Abstract
The Drosophila genome encodes a total of nine Toll and related proteins. The immune and developmental functions of Toll and 18Wheeler (18W) have been analyzed extensively, while the in vivo functions of the other Toll-related proteins require further investigation. We performed transgenic experiments and found that overexpression of Toll-related genes caused different extents of lethality and developmental defects. Moreover, 18w, Toll-6, Toll-7 and Toll-8 often caused related phenotypic changes, consistent with the idea that these four genes have more conserved molecular structure and thus may regulate similar processes in vivo. Deletion alleles of Toll-6, Toll-7 and Toll-8 were generated by targeted homologous recombination or P element excision. These mutant alleles were viable, fertile, and had no detectable defect in the inducible expression of antimicrobial peptide genes except for the Toll-8 mutant had some defects in leg development. The expression of 18w, Toll-7 and Toll-8 mRNA showed wide and overlapping patterns in imaginal discs and the 18w, Toll-8 double and Toll-7, Toll-8 double mutants showed substantially increased lethality. Overall our results suggest that some of the Toll-related proteins, such as 18W, Toll-7 and Toll-8, may have redundant functions in regulating developmental processes.
Keywords: Drosophila, Toll, Toll-6, Toll-7, Toll-8
Introduction
Toll was first identified as a gene that regulated dorsoventral (DV) axis formation in early Drosophila embryos (Anderson et al. 1985; Hashimoto et al. 1988). Further analyses revealed that Toll also regulated innate immune response in larvae and adults (Lemaitre et al. 1996). Toll encodes a single pass transmembrane receptor protein (Hashimoto et al. 1991) and the host protein Spätzle is translated as a precursor and proteolytic cleavage produces an active Spätzle that functions as a ligand for Toll (Morisato & Anderson 1994; Schneider et al. 1994; Weber et al. 2003; Hu et al. 2004). The proteases that control Spätzle activity during DV development include Gastrulation Defective, Snake and Easter (Brennan & Anderson 2004). Meanwhile, separate upstream proteases such as Persephone and Spätzle Processing Enzyme regulate Spätzle during innate immune response (Brennan & Anderson 2004; Jang et al. 2006; Ferrandon et al. 2007). Ligand stimulation of Toll leads to the activation of a highly conserved intracellular signaling pathway. Toll interacts with dMyD88, Tube and Pelle to form the receptor-adaptor complex that ultimately targets the NF-κB-equivalent complex, that is Dorsal, DIF / Cactus in Drosophila (Minakhina & Steward 2006; Ferrandon et al. 2007; Uvell & Engstrom 2007; Pal & Wu 2009). Dorsal and DIF have redundant functions or can substitute each other when expressed in the same tissue (Petersen et al. 1995; Stein et al. 1998; Meng et al. 1999; Rutschmann et al. 2000).
The mammalian counterparts of Toll are known as Toll-like receptors (TLRs). Mammalian genomes have 10–13 different TLR genes and different TLRs are responsible for different pathogen recognition (Kumar et al. 2009). For example, a TLR4 ligand is lipopolysaccharide (LPS) and TLR4 knockout mouse cannot respond to LPS stimulation (Poltorak et al. 1998; Hoshino et al. 1999). The intracellular signaling components for mammalian TLR and Drosophila Toll are well conserved, suggesting that Toll and TLR signaling have a common evolutional origin (Leulier & Lemaitre 2008; Kumar et al. 2009; Pal & Wu 2009). However, a major deviation between insect Toll and mammalian TLRs is the interaction with their ligands. Various TLRs can bind pathogen-associated molecular patterns (PAMPs) either directly or as a receptor complex. Toll, however, interacts with the host protein Spätzle, while pattern recognition in Drosophila involves peptidoglycan recognition proteins (PGRPs) and Gram negative bacteria binding proteins (GNBPs) that act further upstream. Binding of PGRPs/GNBPs with PAMPs leads to activation of a few protease cascades that converge onto Spätzle (Gottar et al. 2006; El Chamy et al. 2008). Therefore, while cytoplasmic signaling is conserved, pathogen recognition in mammals and insects probably evolved independently.
The Drosophila genome encodes eight other proteins that are related to Toll (Tauszig et al. 2000; Ooi et al. 2002). We will refer to these proteins in this report as Toll, 18Wheeler, Toll-3 (MstProx), Toll-4, Toll-5 (Tehao), Toll-6, Toll-7, Toll-8 (Tollo) and Toll-9. Toll has been analyzed extensively as described above, and 18Wheeler (18W) and Toll-8 have been analyzed to some extent. Mutants of 18w showed some defects in innate immune response as well as follicle cell and salivary gland development (Williams et al. 1997; Kambris et al. 2002; Ligoxygakis et al. 2002; Kleve et al. 2006; Kolesnikov & Beckendorf 2007). Toll-8 mutant was first reported to play a role in embryonic central nervous system (CNS) development (Seppo et al. 2003). However, another analysis revealed no developmental phenotype, but instead Toll-8 overexpression affected decapentaplegic (dpp) expression and wing disc development (Kim et al. 2006). Because Toll and TLRs are known to have biological functions in innate immunity, it has been speculated that these other Toll-related proteins in Drosophila may participate in some aspects of innate immune response. However, only Toll and Toll-9 overexpression could lead to significant activation of antimicrobial peptide genes (Tauszig et al. 2000; Ooi et al. 2002; Bettencourt et al. 2004). Sequence comparison of TIR domains showed that the TIR domain of Toll-9 has closer homology with mammalian TLRs while the extracellular domain structure of Toll-9 is more diverged from other Drosophila Toll-related proteins (Imler & Zheng 2004). Meanwhile, the embryonic expression patterns of Toll-related genes suggest that the members of this family may also regulate various developmental processes (Kambris et al. 2002). Moreover, Toll, 18w and Toll-8 have been suggested to function as cell adhesion molecules (Keith & Gay 1990; Eldon et al. 1994; Kim et al. 2006). Further analysis especially at the genetic and molecular levels is needed to understand the in vivo functions of Toll-related proteins in Drosophila innate immunity and development.
We performed gain-of-function and loss-of-function analyses of Toll-related genes in transgenic and mutant flies. Overexpression of Toll and Toll-9 suggested their functional similarities, while 18w, Toll-6, Toll-7 and Toll-8 often showed related phenotypes. Toll-3, Toll-4 and Toll-5 overexpression had no observable phenotypic changes. Analysis of loss of function alleles of Toll-6, Toll-7 and Toll-8 showed that the single mutants were mostly viable with some leg defects. However, the 18w, Toll-8 and Toll-7, Toll-8 double mutant combinations had substantially increased lethality. These data suggest possible interaction or redundant functions among the subgroup of Toll-related genes including 18w, Toll-7 and Toll-8 in Drosophila.
Materials and methods
DNA constructs for transgenic expression
Genomic DNA fragments of Toll-4, Toll-5, Toll-6, Toll-7, Toll-8 and Toll-9 (Ooi et al. 2002) were subcloned into the pUAST vector. Toll and TollD (Toll781Y) cDNA clones into pUAST have been described (Hu et al. 2004). The Toll-3 full length clone contained 3262 bp and included an extra 300 bp 5′ coding sequence and an intron when compared with the earlier predicated Toll-3 coding sequence (Ooi et al. 2002). This Toll-3 full length region represents the updated version in Fly-base and was amplified from wild type genomic DNA and subcloned into pUAST. Coding sequence of 18w and pelle was amplified by polymerase chain reaction (PCR) from genomic DNA or cDNA and subcloned into pUAST. Trangenic fly lines were established by standard procedure. Other fly stocks were obtained from Bloomington stock center if not described.
