Abstract
Analysis of chemical or biomolecular contents in a tiny amount of specimen presents a significant challenge in many biochemical studies and diagnostic applications. In this work, we present a single-layer, optofluidic device for real-time, high-throughput, quantitative analysis of droplet contents. Our device integrates an optical fiber-based, on-chip detection unit with a droplet-based microfluidic unit. It can quantitatively analyze the contents of individual droplets in real-time. It also achieves a detection throughput of 2000 droplets per second, a detection limit of 20 nM, and an excellent reproducibility in its detection results. In a proof-of-concept study, we demonstrate that our device can be used to perform detection of DNA and its mutations by monitoring the fluorescent signal changes of the target DNA/molecular beacon complex in single droplets. Our approach can be immediately extended to a real-time, high-throughput detection of other biomolecules (such as proteins and viruses) in droplets. With its advantages in throughput, functionality, cost, size, and reliability, the droplet-based optofluidic device presented here can be a valuable tool for many medical diagnostic applications.
Keywords: Microfluidics, droplet, optofluidics, DNA analysis, high-throughput detection
Introduction
Droplet-based microfluidics presents great potential for conducting high-throughput chemical and biological assays due to its significant advantages in cost, screening time, and sensitivity.1-7 This technology compartmentalizes reagents into aqueous-in-oil droplets with volumes ranging from femtoliters to nanoliters, as opposed to microliter sample volumes in conventional microfluidic methods. In addition, it allows the aqueous droplets to be separated by an oil phase, thus preventing undesirable contamination and cross-talk between neighboring droplets or between the reagents and channel walls.2-3,5 Moreover, droplet microfluidics allows individual droplet production at kilohertz frequencies and makes it possible to conduct high-throughput assays of complex chemical, biochemical, or pharmaceutical analytical processes in a rapid, automated, and reproducible manner.4
Recently, significant advances have been made in developing droplet microfluidics-based, high-throughput screening methods8-10 for a variety of applications including drug discovery, directed enzyme evolution, bacteria screening, and nucleic acid analysis.4,7,11-13 An effective, droplet-based, screening method will require the following three key functions: 1) encapsulation of materials in droplets, 2) manipulation (moving, splitting, sorting, etc.) of droplets, and 3) detection of products in droplets.1 Thus far, significant progress has been made in realizing the first two functions: researchers have demonstrated encapsulation of various species including cells, microparticles, molecules, and DNA into individual droplets;14-16 they have also achieved varieties of rapid droplet manipulation techniques in a controllable manner, including droplet transportation, fission, fusion, mixing, sorting, trapping, and packaging.17-25 However, limited research has been conducted on developing techniques for rapid and sensitive detection and analysis of droplet contents.10,26-30 In particular, to achieve rapid and quantitative measurements of fast-moving individual droplets often requires expensive instruments and/or sophisticated operation procedures. For example, although droplets can be observed and recorded by a fluorescence microscope equipped with a sensitive CCD camera,4 regular CCD cameras fail in real-time detection of individual droplets moving at a high velocity and rely on detection of the average signal of many droplets. Thus, an expensive fast camera is required to capture data from a single fast-moving droplet.31 Alternatively, confocal fluorescence microscopy has been used to quantify droplet contents in a high-throughput fashion.32,33 Using this method, high-throughput DNA assays have been achieved by characterizing a fluorescence resonance energy transfer (FRET) signal.33 The dependence of the existing droplet-based detection methods on expensive instruments (e.g., fast camera, confocal fluorescence microscope) prevents their widespread applications. We believe that an ideal droplet-based detection method should fulfill the following criteria: 1) low-cost fabrication with easy and automatic operation; 2) real-time and high-throughput individual droplet detection; 3) quantitative and sensitive characterization of droplet contents; and 4) excellent reproducibility.
