Abstract
Glioblastoma multiforme (GBM) is a neurologically debilitating disease that culminates in death 14 to 16 months after diagnosis. An incomplete understanding of how cataloged genetic aberrations promote therapy resistance, combined with ineffective drug delivery to the central nervous system, has rendered GBM incurable. Functional genomics efforts have implicated several oncogenes in GBM pathogenesis but have rarely led to the implementation of targeted therapies. This is partly because many “undruggable” oncogenes cannot be targeted by small molecules or antibodies. We preclinically evaluate an RNA interference (RNAi)–based nanomedicine platform, based on spherical nucleic acid (SNA) nanoparticle conjugates, to neutralize oncogene expression in GBM. SNAs consist of gold nanoparticles covalently functionalized with densely packed, highly oriented small interfering RNA duplexes. In the absence of auxiliary transfection strategies or chemical modifications, SNAs efficiently entered primary and transformed glial cells in vitro. In vivo, the SNAs penetrated the blood-brain barrier and blood-tumor barrier to disseminate throughout xenogeneic glioma explants. SNAs targeting the oncoprotein Bcl2Like12 (Bcl2L12)—an effector caspase and p53 inhibitor overexpressed in GBM relative to normal brain and low-grade astrocytomas—were effective in knocking down endogenous Bcl2L12 mRNA and protein levels, and sensitized glioma cells toward therapy-induced apoptosis by enhancing effector caspase and p53 activity. Further, systemically delivered SNAs reduced Bcl2L12 expression in intracerebral GBM, increased intratumoral apoptosis, and reduced tumor burden and progression in xenografted mice, without adverse side effects. Thus, silencing antiapoptotic signaling using SNAs represents a new approach for systemic RNAi therapy for GBM and possibly other lethal malignancies.
INTRODUCTION
Glioblastoma multiforme (GBM) is the most prevalent and lethal form of malignant brain tumors and considered to be one of the deadliest human cancers (1, 2). Multimodal treatment regimens combining radiation with the DNA alkylating agent temozolomide have only incrementally increased median patient survival by 2.5 months to 14.6 months; recurrence is nearly universal, and salvage therapies to impede further progression are ineffective (3). Disease management is complicated by the coactivation of multiple mitogenic and cell death–inhibitory pathways, resulting in rapid disease progression and intense resistance of tumors toward therapy-induced apoptosis. Coextinction strategies using multiple small molecule–or antibody-based agents, however, are often hampered by drug-drug interactions, systemic toxicity due to pronounced off-target effects, and the emergence of drug resistance (4).
Additional challenges in GBM drug development include ineffective systems for drug delivery to intracerebral tumor elements and the lack of imaging methodologies to quantify intratumoral drug concentrations. Conventional and tailored therapies bound for the central nervous system (CNS) have to negotiate passage through the blood-brain barrier (BBB), the blood–cerebrospinal fluid barrier (BCSF), and the blood-tumor barrier (BTB), and also withstand the substantial dynamic force in the brain caused by CSF flow, brain edema, and tumor mass–related pressure. Moreover, they must be able to disseminate throughout cancerous tissue (5, 6).
RNA interference (RNAi)–based biotherapeutic gene silencing has emerged as a promising approach to target multiple “undruggable” oncogenes implicated in growth, apoptosis, migration, and invasion. The lack of efficient delivery to tumor sites, limited biological activity, and unfavorable safety profile, however, have prevented the implementation of many RNAi-based therapeutics in the clinic (7). Recently, we have developed spherical nucleic acid (SNA) nanoparticle conjugates, which are nanostructures consisting of densely packed, highly oriented small interfering RNA (siRNA) oligonucleotides surrounding an inorganic gold nanoparticle core (8–11) as a platform for gene silencing. SNAs act as single-entity agents capable of simultaneous transfection and gene regulation without the need for auxiliary carriers or cationic transfection agents. SNAs are remarkably stable in physiological environments, resist nuclease degradation, and, in comparison to conventional RNAi delivery platforms, result in more efficient and persistent gene knockdown in cells and tissues without triggering a significant immune response and off-target effects (12–14). SNAs can be cofunctionalized with fluorochromes or gadolinium [Gd(III)]–based magnetic resonance imaging (MRI) contrast agents (15) to track their accumulation in cells and, in this study, in tumors.
To evaluate SNAs as potential anti-glioma therapeutics, we elected the Bcl2Like12 (Bcl2L12) oncogene as a target for SNA-mediated gene silencing. Bcl2L12 localizes to chromosome 19q13, a region frequently amplified in GBM. Furthermore, cell-based assays and expression analyses have identified Bcl2L12 as a putative oncogene with consistent and prevalent mRNA and protein up-regulation in GBM relative to normal brain (16). Mechanistically, Bcl2L12 inhibits apoptosis by neutralizing the activity of effector caspase-3 and caspase-7 downstream of mitochondrial dysfunction and apoptosome activity and blocks the transactivational activity of p53 (17–20), a pivotal tumor suppressor that is frequently deregulated in primary GBM (21, 22).
Given the impact of Bcl2L12 on key nodes of cytoplasmic and nuclear cell cycle and apoptosis signaling cascades, as well as the low-level expression of Bcl2L12 in the adult brain, we describe here an in-depth preclinical characterization of Bcl2L12-targeting SNAs (siL12-SNAs). We report that these nanoconjugates cross the BBB after systemic administration, selectively accumulate in and disseminate throughout tumor tissue, and potently neutralize Bcl2L12 expression, without the use of auxiliary transfection agents, thereby enhancing therapy-induced apoptosis in glioma cells, reducing tumor burden, and significantly delaying cancer progression in glioma-bearing mice, without adverse side effects.
RESULTS
SNAs accumulate in glioma cells and neutralize Bcl2L12 expression
Bcl2L12 is a potent caspase and p53 inhibitor that is overexpressed in the vast majority of human primary GBM specimens, yet is at low or undetectable levels in cells of glial origin, in normal brain surrounding tumor tissue, and in low-grade astrocytoma (16). In addition, our analysis of 343 glioma patients in the Repository of Molecular Brain Neo-plasia Data (REMBRANDT), a Web-based data mining and analysis platform, identified Bcl2L12 as a potential prognostic factor, because GBM patients with high-level overexpression of Bcl2L12 have shorter progression-free survival compared to patients with “intermediate” (0.5- to 4-fold) expression or underexpression of Bcl2L12 [P < 0.001, log-rank P value calculated by REMBRANDT using the Mantel-Haenszel procedure (23)] (fig. S1). Therefore, we decided to explore the use of Bcl2L12-targeted SNAs as a novel GBM therapeutic.
To neutralize Bcl2L12 expression in glioma cells, we first investigated the effectiveness of SNA uptake into human glioma cell lines (U87MG), primary murine cortical astrocytes, and patient- and mouse-derived tumor neurospheres [human (hu) and murine (mu) TNS, respectively] in vitro. Confocal fluorescence microscopy for SNAs conjugated to cyanine5 (Cy5) fluorochromes showed highly efficient cellular uptake (~99% of the cell population) in glioma cell lines, murine cortical astrocytes, and in both huTNS (Fig. 1A) and muTNS (fig. S2, A to C), with widespread diffusion of particles throughout the core of the TNS (Fig. 1A and fig. S2C). Cy5-siRNAs without SNAs were unable to enter cells (fig. S2D). Our previous studies have shown that the accumulation of SNAs depends on the activity of scavenger receptors (SRs), in particular class A (SR-A) (9, 10, 24, 25). In agreement with these studies, inductively coupled plasma mass spectrometry (ICP-MS) revealed reduced accumulation of SNAs in human U87MG cells preincubated with known SR inhibitors, polyinosinic acid (poly I) and fucoidan (fig. S2E).