Generation of Toll-related gene mutants
Toll-6 mutants were generated by imprecise excision of P element from the GS11682 fly strain, obtained from Dr T. Aigaki (Tokyo Metropolitan University). The P element line was crossed with transposase stock and individual w− offsprings were balanced and used for mapping. Deleted regions of the mutant lines were determined by PCR of genomic DNA. The upstream break point of Toll-6ex13 was mapped to approximately 450 bp upstream of the transcription start site. The downstream break point was mapped between 25 and 50 kb from the Toll-6 coding region. There is no known open reading frame (ORF) besides Toll-6 uncovered by these deletions.
We used ends-in knockout system followed by hs-ICreI treatment (Rong et al. 2002) to generate the Toll-7g1−1 deletion allele. A genomic DNA fragment of Toll-7 was modified and mutagenized to introduce two stop codons and an ISceI site into the Toll-7 DNA fragment. Point mutations were introduced using QuickChange Site-Directed Mutagenesis kit (Stratagene). The modified Toll-7 DNA fragment was subcloned into pTV2 vector (Rong et al. 2002). Sequences of primers used for mutagenesis was Toll-7 A: tcg aac cta aaa tgg ctt taa atc cat gga aac tat att gag gcc, Toll-7 B: ggt ggc tcc atg ctg tag cat ggt atc cca gaa. Oligonucleotide of reverse complement sequence for each primer was also synthesized and used for mutagenesis.
Toll-8 mutagenesis started with EY01776 that had a P element insertion located 150 bp upstream of the start codon. Individual offspring that had lost the w+ marker/P element was collected and used for PCR mapping. Toll-858 and Toll-859 were determined to contain deletions. The deleted sequence of Toll-858 was confirmed by sequencing PCR amplified DNA fragment (primers ttcgaacccagctgtttgtg and acggctgaatcttggagatc). It contained a 3068 bp deletion starting from 1508 bp upstream of the start codon and extending downstream, but also contained a 7 bp insertion (cgagcaa) at the break point. The Toll-859 chromosome contains a deletion that uncovers the Toll-8 coding region plus at least 5 kb of upstream and downstream regions but the nearby ORFs CG6888 and Best4 are intact.
Primer sequences for confirmation of genomic deletion in Figure 4 are: P1: forward cac cac cac aac cac caa, reverse aac gga tcc tta cgc cca cag gtt ctt c. P2: forward cat aat aga cac gct cgc act ctc g, reverse cat ttg aaa cct tgt gag gtg gac c. P3: forward gaa cta cac ccc ttg aag gt, reverse ctg tag ttt act cag ctg. P4: forward aac acg gtg cgc ctg cgt ct, reverse cga ttg cgc gag agg atc ag. P5: forward gga cgc tct aat tgg agc tg, reverse ggc cat aga ggt ggt atc tg.
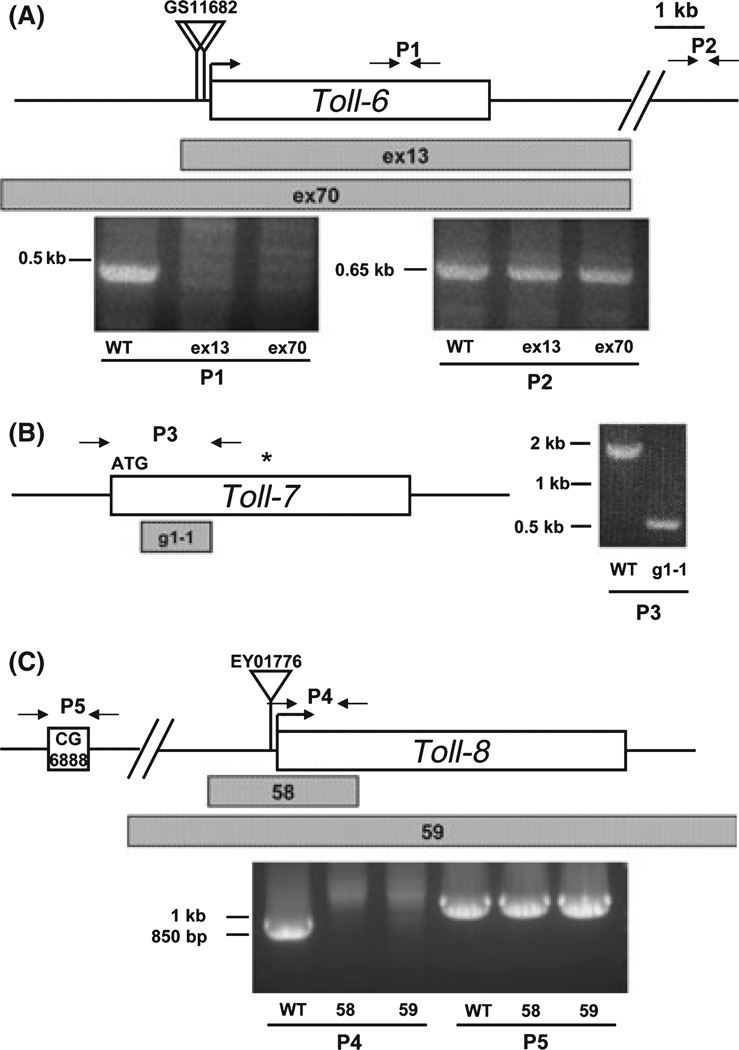
Fig. 4.
Genomic organization and mutant alleles of Toll-6, Toll-7 and Toll-8. The top of each panel shows the wild type genomic organization. The regions deleted are indicated by the shaded bars under each locus and the allele names are as shown. The gel images show the polymerase chain reaction (PCR) products with the primer pairs as shown. Genomic DNA from the indicated alleles are used as templates. Each of the primer pairs (P1 in panel A, P3 in panel B and P4 in panel C) within the coding regions gave positive amplification using wild type DNA but generated no or smaller products when using DNA from the deletion alleles. The control primer pairs (P2 and P5) outside the coding regions yielded the same products when using wild type and mutant DNA.
Quantitative PCR analysis of anti-microbial peptide expression
Adult flies 2–7 days old were infected by septic injury with a mixture of overnight culture of Enterobacter cloecae and Micrococcus luteus. Infected flies and control flies without injury were collected 6 h after infection and kept at −80°C until use. RNA was extracted from the frozen flies by RNeasy mini Plus kit (QIAGEN). Quantitative real time PCR was carried out using High-Capacity cDNA Reverse Transcription kit and Power SYBER Green PCR Master Mix (ABI) with ABI 7500 Real Time PCR System. Primer sets for the antimicrobial peptide genes were synthesized following a previous report (Leulier et al. 2006). The expression level of mRNA was normalized to the rp49 mRNA value in each tissue sample. Three independent experiments were performed and the average and standard error were calculated.
Antibody staining and in situ hybridization of embryos
Toll-859 and Df(3R)Brd15 were balanced with TM6B, AbdB-lacZ and embryos were collected. Homozygous mutant embryos were identified by the lack of β-galactosidase expression originated from the balancer chromosome as detected by X-Gal staining. Rabbit anti-horse radish peroxidase (HRP) was used for immunostaining of embryonic CNS. HRP labeled secondary antibody was used to detect signal with 3,3′-diaminobenzidine tetrachloride (DAB) staining. In situ hybridization was carried out using digoxigenin (DIG) labeled antisense RNA probe. DNA fragments of 18w, Toll-6, Toll-7 and Toll-8 were subcloned and used as templates for in vitro transcription. These probes were tested in embryos to confirm the expression patterns as previously described (Kambris et al. 2002) and then used for imaginal disc staining.
Viability and phenotypic analyses of Toll-related gene mutants
Toll-related gene mutant flies were established as heterozygous, balanced stocks and crossed with each other. The genotypes of offspring were identified and counted based on the presence or absence of the balancer-associated markers. From the number of flies with balancer chromosome markers, the expected number of Toll-related gene mutant or double mutant flies was calculated. The viability of the mutants was expressed as the percentage of hatched homozygotes over expected homozygotes. For malformed leg phenotypic analysis, flies of each genotype were examined and the percentage of flies that had at least one malformed leg was counted and the penetrance of leg phenotype was calculated.
