Abstract
Mucin 13 (MUC13) is a high-molecular-weight transmembrane glycoprotein that is frequently and aberrantly expressed in a variety of epithelial carcinomas, including gastric, colorectal, and ovarian cancers. On the basis of the high expression of MUC13 in cancer cells as well as recent laboratory findings suggesting a malignant phenotype of MUC13-transfected cell lines, the oncogenic potential of MUC13 has emerged. The various functional domains of MUC13 may confer oncogenic potential to MUC13. For example, the bulky extracellular domain with extensive modification with glycan chains may prevent cell–cell and cell–extracellular matrix binding whereas the cytoplasmic tail containing serine and tyrosine residues for potential phosphorylation may participate in cell signaling. MUC13 exhibits the characteristics suitable as an early marker for cancer screening and presents a promising target for antibody-guided targeted therapy.
Introduction
Mucin proteins are known for providing protection and lubrication to epithelial surfaces; in addition, their roles in cell signaling are beginning to be elucidated (1–3). The aberrant expression of mucins, as found in many cancers, is likely associated with cancer biology as alterations in the expression and/or glycosylation patterns of various mucins influence cellular growth, differentiation, transformation, adhesion, invasion, and immune surveillance (1, 4–8). Mucin 13 (MUC13), a transmembrane (TM) mucin, has recently been implicated in cancer development and pathogenesis. Similar to other mucins, MUC13 is characterized by a tandem repeat (TR) domain (the hallmark of mucins) composed of TRs rich in serine and threonine residues that act as glycosylation sites (Fig. 1). MUC13 also contains 3 epidermal growth factor (EGF)-like domains and a cytoplasmic domain containing potential phosphorylation sites, which could play a role in cell signaling. In this review, we describe structural and functional aspects of the newly identified TM mucin, MUC13, and its potential role in cancer pathogenesis (depicted in Figs. 1 and 2).
Figure 1.
Schematic diagram and annotated amino acid sequence of MUC13. Left, a schematic diagram showing the structural features of MUC13 protein. The signal peptide, mucin repeat domain, SEA module, EGF-like domains, TM domain, and the cytoplasmic domain are shown from N-terminal (top) to C-terminal (bottom). Right, the amino acid sequence of MUC13 indicating amino acid residues for predicted posttranslational modification (O-glycosylation, N-glycosylation, and disulfide bonds). The signal peptide, SEA module, and TM sequences are indicated by text that is underlined, bold and underlined, and in italics font, respectively.
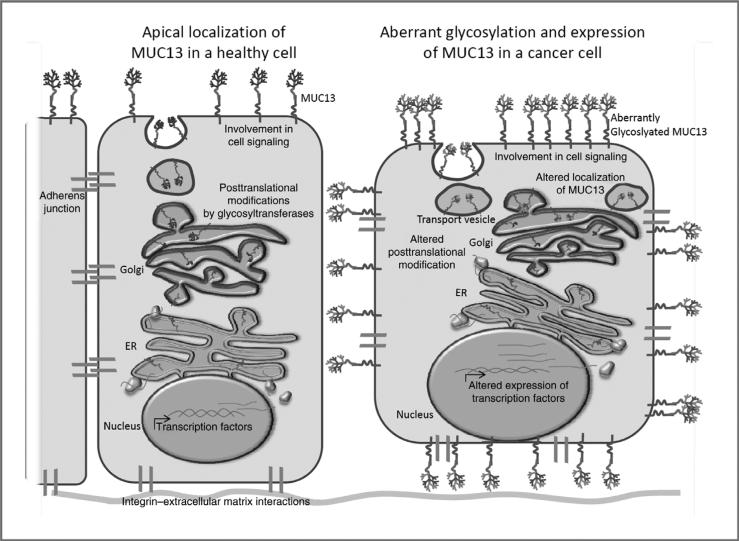
Figure 2.
