Summary
Argonaute proteins (AGOs) are key nuclease effectors of RNA interference (RNAi) [1]. Although purified AGOs can mediate a single round of target-RNA cleavage in vitro, accessory factors are required for short-interfering (si)RNAs loading and to achieve multiple-target turnover [2, 3]. To identify AGO co-factors we immunoprecipitated the C. elegans AGO WAGO-1, which engages amplified small RNAs during RNAi [4]. These studies identified a robust association between WAGO-1 and a conserved Vasa ATPase-related protein RDE-12. rde-12 mutants are deficient in RNAi including viral suppression, and fail to produce amplified secondary siRNAs and certain endogenous siRNAs (endo-siRNAs). RDE-12 co-localizes with WAGO-1 in germline P-granules and in cytoplasmic and peri-nuclear foci in somatic cells. These findings and our genetic studies suggest that RDE-12 is first recruited to target mRNA by upstream AGOs (RDE-1 and ERGO-1) where it promotes small-RNA amplification and/or WAGO-1 loading. Downstream of these events, RDE-12 forms an RNase-resistant (target mRNA-independent) complex with WAGO-1 and may thus have additional functions in target mRNA surveillance and silencing.
Results and Discussion
The VASA homolog RDE-12 is a WAGO-1 interactor
As in other organisms, the upstream events in the C. elegans RNAi response include processing of long dsRNA into siRNAs by the RNase III-related protein, Dicer, loading of siRNAs into an RNase H-related AGO protein, and scanning for target mRNAs by siRNA-mediated base-pairing that precisely positions AGO for target mRNA cleavage [5]. In C. elegans however, the slicer activity of the primary AGO, RDE-1, is not required for silencing [6]. Instead, through mechanisms that are still largely unknown, RDE-1 recruits RNA-dependent RNA polymerase (RdRP) [7, 8]. RdRP then utilizes the target mRNA as a template for the de novo synthesis of secondary siRNAs (termed 22G-RNAs) [4]. 22G-RNAs are then loaded onto secondary AGOs of the “worm-specific AGO” (WAGO) protein family [4]. WAGO proteins lack catalytic residues thought to be essential for target cleavage [9] and thus silencing is thought to involve the recruitment of unknown accessory factors that mediate mRNA turnover.
Given our incomplete understanding of how AGO proteins mediate key events such as RdRP recruitment and mRNA turnover, we sought to identify proteins that interact with worm AGO proteins in vivo. The WAGO-1 protein is abundantly expressed in germline nuage-like structures called P granules [4]. Previous studies suggest that WAGO-1 functions in both exogenous dsRNA-induced (exo-RNAi), as well as several endogenous RNAi pathways [4]. To identify WAGO-1 protein interactors, we immunoprecipitated FLAG-tagged WAGO-1 and analyzed the co-precipitated proteins by SDS-PAGE. This analysis identified a 100 kDa protein that co-precipitated with FLAG∷WAGO-1 (Fig 1A). Tandem mass spectrometry unambiguously identified this protein as F58G11.2, which is predicted to encode a DEAD-box RNA-binding ATPase most similar to Vasa/DDX4 and Belle/DDX3. In addition, F58G11.2 contains two FG (phenylalanine-glycine)-repeat domains that flank the ATPase domain [10]. Because F58G11.2 is required for RNAi (see below), we refer to f58g11.2 as rde-12 (RNAi deficient-12).
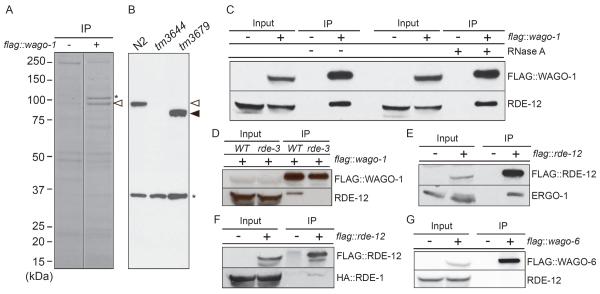
Figure 1.