Here we report an optofluidic34-46 device for high-throughput, real-time, quantitative analysis of droplet contents. Our approach integrates an optical fiber-based detection unit with a droplet-based microfluidic device. By detecting the fluorescence intensity using photomultiplier tubes (PMTs), the contents of individual droplets were accurately analyzed in real-time with a high-throughput format (2000 droplets per second). While being significantly simpler and cheaper, our device can achieve an acceptable performance compared with previous methods, which require expensive equipment such as confocal or fluorescent microscopes coupled to a fast camera.47 In addition, we demonstrate that our device can be used to detect DNA contents in droplets and perform single nucleotide polymorphism (SNP) analysis. SNP analysis is a major diagnostic method for genetic diseases, which is typically limited by sample consumption, non-specific binding, and low reaction efficient in the conventional platform.27 In this work, we demonstrate the detection of the abnormality on the BRCA1 gene which is known to cause breast and ovarian cancer.48
Experiments
Working mechanism
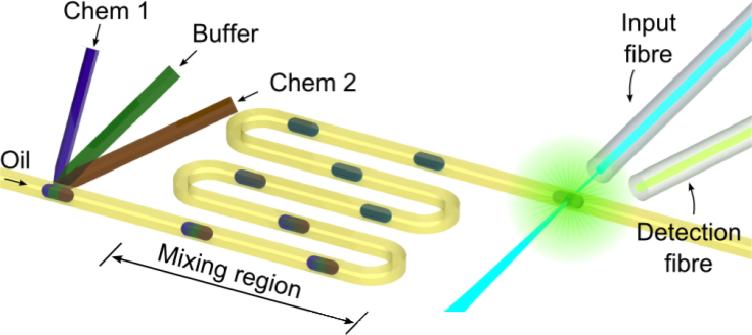
Figure 1 shows a schematic of the droplet-based optofluidic device. This device consists of three components: a droplet-generation unit, a mixing and reaction unit, and an optical fiber-based detection unit. Reagents (Chem 1 and Chem 2) and buffer are loaded from the three-forked inlet and dispersed into the oil flow to generate droplets. Reagents inside each droplet are well mixed during the transportation process through the long curved channel to guarantee complete chemical reaction. Finally, individual droplets are monitored by the fiber-based detection unit in real-time to detect the fluorescence intensity to perform DNA analysis.
Figure 1.
Schematic of the droplet-based optofluidic device composed of a droplet-generation unit, a mixing and reaction unit, and a fiber-based optical detection unit.
Device fabrication
The droplet-based optofluidic device was fabricated using standard soft-lithography and mold-replica techniques in two steps. First, a single-layer pattern of SU8 photoresist (125 μm thick) was developed on a silicon wafer to form the master mold. The width of channel was 200 μm. The length of the serpentine mixing channel depended on the mixing required for the bioreaction under examination. The length of the serpentine mixing channel was 20 mm, 50 mm and 100 mm, for different reactions, respectively. This mold was salinized by vapor of 1H,1H,2H,2H-perfluorooctyl-trichlorosolane (Sigma-Aldrich) to assist polydimethylsiloxane (PDMS) peel-off in the later step. Second, PDMS syrup was poured on the mold and cured in an oven to fabricate the microfluidic channel. Third, the PDMS layer was peeled from the mold, and inlets and outlets were drilled with a Harris Uni-Core punch (0.75 mm) on the PDMA layer. After drilling, the PDMS layer was sealed onto a glass slide to form a microfluidic device. Finally, polyethylene tubes were inserted into the inlets and outlets to connect the microfluidic device to syringe pumps (neMESYS).
Optical setup
The fiber-based optical detection system includes two optical fibers: an input fiber (Thorlabs S405, single-mode, core diameter = 2.9 μm, cladding diameter = 125 μm, N.A. = 0.14) which brings the excitation light from a portable blue laser (488 nm, Innova 300, Coherent) to the detection point, and a detection fiber (Thorlabs AFS105.125Y, multi-mode, core diameter = 105 μm, cladding diameter = 125 μm, N.A. = 0.22) which collects the fluorescent emission from the excited droplets and directs it to an off-chip PMT (Hamamatsu 6780-20). The optical fibers were precisely positioned through guide chambers with a height of 125 μm. As shown in Fig. 2a, the input fiber was aligned perpendicular to the microfluidic channel, and the maximum beam width on the droplet was less than 20 μm thanks to the small numerical aperture of the input fiber. The detection fiber with a much larger numerical aperture and core diameter was fixed at an angle of 27° from the input fiber to maximize the fluorescence detection efficiency and to avoid direct collection of the excitation light. The collected fluorescent emission was filtered by a band-pass filter (532/40 nm) before being fed to the PMT. A home-made control circuit with tunable gain40,49 was developed to drive the PMT. The electronic signals from the PMT were amplified with a high-frequency amplifier (Hamamatsu C6438-01), filtered with an electric signal filter, and recorded by a digital oscilloscope (Tektronix DPO400).