Normal and atypical cellular expression of MUC13. Left, as a TM mucin, theoretically MUC13 is processed through the endoplasmic reticulum (ER) and Golgi apparatus, where posttranslational modifications such as the addition of O- and N-glycosylation occurs and the protein is delivered to the apical cell surface. Right, in cancer cells, MUC13 is localized at basal, lateral, and apical cell surface membranes, which possibly contributes to the loss of cell–cell and cell–ECM binding. Hypothetically, aberrant subcellular localization of MUC13 may alter cell signaling due to interaction with EGFRs. These events could increase tumorigenesis, cell invasion, and metastasis.
Genomic location and expression of MUC13 in normal versus cancer tissues
The human MUC13 gene was originally identified as an orthologue of murine Muc13 (MUC13 for human and Muc13 for other species; ref. 9). MUC13 is located at 3q21.2 and is flanked by genes ITGB5 (β5 integrin) and HEG-1 (Heart of Glass), each transcribed from the reverse strand. An evolutional relationship between HEG and MUC13 genes has been suggested, as they have similar configuration of exons encoding similar domains [TR, EGF receptor (EGFR)-like, and TM domain; ref. 10]. The predominant MUC13 mRNA contains 12 exons (with a final mRNA length of 2.8 kb) and encodes 511 amino acids (11). Although a variety of single nucleotide polymorphisms (SNP) have been identified within the MUC13 gene, their clinical significance has not yet been determined (NCBI: SNPs).
Under normal physiologic conditions, MUC13 mRNA and/or protein have been detected in the large intestine, trachea, kidney, small intestine, gastric epithelium, and esophagus (9). MUC13 protein is normally localized to the apical surface of epithelial cells, as expected by the role in protection and lubrication of the mucosal surface. In general, mucin expression, both secreted and cell membrane associated, provides a protective barrier against colonization by pathogenic bacteria. Using a mouse model, Linden and colleagues show that in response to infection with Clostridium rodentium, the secretion and/or release of many mucins, including Muc13, increases, leading to a depletion of the intracellular stores of mucins (12). MUC13 expression may also be altered in benign conditions of the colon, such as Crohn's disease and ulcerative colitis; however, at this time, additional experimental work is needed. For example, although Moehle and colleagues initially found that MUC13 expression is decreased in colon samples from both Crohn's disease and ulcerative colitis when using gene array analysis of pooled specimens, real-time reverse transcriptase PCR (RT-PCR) assays found that compared with controls, MUC13 expression has a statistically significant upregulation in ulcerative colitis (13). Interestingly, an allelic connection between ulcerative colitis and MUC13-R502 was found; however, the authors suggest that the allelic differences are likely linked to other as of yet unknown genes involved in ulcerative colitis (13). Compared with normal tissues, ovarian, colon, and gastric cancer cells produce a markedly higher amount of MUC13 (protein and/or mRNA, explained in detail below; refs. 14–17). Although aberrant expression of MUC13, including changes in subcellular localization, has been reported, the biological significance of aberrant MUC13 expression has not been fully uncovered and is currently being explored.
Protein structure of MUC13
Overview
The presence of a TR domain is the hallmark feature of the mucin family. The polypeptide backbone of the TR domain acts as a scaffold for a large number of complex O-linked carbohydrate side chains. Consequently, the TR domain constitutes the major spatial part of the MUC13 extracellular domain (ref. 1; Fig. 1). Because of the TR, the extracellular portion of mucins may protrude more than 200 to 2,000 nm above the cell surface and can effectively block cell–cell adhesion and cell adhesion to the extracellular matrix (ECM; Fig. 2). Therefore, the over- or aberrant expression of mucins may be implicated in the exfoliation, dissemination, and invasion of cancer cells (1).
Because of the extensive glycosylation of the TR domain, the molecular weight of matured mucin proteins is much greater than the predicted molecular weight based on the amino acid sequence. Similar to other mucins, the molecular weight of the MUC13 apomucin (the protein “backbone” without glycosylation) is 54.7 kDa whereas the experimental molecular weight (including glycosylation) is approximately 175 kDa (18). In comparison with other membrane-bound mucins, the size of MUC13 is similar to MUC15 and MUC20; however, MUC13 is relatively small compared with MUC1, MUC4, and MUC16 (18, 19). To thoroughly describe the known and presumable functions of MUC13, the following sections describe each domain of MUC13.
