154Ala variant would reduce the ordering of the furanose ring of the natural substrate. However, we instead observed subjectively similar electron density for ribose 5-phosphate in both wild-type and Ser154Ala PPM, and no improvement in ordering of the non-natural dideoxyribose 5-phosphate. Nevertheless, there were fewer enzyme-substrate interactions for only ribose 5-phosphate, thus, we targeted Ser154 for site-saturation mutagenesis via NNK codon degeneracy22.
Figure 2. First shell residues targeted for PPM saturation mutagenesis.
Electron density maps for substrate and Ile195 are |Fo|-|Fc| omit maps calculated in Refmac540,41 after the removal of relevant atoms from each PDB file and contoured at 2σ. Electron density around Ile195 in panels a-c allows comparison of the electron density quality between structures. Hydrogen bonds are shown as dashed lines. Active site residues Ser154, Val158 and Ile195 are highlighted (blue) in the costructures of wild-type PPM with (a) ribose 5-phosphate (gold, PDB entry 3M8Z21) and (b) dideoxyribose 5-phosphate (green). Due to the close proximity (<4.5 Å) of each residue to the bound substrates, all three positions were targeted for saturation mutagenesis. (c) Costructure of Ser154Gly variant with dideoxyribose 5-phosphate (green). The glycine Cα atom shown as a sphere. (d) Overlay of Val158Leu structure (green) with wild-type PPM (gray) with ribose 5-phosphate bound (gold, PDB entry 3M8Z21). A favorable hydrogen bond in the wild-type structure between Arg193 and ribose 5-phosphate induced upon substrate binding is indicated. Putative unfavorable interactions in the Val158Leu variant between the Leu158 side chain and both Arg193 (2.3 Å) and the overlaid ribose 5-phosphate (2.5 Å) prevent ribose 5-phosphate from binding in an orientation optimal for catalysis.
Again using the bioretrosynthetic strategy and monitoring inosine and didanosine production from a coupled reaction with PNP or PNP-46D6, we screened the saturation library against the natural (ribose 5-phosphate) and target non-natural (dideoxyribose 5-phosphate) substrates to compare selectivity changes, and found that the Ser154Gly and Ser154Ala mutations of PPM had the greatest improvements. By including a vast excess of PNP or PNP-46D6 in the reactions, we could calculate accurate kinetic parameters for PPM variants toward both the natural and non-natural substrates (Supplementary Table 1). Comparison of the ratio of catalytic efficiencies for both substrates between each PPM variant and the wild-type enzyme revealed improved substrate selectivity in each enzyme, 49-fold in Ser154Gly and 70-fold in Ser154Ala. Cocrystal structures of the Ser154Gly variant with either ribose 5-phosphate or dideoxyribose 5-phosphate revealed ligand binding similar to that observed in the wild-type and Ser154Ala variant (Fig. 2c and Supplementary Fig. 2e-f). Although the Ser154Ala variant was slightly more selective than the Ser154Gly variant, the latter was chosen as the template for further evolution since the increased active site volume may better accommodate subsequent mutations.
Additional rounds of saturation mutagenesis were performed at Val158 and Ile195 on both the wild-type and Ser154Gly templates. The Val158Leu mutation was found to be the most improved on each template, however no favorable mutation was observed at the Ile195 position. Kinetic analysis via the same bioretrosynthetic strategy using coupled assays showed that the Val158Leu single mutation exhibited a 25-fold increase in KM and 25-fold decrease in kcat for ribose 5-phosphate, but a 2.5-fold decrease in KM and only a slight decrease in kcat for dideoxyribose 5-phosphate (Supplementary Table 1). In total, this single mutation reduced ribose 5-phosphate catalytic efficiency 881-fold such that it was only 1.2-fold greater than that of dideoxyribose 5-phosphate (Supplementary Table 1). The Ser154Gly/Val158Leu double mutant also showed substantially improved dideoxyribose 5-phosphate selectivity (>300-fold), however this benefit came at a severe loss in kcat for both substrates (Supplementary Table 1). Superposition of the structures of wild-type and Val158Leu PPM identified that the mutation introduced steric clash between the Leu158 side chain and both substrate and Arg193, a substrate-orienting residue for ribose 5-phosphate21 (Fig. 2d). These unfavorable interactions would explain the dramatic increase in ribose 5-phosphate KM observed in the Val158Leu variant (Supplementary Table 1) and explains why cocrystallization with both natural and non-natural substrate did not result in the appearance of interpretable electron density (50 datasets).
Directed Evolution of PPM
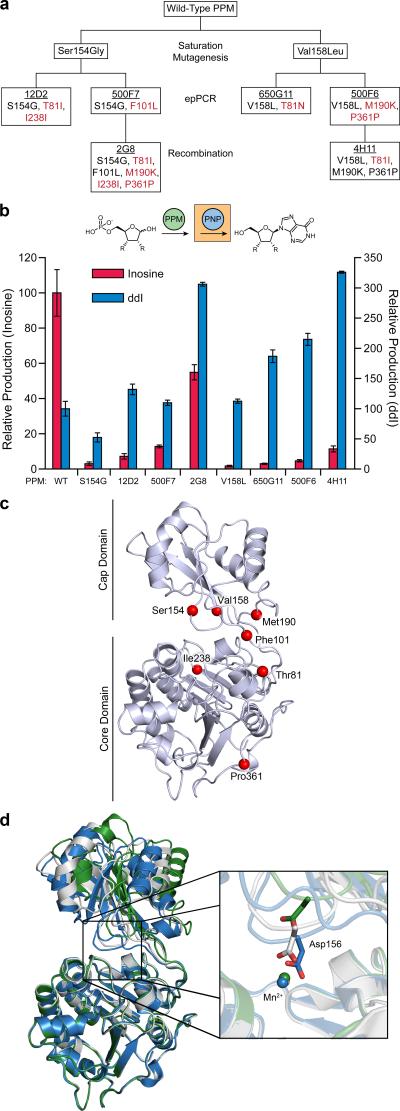
Following the structure-based mutagenesis, additional mutagenesis was required to improve PPM turnover for the target substrate. For this, we performed random mutagenesis by epPCR. Since both the Ser154Gly and the Val158Leu variants had large substrate selectivity changes through distinct and potentially competitive mechanisms, these were separately used as templates for random mutagenesis (Fig. 3a). In the bioretrosynthetic coupled assay, the top two PPM variants from first generation epPCR libraries for each template provided 150-250% higher dideoxyribose 5-phosphate turnover in cell lysate than the respective progenitor (Fig. 3b). These variants were 12D2 (Thr81Ile and Ile238Ile silent mutation) and 500F7 (Phe101Leu) from the Ser154Gly template and 650G11 (Thr81Asn) and 500F6 (Met190Lys and Pro361Pro) from the Val158Leu template (Fig. 3a). Kinetic characterization of the top performing clone in each initial library screen (i.e. 12D2 and 650G11) revealed increased turnover of both substrates, as measured through increased production of nucleosides, accompanied by improvements in selectivity reflected in KM values (Supplementary Table 1). While the Ser154Gly 12D2 variant showed 8.4-fold preference for ribose 5-phosphate, selectivity of the Val158Leu 650G11 variant was entirely reversed, actually favoring dideoxyribose 5-phosphate at 1.4-fold over the natural substrate after >1400-fold change in selectivity (Supplementary Table 1).
Figure 3. Lineage and characterization of PPM variants through generations of evolution.