Results
Transgenic expression of Toll-related genes can lead to various phenotypes
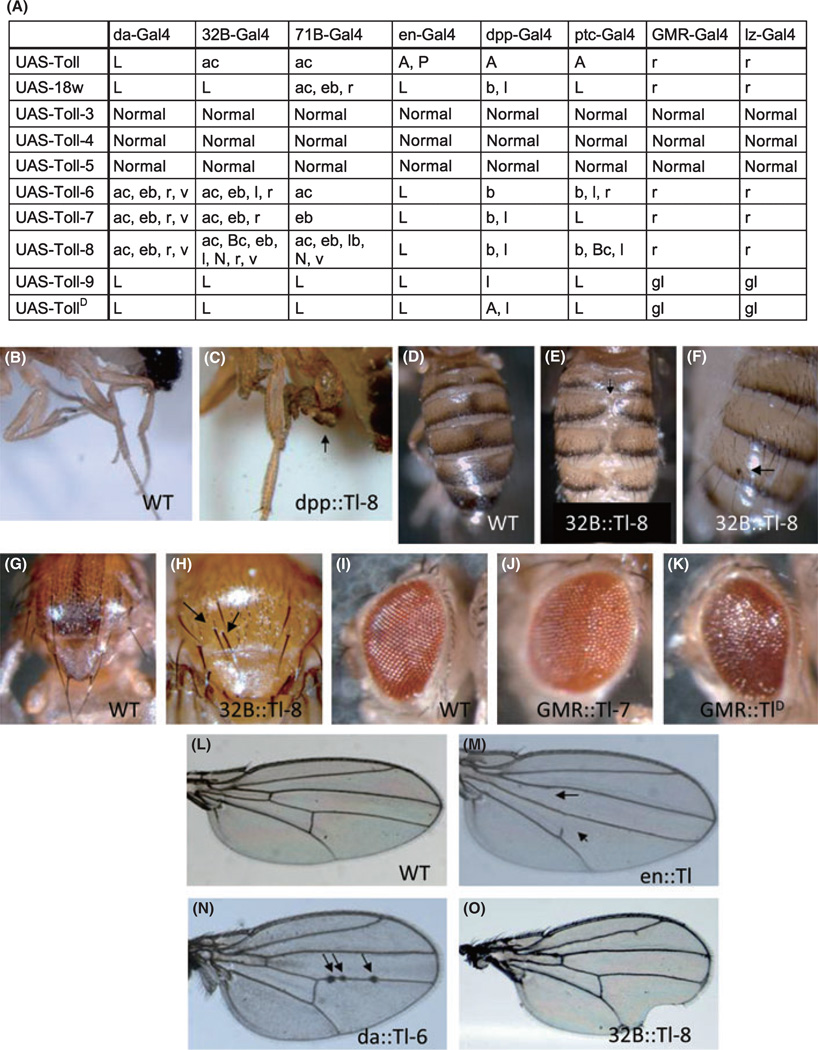
The RNA expression of the Drosophila Toll-related genes in the embryos and in various developmental stages are distinct with some overlap as previously reported (Tauszig et al. 2000; Kambris et al. 2002; Kim et al. 2006). Their functions in most developmental stages, however, have not been studied extensively. To evaluate systematically the possible molecular functions of these Drosophila Toll-related gene products in development, we carried out a transgenic expression assay. The coding sequences were placed under the UAS-vector and transgenic flies were generated. These fly lines were crossed to a series of Gal4 drivers to examine the effect of expression in different tissues (Fig. 1A). We used both ubiquitous and tissue / stage specific drivers. When the ubiquitous expression was driven by the daughterless (da)-Gal4, lethality (L) was observed for Toll, 18w and Toll-9 expressing animals. The penetrance of the lethality was close to 100%. Meanwhile, Toll-6, Toll-7 and Toll-8 showed mild defects of abdominal closure (ac), extra bristle (eb), rough eye (r) and wing vein thickening (v). The penetrance of rough eye phenotype was also close to 100%, while the penetrance of other phenotypes was approximately 5–10%. The expression of Toll-3, Toll-4 and Toll-5 did not cause any lethality or observable morphological defects by the da-Gal4 or any of the other drivers used (Fig. 1A).
Fig. 1.
Transgenic expression of Toll-related genes induces multiple phenotypes. Toll-related genes were expressed under the UAS promoter driven by various Gal4 as indicated in panel A. The panel A table summarizes the phenotypes observed. The abbreviations of the phenotypes are: A, anterior cross vein defect; ac, abdominal closure defect; b, bifurcated wing; Bc, black cell-like phenotype; eb, extra bristle; gl, glazed eye; l, leg defect; L, lethal; lb, loss of bristle; N, notched wing; P, posterior cross vein defect; r, rough eye; v, vein thickening. Panels B–O are representative images of phenotypes in wild type (WT) flies or in transgenic flies expressing the indicated Toll-related gene, as indicated at the bottom of each panel. For example, dpp::Toll-8 is dpp-Gal4 / UAS-Toll-8. TollD was Toll781Y (Hu et al. 2004). Panel C is leg defect phenotype; E is abdominal closure defect; F is black cell-like phenotype; H is extra bristle; J is rough eye; K is glazed eye; M is cross vein defect (arrows); N is vein thickening (arrows); O is notched wing.
We used various drivers intended for expression in imaginal discs. The 32B- and 71B-Gal4 have expression in all imaginal discs. Under these combinations, we observed that Toll-9 and TollD still caused complete lethality while Toll only caused abdominal closure defect. Overexpression of Toll-9 and TollD caused very similar target gene expression profile, likely through the activation of the canonical Toll signaling pathway (Ooi et al. 2002; Bettencourt et al. 2004). This suggests that strong activation of the Toll signaling pathway in imaginal discs is sufficient to kill the animals. Mean-while, similar expression of 18w caused lethality or milder phenotypes consistent with disc developmental defects. The Toll-6, Toll-7 and Toll-8 expression also caused morphological changes in adult tissues derived from various imaginal discs (Fig. 1D–H). The use of engrailed (en)-Gal4 to express 18w, Toll-6, Toll-7, Toll-8, Toll-9 and TollD led to complete lethality. While en is expressed in the posterior compartment of imaginal discs, the gene is also expressed in embryonic stages and in CNS. The lethality may result from the overexpression in other tissues. Wild type Toll overexpression did not induced lethality but anterior cross vein and posterior cross vein were lost in adult wings. We further tested the functions of Toll-related genes by using decapentaplegic (dpp)- and patched (ptc)-Gal4 intended for expression in the more restricted part, anterior–posterior (A–P) boundary of imaginal discs. Under these Gal4 drivers, Toll, Toll-9 and TollD exhibited anterior cross vein missing (A), leg defects (l) or lethality. In comparison, 18w, Toll-6, Toll-7, and Toll-8 exhibited consistent phenotypes of wing bifurcation (b) and leg defects (Fig. 1A,B,C), while lethality was also observed in some combinations. The wing bifurcation phenotype and leg defects had penetrance close to 100%.
Expression of the various Toll-related genes in eye imaginal discs by GMR-Gal4 and lozenge (lz)-Gal4 caused rough eye phenotype. Toll-9 and TollD caused the most severe phenotype so that they became glazed eyes (gl), consistent with severe developmental defects in omatidia. The Toll, 18w, Toll-6, Toll-7, and Toll-8 all caused very mild rough eye phenotype (Fig. 1I–K). All of these eye phenotypes were also close to 100% penetrance.