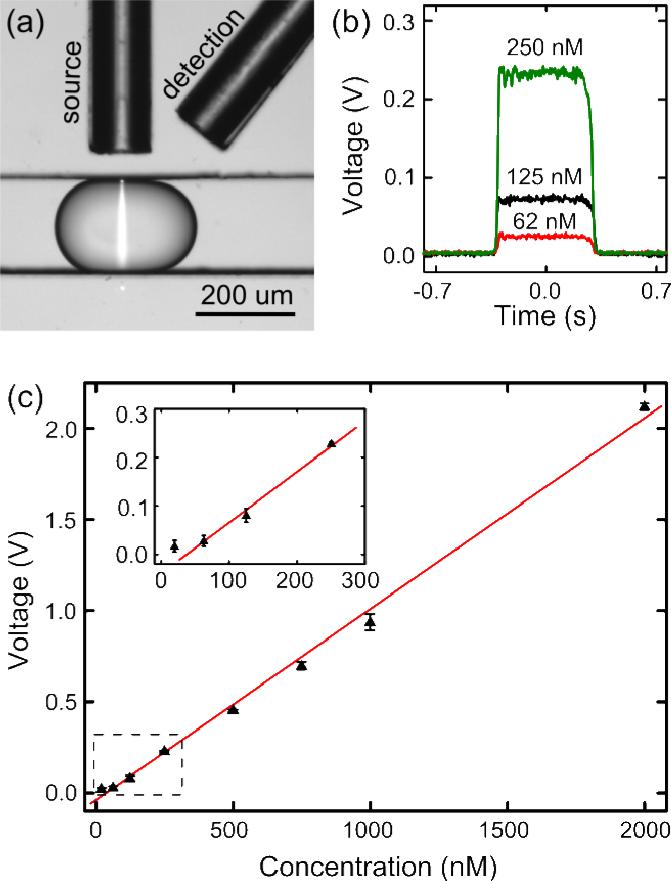
Figure 2.
Fluorescence intensity characterization of the droplet-based optofluidic device. (a) A microscopic snapshot of a droplet excited by a 20 μm-wide laser beam. (b) The electric signal of three droplets with different fluorescent dye concentrations. (c) The calibration curve presenting a linear relationship between the fluorescent dye concentration and the detection voltage.
Sample preparation
Fluorinert® FC-40 (Sigma) with 3 wt% DuPont Krytox 157 FS surfactant (ChemPoint) was used as the oil phase to compartmentalize the aqueous solution into droplets throughout the study. Alexa Fluor® 488 (excitation = 495 nm, emission = 519 nm) conjugated with Dextran (Life Technologies) was diluted into different concentrations ranging from 20 nM to 2 μM as the aqueous phase for quantitative characterization of the fluorescence detection of the device as described in the next section. The molecular beacon (MB), wild-type single-strain DNA BRCA1 (WT), single-mutation single-strain DNA (SM), and control single-strain DNA (CON) were purchased from IDT DNA Technologies and prepared in different concentrations for DNA mutation detection, as summarized in Table 1. The DNase water was purchased from Sigma-Aldrich.
Table 1.
The sequences of molecular beacon (MB) and DNAs used in our experiments. The underlined sequences and lowercase symbol indicate the MB-DNA hybridization sequence and mutation point, respectively.