(a) Lineage tree of PPM variants. Clone name is underlined, with mutations listed below. New mutations accumulated through the indicated method of mutagenesis are shown in red. (b) Comparison of changes in ribose 5-phosphate and dideoxyribose 5-phosphate turnover rate by PPM variants in cell lysate normalized to wild-type PPM, presented as changes in nucleoside production through the two enzyme product-screening assay. Assays were performed in tandem with wild-type PNP or PNP-46D6 using 1 mM ribose 5-phosphate or dideoxyribose 5-phosphate, respectively. Data are mean ± s.d. (n=3). Wild-type PPM turnover is 3.18 ± 0.52 μM min−1 for ribose 5-phosphate and 0.486 ± 0.083 μM min−1 for dideoxyribose 5-phosphate. (c) Location of amino acid substitutions mapped on the structure of wild-type PPM with ribose 5-phosphate bound (PDB entry 3M8Z21). Positions 81, 238 and 361 are located in the core domain, 154 and 158 are located in the cap domain and 101 and 190 are in the hinge region connecting the two domains. (d) Domain movement observed in 4H11 structure. The core and cap domains of the 4H11 variant (green) are related by a different interdomain angle than wild-type PPM in both its active, phosphorylated form (gray, PDB entry 3TWZ42) and the unphosphorylated and unactive form (blue, PDB entry 3TX042). One consequence of the cap domain movement is highlighted in the inset. Asp156 has previously been shown to coordinate an active site Mn2+. However, in the 4H11 structure, the location of this coordinating residue is shifted 4.6 Å away from the Mn2+ and the coordination sphere is instead completed by water molecules. Water molecules are not shown.
To maximize the improvement from this round of evolution, each of these four variants was subjected to random recombination using multiple-site directed mutagenesis. Clones 2G8 in the Ser154Gly lineage and 4H11 of the Val158Leu lineage were the most active recombinants in the screen (Fig. 3a,c). In total, the 2G8 variant showed approximately 300% improved didanosine production while the 4H11 variant demonstrated nearly 325% greater activity than wild-type PPM in cell lysate through the tandem reaction with PNP-46D6 (Fig. 3b). In tracking changes of ribose 5-phosphate activity in cell lysate through PPM evolution, the Ser154Gly and Val158Leu variants each lost greater than 95% of natural substrate activity (Fig. 3b). Successive rounds of evolution recovered a portion of the lost activity, reaching 55% and 11% of the wild-type level for 2G8 and 4H11, respectively.
Overall, most catalytic parameters were similar between the final variant from each lineage (Supplementary Table 1). The one notable exception was in KM values for ribose 5-phosphate. The 2G8 variant showed improved binding (KM of 230 μM) as compared to 4H11 (KM of 2830 μM). The contribution of this difference in KM is reflected in the substrate selectivity of each enzyme; wild-type PPM favors ribose 5-phosphate >1000-fold while selectivity in 4H11 was reduced to a close to equal competence for dideoxyribose 5-phosphate (Supplementary Table 1). To identify the molecular basis for this improvement in selectivity, we determined the structure of this variant. The active site of PPM is located between two domains, with the substrate binding residues on the cap domain and the catalytic residues on the core domain. Interestingly, the 4H11 structure was associated with an interdomain rotation that changed the alignment of the substrate binding and catalytic residues at the active site (Fig. 3d). Moreover, the interdomain rotation results in a more open active site conformation than was previously observed in wild-type structures. This interdomain angle change is likely caused by the mutations at positions 81 and 190, which appear to destabilize important hinge region contacts upon mutation.
Cocrystallization with ribose 5-phosphate or dideoxyribose 5-phosphate did not result in the appearance of clear electron density in the active site. While somewhat complicated by the interdomain twist, manual docking of each substrate into the 4H11 variant was possible by basing ligand position on the location on Ser154, a residue previously identified as important for ribose 5-phosphate binding21 (Fig. 2a, 3c) and located in the cap domain. In this docking mode, the approach of substrate to the catalytic residues is different in the 4H11 variant than it is in the wild-type enzyme. While speculative, this suggests that the observed changes in catalytic parameters in the 4H11 variant may be the result of altered interdomain interactions, which alter presentation of the substrate to the active site during catalysis.
Bioretrosynthetic Step 3: RK
Implementation of a third retro-extension required identification of a progenitor enzyme capable of phosphorylating dideoxyribose to synthesize the non-natural substrate dideoxyribose 5-phosphate for PPM. To this end, we measured didanosine production from dideoxyribose by the evolved PPM and PNP variants in tandem with five candidate kinase progenitor enzymes: E. coli RK23, S. aureus RK24, B. subtilis fructokinase25, E. casseliflavus glycerol kinase26 or B. subtilis hydroxyethylthiazole kinase27. Although didanosine formation was detected in all five systems via tandem enzyme HPLC/MS assay, production was highest in the pathways containing the RK homologs. The E. coli variant slightly outperformed the S. aureus enzyme and is more thoroughly characterized and was therefore selected for use in the full pathway (Supplementary Fig. 3).
Pathway optimization experiments revealed that ATP inhibited PPM at high concentrations (Data not shown). In order to maintain sub-inhibitory concentrations of ATP over extended reaction times, it was necessary to include a cofactor regeneration system along with the kinase progenitor23. We incorporated an ATP recycling system consisting of pyruvate kinase, adenylate kinase and phosphoenolpyruvate to reduce the ATP concentration and prevent loss of cofactor equivalents due to hydrolysis.
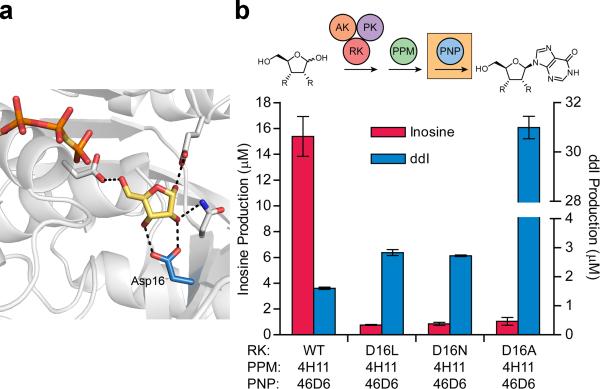
The functional five enzyme system facilitated preliminary mutagenesis of RK. Analysis of an available costructure of E. coli ribokinase with ribose (1GQT28) shows that the active site residue Asp16 interacts with both the C2 and C3 hydroxyls of ribose29 (Fig. 4a). Notably, a residue equivalent to Asp16 is conserved throughout adenosine kinase, ribokinase30, aminoimidazole riboside kinase, ADP-dependent glucokinase31, 6-phosphofructokinase32 and ketohexokinase33 members of the ribokinase-like superfamily, and is present in phosphofructokinase-234 and tagatose-6-phosphate kinase35, in each case appearing to make similar interactions with the respective sugar substrates. As these two hydroxyls are not present in dideoxyribose, we replaced the charged aspartate with non-polar and space filling leucine, the polar isostere asparagine, or alanine (Asp16Leu/Asn/Ala mutants) and measured nucleoside production through the engineered pathway. All three variants reduced inosine formation to ≤1 μM at five minutes (Fig. 4b). The Asp16Leu/Asn mutations provided slight increases in didanosine production, however the Asp16Ala RK (RK-Asp16Ala) exhibited a 20-fold increase in didanosine production compared to wild-type RK, forming 31 μM after 10 h through the three evolved enzymes (Fig. 4b).
Figure 4. Optimization of RK via didanosine production assay.
(a) The interaction between the active site residue Asp16 and bound ribose is highlighted in wild-type E. coli RK (PDB entry 1GQT28). This interaction was targeted for removal in preliminary site directed mutagenesis studies by mutation to leucine, asparagine and alanine (water molecules not shown). (b) RK variants were tested for production of inosine and didanosine from ribose and dideoxyribose, respectively, using the three enzyme engineered pathway and ATP regeneration cycle. Data are mean ± s.d. (n=2).