Signal peptide
Proteins destined for secretion or cell membrane localization enter the endoplasmic reticulum during early events in protein synthesis, move through the Golgi apparatus where glycosylation and trimming of the attached glycan chains occur, and are packaged in vesicles for transport to the cell membrane. This process is initiated by the synthesis of a hydrophobic signal peptide (~20-amino acid sequence) at N-terminus of the protein, which anchors the mRNA–ribosome–signal peptide complex to the endoplasmic reticulum. Once anchored, the signal peptide is cleaved and translation continues with the growing peptide translocated across the endoplasmic reticulum membrane into the endoplasmic reticulum. It is expected that the signal peptide at the N-terminus (amino acids 1–19) of MUC13 (Fig. 1) targets MUC13 for translation into the endoplasmic reticulum. In the endoplasmic reticulum, initial oligosaccharides are added and the protein is transported to the Golgi apparatus for further glycosylation and posttranslational modifications with final delivery to the apical cell membrane of polarized epithelial cells. Mucosal glycosylation can affect a variety of cell behaviors, including proliferation, apoptosis, and response to normal bacterial microflora (20). Altered glycosylation patterns are detected in many types of cancer and may play a role in cancer pathogenesis. Although not thoroughly understood, the aberrant expression pattern of glycosyltransferases and glycosidases within the endoplasmic reticulum and Golgi apparatus of cancer cells likely contributes to the altered glycosylation of mucins (21). Mucin glycosylation is an important and interesting area of mucin biology in need of additional research.
TR domain
The large serine-threonine–rich TR domain, which is present in all mucins, is next to the signal peptide and is located at the N-terminus of matured MUC13 on the cell surface (amino acids 20–170; Fig. 1). There are 10 degenerate TRs (each approximately 10–18 amino acids), with more than 50 predicted glycosylation sites in this domain, which makes this domain a remarkable scaffold on which cells can build oligosaccharide structures (refs. 9, 22; Fig. 1, right). In addition to the many sites for O-glycosylation, there are 7 sites available for N-glycosylation (9). It should be noted that the TR of MUC13 is 150 amino acids long, which is relatively short compared with other membrane-bound mucins, such as MUC4 and MUC16. In some mucins (e.g., MUC2), the length of the TR can vary and is polymorphic (3); however, little is known about MUC13 in this matter.
The glycosylation pattern of the TR domain may have effects on the function of TM mucins, and it can also serve as a potential biomarker for cancer and as a means of targeted cancer therapy. Targeted therapy is an ongoing area of research (recently reviewed by Peracaula and colleagues; see ref. 21) and additional research is needed to determine whether MUC13 epitopes formed by the glycan chains can be a target for antibody-guided targeted cancer therapy. As mentioned earlier, the TR is glycosylated in the endoplasmic reticulum and Golgi apparatus; this a process that is often aberrant in cancer cells, and the altered glycosylation pattern may create unique targets for cancer cells. More work is needed to show that MUC13 glycosylation is altered in cancer cells. In addition, the polarity of epithelial cells is often lost in cancer cells, which could disrupt the appropriate cellular delivery of MUC13 to the apical surface. When the expression of MUC13 is no longer confined to the apical surface, the presence of the large glycosylated TR domain at the lateral and basal surfaces may disrupt binding to adjacent cells or the ECM (see Fig. 2), thus facilitating invasion and metastasis.