| Name | Sequence | Concentration |
|---|---|---|
| MB | 5’- (Alex Fluor® 488) – CC TAG CCC CTA TGT ATG CTC TTT GTT GTG GCT AGG – (BHQ1)-3’ | 2.5 μM |
| WT | 5’-TAA CAC AAC AAA GAG CAT ACA TAG GGT TT-3’ | 1 μM |
| SM | 5’-TAA CAC AAC AAA GAa CAT ACA TAG GGT TT-3’ | 1 μM |
| CON | 5’-CCC TGG GAG GAT CCA AGC TTC CAG TAT C-3’ | 1 μM |
Results and Discussion
Characterization of droplet-based fluorescence detection
We first characterized the fluorescence detection performance of our droplet-based optofluidic device with the commercial Alexa Fluor® 488 fluorescent dye. Both inlets for Chem 1 and Chem 2 were not in use (blocked) during the characterization. The fluorescent solution was injected at different concentrations through the buffer inlet at a flow rate of 0.3 μl min−1 and compartmentalized into uniform droplets (292.8±3.7 μm) by the oil phase with a flow rate of 2 μl min−1. The compartmentalized aqueous droplets were driven through the long curved channel and then monitored by the fiber-based optical detection system in sequence (Fig. 1). Figure 2a shows the microscopic snapshot of a droplet under detection. The droplet was excited by the 488 nm blue laser with a beam width less than 20 μm. The use of a narrow beam significantly reduces excitation time per unit area of the fluorescent solution so that the bleaching effect of the fluorescent could be minimized. The emitted green fluorescence signal from the droplet was collected by the detection fiber and recorded by the digital oscilloscope. Each set of experiments was recorded at a sampling rate of 100 kHz and post-processed using a homemade MATLAB-based program.
Single aqueous-in-oil droplets with a wide range of fluorescent dye concentrations were detected for quantitative characterization. Figure 2b shows the detected optical signals of three representative droplets with different fluorescent dye concentrations (62 nM, 125 nM, and 250 nM). It can be seen that different fluorescent dye concentrations exhibit similar signal waveforms with the same peak width but distinct heights. The identical width of the detected signals proves the uniformity of the generated droplet size by our device, while the distinct peak heights indicate that the fluorescence intensity of each droplet can be quantitatively identified in real-time. The dependency between the fluorescent dye concentration of the droplet and the output detection voltage by our device is displayed in Fig. 2c, with each data point produced from hundreds of uniform droplets. A linear relationship is observed for a large concentration range (20 nM to 2 μM). The small deviation at each data point implies the reproducibility and consistency of our device and the capability to perform fluorescence-based quantitative analyses of droplet contents.
High-throughput fluorescence detection
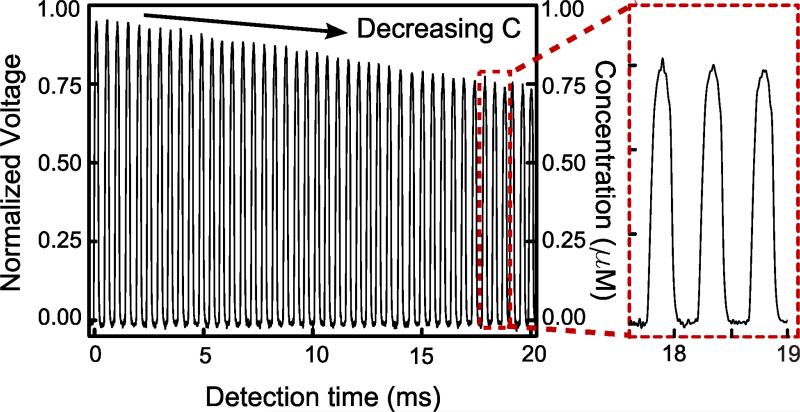
In the previous section we have demonstrated the sensitivity and reproducibility of our droplet-based optofluidic device for fluorescence detection. Here we test the detection throughput of our device. The throughput is majorly limited by the droplet generation process and flow speed. In the throughput-testing setup, the buffer inlet was blocked while the fluorescent solution and DI water were injected through Chem 1 and Chem 2 inlets, respectively. To achieve maximum throughput, the highest droplet production frequency and minimal inter-droplet distance were obtained by adjusting the respective flow rates of the fluorescent solution, DI water, and oil phase. The optimal flow condition of our device was obtained under these conditions: the oil phase had a flow rate of 1500 μl min−1, and the total combined flow rate of 1 μM fluorescent solution and DI water was fixed at 300 μl min−1. The flow rate ratio between the 1 μM fluorescent solution and the DI water defines the concentration of fluorescence in each droplet, which could be varied from 0 M to 1 μM. By continuously increasing the flow rate of DI water from 0 μl min−1 to 300 μl min−1, a concentration gradient of fluorescence was generated across several adjacent droplets. The resolution or “steepness” of the gradient was limited by the response time on the pumps in the system. The droplets were transported through 20 mm channel from the droplet generator to the detection region in around 20 ms, which is long enough to achieve uniform mixing.27 As shown in the processed optical detection signal (Fig. 3), we demonstrate a high detection throughput of 2000 droplets per second (or 0.5 ms per droplet). With such a high throughput, we can perform fast and reliable fluorescence-based detections, such as DNA analysis, by taking the average of the detected signals from thousands of droplets with uniform size and fluorescent concentration. On the other hand, thanks to the consistency of droplet generation and optical detection in our device, we could detect the fluorescence intensity of each droplet with equal size but different fluorescence concentration. To prove the concept, we gradually increased the fluorescent dye concentration in each droplet by steadily increasing the flow rate of the fluorescent dye solution and decreasing that of DI water. After passing through the mixing region, each droplet with various fluorescent dye concentrations was detected and plotted in Fig. 3. This demonstration confirms that our device is capable of detecting fluorescence intensity of each droplet at a throughput of 2000 events per second.