Analysis of enzyme variants in the context of the pathway
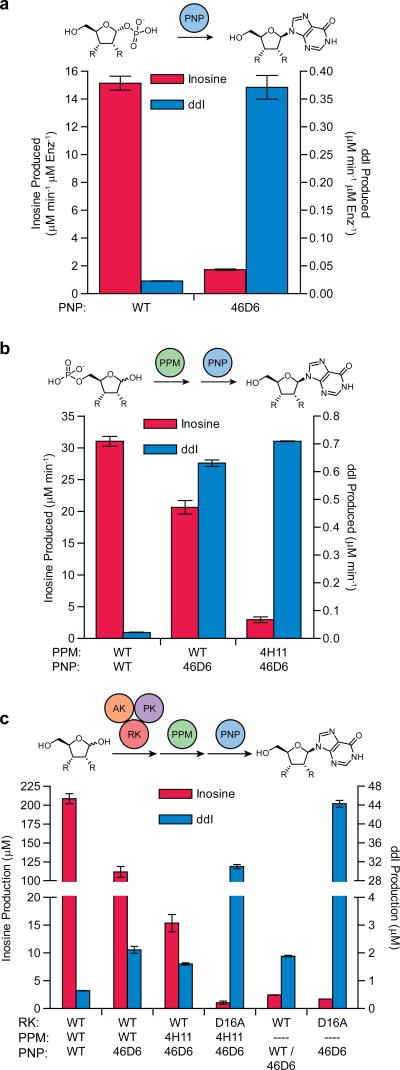
To evaluate the improvements engendered by the separate pathway product selection-based directed evolution experiments, we measured changes in nucleoside production after replacing wild-type enzymes with the engineered counterparts in increasingly longer didanosine biosynthetic pathways. Figure 5a shows the selectivity and turnover improvements resulting from PNP evolution alone, totaling 140-fold change in substrate selectivity and 16-fold increased didanosine production. Extending in vitro nucleoside production to the two enzyme system, the combination of wild-type PPM and wild-type PNP showed a 1420-fold bias for ribose 5-phosphate over the dideoxy substrate (Fig. 5b). Incorporating the evolved PNP variant into the pathway provided a 29-fold increase in didanosine production and a small loss in inosine formation. The effect on inosine biosynthesis was further compounded after pairing the two optimized enzymes, showing a 342-fold total change in substrate selectivity to create a tandem evolved biosynthetic pathway with 32.5-fold improved didanosine production and only a 4-fold preference for the natural substrates.
Figure 5. Selectivity and activity changes in selected variant enzymes as assessed using coupled assays.
Nucleoside production was measured via HPLC/MS analysis (a) from in situ generated sugar 1-phosphates in one step biocatalysis (PNP only), (b) from sugar 5-phosphates in the two-step tandem pathway (PPM and PNP) or (c) from ribose or dideoxyribose in the full biosynthetic pathway (RK, PPM and PNP) or the pathway without PPM. Enzyme variants used in each reaction are listed under bars (dashed line means no variant included). Direct phosphorylation of the sugar C1 position by wild-type RK in panel (c, second from right) was tested in tandem with wild-type PNP for inosine production and PNP-46D6 for didanosine production to allow the best detection of activity. Turnover in panel (a) was normalized to PNP variant concentration and assay length. Production in panel (b) was normalized to incubation time. The full inosine pathway in (c) was incubated for 5 min, while didanosine production was for 10 h. Reactions in (c) without PPM were incubated for 10 h, however inosine production was normalized to 5 min to be directly comparable to production by the full pathway with PPM. Data are mean ± s.d. (n=2).
Measuring nucleoside formation through the complete biosynthetic pathway, the all wild-type pathway efficiently produced inosine, reaching 208 μM in five minutes, while didanosine production was barely detectable with only 0.6 μM ddI formed after 10 h. Introducing the evolved PNP-46D6 and PPM-4H11 variants reduced inosine production by 2-fold and 13.6-fold, respectively, while providing 3-fold and 2.5-fold gains in didanosine production, respectively (Fig. 5c).
Substantial didanosine production gains had been observed in PNP only (16-fold) and PPM-PNP tandem (32.5-fold) systems after enzyme evolution (Fig. 5a and b). However, retro-extension of the pathway with wild-type ribokinase reduced didanosine production, consistent with its low phosphorylation activity on dideoxyribose. This highlighted the need for engineering of ribokinase to increase turnover on the non-natural substrate. Incorporating the RK-Asp16Ala variant into the pathway increased formation of didanosine 50-fold as compared to the pathway with all wild-type enzymes (Fig. 5c). With this improvement, and the ~200-fold reduced production of inosine through the three evolved enzymes, the combination of RK-Asp16Ala, PPM-4H11 and PNP-46D6 resulted in a 9,544-fold total change in nucleoside productivity.
The unexpectedly large increase in productivity in the pathway containing RK-Asp16Ala prompted further investigation of the improved RK. In particular, we considered the possibility of anomeric (C1) phosphorylation. To our knowledge, no enzyme has previously been reported to phosphorylate D-ribose at the 1-position in this or any other superfamily. Excluding PPM-4H11 and assaying RK-Asp16Ala in tandem with PNP-46D6 confirmed this unreported activity of RK. This two enzyme system has the highest didanosine production (44 μM), verified by HPLC/MS comparison to synthetic didanosine standard (Supplementary Fig. 4). This 4.4% yield from dideoxyribose in this two enzyme system was 70-fold greater than the wild-type three enzyme pathway. Notably, the three enzyme pathway including PPM-4H11 demonstrated decreased yields (Fig. 5c), likely due to conversion of ddR1P into dideoxyribose 5-phosphate by the engineered PPM. This has the effect of diverting ribose into the PPM-dependent pathway. Taken together these results suggests that the majority of the RK-Asp16Ala effect on activity in the full pathway was due to increased direct phosphorylation of the anomeric hydroxyl group rather than expected ribose 5-phosphorylation.
Interestingly, our results also demonstrate low level anomeric phosphorylation activity in wild type RK (~1%). However, targeted mutagenesis substantially and selectively improved activity of RK for dideoxyribose. Therefore RK-Asp16Ala potentially provides a unique foundation for further engineering catalytic activity.
DISCUSSION
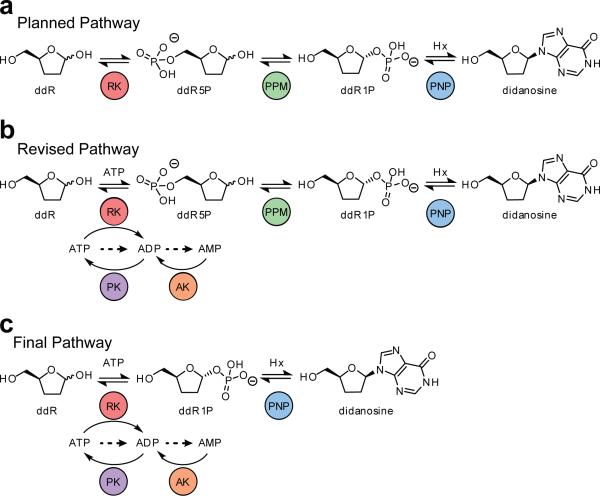
A number of factors account for the current dearth of engineered biosynthetic pathways involving multiple unnatural substrates. For example, individual enzymes in a pathway must be engineered or evolved for optimal intermediate and product formation. However the necessary level of catalytic improvement is unknown prior to a directed evolution campaign, and generalizable methods for the assembly and evolution of unnatural pathways are currently unavailable. For these reasons, we are interested in applying evolutionary theories of metabolic pathway biogenesis to contemporary synthetic biology. Originating in 1945, the retro-evolution hypothesis was the first proposal concerning the evolutionary origin of metabolic pathways. Despite the recognition of the apparent efficiencies of this strategy for artificial pathway construction16,18,36, it has remained untested prior to this study. Initial conceptualization using known enzymatic activities resulted in the planned didanosine pathway shown in Figure 6a. Following directed evolution of the ultimate and penultimate enzymes PNP and PPM, an ATP-recycling system was included to optimize precursor concentrations and yield to allow engineering of the antepenultimate enzyme, RK, again via a didanosine formation assay (Fig. 6b). Finally, RK engineering provided an unanticipated biochemical pathway bypass leading to substantial improvement in didanosine production (Fig. 6c).