The 18w, Toll-6, Toll-7 and Toll-8 can regulate imaginal disc development
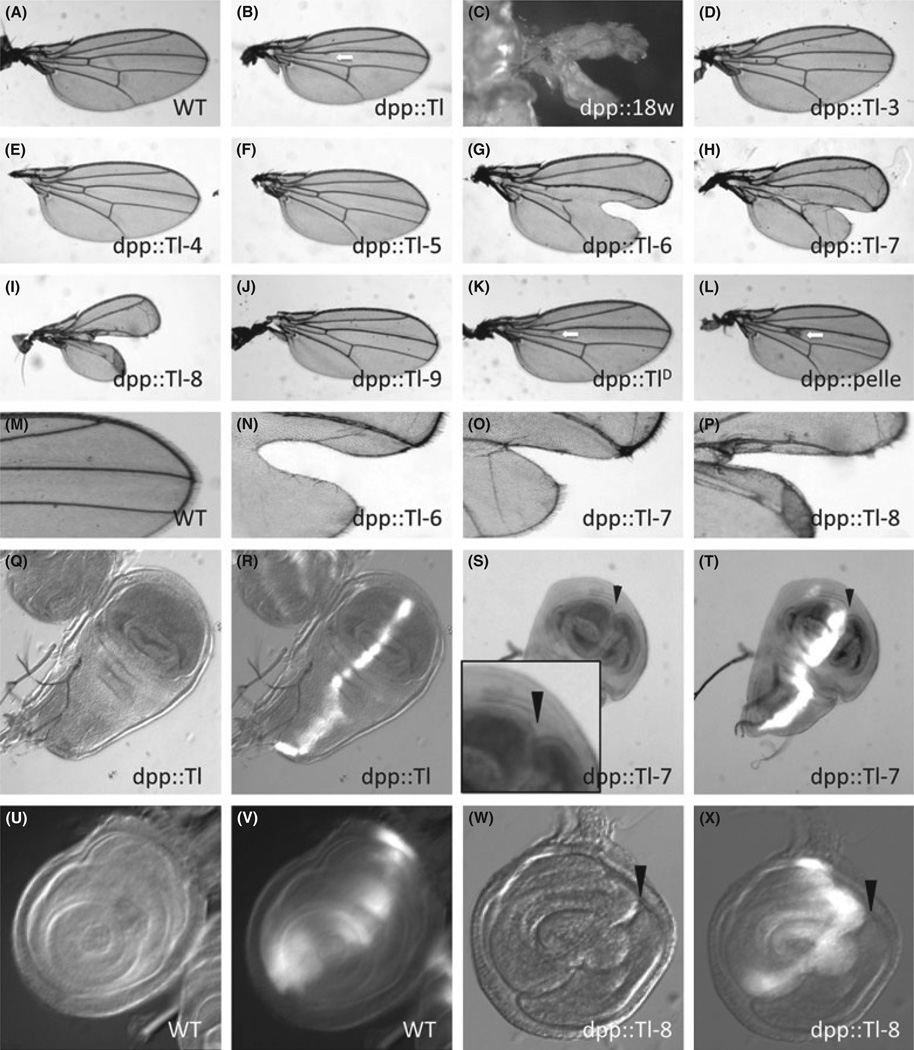
A prominent feature observed from the above experiments is the consistent phenotypic changes in adult tissues developed from imaginal discs expressing 18w, Toll-6, Toll-7, and Toll-8. This group therefore may have a function in imaginal disc development. To investigate this further, we examined the wing and disc phenotypes in adults and larvae. With dpp-Gal4 as the driver, which directs the expression in the A–P boundary, 18w expression led to the most severe phenotype such that only small wings were formed that bifurcated along the A–P boundary (Fig. 2C). Toll-8 caused a similar but milder wing bifurcation phenotype, while Toll-6 and Toll-7 caused mild A–P splitting at the distal end of the wing (Fig. 2G–I). The Toll, TollD and pelle overexpression caused a different and mild phenotype in the wing veins, while Toll-9 did not cause any changes (Fig. 2B,J–L). Therefore, the 18w, Toll-6, Toll-7 and Toll-8 has a specific effect in the formation of A–P boundary when overexpressed by the dpp-Gal4 driver, while other Toll-related genes do not give the same effect.
Fig. 2.
Wing-margin defects induced by the expression of the 18w, Toll-6, -7 and -8. Adult wing preparations are shown in panels A–P. The dpp-Gal4 driver was crossed with UAS lines as indicated at the bottom of each panel. Wild type (WT) was UAS-YFP, and TollD was Toll781Y. Panels M–P are enlarged images of panels A, G, H and I for WT, Toll-6, Toll-7 and Toll-8 overexpression, respectively. The expression of these Toll-related genes induced bifurcated wing phenotype. Overexpression of Toll (panel B) and TollD (panel K) caused the loss of anterior cross vein, while pelle (panel L) overexpression caused thickening of anterior cross vein (white arrows). Panels Q–X are images of imaginal discs that overexpressed the indicated constructs. All of these lines also included the UAS-YFP for visualizing the cells that should express the transgenes. The images in panels Q, S, U, W are light microscopy pictures, while panels R, T, V, X are mergers of light and fluorescent / YFP microscopy pictures. The expression of YFP alone (WT) or together with Toll showed normal disc morphology, while ectopic foldings were observed in Toll-7, -8 overexpressing discs (arroheads in S, T, W, X).
To trace the origin of the wing bifurcation phenotype, we examined the morphology of the imaginal discs of 18w, Toll-6, Toll-7 and Toll-8 overexpressing larvae. We observed abnormal folding at the A–P boundary of the wing and leg imaginal discs (Fig. 2M–T). We performed acridin orange staining to detect cell death but did not observe increased staining in the overexpressing discs (data not shown). It suggests that there may be a change of cell fate or developmental coordination along the A–P boundary of 18w, Toll-6, Toll-7 or Toll-8 expressing cells leading to abnormal folding. This abnormal folding may then have resulted in the adult wing bifurcation phenotype.
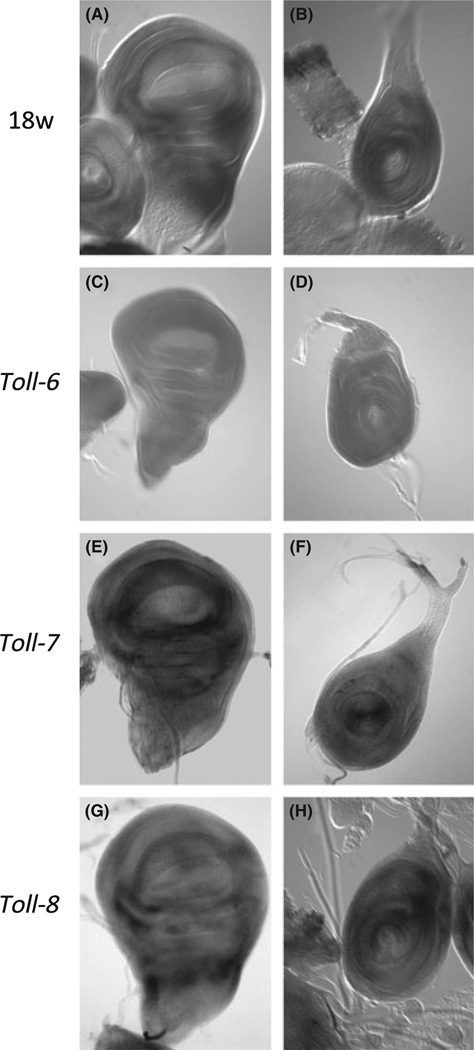
As abnormal phenotypes were observed in the imaginal discs and their derived tissues, we examined the native expression patterns of 18w, Toll-6, Toll-7 and Toll-8 in the imaginal discs by in situ hybridization (Fig. 3). 18w and Toll-8 showed similar expression patterns in the wing and leg discs. RNA signal was observed in the proximal region of the wing discs and anterior compartment of the leg discs. Toll-7 expression in the wing discs was similar to 18w and Toll-8 expression while Toll-7 expression in the leg discs was observed in distal segments. As expression of 18w, Toll-6, Toll-7 and Toll-8 was not observed in the wing pouch, the overexpression phenotype observed in wing was an ectopic expression effect. We could not detect significant expression of Toll-6 in the imaginal discs.