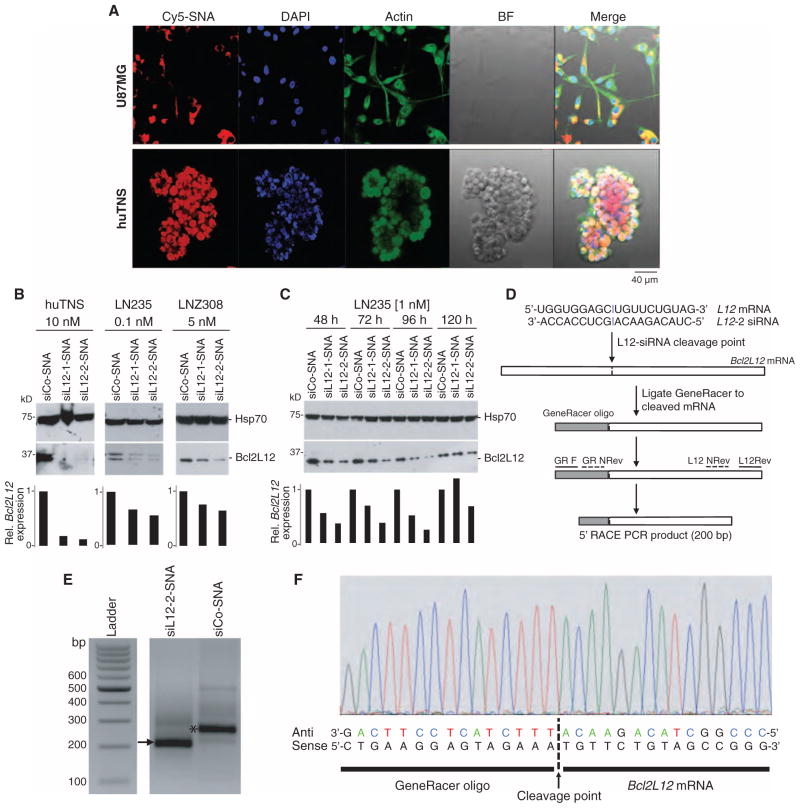
Fig. 1. SNAs penetrate glial cells and down-regulate Bcl2L12 in vitro.
(A) Uptake of Cy5-labeled SNAs (red) into huTNS and the human glioma cell line U87MG. Cells were colabeled with fluorescein isothiocyanate (FITC)–conjugated cytochalasin (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue) to visualize actin filaments and nuclei, respectively. Scale bars, 40 μm. BF, bright field. (B) Effect of Bcl2L12-targeting SNAs siL12-1 and siL12-2 on Bcl2L12 protein levels in huTNS, LN235, and LNZ308 glioma cells relative to control (siCo-SNA) cultures as assessed by Western blot using the noted concentrations of SNAs. Hsp70 served as a loading control. Bar graphs represent a densitometric analysis of Western blots. Band intensity was normalized to Hsp70 and then expressed relative to Bcl2L12 levels in siCo-treated samples. (C) Bcl2L12 knockdown persistance in LN235 cells treated with 1 nM siCo-SNA, siL12-1-SNA, or siL12-2-SNA for 48, 72, 96, and 120 hours as assessed by Western blot. (D) Schematic depicting siL12-2 siRNA cleavage site and subsequent 5′-RLM-RACE PCR. (E) Resulting bands from successive rounds of PCR on the cDNA template using GeneRacer- and Bcl2L12-specific primers in siL12-2-SNA–and siCo-SNA–treated cells. The arrow points to the cleavage product. The asterisk represents a PCR product that is a fragment of Bcl2L12, not ligated to the GeneRacer oligo, and is thus a nonspecific band. (F) DNA sequence chromatogram of resulting PCR product from siL12-2-SNA–treated cells.
To determine whether cellular uptake of SNAs translates into effective neutralization of endogenous Bcl2L12 mRNA and protein expression, we next assessed the knockdown efficacies of siL12-SNAs in glioma cell lines and huTNS. We screened six siL12-SNA constructs targeting different regions of the Bcl2L12 mRNA backbone for their ability to down-regulate endogenous Bcl2L12 levels in comparison to scrambled control sequences (siCo-SNA). Through this process, we identified five conjugates that were able to knock down Bcl2L12 and pursued two of these for subsequent studies: siL12-1-SNA and siL12-2-SNA. Figure S3 provides the synthesis scheme for these siRNA-SNA constructs. SNA synthesis is standardized, so their physicochemical properties are highly reproducible for optimal therapeutic application (table S1). Figure S4 (A and B) displays sequence information and alignment to Bcl2L12 mRNA. These conjugates reduced Bcl2L12 mRNA by 40% and protein expression by up to 90% in patient-derived TNS and transformed glioma cell lines U87MG, LNZ308, and LNZ235 (Fig. 1, B and C, and fig. S4C). SNA concentration as low as 0.1 nM was sufficient to neutralize Bcl2L12 protein levels in LN235 glioma cells (Fig. 1B), comparable to Bcl2L12 knockdown triggered by lipoplex-delivered siL12 oligonucleotides (fig. S4D).
Next, we used 5′ RNA ligand–mediated rapid amplification of complementary DNA (cDNA) ends (5′-RLM-RACE) polymerase chain reaction (PCR) to identify the mRNA cleavage product that results from siL12-2-SNA–triggered, RNA-induced silencing complex (RISC)–mediated RNAi. Upon ligating RNA from siL12-2-SNA–treated U87MG cells to a GeneRacer oligo (Fig. 1D), nested PCR using gene-specific primers amplified a Bcl2L12 mRNA cleavage fragment (Fig. 1E) harboring a 5′ end identical to the predicted cleavage site (Fig. 1F). Thus, siL12-2-SNA–mediated Bcl2L12 gene knockdown was achieved through RNAi.
To verify the functionality of SNA-mediated Bcl2L12 knockdown, we surveyed Bcl2L12-regulated downstream signaling upon SNA administration to glioma cells. Here, SNA-driven knockdown of Bcl2L12 increased effector caspase and p53 activation upon pretreatment of cells with the pan-kinase inhibitor staurosporine, the DNA methylation agent temozolomide, and DNA damage–inducing doxorubicin. SNA-driven Bcl2L12 knockdown increased the levels of active caspase-3 and caspase-7 (Fig. 2, A and B, and fig. S4E), raised p53 protein levels (Fig. 2C), and augmented the protein and mRNA levels of the p53 transcriptional target p21 (Fig. 2, C to E). Bcl2L12-targeting SNAs therefore sensitized glioma cells to therapy-induced apoptosis by enhancing caspase and p53 activities.