Figure 6. Progression of pathway evolution throughout enzyme engineering stages.
(a) The original proposed three enzyme biosynthetic pathway. (b) The full pathway including the ATP regeneration cycle after identifying the kinase progenitor for phosphorylation of dideoxyribose and recognizing inhibition of PPM by nucleotide substrates and products. (c) The two step didanosine biosynthetic pathway as a result of the discovered bypass activity of the engineered RK. Dashed arrows indicate expected degradation products.
Enzyme activities were iteratively improved via mutagenesis, selecting for didanosine formation, and the variants were subsequently substituted back into the intact pathway. This demonstrated selectivity improvements of each engineered enzyme for the non-natural substrate in the context of the full pathway. As shown in Figure 5c, replacing wild-type PNP with evolved PNP (46D6) improved selectivity and didanosine formation. Substitution of the evolved antepenultimate enzyme PPM (4H11) for wild-type PPM resulted in a five-enzyme prototype pathway with further increased selectivity. Finally, exchanging the engineered ribokinase (RK-Asp16Ala) revealed both turnover improvements and an unexpected biochemical bypass with substantial improvement in didanosine production through a shorter biosynthetic pathway. The final improvements in pathway substrate selectivity (8,300-fold) and improvements in turnover for target substrates (70-fold) of this shortened pathway illustrate a further benefit of the retro-evolution hypothesis in vitro.
Concurrently, mutational studies and structural analyses generated new insights into mechanistic enzymology of these enzymes and provided foundational data for future engineering studies. Structural analysis of targeted PPM variants identified residues that modulate selectivity, and directed evolution demonstrated potential for improving turnover in these variants. Intriguingly, the variant with the most improved selectivity toward the non-natural substrate was associated with a large interdomain angle change. This is consistent with recent work with dihydrofolate reductase indicating that during natural evolution, enzymes may alter their reactivity toward alternative substrates by changing their motions37,38.
From a strategic perspective, the bioretrosynthetic method applied here illustrated several advantages and potential constraints when considering future campaigns. Of benefit, the single terminal enzyme screening assay was adaptable towards increasingly tandem reactions and was capable of detecting improvements in enzyme selectivity toward the non-natural substrate, even for the first step in the pathway. The unexpected discovery of a pathway shortening biosynthetic bypass revealed an additional benefit of bioretrosynthesis. In this case, it allows retro-extension from the ultimate enzyme (PNP) rather than the penultimate enzyme (PPM). Importantly, the unexpected detection of 1-phosphorylation regiochemistry was enabled by product monitoring of bioretrosynthesis, and may not have been noticed in a kinase optimization assay. For example, typical UV absorbance based (ribo)kinase assays would indeed have identified apparently improved kinases in this study, as would direct HPLC-MS analysis of phosphorylation products. However, without a functional dideoxyribose phosphate substrate tandem assay optimized for product selection, the pathway shortening 1-phosphorylation activity would have remained unselected as neither assay can distinguish the regiochemistry of phosphorylation (substrate phosphorylation chromatograms shown in Supplementary Fig. 5).
A second advantage is that, as in chemical retrosynthetic analysis17, the product formation focus of bioretrosynthesis potentially minimizes methodological dead-ends during pathway assembly. This aspect is particularly advantageous for enzymes difficult to optimize individually for technical reasons, such as PPM, where the substrate and product are isobaric and chromatographically and spectroscopically indistinguishable.
Several constraints of bioretrosynthesis are shared with directed evolution of pathways through forward concatenation. In both cases it is necessary to optimize precursor and cofactor supply. Substrate or product enzyme inhibition is always a possibility. Most importantly, signal attenuation with increasingly tandem reactions is likely. Indeed, in the present study, it was critical to improve didanosine formation from PNP to provide sufficient signal intensity for accurate and reproducible detection in order to proceed to evolution of the next retro-consecutive step.
A unique constraint of the bioretrosynthetic approach is that it is contingently linear, i.e. enzymes must be improved one at a time in order to take advantage of the economy of product screening or selection. By comparison, in a more traditional, convergent approach, where screening and selection assays are developed for each enzymatic step, multiple enzymes can be optimized simultaneously and then assembled into a complete pathway. Nevertheless, it appears that the assay and substrate requirements for this traditional approach represent a more substantial barrier than individual optimization in practice. Indeed, we are aware of only one reported example of a pathway engineering of this type using unnatural substrates: the atorvastatin side chain11. Moreover, simultaneous evolution for individial enzyme turnover does not necessarily optimize product formation in multi-step sequences. For example, extensive multivariate optimization of individual enzymatic activities in taxadiene and amorphadiene biosynthesis in E. coli was required to generate highest production through the pathway6,39. For these reasons, we propose that the contingent linearity of bioretrosynthetic engineering may be offset by the efficiencies in assay development and post-assembly flux optimization.
Eventual in vivo didanosine production may be realized via additional multiple parameter optimization: improved enzyme turnover and/or substrate selectivity of the pathway enzymes, continued retro-extension to enzymatically generate dideoxyribose, further engineering precursor supply, and optimizing efflux and resistance. Of note, the engineered RK/PNP system we describe may represent a specific solution for the generation of dideoxynucleosides drugs. Direct anomeric phosphorylation of 2- or 3-substituted ribosides (such as would be required for the biosynthesis of the 2’-difluoro nucleoside Gemcitabine, or AZT) is not favored by RK-Asp16Ala. These nucleosides therefore require the development of a PPM generalist activity, as we have discovered herein. Given the breadth of structural diversity of both the sugar and nucleobase moieties in nucleoside analog drugs, both PPM and RK variant enzymes may find complementary application in biocatalytic generation of a variety of compounds within this class of therapeutics.
ONLINE METHODS
Mutant Library Generation
Site-directed and saturation mutagenesis were performed using the QuikChange II mutagenesis kit (Stratagene) with either the wild-type PPM (Genbank Accession Number Q818Z9.1) or the corresponding Ser154Gly template in pET28a+ vector43 or with wild-type ribokinase (Genbank Accession Number ACT45432.1) in pCDFDuet vector23. Template plasmid DNA was digested with DpnI prior to purification of the mutant plasmid by QIAquick PCR Purification Kit (QIAgen, Inc.) and subsequent transformation. Error prone PCR (epPCR) was performed using the GeneMorph II Random Mutagenesis kit (Stratagene, Inc.) using 25 replication cycles and 10 ng/μL template plasmid. The epPCR products were gel purified, digested with NheI/XhoI (New England Biolabs, Inc.) and gel purified before ligation into purified pET28a+ vector that had been similarly digested and treated with alkaline phosphatase (New England Biolabs). Ligations were performed for 15 hours at 4 °C using T4 DNA ligase (Promega, Inc.) and contained 3:1 insert:vector ratio. Pellet Paint Co-Precipitant (Novagen, Inc.) was used to concentrate DNA before transformation. Libraries were generated to create pools of variants possessing 1.5 - 2.5 mutations per gene on average in 10 randomly sequenced clones from each library. Random recombination of epPCR mutations was performed using the QuikChange Multi Site-Directed Mutagenesis kit (Stratagene) where PCR samples contained a single template and a forward primer for each mutation not present at a position on the template, allowing any additional mutations to be incorporated at random in any combination. For example, the 500F6 sample contained primers for the T81I, T81N, F101L and I238I mutations. PCR sample preparation and thermal cycling for recombination of mutations were performed following the recommended protocol by the kit manufacture's manual. Primers used are listed in Supplementary Table 2.