EGF-like domains
The central region of MUC13 contains 3 EGF-like domains (domains 1–3), suggesting that MUC13 may play a role in a signaling cascade. An EGF-like domain consists of 30 to 40 amino acids and contains 6 cysteine residues that form disulfide bonds within the domain (C1–C3, C2–C4, and C5–C6; ref. 23). Subdomains between the conserved cysteines vary in length. The first EGF-like domain (amino acids 177–210) decreases between the TR domain and the SEA (sea urchin sperm protein, enterokinase and agrin) domain (Fig. 1). The second and third EGF-like domains follow the SEA domain (amino acids 326–360 and 367–403). Other TM mucins (MUC3, MUC4, MUC12, and MUC17) contain 1 to 2 EGF-like domains. In comparing the EGF-like domains with sequence information from UniProt, MUC4 and MUC13 have similar spacing patterns between the cysteine residues (similar subdomains). Through its EGF-like domain, the TM mucin rat sialomucin complex (SMC/rat Muc4), has been shown to augment ErbB2/HER2 and ErbB3 signaling by increasing the localization of ErbB2 and ErbB3 to the cell membrane and by suppressing ligand-induced receptor internalization (24). Although the functional role of MUC13 EGF-like domains has not been directly determined, data generated in our laboratory suggest that MUC13 may also be involved in cell signaling, potentially through ErbB2-related pathways (14). At this time, it is not known whether MUC13 and ErbB2 directly interact and additional work is needed to further elucidate the role of MUC13 in cell signaling.
SEA domain
An SEA domain is commonly present in the extracellular portion of dimeric or multimeric membrane-associated proteins. This domain is predicted to provide both the cleavage site and a sequence motif, allowing the 2 subunits of MUC13 to be noncovalently bound at the cell surface. Therefore, it is expected that similar to other mucins, MUC13 will be cleaved during posttranslational processing but the intracellular and extracellular regions will remain noncovalently bound at the cell surface until shedding is initiated through environmental signals involved in outside-to-inside signaling that may alter cell behavior (1). The SEA domain, composed of approximately 120 amino acids of which 80 amino acids (60%) are highly conserved, contains proteolytic cleavage sites and an amino acid sequence motif for noncovalent protein-protein associations (25–27). Of the TM mucins, MUC1, MUC3, MUC12, MUC13, MUC16, and MUC17 have at least one SEA domain (MUC16 has multiple SEA domains that vary in their sequence homology). The majority of these mucins contain the typical GSVV motif, which has been shown to be a site for cleavage (27, 28). Two independent laboratories have published biochemical analysis suggesting that MUC13 is cleaved within the SEA domain (9, 17); however, MUC13 does not have the typical GSVV motif and the amino acid sequence required for the cleavage of MUC13 needs to be determined in future studies. Maeda and colleagues analyzed the solution structure of murine Muc16 and found that, on comparison of the secondary structure-based sequence alignment of the SEA domains, MUC13 does not group with the other SEA containing mucins (27), suggesting that MUC13 may be distinct in function from other mucins.
TM domain
Adjacent to EGF-like domain 3 is a short single-pass TM domain, which anchors MUC13 to the cell membrane (amino acids 421–442; Fig. 1). As explained in detail later (see Biochemical Characterization of MUC13), Williams and colleagues carried out biochemical characterization of MUC13. Their studies suggest that MUC13 is cleaved, resulting in a subunit of MUC13 that contains the cytoplasmic domain, TM domain, the C-terminal EGF-like domain, and at least part of the SEA domain (9). In this regard, independent of protein cleavage and shedding, it is possible that MUC13 could receive extracellular signals via the second and the third EGF-like domains and transduce them through the cytoplasmic domain.
Cytoplasmic tail domain
Following the TM, there is a 69-amino acid long cytoplasmic domain (9, 17). The cytoplasmic domain of MUC13 contains several potential phosphorylation sites (8 serine and 2 tyrosine residues, shown in Fig. 1) and a protein kinase C phosphorylation motif (9), further supporting the hypothesis that MUC13 may be involved in cell signaling pathways through phosphorylation. MUC1 is another TM mucin that is frequently overexpressed and aberrantly glycosylated in cancer. Interestingly, a number of studies have shown that MUC1 signals through a variety of pathways including MAP kinase, β-catenin, p53, and the EGF signaling cascade (18, 29, 30). The extent to which MUC13 plays a role in cell signaling remains to be determined; however, we have observed nuclear localization of MUC13 protein in immunohistochemical staining of ovarian, pancreatic, and colon cancer tissues (unpublished results). With evidence of MUC13 nuclear localization and potential phosphorylation in the cytoplasmic tail, it is very likely that MUC13 plays a significant biological role in oncogenic cellular signaling pathways in a manner similar to MUC1.