Figure 3.
High-throughput, droplet-based fluorescence detection. The left panel plots the normalized output signal and concentration of aqueous-in-oil droplets with slightly increasing fluorescent dye concentrations. The enlarged plot at the right panel depicts the detection throughput at 0.5 ms per droplet (i.e., 2000 droplets per second).
Droplet-based DNA analysis
The sensitivity, reproducibility, and high throughput make our droplet-based optofluidic device an ideal platform for high-throughput screening of biomolecules such as DNA, proteins, and viruses. Here we demonstrate that our device can perform DNA analyses by sensing the fluorescence intensity of complexes of the molecular beacon and DNA in single droplets. The abnormality on BRCA1 gene, known to cause breast and ovarian cancer, was detected in this system. As summarized in Table 1, we prepared wild-type single-strain DNA (WT), single-mutation single-strain DNA (SM), control single-strain DNA (CON), and molecular beacon only (MB) for test. In each experiment, we injected molecular beacon, DNase water, and DNA solutions (WT, SM, CON, or MB) into Chem1, buffer, and Chem 2 inlets at flow rates of 1 μl min-1, 0.05 μl min−1, and 1 μl min−1, respectively. The flow rate of oil phase was kept at 12 μl min−1. All of the reagents were dispersed into 7.5 nL volume droplets at a generation rate of 4 droplets per second. The DNA was fully mixed with the molecular beacon and the reaction between the two was completed during the process of transporting the droplets through the 100 mm long serpentine mixing channel. The transit time of a single droplet through the U shaped structures of the serpentine mixing channel was around 12.5 s. If one were interested in a different bimolecular reaction, the length of the serpentine mixing channel and the flow rates in the system could be adjusted to achieve the desired mixing for the biomolecular reaction of interest. When excited by the blue laser at the optical detection region, the fluorescence emission from the DNA/molecular beacon complex in each droplet was detected and recorded in voltage. Figure 4 shows the output voltage of different sets of experiments. The experiment with molecular beacons only (MB) has the lowest output voltage (0.098 V) and serves as the background noise. The control experiment with mismatched DNA (CON) has a slightly higher output voltage (0.114 V). The wild-type DNA experiment has the highest voltage (0.275 V) while the SM was measured to be slightly lower (0.242 V), indicating that the BRCA1 DNA and its mutation were clearly identified by our device. The relatively high background noise likely resulted from the decomposition of molecular beacon and the absence of a certain metal-ion solution in the buffer, which is consistent with the literarure.50 Nevertheless, our device exhibits high sensitivity and demonstrates that the WT and SM can be distinguished. Our results also prove the robustness of the droplet-based optofluidic device for bioanalysis applications.
Figure 4.
DNA analysis using our droplet-based optofluidic device. The plot presents the output voltage of different fluorescence signals from droplets with different molecular beacon/DNA complexes. The sequences and concentrations of WT, SM, CON and MB are listed in Table 1.