Fig. 3.
Expression patterns of 18w, Toll-6, Toll-7 and Toll-8 in imaginal discs. In situ hybridization was carried out to observe expression patterns of 18w, Toll-6, Toll-7 and Toll-8 mRNA in the imaginal discs. Panels A, C, E, G are wing discs, and panels B, D, F, H are leg discs. The in situ probes used are indicated to the left. The expression of 18w, Toll-7 and Toll-8 is similarly detectable around wing pouch and hinge region (A, E, G). 18w and Toll-8 expression is higher in the anterior segment / left half of leg discs (B, H). Toll-7 expression in the leg disc is higher around the A–P border of tarsal to tibia segment (F). No detectable staining was observed in the imaginal discs for the Toll-6 probe (C, D).
Generation of loss of function alleles of Toll-6, Toll-7 and Toll-8
To further analyze the Toll-6, Toll-7 and Toll-8, we generated loss of function mutant alleles by imprecise excision of P elements (for Toll-6 and Toll-8) or by targeted homologous recombination (for Toll-7). 18w mutant alleles have already been described and analyzed (Eldon et al. 1994; Williams et al. 1997; Ligoxygakis et al. 2002; Kleve et al. 2006; Kolesnikov & Beckendorf 2007), while a small deletion around the 5′ UTR of Toll-8 has been described (Kim et al. 2006). To generate Toll-6 mutants, we used the P element line GS11682, which has two GSV6 vector insertions upstream of the Toll-6 coding region. After crossing with the transposase expression strain, individual lines that had lost the P element were analyzed by PCR of genomic DNA aiming at identifying imprecise excisions. We found that Toll-6ex13 and Toll-6ex70 contained deletions in the Toll-6 coding sequence. The results of PCR analysis using primer pairs P1 and P2 are shown (Fig. 4A). The upstream break point of Toll-6ex13 lies within −449 to +330 bp of the putative transcription start site. The downstream break point lies within 25–50 kb beyond the Toll-6 coding region. No other known gene is reported inside the deleted region of Toll-6ex13. Toll-6ex13 was used for the analyses in this report because Toll-6ex70 has larger deletion and may affect the 5′ neighboring gene CG7255.
We used an ends-in knockout system followed by hs-ICreI treatment to generate a Toll-7 knockout line. In vitro mutagenized Toll-7 coding region was inserted into the pTV2 vector and transgenic fly lines generated were used for genetic crosses to set up the homologous recombination. A correctly recombined line was further crossed with the hs-ICrel line to delete one of the tandem copies. The resulting Toll-7g1−1 has a single copy of mutated Toll-7 gene with a small deletion inside the coding region. This was confirmed by amplifying and sequencing the DNA fragment around the breaking points using the primer pair P3 (Fig. 4B). The PCR product was approximately 1.5 kb smaller than that from the wild type template. The sequencing of this product showed that Toll-7g1#x02212;1 has 1565 bp of deletion, which is from 406 to 1970 bp downstream from the start codon. This deletion causes a frame shift and a new stop codon at codon 140 downstream from the translation start. Further downstream also carries one of our introduced stop codons in the knockout vector (amino acid 1040Q to stop, indicated by * in panel 4B). Thus, Toll-7g1−1 is likely a null mutant.
To generate Toll-8 mutants, we started with the strain EY01776 that had a P element vector inserted 150 bp upstream of the putative start codon. After crossing with a transposase strain, we found two imprecise excision lines, Toll-858 and Toll-859, that had small deletions around the coding region (Fig. 4C). Analysis of the PCR products from primer pairs P4 and P5, plus other PCR mapping encompassing this region, showed that Toll-858 contained a deletion that spanned the 5′ upstream region and part of the coding region. By similar mapping strategy, we found that Toll-859 contained a deletion that uncovered a region extending from further upstream to the region down-stream of the coding sequence. These results demonstrate that the two alleles are likely null mutants.
Phenotypic analyses of the Toll-6, -7, -8 mutant alleles
Both Toll-6ex13 and Toll-6ex70 are largely homozygous viable and fertile (see Fig. 7A). While Toll-7g1−1 homozygotes have very low viability (6%), the Toll-7g1−1 / deficiency showed higher viability (28%), suggesting that the Toll-7g1−1 mutant chromosome may have another mutation that contributes to the lethality. Both Toll-858 and Toll-859 alleles are largely viable and fertile, consistent with previous characterization of a small deletion in Toll-8 5′ UTR (Kim et al. 2006).
Fig. 7.
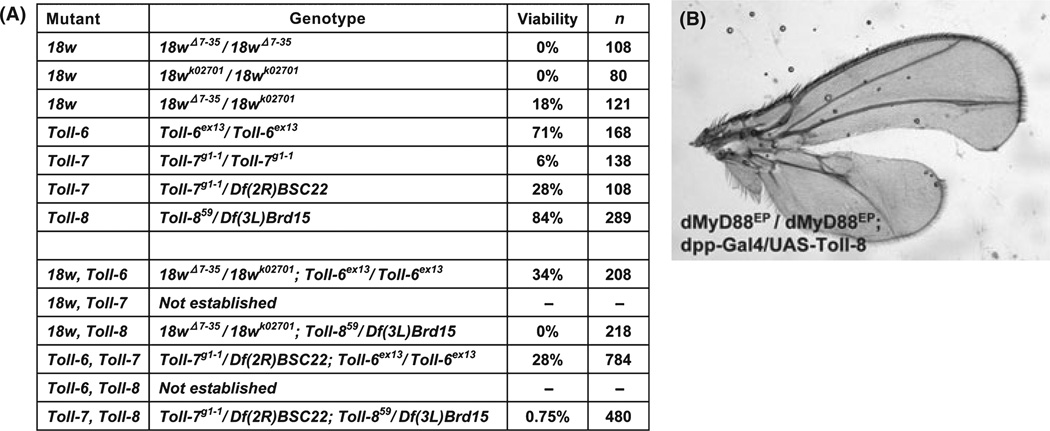
Viability assay of double mutant combinations of Toll-related genes of Drosophila and epistatic analysis of a Toll downstream factor on Toll-8 overexpression phenotype. (A) Viability assay for mutant combinations of 18w, Toll-6, Toll7 and Toll-8. The table shows the total number of flies (n) and the percentage of viable flies of single and double mutants of 18w, Toll-6, Toll-7, and Toll-8. The percent viability is calculated as number of viable homozygotes over the number of expected homozygotes based on the number of other flies hatched. The numbers of viable homozygotes and trans-heterozygotes suggest that 18w and Toll-7 mutant chromosomes have other mutations that cause increased lethality. Therefore, trans-heterozygotes were used whenever possible for double mutant analysis. The 18w, Toll-7 and Toll-6, Toll-8 recombinations were not attempted because the genes are located very close to each other. The combinations of 18w, Toll-8 and Toll-7, Toll-8 showed reduced viability, while 18w, Toll-6 and Toll-6, Toll-7 did not. (B) A dMyD88 allele did not suppress bifurcated wing phenotype. A wing of yw; dMyD88EP2133 / dMyD88EP2133; dpp-Gal4 / UAS-Toll-8 fly is shown. Reduced activity of dMyD88 did not affect bifurcated wing phenotype.