RDE-12 binds to both primary and secondary Argonautes
(A) Identification of FLAG∷WAGO-1 interacting proteins. FLAG∷WAGO-1 immune complexes from N2 wild-type (−) and flag∷wago-1 transgenic worms (+) were resolved on a denaturing polyacrylamide gel, and proteins were visualized by colloidal blue staining. FLAG∷WAGO-1 is indicated with an asterisk, and a prominent 100 kDa interacting protein is indicated with an open arrow head.
(B) Specificity of RDE-12 antisera. Immunoblot analysis with RDE-12-specific antisera on extracts from wild-type (N2), rde-12(tm3644) and rde-12(tm3679) animals. The open and closed arrowheads indicate the expected mobility of RDE-12 produced in wild-type and tm3679 animals, respectively. The asterisk indicates a background band.
(C–G) RDE-12 interactions with AGO proteins. Immunoblot analyses of FLAG IP experiments to monitor RDE-12 interactions with (C) FLAG∷WAGO-1 in the presence (+) or absence (−) of RNase A, (D) FLAG∷WAGO-1 in wild type, WT and rde-3 mutant, (E) ERGO-1, (F) HA∷RDE-1 and (G) FLAG∷WAGO-6 lysates. flag transgenic lysates are indicated by a (+) in the headings above the blots. The protein blotted by the antibody probes used in each experiment is indicated to the right of each blot.
RDE-12-specific antisera recognized a 100 kDa protein in wild-type that is absent or truncated in rde-12 mutant animals (Fig 1B). Consistent with our WAGO-1 mass-spectrometry findings, RDE-12 protein co-immunoprecipitated FLAG∷WAGO-1 (Fig 1C). The interaction between WAGO-1 and RDE-12 was resistant to RNase A treatment, suggesting that the interaction is not bridged by RNA. Interestingly, we found that WAGO-1 and RDE-12 do not co-immunoprecipitate (IP) in rde-3 mutants (Fig. 1D), where the majority of both endo- and exo-secondary siRNAs are absent [4, 11]. RDE-12 IP followed by multidimensional protein identification technology (MudPIT) analysis detected WAGO-1 and a primary AGO, ERGO-1 [12], but failed to detect other AGO proteins (Data Not Shown). The ERGO-1 interaction was confirmed by co-IP/immunoblot analysis with a rescuing FLAG∷RDE-12 (Fig. 1E). FLAG∷RDE-12 IP also co-precipitated HA∷RDE-1 (Fig. 1F) suggesting that RDE-12 and RDE-1 may also interact. Finally, RDE-12 failed to interact in IP/immunoblot analysis with FLAG∷WAGO-6, another cytoplasmic WAGO (Fig. 1G).
rde-12 mutants are partially defective in RNAi
To examine the function of rde-12, we obtained deletion alleles (tm3644 and tm3679) from the National BioResource Project for the Experimental Animal “Nematode C. elegans” [13]. Immunoblot analysis failed to detect a protein in rde-12(tm3644) extracts, and detected a protein of lower molecular weight in extracts from rde-12(tm3679) (Fig. 1B). Animals homozygous for both alleles were viable and showed no obvious developmental defects. In dsRNA feeding assays with several triggers, both rde-12 mutant strains were strongly, but not completely, resistant to RNAi targeting the muscle-specific gene unc-22 and the essential gene cpsf-2 (Fig. 2B). This incomplete RNAi deficit was more apparent in assays targeting the germline gene pos-1. We found that 63% of rde-12(tm3644) and 89% of rde-12(tm3679) embryos were sensitive to pos-1(RNAi) (Fig. 2B). Together with the immunoblot analysis, these findings suggest that tm3644 is a stronger, likely null allele, while tm3679 may retain partial function.