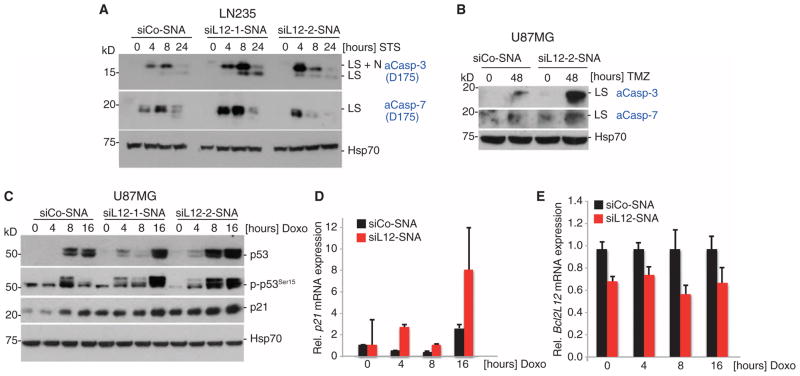
Fig. 2. Bcl2L12-specific SNAs promote apoptotic signaling in glioma cells.
(A and B) Bcl2L12 knockdown by siL12-SNAs sensitizes cells to apoptosis (measured by caspase-3 and caspase-7 cleavage) upon treatment with staurosporine (STS) and temozolomide (TMZ). LS, large subunit; LS+N, large subunit plus N-peptide. (C) Effect of Bcl2L12 knockdown on p53 protein stability, phosphorylated p53 (p-p53Ser15), and the p53 target p21 upon doxorubicin (Doxo) treatment for the indicated times. (D) p53 target gene induction of p21 on the mRNA level as measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR). (E) Bcl2L12 mRNA knockdown was assessed in parallel and expressed as fold change relative to siCo-SNA–treated cells at 0, 4, 8, and 16 hours after application of doxorubicin. Data are means ± SD (n = 3).
SNAs disseminate throughout glioma tissue upon local and systemic administration
After in vitro studies, we assessed whether SNAs could penetrate glioma tissue in vivo using local (intracranial) and systemic (intravenous) administration. Glioma-bearing severe combined immunodeficient (SCID) mice received intracranial injections of Cy5- or gadolinium [Gd(III)]–labeled SNAs, and SNA distribution was studied by ICP-MS, MRI, and confocal fluorescence. (Note that Gd-SNAs are conjugated to DNA instead of siRNA; however, we have not observed differences in uptake efficiency between DNA-based SNAs and siRNA-based SNAs.) ICP-MS analysis indicated 10-fold greater accumulation of SNAs in tumor versus nontumor brain regions (Fig. 3A). This selective intratu-moral SNA accumulation may be due to the enhanced permeability and retention (EPR) effect (26). Correspondingly, MRI studies using Gd(III)-SNAs (Fig. 3B) revealed pervasive intratumoral dissemination (Fig. 3C). Here, gold and Gd(III) distribution within corresponding coronal brain sections localized to tumor elements, as determined by laser ablation (LA)–ICP–MS analysis (Fig. 3D). Confocal fluorescence of serial coronal brain sections further revealed effective dispersion of Cy5-SNAs in tumor tissue (Fig. 3E and movie S1). This further substantiated intratumoral dispersion of SNA conjugates and demonstrated that the tumor-associated Gd(III)-SNA signal resulted from predominant accumulation of conjugates within the extravascular tumor parenchyma.
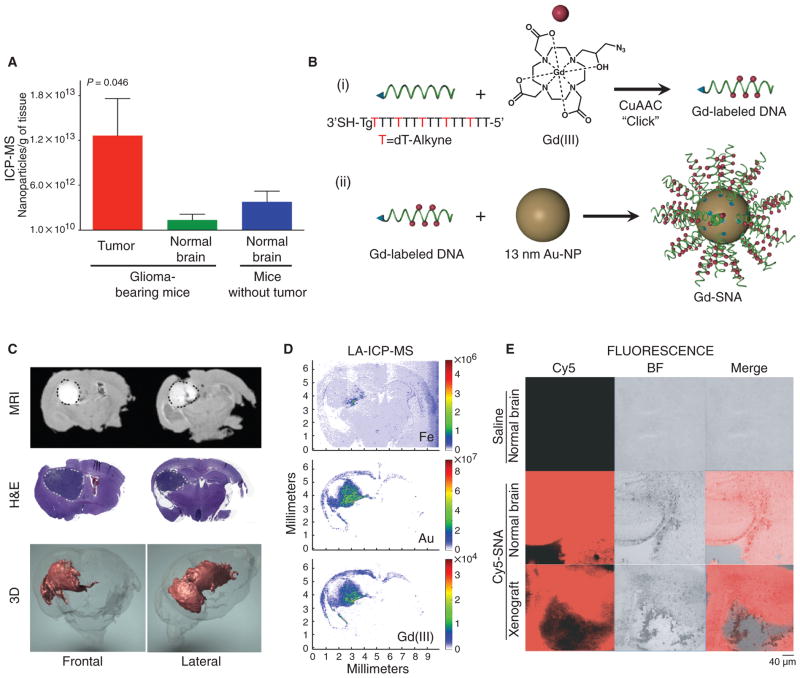
Fig. 3. SNAs disseminate throughout glioma tissue.
(A) ICP-MS quantification of SNA uptake into orthotopic U87MG tumor and adjacent normal tissue after 48 hours. Data are means ± SEM (n = 3 glioma-bearing mice, n = 5 normal mice). P value was calculated with one-way analysis of variance (ANOVA). (B) Schematic overview of the synthesis of Gd(III)-functionalized SNAs. (i) Gd(III)-SNA conjugates were prepared from alkyne-modified T bases and azide-labeled Gd(III) complexes through click chemistry. (ii) Gd(III)-conjugated DNA was then functionalized onto gold nanoparticle (Au-NP) surface to form Gd(III)-SNA following the same procedure as fig. S3, except without oligoethylene glycol/polyethylene glycol (PEG) backfill. (C) MR images of tumor-bearing mouse brains injected intracranially with Gd(III)-SNAs. Two representative coronal sections imaged 24 hours after Gd(III)-SNA injection (upper panel) show localization of Gd(III)-SNA within the intracerebral lesion. Gd(III) signal is white and outlined in black dotted line. Also shown are corresponding hematoxylin and eosin (H&E) sections showing the location of the tumor (darker purple, outlined in light gray dotted line), and three-dimensional (3D) reconstruction of MR images [Gd(III) signal in red]. See movie S1. (D) LA-ICP-MS shows localization of Au, Fe, and Gd(III) contents in coronal brain sections of mice injected intracranially with Gd(III)-SNAs. The heat map indicates the relative amount of the element detected in the tissue. (E) Confocal fluorescence microscopy of coronal brain sections derived from tumor-bearing and non–tumor-bearing mice injected with saline or Cy5-SNAs. Representative sections are shown for n = 5 mice.
Next, we tested whether SNAs could cross the BBB/BTB to target intracranial lesions in an in vitro BBB model and in xenogeneic explants in vivo (Fig. 4 and fig. S5). For the in vitro study, we used a previously established model (27) consisting of a noncontact coculture of human primary brain microvascular endothelial cells (huBMECs) coating a semipermeable filter insert with human astrocytes cultivated in a cell culture well on the opposite side of the filter (Fig. 4A). For this model, the conductivity and composition of tight junctions have been shown to recapitulate an in vivo BBB (28, 29). Because the astrocytes were physically separated from the huBMECs, we were able to independently monitor SNA uptake into the endothelial and, upon transcytosis, the astrocytic compartment by confocal fluorescence microscopy. At early stages of the experiment, Cy5.5-SNAs accumulated within huBMECs when added on top of the endothelial layer and, upon passage through the huBMEC layer and filter, rapidly entered the astrocytes (Fig. 4A). Similar to SNA uptake into cell monolayers (Fig. 1A and fig. S2), accumulation of SNAs in astrocytes was blocked by pretreatment of the huBMECs with poly I, suggesting that SRs may play a role in BBB transcytosis (10, 24).