PPM Library Screening
Colonies were picked to 96-well round bottom plates containing 100 μL LB medium with 50 μg/mL kanamycin and grown for 24 h at 37 °C with shaking at 200 rpm. Plates held four wells of negative control (E. coli with empty pET28a+ vector), four wells of positive control (vector with template gene) and 88 wells of the mutant library. For saturation mutagenesis libraries (176 clones), plates were replicated to 100 μL and 150 μL LB medium (wild-type PPM template) or 50 μL and 150 μL LB medium (Ser154Gly PPM template) and grown for an additional 24 h. The 50 μL cultures of Ser154Gly PPM were induced by addition of 50 μL LB medium containing 50 μg/mL kanamycin and 2 mM IPTG and grown for an additional 24 h. Cells were collected by centrifugation at 1600 rcf and the broth was removed by inversion before storing the plates at −80 °C until ready for assay. Initial 100 μL overnight plates of the epPCR and recombination libraries (1056 and 96 clones, respectively) were copied to 50 μL LB medium with kanamycin and grown for an additional 24 h before collecting the cells by centrifugation.
PPM activity was determined in tandem assays with purine nucleoside phosphorylase where hypoxanthine consumption was measured by xanthine oxidase at endpoints using a typical hypoxanthine detection assay44. Thawed cell pellets were resuspended in 100 – 200 μL lysis mix containing 0.1 mM MnCl2, 0.25 mg/mL egg white lysozyme (Sigma) and DNase I (Sigma) in 25 mM Tris-HCl, pH 8 and incubated for 10 min at 25 °C before undergoing a single freeze/thaw cycle from −80 °C to a 37 °C incubator for 40 – 60 min. After centrifugation at 1600 rcf, 25 μL of the clarified cell lysate was transferred to a 96-well flat bottom plate and 55 μL of an assay mix was added to initiate the reaction. Final concentrations of components in 80 μL reactions for the ribose 5-phosphate screen were 0.1 mM MnCl2, 5 μM PNP, 1 μM glucose 1,6-bisphosphate (Sigma), 600 μM hypoxanthine (Sigma) and 1 mM ribose 5-phosphate (Sigma). Conditions in 80 μL reactions for the dideoxyribose 5-phosphate screen were the same but with 10 μM hPNP-46D6 variant and 1-2 mM dideoxyribose 5-phosphate. Assays were incubated at room temperature for 12 min (ribose 5-phosphate) or from 50-90 min (dideoxyribose 5-phosphate) before being quenched by addition of either 30 μL 1 M HCl (ribose 5-phosphate) or 30.5 μL 1 M NaOH (dideoxyribose 5-phosphate). After a minimum of 30 min, the solution was neutralized using 30 μL 1 M NaOH or 29.5 μL 1 M HCl before addition of 35 μL developing solution containing 0.2% Triton X-100, 7.5 mM iodonitrotetrazolium chloride and xanthine oxidase in 25 mM Tris-HCl, pH 8. Hypoxanthine consumption was determined by measuring absorbance of the colored formazan at 546 nm and normalized to percent activity in comparison to the positive and negative controls. The top ~45 hits from each primary screen were regrown from fresh transformants and retested in duplicate to validate activity. The best 4 - 9 hits after validation were screened in a tertiary assay in duplicate under the same conditions as above with 500, 1000 and 2000 μM dideoxyribose 5-phosphate to select the top hit from each round. The Ser154Gly variant isolated through saturation mutagenesis contained a codon mutation from TCT to GGG and the Val154Leu variant possessed a codon mutation from GTA to CTG in the respective mutagenic primers listed in Supplementary Table 2. Mutations gained through epPCR and used in the recombination mutagenesis steps are indicated in the primers given for the recombination protocol in Supplementary Table 2.
Libraries containing the top hits from each round were freshly prepared to directly compare activity changes. Plates containing 100 μL cultures of colonies hosting empty pET28a+, wild-type PPM, Ser154Gly, 12D2, 500F7, 2G8, Val158Leu, 650G11, 500F6 and 4H11 were grown for 24 h and replicated to 50 μL to grow an additional 24 h before collecting the cells. Cell pellets were lysed in 200 or 150 μL lysis mix and assayed with 1 mM ribose 5-phosphate or dideoxyribose 5-phosphate, respectively, as described above. Reactions were incubated for 10 – 90 min (ribose 5-phosphate) or 45 – 60 min (dideoxyribose 5-phosphate) before being quenched with 30.5 μL 1 M NaOH then neutralized and developed as described above. Turnover was normalized by incubation length then to activity of wild-type PPM.
Expression and Purification
Plasmids containing PPM variants, PNP variants19, RK variants, adenylate kinase or pyruvate kinase23 were transformed into Escherichia coli BL21(DE3) and grown at 37 °C in LB medium supplemented with 50 μg/mL kanamycin or 50 μg/mL streptomycin (RK variants) and induced with 1 mM IPTG for 3 – 6 h after OD600 had reached 0.5 – 0.6. Plasmids containing Staphylococcus aureus ribokinase24, Bacillus subtilis fructokinase25, Enterococcus casseliflavus glycerol kinase26 and Bacillus subtilis hydroxyethylthiazole kinase27 were transformed into E. coli BL21(DE3) and grown at 37 °C in LB or 2xYT medium (hydroxyethylthiazole kinase only) supplemented with 50 μg/mL ampicillin and were induced with 0.5 – 2 mM IPTG for 20 h at 16 °C using the published protocols. E. casseliflavus glycerol kinase and B. subtilis hydroxyethylthiazole kinase were also cotransformed with pREP4 plasmid and maintained in culture using 25 μg/mL kanamycin. Cell pellets were resuspended in Buffer A (50 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole, pH 7.4) and disrupted by passing through a French Pressure cell (1400 psi). The clarified lysate was applied to a HisTrap FF Ni affinity column (GE Healthcare, Inc.) and washed with 10% Buffer B (Buffer A with 500 mM imidazole). Protein was eluted by a linear gradient from 10% Buffer B to 60% Buffer B, before a step to 100% Buffer B to fully elute the column. Purified enzymes were concentrated, desalted and exchanged into 25 mM Tris-HCl, pH 8 or 5 mM MgCl2 in 25 mM Tris-HCl, pH 8 (all kinase variants) before storage at −80 °C. Hydroxyethylthiazole kinase purification buffers also contained 2 mM β-mercaptoethanol and both hydroxyethylthiazole kinase and fructokinase were exchanged into Tris-HCl and MgCl2 buffer containing 2mM DTT before storage. Enzyme concentrations were determined using the BCA Protein Assay Kit (Thermo Scientific, Inc.). Xanthine oxidase was purified from raw bovine milk using previously reported protocols45.
PPM Kinetic Assays
PPM variant activity was measured in a tandem assays with catalytic excess of either PNP or hPNP-46D6 in the presence of hypoxanthine to produce (dideoxy)inosine. Nucleoside produced in the assay was separated from other reaction components using a Luna Phenyl-Hexyl column (4.6 X 250 mm, Phenomenex) and an isocratic flow of 1.0 mL/min of 10 mM ammonium acetate in 95% water:5% acetonitrile, pH 6. Nucleosides were analyzed on a TSQ Quantum Access triple quadrupole electrospray ionization-LC/MS (Thermo, Inc.) using selected reaction monitoring fragmentation to the free nucleobase (inosine [M+H]+ 269 m/z and didanosine [M+H]+ 237 m/z transition to hypoxanthine [M+H]+ 137 m/z) with 2-deoxyguanosine (Aldrich) as the internal standard ([M+H]+ 268 m/z to guanine [M+H]+ 152 m/z). Nitrogen was used for both the auxiliary and sheath gases and was set to 45 units and 30 units, respectively. The following instrument parameters were used: source voltage 4.5 kV; vaporizer temperature 0 °C; capillary temperature 270 °C; tube lens 101 V; skimmer offset −5 V; collision energy −10 V. Data acquisition and analysis were conducted with Thermo Xcalibur software, version 2.1.