Next, we calculated the signal-to-noise ratio (SNR) as an indicator of detection capability of our device. The SNR is defined as
| (1) |
where I and V are intrinsic fluorescence intensity and detected voltage, respectively, and subscripts b, c, and o denote molecular beacon only (background: MB), molecular beacon in the presence of the DNA target (WT or SM), and molecular beacon in the presence of the negative-control DNA (CON), respectively. The calculated SNR for WT and SM is 11.06 and 9.0, respectively. The SNR of single mutation hybridization is 18.6% smaller than that of the wild-type hybridization and can be used to analyze the DNA mutations. The demonstration presented in this section shows that our droplet-based, optofluidic device has the capability to quantitatively detect DNA and its mutations.
Conclusions
In summary, we have successfully developed a droplet-based optofluidic device for high-throughput fluorescence detection, which integrates droplet generation, on-chip mixing, and fiber-based optical detection into a single-layered chip. We show that our device can quantitatively sense the fluorescent emission from droplets with a detection limit of 20 nM of fluorescent dye concentration (Alexa Fluor® 488 dye). A detection throughput of 2000 droplets per second was achieved with excellent fluorescence intensity identification. Compared to the existing droplet-based, high-throughput detection methods (confocal or fluorescence microscopy equipped with a fast camera), our optofluidic approach possesses the advantages of low cost, less fluorescence bleaching, real-time single droplet detection, and a low detection limitation. Moreover, we demonstrated that the device can detect single nucleotide mutations in DNA, making it an ideal platform for fast and cost-effective breast and ovarian cancer diagnosis. This technique will be useful in many chemical, biochemical, and pharmaceutical analytical processes such as immunoassays and cancer diagnosis.51-58
Acknowledgement
The authors would like to thank Matt Jaffe, Joey Rufo, Dr. Danqi Chen, Dr. Yijun Deng, and Dr. Zhenxin (Peter) Hu for helpful discussions. This research was supported by the National Institutes of Health (NIH) Director's New Innovator Award (1DP2OD007209-01), National Science Foundation, and the Penn State Center for Nanoscale Science (MRSEC). Components of this work were conducted at the Penn State node of the NSF-funded National Nanotechnology Infrastructure Network (NNIN).
Footnotes
Supplementary Information. Additional information and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Reference
- 1.Chiu DT, Lorenz RM, Jeffries GD. M. Analytical Chemistry. 2009;81:5111. doi: 10.1021/ac900306q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo MT, Rotem A, Heyman JA, Weitz DA. Lab on a Chip. 2012;12:2146. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 3.Theberge A, Courtois F, Schaerli Y, Fischlechner M, Abell C, Hollfelder F, Huck W. Angewandte Chemie International Edition. 2010;49:5846. doi: 10.1002/anie.200906653. [DOI] [PubMed] [Google Scholar]