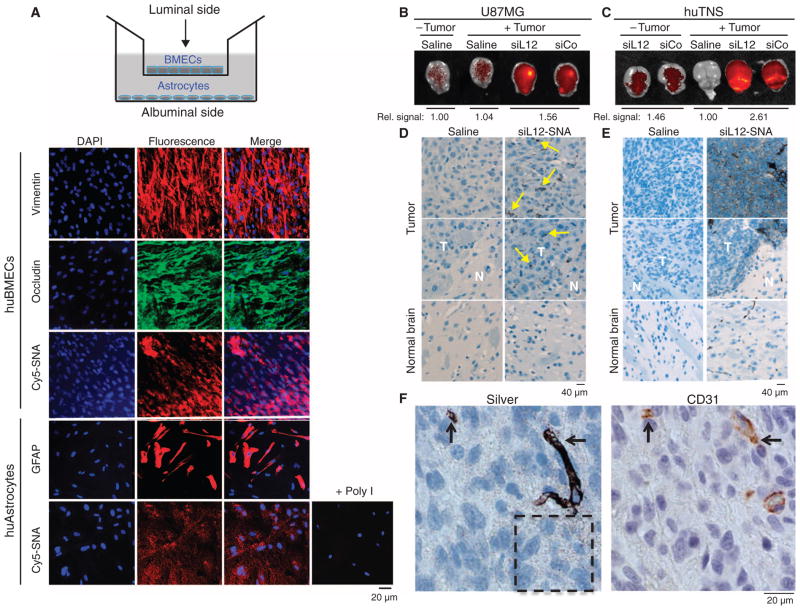
Fig. 4. SNAs cross the BBB/BTB and selectively accumulate in glioma tissue.
(A) Noncontact in vitro BBB model using a coculture of huBMECs and human astrocytes. Representative confocal fluorescence microscopy images demonstrate Cy5.5-SNA (red) distribution in endothelial and astrocytic cells. Endothelial and astrocytic cells stained positively for occludin (a marker for tight junctions) and glial fibrillary acidic protein (GFAP), respectively. Anti-vimentin and DAPI stained cytoplasm and nuclei, respectively. (B and C) IVIS analysis of brains with or without U87MG (B) or huTNS (C) tumors 48 hours after systemic delivery of saline or Cy5.5-SNAs. SNA accumulation is indicated by increased fluorescence (yellow). Quantification of radiant efficiency is shown as relative signal amount under the images. (D and E) Selective accumulation of Cy5.5-SNAs within U87MG (D) or huTNS (E) tumors, but not normal brain elements. Coronal brain sections were silver-stained and counterstained with hematoxylin. Yellow arrows indicate sites of SNA accumulation, which appear as dark brown regions. T, tumor; N, normal brain. (F) huTNS-derived accumulated SNAs in vascular and extravascular tumor elements as revealed by silver staining and anti-CD31 immunohistochemistry on adjacent coronal sections. Arrows point to areas of SNA and CD31 colocalization. Hashed box shows SNAs that extravasated into the tumor parenchyma.
To study the BBB penetration of SNAs in vivo, we injected glioma-bearing mice intravenously with Cy5.5-SNAs and monitored them by near-infrared fluorescence (NIRF) using an in vivo imaging system (IVIS). Consistent with SNA penetration of intracerebral gliomas upon local delivery, analysis of intracranial SNA distribution by IVIS (Fig. 4, B and C) and subsequent histological analysis using hematoxylin and silver stainings (Fig. 4, D and E) confirmed higher accumulation of SNAs in tumor versus normal brain tissue; specifically, quantification of radiant efficiency in excised brains using Living Image software revealed 1.8-fold higher signal in huTNS tumor–bearing mice than in mice that received a sham tumor inoculation. High-resolution imaging of silver-stained brain sections revealed a speckled, cytoplasmic distribution of SNAs within the extravascular tumor parenchyma (hashed box in Fig. 4F), together with robust localization of SNAs to CD31+ endothelial cells (Fig. 4F).
Selective SNA infiltration of the glioma mass in comparison to adjacent noncancerous brain tissue was further confirmed by ICP-MS analysis (fig. S5A) and is likely due to a functionally compromised BBB/BTB in glioma-bearing mice (30) and the EPR effect of the glioma-associated vasculature. Specifically, SNA accumulation in brain tissue of non–tumor-bearing mice was extensive, with about 1 × 1010 SNA particles per gram of tissue. Tumor and adjacent normal brain in glioma-bearing mice, however, revealed an almost three orders of magnitude higher SNA accumulation (fig. S5A), suggesting that BBB/BTB compromise is a major driver of selective SNA accumulation in glioma-bearing brain tissue. Increased BBB/BTB permeability in tumor-bearing mice was confirmed by Evans blue (a BBB-impermeable dye) extravasation, documenting dye accumulation selectively in areas of tumor growth as early as 7 days after tumor cell implantation (fig. S6). Bio-distribution analysis revealed that up to 1% of the total SNA amount injected was found within the tumor, with most of the SNAs accumulating in the liver and spleen 24 hours after the last injection (fig. S5B).
Pharmacokinetic analyses (fig. S7A) showed that the circulation time for the bulk of a single dose was short, with more than 90% distributing to tissues in the first 5 min after intravenous injection (half-life for distribution, αt1/2, ~1 min; fig. S7B). The elimination phase for these particles was much slower (half-life for elimination, βt1/2, ~8.5 hours) (fig. S7B), with low levels of SNAs in the blood through the 72-hour course of the pharmacokinetics study. Similar to glioma-bearing mice, tissue distribution in healthy animals was primarily in the liver and spleen, with small accumulation in the lungs, kidneys, heart, brain, and intestines (fig. S8). By 72 hours after injection, the liver contained most of the remaining gold, and the gold in other tissues was reduced to near background levels (fig. S8).
SNAs are nontoxic in rodents
We next evaluated the potential toxicity of Bcl2L12-targeting SNAs. To begin to determine potential adverse side effects of SNAs, we looked for induction of inflammatory cytokines in blood samples collected from xenografted mice treated with siCo-SNA or siL12-2-SNA in comparison to blood samples from saline-treated control mice 21 days after cell inoculation. These animals received seven injections of SNAs, given every other day, for a total dose of ~10 mg/kg (siRNA/body weight). We did not observe any significant differences between the groups using one-way ANOVA followed by Dunn’s multiple comparison test (table S2).
In addition, we administered siL12-2-SNAs (10 mg/kg) or an equivalent volume of phosphate-buffered saline (PBS) as a single dose intravenously into Sprague-Dawley rats. Assessment of toxicity was based on mortality, clinical signs, body weights, blood chemistry, complete blood counts, and detailed histopathology. We did not observe siL12-2-SNA–related differences in histology (fig. S9) or body weight (table S3) 1 or 14 days after SNA administration. Complete blood counts (hematology) and blood chemistry panels were within normal range (table S3), confirming the absence of acute or long-term toxicity at therapeutic dosages. In addition, histopathological evaluation of major organs failed to detect abnormal lesions or signs of pathological processes, and findings were consistent with organ structures in clinically normal, age- and strain-matched rats (fig. S9).