All reactions were performed in 100 μL volumes in 96-well plates. Wild-type or variant PPM was activated at a concentration 10-fold higher than that used in the assay by incubation for 10 min at room temperature in 25 mM Tris-HCl and 0.1 mM MnCl2 with either 5 μM (wild-type PPM) or 10 μM (variant PPM) glucose 1,6-bisphosphate then held at 4 °C until assayed. Ribose 5-phosphate activity assays contained 0.1 mM MnCl2, 5 μM PNP, 600 μM hypoxanthine and 0 – 500 μM, 0 – 1000 μM or 0 – 4000 μM ribose 5-phosphate in 25 mM Tris-HCl, pH 8. Assays for activity on dideoxyribose 5-phosphate contained 0.1 mM MnCl2, 10 μM hPNP-46D6, 600 μM hypoxanthine and 0 – 5000 μM dideoxyribose 5-phosphate in 25 mM Tris-HCl, pH 8. PPM variant concentrations ranged from 0.02 – 1 μM for ribose 5-phosphate assays and 0.15 – 1 μM for dideoxyribose 5-phosphate assays. Reactions were initiated by 10 μL addition of the sugar 5-phosphate substrate to 90 μL mix containing all other components and were incubated for 2 – 8 min at room temperature before being quenched by addition of 5 μL 2 M NaOH. After 30 min, 5 μL of 2 M HCl/1 M CaCl2 was added to neutralize the mixture and the assay plate was centrifuged to pellet the precipitates. A 40 μL aliquot of each sample was combined with 10 μL of 50 μM 2-deoxyguanosine internal standard to prepare the sample for LC/MS analysis. Inosine and didanosine formation was quantified by relative peak area of analyte to a 10 μM 2-deoxyguanosine internal standard in comparison to a standard curve made using authentic inosine (Acros Organics) and didanosine (3B Pharmachem (Wuhan) International Co. Ltd.). Retention times were approximately 5 min for inosine, 6.2 min for 2-deoxyguanosine and 14 min for didanosine.
Characterization of dideoxyribose activity of kinase enzymes
Didanosine production by wild-type E. coli ribokinase23, S. aureus ribokinase24, B. subtilis fructokinase25, E. casseliflavus glycerol kinase26 or B. subtilis hydroxyethylthiazole kinase27 was screened to determine the most active enzyme. Reactions contained 0.6 mM MnCl2, 0.6 mM MgCl2, 30 mM KCl, 10 μM hPNP-46D6, 10 μM 4H11 PPM, 100 μM above kinase, 5 μM adenylate kinase, 5 μM pyruvate kinase, 15 μM glucose 1,6-bisphosphate, 3 mM hypoxanthine, 1 mM ATP, 2 mM phosphoenolpyruvate and 1 mM dideoxyribose in 25 mM Tris, pH 8. Reactions were initiated by addition of dideoxyribose and incubated 22 h at room temperature. Aliquots of 250 μL were quenched by loading onto Oasis® HLB (3 mL, 60 mg) solid-phase extraction cartridges (Waters) preconditioned with 3 mL methanol and 3 mL water. Loaded cartridges were then washed with 1 mL water and nucleosides were eluted with 1.5 mL methanol46. The methanol fractions were evaporated to dryness and the remaining residue was reconstituted in 50 μL water and LC/MS samples were prepared as previously described.
In vitro production of inosine and didanosine
Nucleoside production turnover through the one step (PNP only), two step (PPM and PNP) and full three step (RK, PPM and PNP) pathways was measured using purified enzymes. For the one step pathway, sugar 1-phosphate was generated in situ from (dideoxy)ribose 5-phosphate by preincubation with either wild-type PPM or 4H11 PPM. The PPM variant (5.27 μM) was activated in the presence of 0.11 mM MnCl2, 5.27 μM glucose 1,6-bisphosphate and 3.34 mM hypoxanthine in 25 mM Tris, pH 8 for 15 min in a volume of 900 μL before addition of 50 μL 50 mM sugar 5-phosphate (2.63 mM final) and incubated for 4 h to create the sugar 1-phosphates. Each reaction was then passed through a 10 kDa molecular weight cutoff filter at 4 °C to remove PPM and aliquoted to 95 μL portions on a 96-well plate before 5 μL of 0.4 μM PNP variant (for inosine production) or 10 μM PNP variant (for didanosine production) was added to initiate the reaction. The final reaction performed in duplicate contained 20 nM or 500 nM PNP variant, 0.1 mM MnCl2 and 3 mM hypoxanthine in 25 mM Tris, pH 8 with equilibrium concentrations of (dideoxy)ribose 5-phosphate and (dideoxy)ribose 1-phosphate and trace amounts of glucose 1,6-bisphosphate. Assays were incubated for 5 min (for ribose 1-phosphate) and 20 min (for dideoxyribose 1-phosphate) before being quenched by addition of 5 μL 2 M NaOH then neutralized after 30 min by addition of 5 μL 2 M HCl/1 M CaCl2. Assay plates were centrifuged to pellet the precipitates and samples were prepared for LC/MS analysis as described above.
Catalysis via the two step pathway was performed on a 96-well plate in 100 μL reactions in duplicate comparing nucleoside production through multiple pairs of PPM and PNP variants from sugar 5-phosphates. Reactions were initiated by addition of 10 μL of 10 mM sugar 5-phosphate to 90 μL of assay mix containing all other reaction components. Final reaction conditions were 0.1 mM MnCl2, 0.5 μM glucose 1,6-bisphosphate, 0.1 μM PPM variant, 2 μM PNP variant, 2 mM hypoxanthine and 1 mM sugar 5-phosphate. Reactions were incubated for 5 min (ribose 5-phosphate) and 20 min (dideoxyribose 5-phosphate) before quenching, neutralizing and preparing samples for LC/MS as described above.
Nucleoside production through the three step pathway and ATP regeneration cycle was performed as follows. Conditions for inosine production were 0.6 mM MnCl2, 0.6 mM MgCl2, 30 mM KCl, 1 μM PNP variant, 0.1 μM PPM variant, 1 μM RK variant, 5 μM adenylate kinase, 5 μM pyruvate kinase, 0.5 μM glucose 1,6-bisphosphate, 0.5 mM ATP and 1 mM ribose. Reactions for didanosine production were as listed above with 10 μM PNP variant, 10 μM PPM variant, 100 μM RK variant, 15 μM glucose 1,6-bisphosphate and 1 mM dideoxyribose. Duplicate assays were initiated by addition of ribose or dideoxyribose and incubated at room temperature for 5 min (ribose) or 10 hr (dideoxyribose). Aliquots of 250 μL were quenched using Oasis® HLB cartridges as described above. The dried methanol fraction was reconstituted in 250 μL water (ribose pathway) or 50 μL water (dideoxyribose pathway) and LC/MS samples were prepared as described. Reactions for production of nucleosides without PPM were performed identically without the addition of the PPM variant and were incubated for 10 h before quenching. Samples were prepared for LC/MS as previously described. Turnover in the reaction mix without the sugar substrate incubated for the same time were subtracted from the final turnover measurements.
RK-Asp16Ala Product Assay
Sugar phosphate reaction products of RK-Asp16Ala and dideoxyribose were analyzed via HPLC-MS using a Hypercarb porous graphite column (3 X 50 mM, ThermoFisher Scientific) and an isocratic flow of 0.2 mL/min of 20 mM ammonium acetate, pH 6 and 0.1 % diethylamine21. Sugar phosphates were analyzed on a TSQ Quantum Access triple quadrupole electrospray ionization-LC/MS (Thermo, Inc.) on full scan mode. Traces shown in Supplementary Figure 5 are filtered for the sugar phosphate diethylammonium adduct (dideoxyribose 1-phosphate and dideoxyribose 5-phosphate [M+C4H12N]+ 272 m/z). Nitrogen was used for both the auxiliary and sheath gases and was set to 10 and 54 units, respectively. The following instrument parameters were used: source voltage 4.5 kV; vaporizer temperature 280 °C; capillary temperature 270 °C; tube lens −138 V; skimmer offset −5 V, collision energy −10 V. Data acquisition and analysis were conducted with Thermo Xcalibur software, version 2.1.