- 4.Song H, Chen DL, Ismagilov RF. Angewandte Chemie-International Edition. 2006;45:7336. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teh SY, Lin R, Hung LH, Lee AP. Lab on a Chip. 2008;8:198. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 6.Dittrich PS, Tachikawa K, Manz A. Analytical Chemistry. 2006;78:3887. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 7.Neuzil P, Giselbrecht S, Lange K, Huang TJ, Manz A. Nature Reviews Drug Discovery. 2012;11:620. doi: 10.1038/nrd3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14195. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granieri L, Baret JC, Griffiths AD, Merten CA. Chemistry & Biology. 2010;17:229. doi: 10.1016/j.chembiol.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Kiss MM, Ortoleva Donnelly L, Beer NR, Warner J, Bailey CG, Colston BW, Rothberg JM, Link DR, Leamon JH. Analytical Chemistry. 2008;80:8975. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rane TD, Zec HC, Puleo C, Lee AP, Wang TH. Lab on a Chip. 2012;12:3341. doi: 10.1039/c2lc40537g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang TH, Peng YH, Zhang CY, Wong PK, Ho CM. Journal of the American Chemical Society. 2005;127:5354. doi: 10.1021/ja042642i. [DOI] [PubMed] [Google Scholar]
- 13.Meserve D, Wang ZH, Zhang DD, Wong PK. Analyst. 2008;133:1013. doi: 10.1039/b804853c. [DOI] [PubMed] [Google Scholar]
- 14.He M, Edgar JS, Jeffries GDM, Lorenz RM, Shelby JP, Chiu DT. Analytical Chemistry. 2005;77:1539. doi: 10.1021/ac0480850. [DOI] [PubMed] [Google Scholar]
- 15.Clausell Tormos J, Lieber D, Baret JC, El Harrak A, Miller OJ, Frenz L, Blouwolff J, Humphry KJ, Koster S, Duan H, Holtze C, Weitz DA, Griffiths AD, Merten CA. Chemistry & Biology. 2008;15:427. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Edd JF, Di Carlo D, Humphry KJ, Koster S, Irimia D, Weitz DA, Toner M. Lab on a Chip. 2008;8:1262. doi: 10.1039/b805456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link DR, Anna SL, Weitz DA, Stone HA. Physical Review Letters. 2004:92. doi: 10.1103/PhysRevLett.92.054503. [DOI] [PubMed] [Google Scholar]
- 18.Zeng SJ, Pan XY, Zhang QQ, Lin BC, Qin JH. Analytical Chemistry. 2011;83:2083. doi: 10.1021/ac1028775. [DOI] [PubMed] [Google Scholar]
- 19.Fidalgo LM, Abell C, Huck WTS. Lab on a Chip. 2007;7:984. doi: 10.1039/b708091c. [DOI] [PubMed] [Google Scholar]
- 20.Huebner A, Bratton D, Whyte G, Yang M, deMello AJ, Abell C, Hollfelder F. Lab on a Chip. 2009;9:692. doi: 10.1039/b813709a. [DOI] [PubMed] [Google Scholar]
- 21.Liu K, Ding HJ, Liu J, Chen Y, Zhao XZ. Langmuir. 2006;22:9453. doi: 10.1021/la061729+. [DOI] [PubMed] [Google Scholar]
- 22.Guo F, Ji XH, Liu K, He RX, Zhao LB, Guo ZX, Liu W, Guo SS, Zhao XZ. Applied Physics Letters. 2010;96:193701. [Google Scholar]
- 23.Surenjav E, Priest C, Herminghaus S, Seemann R. Lab on a Chip. 2009;9:325. doi: 10.1039/b808160c. [DOI] [PubMed] [Google Scholar]
- 24.Watson MWL, Abdelgawad M, Ye G, Yonson N, Trottier J, Wheeler AR. Analytical Chemistry. 2006;78:7877. doi: 10.1021/ac0613378. [DOI] [PubMed] [Google Scholar]
- 25.Jebrail MJ, Wheeler AR. Analytical Chemistry. 2009;81:330. doi: 10.1021/ac8021554. [DOI] [PubMed] [Google Scholar]
- 26.Roman GT, Wang M, Shultz KN, Jennings C, Kennedy RT. Analytical Chemistry. 2008;80:8231. doi: 10.1021/ac801317t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh ATH, Pan PJH, Lee AP. Microfluidics and Nanofluidics. 2009;6:391. [Google Scholar]
- 28.Walter A, Marz A, Schumacher W, Rosch P, Popp J. Lab on a Chip. 2011;11:1013. doi: 10.1039/c0lc00536c. [DOI] [PubMed] [Google Scholar]
- 29.Chan KLA, Niu X, de Mello AJ, Kazarian SG. Lab on a Chip. 2010;10:2170. doi: 10.1039/c004246c. [DOI] [PubMed] [Google Scholar]
- 30.Liu SJ, Gu YF, Le Roux RB, Matthews SM, Bratton D, Yunus K, Fisher AC, Huck WTS. Lab on a Chip. 2008;8:1937. doi: 10.1039/b809744e. [DOI] [PubMed] [Google Scholar]
- 31.Cecchini MP, Hong J, Lim C, Choo J, Albrecht T, Demello AJ, Edel JB. Analytical Chemistry. 2011;83:3076. doi: 10.1021/ac103329b. [DOI] [PubMed] [Google Scholar]