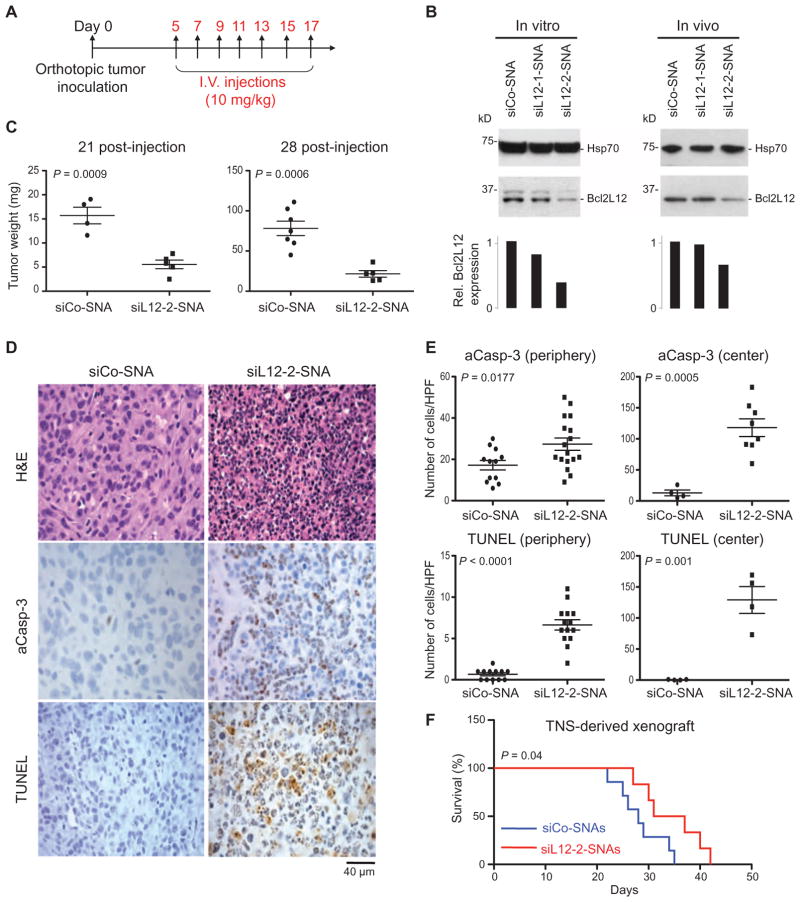
SNAs silence Bcl2L12 and reduce tumor burden in xenografted mice
To assess the impact of siL12-2-SNAs on GBM progression in vivo, we determined Bcl2L12 protein knockdown, tumor burden, histology, and survival of xenografted mice upon intravenous administration of siL12-2-SNAs. Animals were treated per the scheme in Fig. 5A. Bcl2L12-targeting SNAs triggered intraglioma mRNA (26%) and protein (40%) knockdown relative to siCo-SNAs (Fig. 5B) and impaired tumorigenicity as measured by reduced tumor burden in U87MG-derived xenogeneic mice (Fig. 5C). Bcl2L12 knockdown significantly increased intratumoral apoptosis, as evidenced by enhanced levels of DNA strand breaks and active caspase-3 (Fig. 5, D and E). This cellular-level effect elicited by siL12-2-SNAs translated into a significant improvement in survival in a huTNS-derived xenogeneic GBM model (Fig. 5F). This model recapitulated important phenotypic characteristics of the human disease, including hypercellularity, micro-vascular proliferation, extensive necrosis, and diffuse and widespread invasion, including the presence of single invasive cells separated from the bulk tumor (fig. S10).
Fig. 5. Systemic administration of SNAs reduces intratumoral Bcl2L12 and decreases tumor burden in mice.
(A) Scheme of cell and SNA injections. Mice were orthotopically injected with either U87MG or huTNS cells for tumor burden, histology, and survival analyses (day 0) followed by seven intravenous (I.V.) injections of siCo- or siL12-SNAs. (B) Bcl2L12 knockdown in U87MG-derived intracranial tumors treated with siL12-SNAs. Hsp70 was used as a loading control. Histograms represent a densitometric analysis of the Western blots. The intensity of Bcl2L12 bands was normalized to Hsp70 and ex-pressedasrelativeBcl2L12 expression. (C) Weight of xenografted tumors 21 and 28 days after SNA injection. Data points display tumor weight from individual mice. P value was calculated with two-tailed Student’s t test. (D and E) Intratumoral apoptosis in mice injected with siL12-2-SNA. (E) The amount of activated caspase-3 (aCasp-3) and terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) was quantified from (D) in peripheral and central tumor regions. Data points are the number of stained cells per field. Bars are means ± SEM. P values were calculated with two-tailed Student’s t test. HPF, high-power field. (F) Kaplan-Meier survival curves of mice with TNS-derived xenografts treated with siL12-2-SNA (n = 6) or siCo-SNA (n = 7). P value was calculated with the Mantel-Cox test.
DISCUSSION
Anticancer drugs must disseminate throughout tumor parenchyma and accumulate in therapeutic concentrations to effectively inhibit neoplastic growth. Intratumoral pressure gradients, abnormal tumor-associated vasculature, and, perhaps most importantly, the BBB serve as physical and physiological barriers for drug delivery to brain tumors. Tight junctions, lack of fenestration, and a dense layer of pericytes characterize endothelial cells of the BBB. Together with high electrical resistance and abundant expression of efflux transporters, anticancer compounds are forced back into systemic circulation, and these structural barriers prevent the extravasation of drugs into the CNS (31). Indeed, most chemotherapeutics for brain cancers are delivered orally or intravenously, and their inadequate BBB penetration limits drug efficacy (6). Additional approaches have focused on local drug delivery via implanted polymers, catheters, or convection-enhanced delivery (CED). These locally delivered agents exhibit limited intratumoral dispersion, require surgery for wafer and catheter implementation, and, in the case of CED, depend on high infusion rates that may be associated with increased risk of neuro-toxicity caused by elevated intracranial pressure (6).
Intraglioma delivery of siRNA is particularly challenging because RNA is rapidly degraded by nucleases and entrapped in endosomes and inadequately penetrates extravascular tumor tissue beyond perivascular regions. Such low-level penetration stems from poor blood perfusion of tumor elements and high interstitial pressure (31, 32). In contrast, we show that SNAs penetrate the BBB/BTB and disseminate throughout extravascular glioma tissue. Consequently, SNA technology represents an important therapeutic paradigm for systemically targeting undruggable oncoproteins that are important for GBM progression and therapy responses via passive, EPR-mediated tumor targeting. Furthermore, the evaluation of intratumoral drug delivery, concentration, and tissue dissemination are critical for the (pre)clinical assessment of antineoplastic drugs (31, 32). Here, we showed that intratumoral SNA levels and distribution could be precisely measured by LA-ICP-MS in resected tissue, allowing one to track the biodistribution and tissue distribution of the SNA–gold nanoparticle therapeutic.
Notably, cellular uptake and possibly BBB penetration depend on SR-mediated endocytosis (10, 24, 25). In agreement with our previous studies using pharmacological SR inhibitors (10), we found that inhibition of SR-A with poly I and fucoidan blocked SNA uptake into glioma cells and BBB transcytosis. The mechanism of cellular uptake of SNAs into GBM cells and tumors is not yet clear; however, our previous studies suggest that SNA uptake is dependent on the 3D architecture of the conjugates because hollow, gold-free SNAs, but not single-stranded DNAs, effectively compete with SNAs to block their uptake into cells (25). In addition, we discovered that RNAi-mediated knockdown of SR-A reduced the accumulation of SNAs in cells and, consequently, SNAs chemically engaged with SR-A and lipid raft proteins, suggesting that SR-A and lipid rafts are important factors mediating SNA endocytosis (25). SRs are highly expressed within endothelial cells that make up the BBB, suggesting that the robust penetration of SNAs into brain and tumor elements that we observed may involve SR-dependent transcytosis (33, 34). This observation has potential implications for the delivery of SNAs to the CNS to treat other neurological diseases and CNS malignancies.