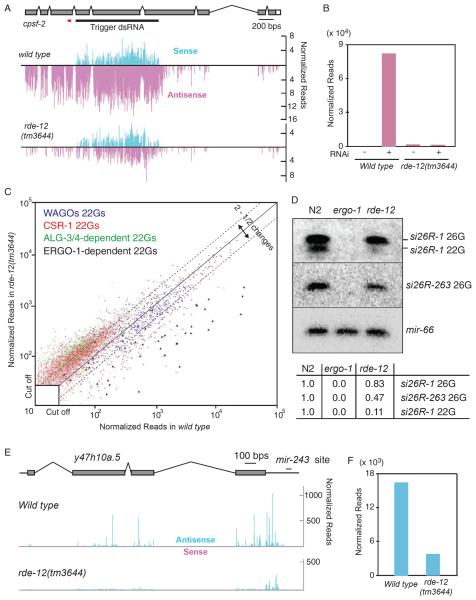
Figure 2.
RDE-12 is a DEAD-box RNA ATPase required for RNAi and viral infection
(A) Schematic diagram of the predicted domain structure of RDE-12 protein. The approximate positions of eleven conserved motifs (F-VI) found in the RNA helicase domain are indicated (black boxes), as are two FG/GF-Rich domains (gray boxes). The region deleted by a putative null allele rde-12(tm3644) and by a less severe deletion allele rde-12(tm3679) are indicated by the black bars above the diagram. An alignment of domain I from RDE-12 and related C. elegans (CE), Drosophila melanogaster (DM), and homo sapiens (HS) proteins is provided below the diagram. Invariant amino acids are indicated with an asterisk (below), and the conserved lysine mutated in RDE-12(K429A) is indicated in bold.
(B) rde-12(tm3644) is deficient in RNAi. Wild type, rde-12(tm3644 and tm3679) and transgenic animals (as indicated) were assayed for sensitivity to RNAi by feeding. The percent of unc-22 twitching/paralyzed, the cpsf-2 sterile, or the pos-1 dead embryo phenotypes among (n) animals analyzed is shown.
(C) rde-12 mutants are sensitive to virus infection. RT-qPCR analysis to measure the relative levels of Orsay virus RNA in N2 wild-type (WT), rde-1(ne300), rde-12(tm3644 and tm3679) animals infected with Orsay virus. The fold-change relative to wild-type infected worms is indicated in log2 scale.
(D) rde-12 is not required for production and/or RDE-1-loading of primary siRNAs. Northern blots and immunoblots were probed for unc-22 siRNA and HA∷RDE-1 respectively. RNA and protein extracts were prepared from wild-type and rde-12(tm3644) mutant animals before (Input) and after IP with HA∷RDE-1 (HA IP). The presence (+) or absence (−) of a transgene driving an unc-22 hairpin dsRNA trigger is indicated. (E) Secondary siRNAs fail to accumulate in rde-12 mutants. Northern blot analysis of small RNAs isolated from wild-type and rde-12(tm3644) mutant animals fed with bacteria expressing cpsf-2 dsRNA (+) or a control feeding vector (−). A cpsf-2-specific probe located upstream of trigger sequence (red bar below the gene structure in Fig. 3A) was used to detect cpsf-2 22G-RNAs. An f37d6.3-specific probe was used to detect rde-12-independent endo-22G-RNAs.
The rde-12(tm3644) RNAi-deficient phenotype was rescued by both FLAG-or GFP-tagged RDE-12 transgenes (Fig. 2B). Interestingly, the overexpression of rde-12 in muscle from the myo-3 promoter not only rescued the Rde phenotype of tm3644 but also enhanced the sensitivity of the transgenic animals to unc-22 RNAi. Wild-type, non-transgenic, animals exposed to unc-22(RNAi) produced 100% twitching progeny, but only 14% (n=36) showed the most severe paralyzed twitching phenotype. Strikingly, the percentage of paralyzed animals increased to 72% (n=29) for myo-3∷flag∷rde12 transgenic animals among 100% twitching progeny, suggesting that the overexpression of RDE-12 enhances RNAi in the muscles.