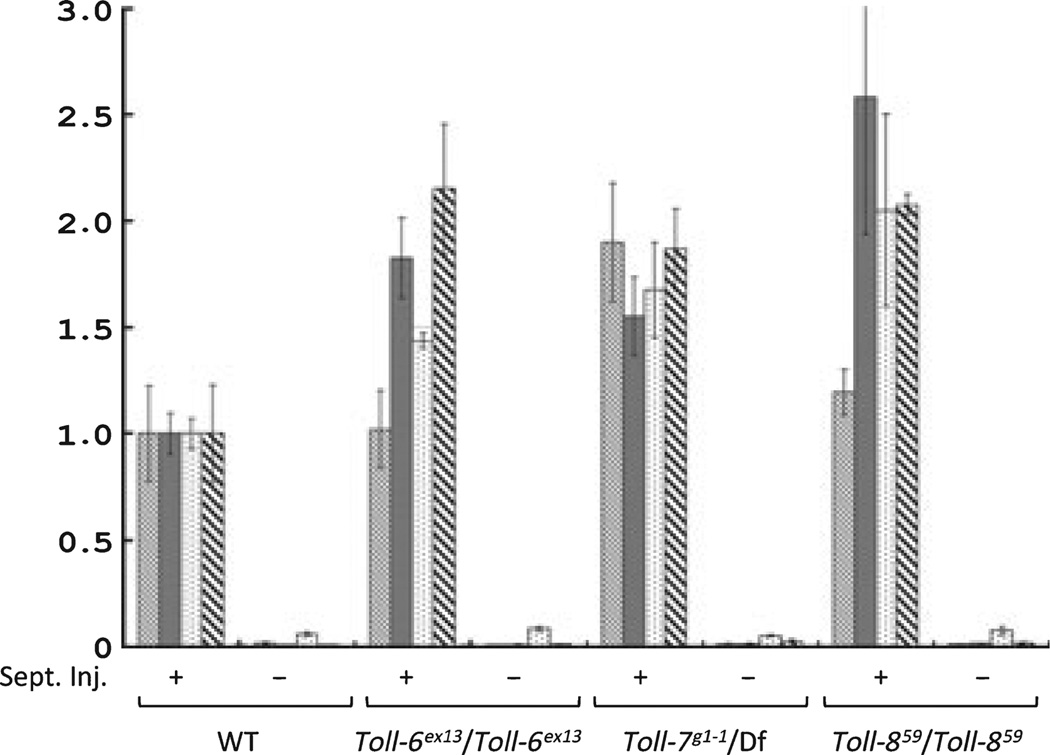
Because Toll-related proteins have been speculated to have innate immune functions, we examined the induction of antimicrobial peptide genes using the viable adult mutant flies. Septic injury of wild type flies causes high level expression of four different antimicrobial peptide genes (Fig. 5). All Toll-6, Toll-7, and Toll-8 mutants exhibited similar inducible expression of these four antimicrobial peptide genes. We also examined the viability of these mutant flies after septic injury and the viability was similar to wild type flies, which suggests that the overall innate immune response against bacteria does not have a severe defect in these mutant flies (data not shown). These results suggest that a main branch of innate immune response does not involve the individual function of these three Toll-related genes.
Fig. 5.
Normal induction of antimicrobial peptide genes in Toll-6, -7 and -8 mutants. Adult flies of the genotypes as indicated were collected and subject to septic injury. Antimicrobial peptide gene expression was measured by quantitative reverse transcription–polymerase chain reaction using gene-specific primers as indicated. The expression level was first normalized by the rp49 expression level in each sample. The value of each antimicrobial peptide gene in the wild type background after septic injury was then set to 1, and the expression levels in flies without septic injury and in other genetic backgrounds were calculated relative to this level of 1 for each gene separately. The plot as shown is an average of three independent experiments. Error bars represent standard error. AttA is AttacinA ( ); Dipt is Diptericin (
); Dipt is Diptericin ( ); Drs is Drosomycin (
); Drs is Drosomycin ( ); Mtk is Metchnikowin (
); Mtk is Metchnikowin ( ). The inducibility of the antimicrobial peptide genes tested remains high in the mutants as in the wild type flies.
). The inducibility of the antimicrobial peptide genes tested remains high in the mutants as in the wild type flies.
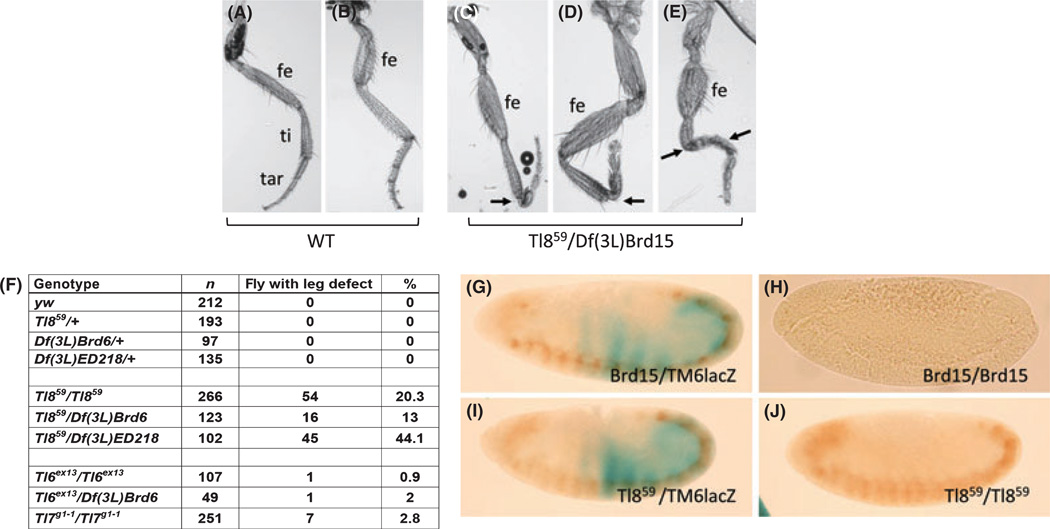
A detectable phenotypic change was that Toll-8 mutant flies exhibited a slight defect in adult legs (Fig. 6A–E). These mutant flies had a normal number of segments but the legs were bent between tibia and tarsus (Fig. 6C–E, arrows) and thus twisted towards other directions. Flies with stronger phenotype exhibited malformed femur and double bending in both tibia and tarsus (Fig. 6E). The penetrance of this leg phenotype is 20–44% of the Toll-8 mutant flies, depending on the mutant combination (Fig. 6F). The Toll-6, and Toll-7 mutants have much lower penetrance of the leg phenotype that might be a genetic background effect and therefore not significant. A previous report showed that some allelic combinations of 18w also showed leg developmental defects (Eldon et al. 1994). The mild leg defect observed in Toll-8 mutants is consistent with some of the imaginal disc-derived phenotypes induced by the overexpression experiments described above.
Fig. 6.
Toll-8 mutants have defects in adult legs but not in embryonic central nervous system (CNS). (A–B) Wild type legs showing normal arrangement of different segments, from femur (fe) to tibia (ti) and tarsus (tar). (C–E) Malformed legs from the Toll-859 / deficiency mutant flies. Most segments have similar length but the first tarsal segment is frequently bent as indicated by the arrows. The distal end of the leg in panel D is lost. Panel E is a more severely malformed leg, in which bending occurs in both tibia and tarsal segments as indicated by the arrows and the femur is malformed. The whole leg is therefore twisted. (F) A table summarizing the penetrance of malformed leg phenotype. Flies with one or more malformed leg were counted. A high percentage (13–44%) of Toll-8 mutants have such developmental defects. Toll-6 (1%) and Toll-7 (3%) mutants have very low penetrance of this phenotype. (G–J) CNS morphology was visualized by anti-horse radish peroxidase (HRP) staining (brown). The balancer chromosome contained a lacZ transgene and the embryos that lacked β-galactosidase expression (blue staining) were homozygous mutants for Toll-8. The genotypes of the embryos were G: Df(3R)Brd15 / TM6B AbdB-lacZ, H: Df(3R)Brd15 / Df(3R)Brd15, I: Toll-859 / TM6B AbdB-lacZ, J: Toll-859 / Toll-859. The Df(3R)Brd15 homozygous embryos lost anti-HRP antigen but Toll-8 homozygous mutant embryos showed normal anti-HRP antigen.