Other than successful intravenous delivery and penetration of BBB, SNA conjugates triggered efficient, specific, and persistent Bcl2L12 gene knockdown, which translated into a significant reduction in tumor burden in mice without adverse effects or toxicity. SNAs have an ion cloud associated with the high-density oligonucleotide shell and steric inhibition at the particle surface, which creates a unique micro-environment that inhibits enzymatic nucleic acid degradation and, in turn, results in an increased stability of siRNAs (8, 9, 13) and potentially longer therapeutic lifetimes.
Although the SNA platform enables specific genetic knockdown upon systemic delivery, their efficacy could be further improved by potentially increasing circulation times and accumulation in targeted tissues. One way to increase targeting is through the use of conjugated monoclonal antibodies, an avenue that has shown promise in vitro for Her2 in breast cancer cells (35) but has yet to be evaluated in vivo. Additionally, circulation time and SNA stability could be extended through small adjustments to the physical properties of SNAs, for example, the length of the spacer between the siRNA and the gold and/or the amount and length of PEG used for backfill. Finally, this proof-of-concept study has explored the effect that neutralization of a single oncogene has on an overall disease state. Targeting multiple oncogenes, such as driving mutations of GBM implicated in aberrant growth factor receptor signaling, may be possible with single SNA constructs composed of multiple siRNA reagents.
Finally, the SNA platform provides a tool for discovery science to validate additional genes implicated in GBM pathogenesis as therapeutic targets, including receptor tyrosine kinases and downstream RAS-MAPK (mitogen-activated protein kinase) and PI3K (phosphatidylinositol 3-kinase)–AKT–mTOR (mammalian target of inhibitor) signaling, additional antiapoptotic members of the Bcl-2 protein family, genes implicated in metabolic control of GBM progression, and microRNAs. Current strategies focusing on viral delivery of short hairpin RNAs (shRNAs) are often time-consuming, require manipulation of cells in culture, and necessitate extensive optimization of shRNA induction in cell and animal models. Thus, SNA-based systemic delivery of siRNAs may represent a more effective way to target cancer genes in cell-based and in tumor regression studies in vivo.
In summary, this work provides proof of concept that systemically administered SNAs can cross the BBB/BTB in GBM cell and mouse models, infiltrate tumor parenchyma, silence genetic lesions of established GBM in vitro and in vivo, and effectively reduce tumor burden. More generally, this work establishes SNAs as a promising new class of therapeutic gene regulation agents capable of treating disease through systemic injection. Systemic delivery of Bcl2L12-specific SNAs to intracranial lesions exhibited both selectivity and favorable toxicity and pharmacokinetic profiles that may enable siRNA delivery to GBM patients. Bcl2L12 may also be a good target for other cancers such as melanoma (36) and non-CNS malignancies, considering its implication as a bio-marker for early-stage gastric cancer and colon cancer, and for the prediction of short-term relapse in nasopharyngeal carcinoma (37–39). Toward translation, further development of the SNA platform will be required, including optimization of circulation times, two-species toxicity studies, and efficacy studies in additional models of human GBM. Together, SNAs harness the great promise of biotherapeutic gene silencing as a personalized medicine approach to neutralize many genes, including undruggable oncogenes, and can overcome some of the major challenges associated with CNS-directed drug delivery and RNAi-based therapy.
MATERIALS AND METHODS
Study design
This study aimed to evaluate SNA nanoparticle conjugates as an RNAi-based therapy for glioblastoma. The ability of Bcl2L12-specific SNAs (siL12-SNAs) to enter multiple cell types, penetrate tumor and normal tissue, neutralize target gene expression, and affect downstream apoptotic signaling was determined both in vitro and in vivo. SNA uptake into cells or tissues was assessed qualitatively and quantitatively with microscopy, MRI, ICP-MS, and IVIS. The subsequent impact on Bcl2L12 expression and downstream apoptotic signaling was determined by Western blotting and qRT-PCR. Additionally, the pharmacokinetics and toxicity of SNAs were evaluated in both rats and mice. For animal studies to determine SNA efficacy in vivo, the number of mice per group was calculated with a power of 0.8, and animals were randomized by weight before tumor inoculation with Studylog’s stratified sampling randomization to establish a P value of 0.99 between groups. Animals were then further randomized to different treatment groups within each cage, and ear tags were used to identify mice so that researchers could remain blinded to which treatment each animal had received. All experiments were conducted under an approved protocol of the Institutional Animal Care and Use Committee of Northwestern University.
SNA synthesis
RNA synthesis, gold nanoparticle functionalization and characterization, and synthesis of Gd(III)-SNAs are provided in the Supplementary Materials.
In vitro SNA administration
SNAs were added directly to the medium of subconfluent glioma and a patient-derived TNS line at varying concentrations (0.1 to 10 nM). To visualize SNA uptake, we plated the cells on chamber slides and treated them with SNAs for 12 to 16 hours. Cells were counterstained with DAPI and FITC-labeled cytochalasin to visualize nuclei and actin filaments, respectively, and subjected to confocal fluorescence microscopy (n = 3 cultures per cell line). To assess functionality of SNA-mediated knockdown of Bcl2L12, we treated the cells with the pan-specific kinase inhibitor staurosporine (0.5 μM, Sigma), temozolomide (100 μM, Sigma), or doxorubicin (0.4 μg/ml, Sigma) for the indicated time points, and we collected protein lysate or total cellular RNA. The degree of apoptosis was determined by Western blotting for cleaved caspase-3 and caspase-7 in response to staurosporine treatment, by Western blotting for p53, p-p53Ser16, and p21, and by qPCR for p21 in response to doxorubicin treatment (Supplementary Methods).
In vitro BBB model
The in vitro BBB model was adapted from (27) with modifications. huBMECs (ScienCell) were maintained in fibronectin-coated T-75 flasks in endothelial cell medium (ScienCell), and primary human astrocytes (ScienCell) were cultivated in Dulbecco’s modified Eagle’s medium and 10% fetal bovine serum. Primary human astrocytes (1 × 105) were plated in 24-well plates and allowed to grow for 4 days. huBMECs (4 × 105) were plated onto the upper side of 6.4-mm collagen I–coated, 0.4-μm pore size cell culture inserts (BD Biosciences) and were transferred to the 24-well plates containing the primary astrocytes. huBMECs and human astrocytes were cocultured for 12 days to allow for formation of the in vitro BBB. Cy5.5-labeled SNAs (5 nM) were then added into the insert well and incubated for 24 hours, and penetration across the huBMECs layer and into primary astrocytes below was followed by fluorescence microscopy. Cells on coverslips or collagen-coated inserts were washed in PBS and fixed in 4% paraformaldehyde for 5 min at 37°C, blocked for 1 hour at 37°C in 5% bovine serum albumin, and incubated overnight with primary anti-GFAP (Cell Signaling, 1:100), anti-occludin (Santa Cruz Biotechnology, 1:200), or anti-vimentin antibodies (Thermo Fisher Scientific, 1:400).