- 32.Jeffries GDM, Lorenz RM, Chiu DT. Analytical Chemistry. 2010;82:9948. doi: 10.1021/ac102173m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srisa Art M, deMello AJ, Edel JB. Analytical Chemistry. 2007;79:6682. doi: 10.1021/ac070987o. [DOI] [PubMed] [Google Scholar]
- 34.Psaltis D, Quake SR, Yang CH. Nature. 2006;442:381. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 35.Fan XD, White IM. Nature Photonics. 2011;5:591. doi: 10.1038/nphoton.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu AQ, Huang HJ, Chin LK, Yu YF, Li XC. Analytical and Bioanalytical Chemistry. 2008;391:2443. doi: 10.1007/s00216-008-1878-2. [DOI] [PubMed] [Google Scholar]
- 37.Greenbaum A, Sikora U, Ozcan A. Lab on a Chip. 2012;12:1242. doi: 10.1039/c2lc21072j. [DOI] [PubMed] [Google Scholar]
- 38.Chiou PY, Ohta AT, Wu MC. Nature. 2005;436:370. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- 39.Mao XL, Lin SCS, Dong C, Huang TJ. Lab on a Chip. 2009;9:1583. doi: 10.1039/b820138b. [DOI] [PubMed] [Google Scholar]
- 40.Lapsley MI, Chiang IK, Zheng YB, Ding X, Mao X, Huang TJ. Lab on a Chip. 2011;11:1795. doi: 10.1039/c0lc00707b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erickson D, Sinton D, Psaltis D. Nature Photonics. 2011;5:583. [Google Scholar]
- 42.Mao X, Stratton ZI, Nawaz AA, Lin SCS, Huang TJ. Biomicrofluidics. 2010;4:43007. doi: 10.1063/1.3497934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao X, Waldeisen JR, Juluri BK, Huang TJ. Lab on a Chip. 2007;7:1303. doi: 10.1039/b708863a. [DOI] [PubMed] [Google Scholar]
- 44.Huang H, Mao X, Lin SCS, Kiraly B, Huang Y, Huang TJ. Lab on a Chip. 2010;10:2387. doi: 10.1039/c005071g. [DOI] [PubMed] [Google Scholar]
- 45.Mao X, Nawaz AA, Lin SCS, Lapsley MI, Zhao Y, McCoy JP, El Deiry WS, Huang TJ. Biomicrofluidics. 2012;6:24113. doi: 10.1063/1.3701566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao X, Lin SCS, Lapsley MI, Shi J, Juluri BK, Huang TJ. Lab on a Chip. 2009;9:2050. doi: 10.1039/b822982a. [DOI] [PubMed] [Google Scholar]
- 47.Rane TD, Puleo CM, Liu KJ, Zhang Y, Lee AP, Wang TH. Lab on a Chip. 2010;10:161. doi: 10.1039/b917503b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shattuck Eidens D, McClure M, Simard J, et al. JAMA. 1995;273:535. [PubMed] [Google Scholar]
- 49.Mao XL, Luo Y, Dai ZP, Wang KY, Du YG, Lin BC. Analytical Chemistry. 2004;76:6941. doi: 10.1021/ac049270g. [DOI] [PubMed] [Google Scholar]
- 50.Yao G, Tan WH. Analytical Biochemistry. 2004;331:216. doi: 10.1016/j.ab.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Karns K, Herr AE. Analytical Chemistry. 2011;83:8115. doi: 10.1021/ac202061v. [DOI] [PubMed] [Google Scholar]
- 52.Kim D, Karns K, Tia SQ, He M, Herr AE. Analytical Chemistry. 2012;84:2533. doi: 10.1021/ac3000013. [DOI] [PubMed] [Google Scholar]
- 53.Mao X, Huang TJ. Lab on a Chip. 2012;12:1412. doi: 10.1039/c2lc90022j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang S, Guo F, Kiraly B, Mao X, Lu M, Leong KW, Huang TJ. Lab on a Chip. 2012;12:2097. doi: 10.1039/c2lc90046g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin SCS, Mao X, Huang TJ. Lab on a Chip. 2012;12:2766. doi: 10.1039/c2lc90076a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang TJ, Liu M, Knight LD, Grody WW, Miller JF, Ho CM. Nucleic Acids Research. 2002;30:e55. doi: 10.1093/nar/gnf054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi J, Yazdi S, Lin SCS, Ding X, Chiang I, Sharp K, Huang TJ. Lab on a Chip. 2011;11:2319. doi: 10.1039/c1lc20042a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziober BL, Mauk MG, Falls EM, Chen Z, Ziober AF, Bau H. Head Neck-J Sci Spec. 2008;30:111. doi: 10.1002/hed.20680. [DOI] [PubMed] [Google Scholar]