Reactions were performed in 250 μL volumes containing 0.6 mM MnCl2, 0.6 mM MgCl2, 30 mM KCl, 100 μM RK-Asp16Ala, 5 μM pyruvate kinase, 5 μM adenylate kinase, 0.5 mM ATP and 2 mM phosphoenolpyruvate in 25 mM Tris-HCl, pH 8. Reactions were initiated by addition of dideoxyribose to 1 mM and incubated at room temperature before quenching 50 μL aliquots in 50 μL methanol after 2, 4 and 9 hr. Quenched reactions were held on ice for at least 10 min before being centrifuged at 4 °C and analyzed by LC-MS. A dideoxyribose 1-phosphate standard was enzymatically prepared through phosphorolysis of didanosine by hPNP-46D6. A 250 μL reaction containing 5 mM KH2PO4, 100 μM didanosine and 10 μM hPNP-46D6 in 25 mM Tris-HCl, pH 8 was incubated for 1 h at room temperature before being quenched as described above. The coinjection sample analyzed contained equal volumes of the RK-Asp16Ala reaction and the dideoxyribose 1-phosphate standard mixed prior to analysis by LCMS. Synthesis of the dideoxyribose 5-phosphate standard is described in the Supplementary Note.
Crystallization, data collection, and structure determination of wild-type and variant PPM
Purification and preparation of the PPM variants for crystallography was performed as previously described43. Crystals of wild-type PPM, the Ser154Ala variant, and the Ser154Gly variants were all grown using the hanging-drop vapor diffusion method by combining 1 μL of protein solution (12 mg/ml enzyme in 1 mM MnCl2, 25 mM Tris-HCl, pH 7.4 unless otherwise noted) and 1 μL reservoir solution and incubating drops over the reservoir solution at 18 °C while crystals of the Val158Leu and 4H11 variants grew after combining 2 μL protein solution and 2 μL reservoir solution. Crystals of wild-type PPM grew over a reservoir solution containing 13% polyethylene glycol 3350, 50 mM MnCl2, 75 mM NH4CH3COO, and 100 mM Bis-Tris, pH 5.5. Crystals of the Ser154Ala variant grew over a reservoir solution containing 17% polyethylene glycol 3350, 50 mM MnCl2, 50 mM NH4CH3COO, and 100 mM Bis-Tris, pH 5.5. Crystals of the Ser154Gly variant grew over a reservoir solution containing 17% polyethylene glycol 3350, 50 mM MnCl2, 50 mM NH4CH3COO, and 100 mM Bis-Tris, pH 5.5. Crystals of the Val158Leu variant grew over a reservoir solution containing 20% polyethylene glycol 3350, 50 mM MnCl2, 50 mM NH4CH3COO, and 100 mM Bis-Tris, pH 5.45. Crystals of the 4H11 variant (20 mg/ml enzyme in 1 mM MnCl2, 25 mM Tris-HCl, pH 7.4) grew over a reservoir solution containing 25% polyethylene glycol 3350, 50 mM (NH4)2SO4, and 100 mM Bis-Tris, pH 5.45. Co-crystals of the Ser154Ala or Ser154Gly variants with ribose 5-phosphate were prepared by soaking fully formed crystals in a solution that contained all components of the reservoir solution and 5 mM ribose 5-phosphate for 30 minutes. Co-crystals of PPM variants with dideoxyribose 5-phosphate were prepared by soaking fully formed crystals in a solution that contained all components of the reservoir solution and 10 mM dideoxyribose 5-phosphate for 30 minutes. All crystals were cryoprotected with a solution that was 70% v/v reservoir solution and 30% glycerol before flash cooling by plunging into liquid nitrogen.
Data were collected at a temperature of 100 K using a wavelength of 0.979 Å and a MarMosaic225 CCD detector at the Advanced Photon Source (Argonne, IL) LS-CAT beamline 21-ID-G (Supplementary Tables 3, 4 and 5). Data were processed and scaled using the HKL suite of programs47. Space group and unit cell information for all crystals is listed in Supplementary Tables 3, 4 and 5. For wild-type PPM and the Ser154Ala and Ser154Gly variants, initial phases were determined by subjecting the structure of wild-type B. cereus PPM (PDB entry 3M8W21) to rigid body refinement using CNS48. For the Val158Leu and 4H11 variants, initial phases were obtained by molecular replacement with Molrep49 of the CCP4 Suite40, using PDB entries 3UN3 and 3M8Z, respectively. The model was refined using iterative rounds of model building in COOT50 and refinement in CNS48 and Refmac540,41 using Translation/Libration/Screw (TLS) refinement51. Ramachandran statistics are reported in Supplementary Table 7. Figures 2, 3c, d, 4 and Supplementary Figure 2 were made using PyMOL (Delano Scientific). RMSD calculations were performed using DaliLite52.
Supplementary Material
Acknowledgements
We thank V. Phelan, K. McCulloch, and C. Goodwin for assistance with data acquisition. We also thank Jianye Zang (Chinese Academy of Sciences), Andrzej Joachimiak (Argonne National Laboratory and University of Chicago), Josef Deutscher (CNRS) and Steven Ealick (Cornell University) for expression plasmids used in this study. This work was supported by T32 GM065086 and the D. Stanley and Ann T. Tarbell Endowment fund (W.R.B.), 5T32GM008320 and an NSF individual graduate fellowship DGE:0909667 (C.A.S.), T32NS07491 and 5T32GM008320 (T.D.P.), T90 DA022873 (D.P.N.), NIH Grant GM079419 (T.M.I.), GM077189 (B.O.B), and the Vanderbilt Institute of Chemical Biology.
Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Footnotes
Author Contributions
B.O.B supervised bioretrosynthetic studies and T.M.I. supervised X-ray crystallographic work. W.R.B. and D.P.N. designed assays. W.R.B performed assays, screened mutagenesis libraries, performed kinetic characterization, and tested enzymes in the biosynthetic pathway studies. C.A.S. determined the crystal structures of Val158Leu and 4H11 PPM variants. T.D.P. determined crystal structures of wild-type PPM and the Ser154Ala and Ser154Gly variants. D.P.N. established initial synthesis routes of dideoxyribose and dideoxyribose 5-phosphate. W.R.B., C.A.S., T.M.I and B.O.B. wrote this paper.
Competing financial interests
These authors declare no competing financial interests.
Accession codes. The atomic coordinates and structure factors have been deposited under accession codes 4LR7, 4LR8, 4LR9, 4LRA, 4LRB, 4LRC, 4LRD, 4LRE and 4LRF.