Tumor inoculation and in vivo efficacy studies
All animals were used under an approved protocol of the Institutional Animal Care and Use Committee of Northwestern University. To implant tumors, we suspended U87MG and a patient-derived TNS line in Hanks balanced salt solution. Each mouse was anesthetized and placed in a stereotaxic frame, and the surgical area was cleaned with alcohol and Betadine. For these studies, the cells were implanted in ~7-week-old female CB17 SCID mice (Taconic Farms). An incision was made in the scalp, and then a 0.7-mm burr hole was created in the skull with a microsurgical drill 2 mm lateral right of the sagittal suture and 0.5 mm posterior of bregma. A Hamilton syringe was loaded with either 1 × 105 U87MG cells or 2 × 105 huTNS cells and inserted 3.5 mm into the brain. The cells were implanted over a period of 5 min, and the needle was left in place for 1 min before withdrawing the syringe. After surgery, the skin was closed with sutures.
To assess therapeutic efficacy, we administered 500 nM SNAs via the tail vein at a volume that allowed for 1.4 mg/kg (RNA/mouse weight) per injection, with injections performed every other day (Fig. 5A). In the U87MG trial, the mice received five injections, for a total siRNA dose of 7 mg/kg. In the TNS trial, the mice received seven injections, for a total siRNA dose of 9.8 mg/kg.
SNA content in tissues
ICP-MS, MRI and 3D movie reconstruction, LA-ICP-MS, NIRF imaging, and silver staining are provided in the Supplementary Materials.
Statistical analysis
REMBRANDT, a clinical genomics database available at https://caintegrator.nci.nih.gov/rembrandt/, was used to calculate the effect of Bcl2L12 expression on overall GBM patient survival in fig. S1. P values were calculated by the log-rank test. For further details on statistical analyses, see (23). Two-tailed t tests were used to calculate statistical significance for fig. S2E and Fig. 5 (C and E). Statistical significance for Fig. 3A was calculated with one-way ANOVA. Statistics for overall survival in mouse studies (Fig. 5F) was calculated with the log-rank (Mantel-Cox) test. Statistical significance was established at P < 0.05.
Supplementary Material
Acknowledgments
We thank H. Zheng (Cold Spring Harbor Laboratory) for providing muTNS. We also thank H. Bradshaw and K. Sidell for technical support. We acknowledge the Cell Imaging Facility (CIF), the Mouse Histology and Phenotyping Laboratory (MHPL), the Center for Advanced Molecular Imaging (CAMI), the Comprehensive Metabolic Core, and the Quantitative Bioelemental Imaging Center (QBIC) at Northwestern University.
Funding: The research was supported by the Center for Cancer Nanotechnology Excellence initiative of the NIH (U54 CA151880 to C.A.M., A.H.S., and T. J. Meade), National Institute of Arthritis and Musculoskeletal and Skin Diseases/NIH grants R01AR060810 and R21AR062898 (to C.A.M.), the Dixon Translational Research Grants Initiative of the Northwestern Memorial Foundation (to C.A.M. and A.H.S.), the James S. MacDonnell 21st Century Initiative and the Coffman Charitable Trust (to A.H.S.), and National Institute of Biomedical Imaging and Bioengineering/NIH R01EB005866-06 (to T. J. Merkel). The research was also supported by the NIH National Cancer Institute (NCI) under Award Number F32CA171949 to E.S.D. The American Cancer Society supported C.H.K. with Postdoctoral Fellowship PF-11-107-01-CDD, and E.S.D., C.H.K., and T. J. Merkel were supported by the Northwestern University International Institute of Nanotechnology. S.A.J. received funding from the Malkin Scholars Programs of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University, and NCI/NIH training grant T32CA09560. MRI was performed on the 9.4-T Bruker BioSpec system purchased with the support of National Center for Research Resources 1S10RR023707-01. CAMI, CIF, and MHPL are all supported by NCI Cancer Center Support Grant P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center, and QBIC is supported by NASA Ames Research Center NNA06CB93G. This work was also supported, in part, by the Defense Advanced Projects Agency Cooperative Agreement No. HR0011-13-2-0018.
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/5/209/209ra152/DC1
Methods
Fig. S1. Bcl2L12 is a GBM oncogene.
Fig. S2. SNA uptake into murine astrocytes and muTNS depends on scavenger receptors.
Fig. S3. Synthesis scheme for SNAs.
Fig. S4. Evaluation of additional SNAs targeting Bcl2L12.
Fig. S5. Biodistribution of SNAs in glioma-bearing mice after systemic administration.
Fig. S6. The BBB/BTB is compromised in tumor-bearing mouse brains.
Fig. S7. Pharmacokinetics of SNAs.
Fig. S8. Biodistribution of SNAs in tumor-free mice.
Fig. S9. Histological evaluation of siL12-2-SNAs in major tissues.
Fig. S10. Histological analysis of huTNS-derived mouse tumors.
Table S1. Characterization of SNAs.
Table S2. Evaluation of serum cytokines in mice.
Table S3. Body weight and blood chemistry after systemic SNA administration to Sprague-Dawley rats.
Movie S1. 3D reconstruction of MR images after intracranial injection of Gd(III)-functionalized SNAs. References (40–42)
Author contributions: Designed the experiments: S.A.J., E.S.D., C.H.K., C.A.M., and A.H.S. Synthesized and characterized the nanomaterials: E.S.D., C.H.K., J.I.C., W.L.D., A.W.S., M.W.R., D.A.G. and T. J. Meade Conducted in vitro experiments: S.A.J., C.H.K., J.P.L., and D.A.G. Conducted in vivo experiments: S.A.J., E.S.D., C.H.K., L.A.H., J.P.L., and F.M.K. Performed data analysis: S.A.J., E.S.D., C.H.K., L.A.H., and J.P.L. Obtained ICP data: C.H.K., A.J.L., and P.C.P. Obtained pharmacokinetic and toxicology data: T. J. Merkel Prepared the manuscript: S.A.J., E.S.D., C.H.K., C.A.M., and A.H.S. All authors edited the manuscript.
Competing interests: D.A.G., C.A.M., W.L.D., A.H.S., and P.C.P. have interest in AuraSense Therapeutics, which develops SNA-based technologies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the sponsors or government, and no official endorsement should be inferred.
Data and materials availability: SNA conjugates and Bcl2L12-related biologicals are available upon request from C.A.M. and A.H.S. with an executed materials transfer agreement.