To determine whether the ATPase activity in RDE-12 is required for its function, we generated an rde-12 transgene construct bearing a lesion within motif I in the conserved lysine residue (K429A), which is required for ATP hydrolysis in DEAD-box proteins [14] (Fig. 2A). Based on GFP fluorescence (Fig. S1A) and by immunoblotting with FLAG-specific antibodies (Data Not Shown), we found that the expression of RDE-12(K429A) was identical to the expression of similarly tagged wild-type constructs. Importantly, we found that RDE-12(K429A) was unable to rescue the RNAi deficiency of rde-12(tm3644) (Fig. 2B), indicating that the ATPase activity of RDE-12 is essential for function.
rde-12 mutants are sensitive to viral infection
The exo-RNAi pathway in C. elegans is an antiviral pathway [15, 16]. To ask if rde-12 mutants are defective in viral-suppression, we exposed animals to Orsay virus [17]. We found that both rde-12 mutant strains (tm3644 and tm3679) exhibited dramatically increased viral-RNA levels relative to wild-type, comparable in the case of tm3644 to the levels observed in rde-1 mutants (Fig. 2C). These findings suggest that RDE-12 functions in the antiviral response in C. elegans.
Primary and secondary Argonaute pathways are uncoupled in rde-12 mutants
During exoRNAi, RDE-1 engages primary sense and antisense siRNAs produced by the Dicer-mediated processing of the dsRNA trigger [9]. Once RDE-1 engages target mRNAs, secondary siRNAs, which are exclusively antisense, are produced by RdRP-dependent amplification [9, 18, 19]. To identify which step(s) of the RNAi pathway is compromised in rde-12 mutants, we examined the level of primary and secondary siRNAs in mutant animals exposed to RNAi. To ask if the production of primary siRNA and loading onto RDE-1 requires RDE-12 activity, we examined animals expressing a short hairpin RNA targeting the unc-22 gene [19]. We found that the total level of siRNA produced from this transgene was similar in wild-type and rde-12 mutant animals (Fig. 2D). Furthermore, northern-blot analysis of RDE-1 IP indicated that a similar, or higher, level of unc-22 siRNA was loaded onto RDE-1 in the rde-12 mutant extracts compared to wild type (Fig. 2D).
To ask if RDE-12 is required for RdRP-dependent secondary siRNAs, we exposed animals to cpsf-2 dsRNA and used northern-blot analysis to detect secondary siRNAs that accumulate immediately upstream of the trigger dsRNA region in wild-type animals. Strikingly, this analysis revealed that secondary siRNAs were strongly depleted in rde-12 mutants (Fig. 2E). Deep sequencing of small RNAs from cpsf-2(RNAi) animals revealed a robust accumulation of primary siRNAs in rde-12 mutants: sense siRNA levels under the trigger dsRNA region were similar in rde-12 and wild-type animals (compare upper and lower graphs in Fig. 3A). However, antisense small-RNA levels within this region and upstream of the trigger were reduced dramatically in rde-12 mutants (Fig. 3A and 3B), consistent with a defect in secondary siRNA biogenesis. Taken together, these findings are consistent with a role for RDE-12 downstream of primary siRNA biogenesis and RDE-1 loading, but upstream of secondary siRNA (22G-RNA) production and/or WAGO loading.
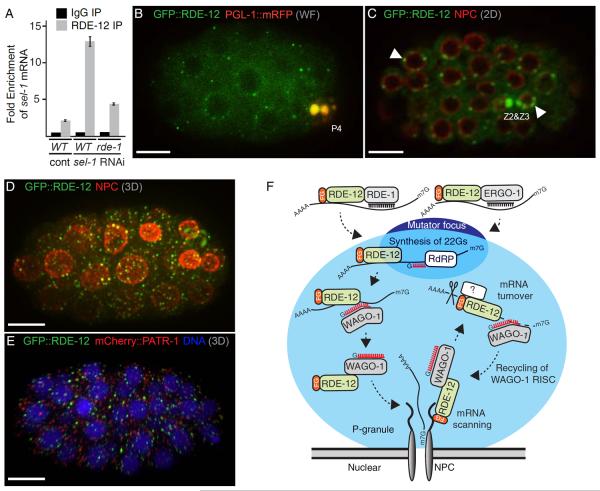
Figure 3.