References
- 1.Savile CK, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 2.Liang J, et al. Development of a biocatalytic process as an alternative to the (−)-DIP-Cl-mediated asymmetric reduction of a key intermediate of montelukast. Org. Process. Res. Dev. 2010;14:193–198. [Google Scholar]
- 3.Gao X, et al. Directed evolution and structural characterization of a simvastatin synthase. Chem. Biol. 2009;16:1064–1074. doi: 10.1016/j.chembiol.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 5.Paddon CJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 6.Ajikumar PK, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito T, et al. Deciphering pactamycin biosynthesis and engineered production of new pactamycin analogues. Chembiochem. 2009;10:2253–2265. doi: 10.1002/cbic.200900339. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M-Q, et al. Optimizing natural products by biosynthetic engineering: Discovery of nonquinone Hsp90 inhibitors. J. Med. Chem. 2008;51:5494–5497. doi: 10.1021/jm8006068. [DOI] [PubMed] [Google Scholar]
- 9.Niu W, Molefe MN, Frost JW. Microbial synthesis of the energetic material precursor 1,2,4-butanetriol. J. Am. Chem. Soc. 2003;125:12998–12999. doi: 10.1021/ja036391+. [DOI] [PubMed] [Google Scholar]
- 10.Yim H, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 11.Ma SK, et al. A green-by-design biocatalytic process for atorvastatin intermediate. Green Chem. 2010;12:81. [Google Scholar]
- 12.Pinheiro E, et al. Examining the production costs of antiretroviral drugs. Aids. 2006;20:1745–1752. doi: 10.1097/01.aids.0000242821.67001.65. [DOI] [PubMed] [Google Scholar]
- 13.Medema MH, van Raaphorst R, Takano E, Breitling R. Computational tools for the synthetic design of biochemical pathways. Nature Rev. Microbiol. 2012;10:191–202. doi: 10.1038/nrmicro2717. [DOI] [PubMed] [Google Scholar]
- 14.Eriksen DT, Lian J, Zhao H. Protein design for pathway engineering. J. Struct. Biol. 2014;185:234. doi: 10.1016/j.jsb.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horowitz NH. On the evolution of biochemical syntheses. Proc. Natl. Acad. Sci. U.S.A. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachmann BO. Biosynthesis: Is it time to go retro? Nat. Chem. Biol. 2010;6:390–393. doi: 10.1038/nchembio.377. [DOI] [PubMed] [Google Scholar]
- 17.Corey EJ. The logic of chemical synthesis - Multistep synthesis of complex natural carbogenic molecules. Angew. Chem. Int. Ed. Engl. 1991;30:455–465. [Google Scholar]
- 18.Turner NJ, O'Reilly E. Biocatalytic retrosynthesis. Nat. Chem. Biol. 2013;9:285–288. doi: 10.1038/nchembio.1235. [DOI] [PubMed] [Google Scholar]
- 19.Nannemann DP, Kaufmann KW, Meiler J, Bachmann BO. Design and directed evolution of a dideoxy purine nucleoside phosphorylase. Protein Eng. Des. Sel. 2010;23:607–616. doi: 10.1093/protein/gzq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamamoto T, Noguchi T, Midorikawa Y. Phosphopentomutase of Bacillus stearothermophilus TH6-2: The enzyme and its gene ppm. Biosci. Biotechnol. Biochem. 1998;62:1103–1108. doi: 10.1271/bbb.62.1103. [DOI] [PubMed] [Google Scholar]
- 21.Panosian TD, et al. Bacillus cereus phosphopentomutase is an alkaline phosphatase family member that exhibits an altered entry point into the catalytic cycle. J. Biol. Chem. 2011;286:8043–8054. doi: 10.1074/jbc.M110.201350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patrick WM, Firth AE, Blackburn JM. User-friendly algorithms for estimating completeness and diversity in randomized protein-encoding libraries. Protein Eng. Des. Sel. 2003;16:451–457. doi: 10.1093/protein/gzg057. [DOI] [PubMed] [Google Scholar]
- 23.Scism RA, Bachmann BO. Five-component cascade synthesis of nucleotide analogues in an engineered self-immobilized enzyme aggregate. Chembiochem. 2010;11:67–70. doi: 10.1002/cbic.200900620. [DOI] [PubMed] [Google Scholar]
- 24.Li J, et al. Crystal structure of Sa239 reveals the structural basis for the activation of ribokinase by monovalent cations. J. Struct. Biol. 2012;177:578–582. doi: 10.1016/j.jsb.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Nocek B, et al. Structural studies of ROK fructokinase YdhR from Bacillus subtilis: Insights into substrate binding and fructose specificity. J. Mol. Biol. 2011;406:325–342. doi: 10.1016/j.jmb.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charrier V, et al. Cloning and sequencing of two Enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. J. Biol. Chem. 1997;272:14166–14174. doi: 10.1074/jbc.272.22.14166. [DOI] [PubMed] [Google Scholar]
- 27.Campobasso N, Mathews II, Begley TP, Ealick SE. Crystal structure of 4-methyl-5-β-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 angstrom resolution. Biochem. 2000;39:7868–7877. doi: 10.1021/bi0000061. [DOI] [PubMed] [Google Scholar]
- 28.Andersson CE, Mowbray SL. Activation of ribokinase by monovalent cations. J. Mol. Biol. 2002;315:409–419. doi: 10.1006/jmbi.2001.5248. [DOI] [PubMed] [Google Scholar]
- 29.Sigrell JA, Cameron AD, Jones TA, Mowbray SL. Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 angstrom resolution: Insights into a new family of kinase structures. Structure. 1998;6:183–193. doi: 10.1016/s0969-2126(98)00020-3. [DOI] [PubMed] [Google Scholar]
- 30.Datta R, et al. Homology-model-guided site-specific mutagenesis reveals the mechanisms of substrate binding and product-regulation of adenosine kinase from Leishmania donovani. Biochem. J. 2006;394:35–42. doi: 10.1042/BJ20051513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Dougherty M, Downs DM, Ealick SE. Crystal structure of an aminoimidazole riboside kinase from Salmonella enterica: Implications for the evolution of the ribokinase superfamily. Structure. 2004;12:1809–1821. doi: 10.1016/j.str.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 32.Currie MA, et al. ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3 -Structure determination and biochemical characterization of PH1645. J. Biol. Chem. 2009;284:22664–22671. doi: 10.1074/jbc.M109.012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trinh CH, Asipu A, Bonthron DT, Phillips SE. Structures of alternatively spliced isoforms of human ketohexokinase. Acta Crystallogr. D Biol. Crystallogr. 2009;65:201–211. doi: 10.1107/S0907444908041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabrera R, et al. The crystal complex of phosphofructokinase-2 of Escherichia coli with fructose-6-phosphate - Kinetic and structural analysis of the allosteric ATP inhibition. J. Biol. Chem. 2011;286:5774–5783. doi: 10.1074/jbc.M110.163162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miallau L, Hunter WN, McSweeney SM, Leonard GA. Structures of Staphylococcus aureus D-tagatose-6-phosphate kinase implicate domain motions in specificity and mechanism. J. Biol. Chem. 2007;282:19948–19957. doi: 10.1074/jbc.M701480200. [DOI] [PubMed] [Google Scholar]
- 36.Sutherland JD, Wilson EJ, Wright MC. Directed evolution of novel biosynthetic pathways: Growth of an Escherichia coli proline auxotroph on Δ(1)-pyrroline-2-carboxylic acid. Bioorg. Med. Chem. Lett. 1993;3:1185–1188. [Google Scholar]
- 37.Bhabha G, et al. Divergent evolution of protein conformational dynamics in dihydrofolate reductase. Nat. Struct. Mol. Biol. 2013;20:1243–1249. doi: 10.1038/nsmb.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfleger BF, Pitera DJ, D Smolke C, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 40.Bailey S. The CCP4 Suite - Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Iverson TM, Panosian TD, Birmingham WR, Nannemann DP, Bachmann BO. Molecular differences between a mutase and a phosphatase: Investigations of the activation step in Bacillus cereus phosphopentomutase. Biochem. 2012;51:1964–1975. doi: 10.1021/bi201761h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panosian TD, Nannemann DP, Bachmann BO, Iverson TM. Crystallization and preliminary X-ray analysis of a phosphopentomutase from Bacillus cereus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:811–814. doi: 10.1107/S1744309110017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.deGroot H, Noll T. Enzymic determination of inorganic phosphates, organic phosphates and phosphate-liberating enzymes by use of nucleoside phosphorylase-xanthine oxidase (dehydrogenase)-coupled reactions. Biochem. J. 1985;230:255–260. doi: 10.1042/bj2300255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ball EG. Xanthine oxidase: Purification and properties. J. Biol. Chem. 1939;128:51–67. [Google Scholar]
- 46.Bezy V, Morin P, Couerbe P, Leleu G, Agrofoglio L. Simultaneous analysis of several antiretroviral nucleosides in rat-plasma by high-performance liquid chromatography with UV using acetic acid/hydroxylamine buffer - Test of this new volatile medium-pH for HPLC-ESIMS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;821:132–143. doi: 10.1016/j.jchromb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 49.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 52.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.