REFERENCES AND NOTES
- 1.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, Brennan C, Ligon KL, Furnari F, Cavenee WK, Depinho RA, Chin L, Hahn WC. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 4.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–79. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Stukel JM, Caplan MR. Targeted drug delivery for treatment and imaging of glioblastoma multiforme. Expert Opin Drug Deliv. 2009;6:705–718. doi: 10.1517/17425240902988470. [DOI] [PubMed] [Google Scholar]
- 6.Zhan C, Lu W. The blood-brain/tumor barriers: Challenges and chances for malignant gliomas targeted drug delivery. Curr Pharm Biotechnol. 2012;13:2380–2387. doi: 10.2174/138920112803341798. [DOI] [PubMed] [Google Scholar]
- 7.Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med. 2009;1:142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutler JI, Auyeung E, Mirkin CA. Spherical nucleic acids. J Am Chem Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 9.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA–nanoparticle conjugates. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nano-particles. Bioconjug Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 12.Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA. Cellular response of polyvalent oligonucleotide-gold nanoparticle conjugates. ACS Nano. 2010;4:5641–5646. doi: 10.1021/nn102228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA nano-particle conjugates stabilize nucleic acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng D, Giljohann DA, Chen DL, Massich MD, Wang XQ, Iordanov H, Mirkin CA, Paller AS. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci USA. 2012;109:11975–11980. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y, Xu X, MacRenaris KW, Zhang X, Mirkin CA, Meade TJ. Multimodal gadolinium-enriched DNA–gold nanoparticle conjugates for cellular imaging. Angew Chem Int Ed Engl. 2009;48:9143–9147. doi: 10.1002/anie.200904666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stegh AH, Kim H, Bachoo RM, Forloney KL, Zhang J, Schulze H, Park K, Hannon GJ, Yuan J, Louis DN, DePinho RA, Chin L. Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes Dev. 2007;21:98–111. doi: 10.1101/gad.1480007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stegh AH, Brennan C, Mahoney JA, Forloney KL, Jenq HT, Luciano JP, Protopopov A, Chin L, Depinho RA. Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev. 2010;24:2194–2204. doi: 10.1101/gad.1924710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stegh AH, DePinho RA. Beyond effector caspase inhibition: Bcl2L12 neutralizes p53 signaling in glioblastoma. Cell Cycle. 2011;10:33–38. doi: 10.4161/cc.10.1.14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stegh AH, Chin L, Louis DN, DePinho RA. What drives intense apoptosis resistance and propensity for necrosis in glioblastoma? A role for Bcl2L12 as a multifunctional cell death regulator. Cell Cycle. 2008;7:2833–2839. doi: 10.4161/cc.7.18.6759. [DOI] [PubMed] [Google Scholar]
- 20.Stegh AH, Kesari S, Mahoney JE, Forloney KL, Protopopov A, Louis DN, Chin L, DePinho RA. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc Natl Acad Sci USA. 2008;105:10703–10708. doi: 10.1073/pnas.0712034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson AM, Rich A, Krieger M. Polynucleotide binding to macrophage scavenger receptors depends on the formation of base-quartet-stabilized four-stranded helices. J Biol Chem. 1993;268:3546–3554. [PubMed] [Google Scholar]
- 25.Choi CHJ, Hao L, Narayan SP, Auyeung E, Mirkin CA. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc Natl Acad Sci USA. 2013;110:7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda H, Greish K, Fang J. The EPR effect and polymeric drugs: A paradigm shift for cancer chemotherapy in the 21st century. Adv Polym Sci. 2006;193:103–121. [Google Scholar]
- 27.Boveri M, Berezowski V, Price A, Slupek S, Lenfant AM, Benaud C, Hartung T, Cecchelli R, Prieto R, Dehouck MP. Induction of blood-brain barrier properties in cultured brain capillary endothelial cells: Comparison between primary glial cells and C6 cell line. Glia. 2005;51:187–198. doi: 10.1002/glia.20189. [DOI] [PubMed] [Google Scholar]
- 28.Cecchelli R, Dehouck B, Descamps L, Fenart L, Buée-Scherrer V, Duhem C, Lundquist S, Rentfel M, Torpier G, Dehouck MP. In vitro model for evaluating drug transport across the blood–brain barrier. Adv Drug Deliv Rev. 1999;36:165–178. doi: 10.1016/s0169-409x(98)00083-0. [DOI] [PubMed] [Google Scholar]
- 29.Culot M, Lundquist S, Vanuxeem D, Nion S, Landry C, Delplace Y, Dehouck MP, Berezowski V, Fenart L, Cecchelli R. An in vitro blood-brain barrier model for high throughput (HTS) toxicological screening. Toxicol In Vitro. 2008;22:799–811. doi: 10.1016/j.tiv.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Abbott NJ, Rönnbäck L, Hansson E. Astrocyte–endothelial interactions at the blood–brain barrier. Nat Rev Neurosci. 2006;7:41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 31.Allhenn D, Boushehri MA, Lamprecht A. Drug delivery strategies for the treatment of malignant gliomas. Int J Pharm. 2012;436:299–310. doi: 10.1016/j.ijpharm.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 32.Blakeley J. Drug delivery to brain tumors. Curr Neurol Neurosci Rep. 2008;8:235–241. doi: 10.1007/s11910-008-0036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackic JB, Stins M, McComb JG, Calero M, Ghiso J, Kim KS, Yan SD, Stern D, Schmidt AM, Frangione B, Zlokovic BV. Human blood-brain barrier receptors for Alzheimer’s amyloid-beta 1–40. Asymmetrical binding, endocytosis, and transcytosis at the apical side of brain micro-vascular endothelial cell monolayer. J Clin Invest. 1998;102:734–743. doi: 10.1172/JCI2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goti D, Hrzenjak A, Levak-Frank S, Frank S, van der Westhuyzen DR, Malle E, Sattler W. Scavenger receptor class B, type I is expressed in porcine brain capillary endothelial cells and contributes to selective uptake of HDL-associated vitamin E. J Neurochem. 2001;76:498–508. doi: 10.1046/j.1471-4159.2001.00100.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang K, Hao L, Hurst SJ, Mirkin CA. Antibody-linked spherical nucleic acids for cellular targeting. J Am Chem Soc. 2012;134:16488–16491. doi: 10.1021/ja306854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gartner JJ, Parker SCJ, Prickett TD, Dutton-Regester K, Stitzel ML, Lin JC, Davis S, Simhadri VL, Jha S, Katagiri N, Gotea V, Teer JK, Wei X, Morken MA, Bhanot UK, Chen G, Elnitski LL, Davies MA, Gershenwald JE, Carter H, Karchin R, Robinson W, Robinson S, Rosenberg SA, Collins FS, Parmigiani G, Komar AA, Kimchi-Sarfaty C, Hayward NK, Margulies EH, Samuels Y NISC Comparative Sequencing Program. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci USA. 2013;110:13481–13486. doi: 10.1073/pnas.1304227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fendri A, Kontos CK, Khabir A, Mokdad-Gargouri R, Scorilas A. Bcl2L12 is a novel biomarker for the prediction of short-term relapse in nasopharyngeal carcinoma. Mol Med. 2011;17:163–171. doi: 10.2119/molmed.2010.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Florou D, Papadopoulos IN, Scorilas A. Molecular analysis and prognostic impact of the novel apoptotic gene Bcl2L12 in gastric cancer. Biochem Biophys Res Commun. 2010;391:214–218. doi: 10.1016/j.bbrc.2009.11.034. [DOI] [PubMed] [Google Scholar]
- 39.Kontos CK, Papadopoulos IN, Scorilas A. Quantitative expression analysis and prognostic significance of the novel apoptosis-related gene Bcl2L12 in colon cancer. Biol Chem. 2008;389:1467–1475. doi: 10.1515/BC.2008.173. [DOI] [PubMed] [Google Scholar]
- 40.Mastarone DJ, Harrison VSR, Eckermann AL, Parigi G, Luchinat C, Meade TJ. A modular system for the synthesis of multiplexed magnetic resonance probes. J Am Chem Soc. 2011;133:5329–5337. doi: 10.1021/ja1099616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nature Phys Sci. 1973;241:20–22. [Google Scholar]
- 42.Demers LM, Mirkin CA, Mucic RC, Reynolds RA, III, Letsinger RL, Elghanian R, Viswanadham G. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nano-particles. Anal Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.