rde-12 is required for 22G-RNA accumulation in the RDE-1 and ERGO-1 pathways
(A, B) rde-12 is required for secondary 22G-RNA accumulation during exo-RNAi. (A) Genome browser view showing the cpsf-2 gene structure aligned above deep-sequencing read-density plots of the cpsf-2 small-RNAs from wild-type and rde-12(tm3644) mutant strains exposed to cpsf-2(RNAi). Sense reads (light blue) are indicated above the line, and anti-sense reads (pink) are indicated below the line. The height of each bar indicates the frequency (log2 scale) of small-RNAs that initiate at that genomic coordinate. The black bar below the gene structure indicates the location of the cpsf-2 trigger dsRNA. The red bar below the gene structure indicates the probe used in Fig. 2E. (B) Bar graph analysis of the data in (A) indicating the normalized numbers of antisense siRNA reads mapping outside of the dsRNA trigger in wild-type and rde-12(tm3644) mutant animals with (+) or without (−) exposure to cpsf-2 dsRNA.
(C, D) RDE-12 is required for ERGO-1 pathway 22G-RNAs. (C) Scatter plot analysis of endo-siRNAs in wild-type and rde-12(tm3644) mutants. Each point represents a previously defined locus (or gene) targeted by 22G-RNAs that depend on each of four pathways as indicated by the color code. Deviations from the diagonal indicate, on a log10 scale, the enrichment (above the line) or depletion (below the line) in rde-12(tm3644) mutants relative to wild-type animals. The minimal read cutoff was 25 in this analysis. (D) Northern blot analysis of ERGO-1 pathway primary, 26G-, and secondary, 22G-RNAs, in wild-type (N2), ergo-1(tm1890), and rde-12(tm3644) mutants. The blot was analyzed with specific probes to detect si26R-1 and its corresponding 22G-RNA, si26R-263, and the micro-RNA mir-66 (as indicated). Quantification of the relative amounts of 22G- and 26G-RNAs normalized to miR-66 in rde-12 mutants compared to wild type is shown below.
(E, F) rde-12 is required for RDE-1-dependent endo-siRNAs targeting y47h10a.5. (E) Genome browser view showing y47h10a.5 aligned above deep-sequencing read-density plots. Small-RNA distributions in wild-type and rde-12(tm3644) mutant strains are indicated on a linear scale. Antisense reads are indicated in light blue and sense reads (though largely absent) in pink. The location of the mir-243 target site (which triggers secondary 22G-RNA accumulation) is indicated above the 3'UTR of the y47h10a.5 gene. (F) Bar graph analysis of the data in (E) indicating the normalized total numbers of antisense siRNA reads mapping to the y47h10a.5 gene in wild-type and rde-12(tm3644) mutants.
We also analyzed endo-siRNA accumulation in rde-12 mutants. A variety of small RNA species are produced in at least seven different AGO pathways in C. elegans [4, 12, 20–27]. We found that the majority of small RNA species in these pathways were largely unaffected in rde-12 mutants (Fig. 3C and Data Not Shown). In contrast, although the 26G-RNAs that bind to ERGO-1 were largely unaffected, we found that ERGO-1-dependent 22G-RNAs were strongly depleted in rde-12 mutant animals (Fig. 3C, 3D, S2 and Table S1). Similarly, the only verified endogenous RDE-1 target, y47h10a.5 [22], was strongly depleted of secondary siRNAs in rde-12 mutants (Fig. 3E, F and Table S1). These findings suggest that RDE-12 is required for the production or stability of 22G-RNAs downstream of RDE-1 and ERGO-1.
RDE-12 is recruited to the target RNA in an RDE-1-dependent manner
Although sequence-specific RNA binding has not been demonstrated for the majority of DEAD-box proteins, they can be recruited to specific target RNAs by interacting with sequence-specific RNA-binding co-factors [28]. We therefore asked if RDE-12 is recruited to target mRNA during RNAi. To test this possibility, we immunoprecipitated RDE-12 from animals exposed to dsRNA targeting the abundantly-expressed sel-1 mRNA and then measured sel-1 mRNA in the RDE-12 immune complexes by quantitative PCR after reverse transcription (RT-qPCR) using primers upstream of the trigger dsRNA. This analysis revealed a sel-1(RNAi)-dependent interaction between RDE-12 and sel-1 mRNA. Despite the fact that sel-1 mRNA levels were reduced by 71% by sel-1(RNAi), we observed a 6-fold enrichment of sel-1 mRNA in RDE-12 co-IP assays relative to levels detected in experiments on control animals exposed to empty vector (Fig. 4A). The interaction between RDE-12 and the sel-1 target-mRNA was reduced in rde-1(ne300) mutants (Fig 4A). Taken together with our AGO co-IP studies, these findings support the idea that RDE-1 (and perhaps ERGO-1) interacts with and recruits RDE-12 to target mRNA.