A previous report suggested that Toll-8 had a function in CNS because a deficiency chromosome exhibited loss of anti-HRP-staining in CNS, which was rescued by Toll-8 expression (Seppo et al. 2003). Therefore, we performed anti-HRP antibody staining in our Toll-8 null mutant embryos (Fig. 6G–J). However, the anti-HRP antigen expression was normal in these mutant embryos (Fig. 6I,J), contrary to the total loss of staining in the previously described deficiency Df(3L)Brd15 (Fig. 6G,H). This result suggests that loss of Toll-8 function is not the only reason for the loss of anti-HRP staining in TM3 and deficiencies of the 71C region.
Double mutant analysis suggests possible redundant functions among 18w, Toll-7 and Toll-8
Single mutants of Toll-6, Toll-7 and Toll-8 did not show significant phenotype and are mostly viable, thus we generated double mutant combinations within this subgroup to further examine their developmental requirement. The 18w, Toll-6 double mutant did not show significant change of viability when compared with the 18w single mutant, and therefore did not imply functional redundancy. We did not attempt to make the 18w, Toll-7 double mutant because both genes are located in the map region 56E-F. The 18w, Toll-8 double mutant, however, exhibited significantly lower viability (0%) when compared with the 18w (18%) or Toll-8 (84%) mutant alone. Double mutant embryos for 18w and Toll-8 died at late embryonic to early larval stage. We examined cuticle preparations of these double mutant embryos. Even though 18w and Toll-8 RNA are expressed as stripes in the embryos (Kambris et al. 2002), we could not detect any segmentation defect in the double mutant embryos (data not shown). These results suggest that 18w and Toll-8 have functional redundancy or interaction for some yet to be identified developmental processes.
In comparison, the Toll-6, Toll-7 double mutant exhibited the same lethality as the Toll-7 mutant (28%). We did not make the Toll-6, Toll-8 double mutant for the same reason that both are located in the 71C region. Meanwhile, Toll-7, Toll-8 double mutants showed strongly reduced viability (<1%). Thus, Toll-7 and Toll-8 may also have redundant functions in development. The Toll-7, Toll-8 double mutant embryos also died at late embryonic to early larval stage and no specific morphological defects were observed. Our results support the idea that 18w, Toll-7 and Toll-8 have closely related functions in vivo.
Discussion
Examination of the overexpression phenotypes leads us to postulate that the Drosophila Toll-related genes may be divided into subgroups, which have different functions. Previous reports have shown that Toll and Toll-9 can activate the canonical Toll signaling pathway (Ooi et al. 2002; Bettencourt et al. 2004). While our results are consistent that TollD and Toll-9 overexpression induced almost the same phenotype, the functions of Toll and Toll-9 are not identical. For example, Toll and TollD overexpression caused loss of anterior cross vein. However, we did not observe the same phenotype by overexpressing Toll-9 or pelle. As Toll-9 or pelle overexpression is sufficient to stimulate the Toll signaling pathway and antimicrobial peptide gene expression, the anterior cross vein phenotype induced by Toll and TollD may depend on other functions of Toll protein, such as being an adhesion molecule through the extracellular leucine-rich repeats (Keith & Gay 1990).
We could not find any phenotype by overexpressing Toll-3, -4 or -5. As the Toll-5 protein has the highest sequence homology with Toll and overexpression of a constitutively active form of Toll-5 could weakly activate the Toll signaling pathway (Tauszig et al. 2000; Imler & Hoffmann 2001; Luo et al. 2001; Ooi et al. 2002), it was expected that Toll-5 overexpression shows similar phenotype with Toll overexpression. Surprisingly, our overexpression of Toll-5 in transgenic flies did not show any phenotype (Figs 1, 7A). Toll-3 and Toll-4 do not have mosquito or moth counterparts (Christophides et al. 2002; Cheng et al. 2008), suggesting that they have evolved independently in fly. As overexpression of Toll-3, Toll-4 and Toll-5 did not show any phenotype, we did not analyze these genes further in this study.
Our gain-of-function studies suggest that 18w together with Toll-6, Toll-7 and Toll-8 to form a subgroup which has similar functions, and loss-of-function analyses provide some support for this model. In particular there is increased lethality of the double mutant combinations of 18w, Toll-8 and Toll-7, Toll-8. However, the Toll-6 mutant did not show significant phenotype, and the 18w, Toll-6 and Toll-7, Toll-6 double mutants did not show more severe phenotype or lethality. Other analyses may help to unveil the Toll-6 functions. Furthermore, we have not yet been able to examine other double mutant combinations, as the short distance between 18w and Toll-7 (290 kb) and between Toll-6 and Toll-8 (100 kb) makes it difficult to generate the double mutants by meiotic recombination. RNAi should be useful method to overcome such difficulty but RNAi lines that we tested did not efficiently knockdown the expression of these genes (data not shown). Further analysis of the reason for the lethality of the double mutants may shed light on the mechanism and developmental function of Toll-related proteins.
Although Toll-8 expression could rescue the loss of anti-HRP staining in the TM3 balancer chromosome and 71C locus deficiencies (Seppo et al. 2003), our Toll-8 null mutant alleles showed normal anti-HRP pattern in embryonic CNS (Fig. 6). This is consistent with the biochemical analysis that suggests Toll-8 is not directly involved in sugar chain synthesis (Aoki et al. 2007). It is not yet clear why Toll-8 expression could rescue loss of anti-HRP antigen phenotype in the previous report. One possibility is that Toll-8 and Toll-6 are both deleted in these mutants and that either gene can rescue the defect because they have redundant functions.
Because none of the observed phenotypes related to the 18w subgroup match the loss of function phenotypes of the Toll pathway signaling components, the 18w subgroup may use a separate signaling pathway or may function as transmembrane adhesion molecules. A previous report showed that overexpression of Toll or Toll-8 can induce homotypic cell aggregation, suggesting that they can act as cell adhesion molecules (Keith & Gay 1990; Kim et al. 2006). We examined whether downstream signaling components of the Toll pathway are required for 18w subgroup function. Toll-8 was overexpressed by dpp-Gal4 in the dMyd88EP2133 background (Fig. 7B) or by ptc-Gal4 in the pelle2 / pelle7 trans-heterozygote background (data not shown). We could not observe suppression of Toll-8 overexpression phenotype, therefore implying that downstream components of the Toll pathway is not needed for 18w subgroup function.
The mosquito Anopheles gambie genome sequence showed that there were orthologues of Toll-6, Toll-7 and Toll-8, but 18w orthologue was not found (Christophides et al. 2002). The genome of the silk moth Bombyx mori has three Toll genes that are not clear orthologues but related to 18w or Toll-7 (Cheng et al. 2008). The recently available genome sequence of the water flea Daphnia pulex reveals orthologues of Toll-8 (McTaggart et al. 2009). These comparisons suggest that an ancestral gene of the 18w subgroup was present before insect and crustacean diverged, and this gene was duplicated and evolved independently in the individual genome to give rise to a different number of genes within this subgroup. Though overall structure is common between the Toll-related proteins of Drosophila, the four Drosophila Toll-related proteins in the 18w subgroup have some common characters, such as longer leucine-rich repeats in their extracellular domain and longer stretches of amino acids extended beyond the conserved TIR domain in the cytoplasmic domain (Imler & Hoffmann 2001). These structural similarities and our functional analyses reported here support the notion that these four Drosophila genes are more evolutionarily related.