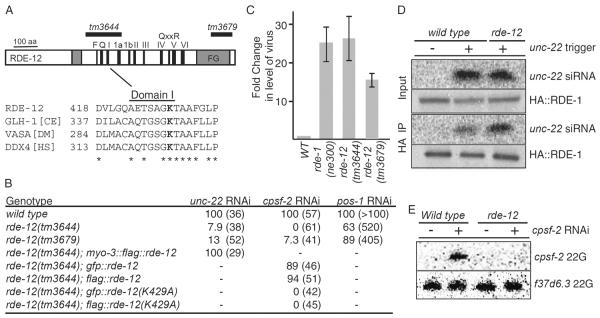
Figure 4.
RDE-12 binds target mRNA during RNAi and localizes to cytoplasmic and perinuclear foci
(A) RDE-12 associates with target mRNA during RNAi. Bar graph showing RT-qPCR analysis of sel-1 mRNA recovered from rabbit IgG or RDE-12 IP from wild-type or rde-1 mutant lysates after control (cont) or sel-1(RNAi) (as indicated).
(B) Fluorescence micrograph showing GFP::RDE-12 (green) is visible throughout the cytoplasm and in cytoplasmic and peri-nuclear foci, while PGL-1∷mRFP (red) [29] is localized exclusively in the germline blastomere P4 (indicated). Co-localization in P4 appears (yellow) in this merged image.
(C and D) Confocal images of GFP∷RDE-12 (green) and nuclear pore complexes (NPC, red) stained with GFP and nucleoporin-specific antibody mAb414, respectively. A single confocal section of a ~50-cell stage embryo (C) shows cytoplasmic and peri-nuclear foci of GFP∷RDE-12 in somatic cells and enriched in the two primordial germ cells (Z2 and Z3, indicated). Arrowheads show examples of peri-nuclear localization of GFP∷RDE-12 in somatic and germ cells. A 3D projection of confocal images from a 24-cell stage embryo is shown in (D).
(E) GFP∷RDE-12 fails to co-localize with PATR-1. A 3D projection of confocal images from a 28-cell embryo showing GFP∷RDE-12 (green), mCherry∷PATR-1 (red), and DNA (blue). Scale bars: 10 μm (B–E).
(F) Schematic model illustrating potential functions and interactions for RDE-12 during RNAi. A nuclear envelope and pore (NPC) are drawn at the base of the diagram with a P-granule (light blue sphere) docked above. A mutator focus (dark blue sphere) is located in the top part of P-granule, where secondary siRNAs are produced. RDE-12 is shown engaged at the pore through its FG domains. Dashed arrows indicate potential functions for RDE-12.
RDE-12 localizes to germline P-granules and to similar, though smaller, peri-nuclear cytoplasmic foci in somatic cells
A previous study using a C-terminally, GFP-tagged transgene showed that RDE-12∷GFP localizes to germline P-granules and co-localizes with nuclear pore components [10]. We confirmed the P-granule localization and nuclear pore association of RDE-12 using GFP∷RDE-12 (Fig. 4B–D and S1A). GFP::RDE-12 localized throughout the cytoplasm and in peri-nuclear foci, and co-localized prominently with P-granules (PGL-1∷mRFP [29]) in the germline (Fig. 4B and Fig. S1A).
Confocal microscopy revealed that peri-nuclear GFP∷RDE-12 foci in somatic cells sometimes overlap with nuclear pores stained with a nucleoporin-specific antibody (Fig. 4C and 4D, Movies S1 and S2). The cytoplasmic (non-peri-nuclear) RDE-12 foci in somatic cells appear similar to cytoplasmic processing bodies, or P bodies, which function in mRNA turnover [30]. However, we found that GFP∷RDE-12 did not significantly co-localize with the P-body marker mCherry∷PATR-1 [31] (Fig. 4E). We speculate that the somatic GFP∷RDE-12 foci represent structures that are analogous to, albeit smaller than, germline P-granules.
A recent study identified distinct cytoplasmic structures termed “mutator foci” [32] that are adjacent to, or partially overlap with, P-granules and contain many factors required for the amplification of secondary siRNAs, including the cellular RdRP RRF-1 and the beta-nucleotidyltransferase RDE-3 [32]. We noted that GFP∷RDE-12, like other P-granule components, partially overlapped with mutator foci (Fig. S1B). The germline localization of RDE-12 was similar to GFP∷WAGO-1 [4], suggesting that the physical association of RDE-12 and WAGO-1 may occur in P-granules. However, localization of GFP∷WAGO-1 in P-granules is not dependent on RDE-12 (Data Not Shown).
Here we have identified the Vasa homolog RDE-12 as an AGO-interacting protein required for RNAi in C. elegans. The interaction between Vasa-related RNA-binding ATPases and Argonautes appears to be evolutionally conserved. In Drosophila, Vasa has been reported to interact with Aubergine (Aub), a Piwisubfamily protein that binds germline piRNAs [33], while Belle, a close paralog of Vasa interacts with Drosophila Argonaute2 (Ago2) and appears to function with AGO2 in promoting chromosome segregation [34]. MVH, a mouse homolog of Vasa associates with Piwi Argonautes, Miwi and Mili [35]. Although Vasa and Belle have been implicated in the piRNA, endo-siRNA and miRNA pathways [36–38], their specific biochemical functions in these pathways remain unclear.
We found that RDE-12 is required downstream of the primary AGOs RDE-1 and ERGO-1 for the accumulation of RdRP-dependent 22G-RNAs. Previous reports have shown that the RDE-10/RDE-11 complex is also essential for the amplification of secondary siRNA in both pathways [39, 40]. Perhaps, once loaded on the target mRNA, RDE-10/11 and RDE-12 signal downstream to recruit RdRP or other factors, such as RDE-3, that ensure efficient secondary small-RNA (22G-RNA) accumulation (see model, Fig. 4F). We identified RDE-12 as a protein that interacts robustly with WAGO-1, a secondary AGO that engages 22G-RNAs downstream of RdRP amplification. Interestingly, the association of RDE-12 with WAGO-1 required RDE-3 activity, raising the possibility that the association of RDE-12 with WAGO-1 may occur on target mRNAs after 22G-RNA biogenesis and/or WAGO-1 loading. One attractive possibility is that RDE-12 functions on the target mRNA to promote loading of nascent 22G-RNAs onto WAGO-1. Perhaps RDE-12 dissociates from the target mRNA together with the newly loaded WAGO-1/22G-RNA RISC and, through its FG-repeat domains, positions WAGO-1 RISC in close proximity to the nuclear pore, where it is thus poised to scan mRNAs exiting the nucleus (Fig. 4F).
WAGO-1, like other WAGOs lacks key metal-coordinating residues in its RNase H-related PIWI domain [9], raising important questions regarding how WAGO-1 promotes silencing and whether WAGO-1 RISC is recycled for subsequent targeting events. It is possible that RDE-12 also functions in one or both of these downstream activities (Fig. 4F). Further analysis of how RDE-12 binds to target mRNA and interacts with WAGO-1 during RNAi should shed light on the fascinating role of this and other DEAD-box proteins in AGO-mediated mRNA surveillance.
Experimental Procedures
Genetics
All C. elegans strains were derived from the Bristol N2 strain and cultured as described [41]. The strains used in this study are listed in Supplemental Information.
RDE-12 antibodies
RDE-12 antibodies were affinity purified from rabbit antiserum raised against a synthetic peptide corresponding to residues 12–29 (GREYHDDRSNRDHRHGNG) of RDE-12.