Acknowledgments
We thank Bloomington stock center for fly strains. We thank Masako Wada, Mari Motegi, Noriko Tamura and Xiaodi Hu for their technical assistance and Michiaki Yamada and Harutaka Endo for their assistance in mutagenesis and analysis. The critical reading of the manuscript by Shin Sugiyama is greatly appreciated. The work in YTI laboratory was supported by an NIH grant (GM53269). YTI is a member of the UMass Diabetes Endocrinology Research Center (DK32520) and core resources supported by the DERC grant were also used. The work in Nagoya University was supported by grants from the Ministry of Education, Cultures, Sports, Science and Technology (MEXT) of Japan.
References
- Anderson KV, Jurgens G, Nusslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell. 1985;42:779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J. Biol. Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- Bettencourt R, Tanji T, Yagi Y, Ip YT. Toll and Toll-9 in Drosophila innate immune response. J. Endotoxin Res. 2004;10:261–268. doi: 10.1179/096805104225004897. [DOI] [PubMed] [Google Scholar]
- Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- Cheng TC, Zhang YL, Liu C, Xu PZ, Gao ZH, Xia QY, Xiang ZH. Identification and analysis of Toll-related genes in the domesticated silkworm, Bombyx mori. Dev. Comp. Immunol. 2008;32:464–475. doi: 10.1016/j.dci.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat. Immunol. 2008;9:1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldon E, Kooyer S, D’Evelyn D, Duman M, Lawinger P, Botas J, Bellen H. The Drosophila 18 wheeler is required for morphogenesis and has striking similarities to Toll. Development. 1994;120:885–899. doi: 10.1242/dev.120.4.885. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Matskevich AA, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C, Gerttula S, Anderson KV. Plasma membrane localization of the Toll protein in the syncytial Drosophila embryo: importance of transmembrane signaling for dorsal-ventral pattern formation. Development. 1991;111:1021–1028. doi: 10.1242/dev.111.4.1021. [DOI] [PubMed] [Google Scholar]
- Hashimoto C, Hudson KL, Anderson KV. The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- Hu X, Yagi Y, Tanji T, Zhou S, Ip YT. Multimerization and interaction of Toll and Spatzle in Drosophila. Proc. Natl. Acad. Sci. U S A. 2004;101:9369–9374. doi: 10.1073/pnas.0307062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imler JL, Hoffmann JA. Toll receptors in innate immunity. Trends Cell Biol. 2001;11:304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- Imler JL, Zheng L. Biology of Toll receptors: lessons from insects and mammals. J. Leukoc. Biol. 2004;75:18–26. doi: 10.1189/jlb.0403160. [DOI] [PubMed] [Google Scholar]
- Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, Kambris Z, Brun S, Hashimoto C, Ashida M, Brey PT, Lee WJ. A Spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev. Cell. 2006;10:45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Kambris Z, Hoffmann JA, Imler JL, Capovilla M. Tissue and stage-specific expression of the Tolls in Drosophila embryos. Gene Expr. Patterns. 2002;2:311–317. doi: 10.1016/s1567-133x(02)00020-0. [DOI] [PubMed] [Google Scholar]
- Keith FJ, Gay NJ. The Drosophila membrane receptor Toll can function to promote cellular adhesion. EMBO. J. 1990;9:4299–4306. doi: 10.1002/j.1460-2075.1990.tb07878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chung S, Yoon J, Choi KW, Yim J. Ectopic expression of Tollo/Toll-8 antagonizes Dpp signaling and induces cell sorting in the Drosophila wing. Genesis. 2006;44:541–549. doi: 10.1002/dvg.20245. [DOI] [PubMed] [Google Scholar]
- Kleve CD, Siler DA, Syed SK, Eldon ED. Expression of 18-wheeler in the follicle cell epithelium affects cell migration and egg morphology in Drosophila. Dev. Dyn. 2006;235:1953–1961. doi: 10.1002/dvdy.20820. [DOI] [PubMed] [Google Scholar]
- Kolesnikov T, Beckendorf SK. 18 wheeler regulates apical constriction of salivary gland cells via the Rho-GTPase-signaling pathway. Dev. Biol. 2007;307:53–61. doi: 10.1016/j.ydbio.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem. J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Leulier F, Lemaitre B. Toll-like receptors – taking an evolutionary approach. Nat. Rev. Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol. Cell. Biol. 2006;26:7821–7831. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis P, Bulet P, Reichhart JM. Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defense of Drosophila. EMBO Rep. 2002;3:666–673. doi: 10.1093/embo-reports/kvf130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Shen B, Manley JL, Zheng L. Tehao functions in the Toll pathway in Drosophila melanogaster: possible roles in development and innate immunity. Insect Mol. Biol. 2001;10:457–464. doi: 10.1046/j.0962-1075.2001.00284.x. [DOI] [PubMed] [Google Scholar]
- McTaggart SJ, Conlon C, Colbourne JK, Blaxter ML, Little TJ. The components of the Daphnia pulex immune system as revealed by complete genome sequencing. BMC Genomics. 2009;10:175. doi: 10.1186/1471-2164-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Khanuja BS, Ip YT. Toll receptor-mediated Drosophila immune response requires Dif, an NF-κB factor. Genes Dev. 1999;13:792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhina S, Steward R. Nuclear factor-kappa B pathways in Drosophila. Oncogene. 2006;25:6749–6757. doi: 10.1038/sj.onc.1209940. [DOI] [PubMed] [Google Scholar]
- Morisato D, Anderson KV. The spätzle gene encodes a component of the extracellular signaling pathway establishing the dorsal-ventral pattern of the Drosophila embryo. Cell. 1994;76:677–688. doi: 10.1016/0092-8674(94)90507-x. [DOI] [PubMed] [Google Scholar]
- Ooi JY, Yagi Y, Hu X, Ip YT. The Drosophila Toll-9 activates a constitutive antimicrobial defense. EMBO Rep. 2002;3:82–87. doi: 10.1093/embo-reports/kvf004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Wu LP. Pattern recognition receptors in the fly: lessons we can learn from the Drosophila melanogaster immune system. Fly. 2009;3:121–129. doi: 10.4161/fly.8827. [DOI] [PubMed] [Google Scholar]
- Petersen UM, Bjorklund G, Ip YT, Engstrom Y. The dorsal-related immunity factor Dif, is a sequence-specific trans-activator of Drosophila Cecropin gene expression. EMBO. J. 1995;14:3146–3158. doi: 10.1002/j.1460-2075.1995.tb07317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tl. 1998;4 doi: 10.1126/science.282.5396.2085. **(gene), Science–282. [DOI] [PubMed] [Google Scholar]
- Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- Schneider DS, Jin Y, Morisato D, Anderson KV. A processed form of the Spätzle protein defines dorsal-ventral polarity in the Drosophila embryo. Development. 1994;120:1243–1250. doi: 10.1242/dev.120.5.1243. [DOI] [PubMed] [Google Scholar]
- Seppo A, Matani P, Sharrow M, Tiemeyer M. Induction of neuron-specific glycosylation by Tollo/Toll-8, a Drosophila Toll-like receptor expressed in non-neural cells. Development. 2003;130:1439–1448. doi: 10.1242/dev.00347. [DOI] [PubMed] [Google Scholar]
- Stein D, Goltz JS, Jurcsak J, Stevens L. The Dorsal-related immunity factor (Dif) can define the dorsal-ventral axis of polarity in the Drosophila embryo. Development. 1998;125:2159–2169. doi: 10.1242/dev.125.11.2159. [DOI] [PubMed] [Google Scholar]
- Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc. Natl. Acad. Sci. USA. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uvell H, Engstrom Y. A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet. 2007;23:342–349. doi: 10.1016/j.tig.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelievre E, Gascan H, Ray KP, Morse MA, Imler JL, Gay NJ. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat. Immunol. 2003;4:794– 800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- Williams MJ, Rodriguez A, Kimbrell DA, Eldon ED. The 18-wheeler mutation reveals complex antibacterial gene regulation in Drosophila host defense. EMBO. J. 1997;16:6120–6130. doi: 10.1093/emboj/16.20.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]