Immunoblot analysis
Antibodies used for immunoblotting include anti-RDE-12 (1/1000), anti-ERGO-1 (1/1000; [12]), anti-PRG-1 (1/1000; [26]), anti-CSR-1 (1/1000; [20]), anti-FLAG (1/1000; M2, Sigma), and anti-HA (1/4000; ab9110, Abcam).
RNA and Northern blot analysis
RNA purification and Northern blot analysis were performed as described [4, 42].
Immunostaining
Embryos were dissected from worms directly on poly-L-lysine-coated glass slides and fixed as described (http://www.wormatlas.org/images/earlyembstaining.pdf).
Supplementary Material
Highlights
Vasa ATPase-related protein RDE-12 promotes RNAi and virus suppression in C.elegans
RDE-12 is recruited to target mRNA by primary Ago RDE-1 in RNAi pathway
RDE-12 promotes secondary siRNA biogenesis and associates with secondary Ago WAGO-1
RDE-12 localizes to P-granules in germline and novel peri-nuclear foci in soma
Acknowledgments
We thank Ho Yi Mak for sharing information prior to publication. We thank Paul Furcinitti for creating and processing confocal images, Shohei Mitani for rde-12 deletion alleles, Hsin-Yue Tsai for help with RNA IP, and James Moresco and John Yates III for MudPIT analysis. We are grateful to the members of the Mello lab for input and discussion, especially Wen Tang and HengChi Lee for their technical help with Orsay virus assays and IP/Immunoblot, respectively, and Darryl Conte for comments on the text and figures. Some strains used in this study were obtained from the Caenorhabditis Genetics Center. M. Seth is a Howard Hughes Medical Institute International Student Research Fellow. This work was supported by NIH grant (GM058800) to C.C.M. C.C.M. is a Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Number Illumina data are available from GEO under the accession number GSE54396.
Supplemental Information Supplemental Information includes Supplemental Experimental Procedures, Supplemental Figures, Supplemental Movies, and Supplemental Table.
References
- 1.Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb Perspect Biol. 2011;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 3.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 4.Gu W, Shirayama M, Conte D, Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:207–211. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- 7.Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 8.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 9.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 10.Sheth U, Pitt J, Dennis S, Priess JR. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 2010;137:1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, Mello CC. A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol. 2005;15:378–383. doi: 10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitani S. Nematode, an experimental animal in the national BioResource project. Exp Anim. 2009;58:351–356. doi: 10.1538/expanim.58.351. [DOI] [PubMed] [Google Scholar]
- 14.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 15.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009;5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 19.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 20.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Correa RL, Steiner FA, Berezikov E, Ketting RF. MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000903. doi: 10.1371/journal.pgen.1000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr., Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniar JM, Fire AZ. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr Biol. 2011;21:449–459. doi: 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 28.Young C, Karbstein K. Analysis of cofactor effects on RNA helicases. Methods Enzymol. 2012;511:213–237. doi: 10.1016/B978-0-12-396546-2.00010-3. [DOI] [PubMed] [Google Scholar]
- 29.Wolke U, Jezuit EA, Priess JR. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development. 2007;134:2227–2236. doi: 10.1242/dev.004952. [DOI] [PubMed] [Google Scholar]
- 30.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol. 2008;323:76–87. doi: 10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Phillips CM, Montgomery TA, Breen PC, Ruvkun G. MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 2012;26:1433–1444. doi: 10.1101/gad.193904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev. 2008;125:865–873. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pek JW, Kai T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci U S A. 2011;108:12007–12012. doi: 10.1073/pnas.1106245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. 2010;285:8148–8154. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H, Zhang Y, Vallandingham J, Li H, Florens L, Mak HY. The RDE-10/RDE-11 complex triggers RNAi-induced mRNA degradation by association with target mRNA in C. elegans. Genes Dev. 2012;26:846–856. doi: 10.1101/gad.180679.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C, Montgomery TA, Fischer SE, Garcia SM, Riedel CG, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr Biol. 2012;22:881–890. doi: 10.1016/j.cub.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr., Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.