Biochemical characterization of MUC13
It is biochemically appropriate to consider that MUC13 consists of 2 distinct subunits, an extracellular α-subunit (consisting of the TR domain, 1 EGF-like domain, and a portion of the SEA domain) and a β-subunit (consisting of a portion of the SEA domain, 2 EGF-like domains, the TM domain, and the cytoplasmic tail, shown in Fig. 1). As described earlier, the SEA domain is predicted to contain a cleavage site and, although the amino acid sequence is unknown, there is biochemical evidence that MUC13 undergoes cleavage (9, 17). Williams and colleagues carried out a comprehensive Western blot analysis of MUC13 protein expressed in 2 cancer cell lines by using a polyclonal MUC13 antibody (9). In this experiment, under nonreducing conditions, MUC13 migrated as a 47-kDa band plus a 93-kDa band homodimer. In contrast, under reducing conditions, MUC13 appeared as a 58-kDa single band. The size difference observed with different conditions can be explained by the denaturation of MUC13 in the reducing condition accompanied by cleavage of intrastrand disulfide bonds, which created a slower migrating conformation of MUC13 (9). However, when mild detergents and reducing conditions were used, MUC13 appeared as a larger size protein with an intense band at 72 kDa and a weak band at 120 kDa. These larger sizes of MUC13 protein are likely noncovalently bound MUC13 α- and β-subunits (that did not dissociate under mild denaturing conditions). To examine N-glycosylation, Williams and colleagues also showed specific bands on immunoprecipitation with anti-MUC13 polyclonal antibodies and probing with lectins specific for N-glycosylated sites. In addition, they observed the appearance of smaller bands (49 and 38 kDa) after treatment with PNGase F (also known as N-glycosidase F, an amidase that cleaves between the innermost GlcNAc and asparagine residues of complex oligosaccharides from N-linked glycoproteins). Together, these data imply that MUC13 contains disulfide bonds and O- and N-linked glycosylation and may exist as a homodimer of β-subunits.
MUC13 has also been characterized with monoclonal antibodies (17). These experiments suggest the presence of alternative O- and N-glycosylation patterns as well as cleavage of MUC13 within the SEA domain. Shimamura and colleagues, using 2 monoclonal antibodies and a third polyclonal antibody to the TR, EGF-like domain 2, and cytoplasmic tail, respectively, show that depending on the extent of O- and N-glycosylation, MUC13 appears as either a 120- or 80-kDa protein (17). The β-subunit appears as a 35-kDa band and would be potentially important for cell signaling. The cleaved α-subunit was not detected from cell culture supernatant, possibly because of the low affinity of the antibody to the highly glycosylated form of the α-subunit. We also detected glycosylated and nonglycosylated forms of recombinant MUC13-Fc fusion protein in 293T cells by using a newly developed anti-MUC13 monoclonal antibody (unpublished data).
Aberrant expression of MUC13
Colon cancer
Walsh and colleagues studied the expression of MUC13 in various stages of colon cancer (16). In normal colon, MUC13 was detected as a thin layer on the apical surface of glands; however, MUC13 was highly expressed in most of the colon tumors, with 81% of well-differentiated adenocarcinomas exhibiting strong MUC13 staining. Mucinous tumors expressed MUC13, but at a lower level, based on the lower staining intensity than adenocarcinomas (50% vs. 81%, indicating strong staining). Although not statistically significant, there was a trend toward poorer survival in patients with tumors showing basolateral MUC13 expression. In contrast to these results, Packer and colleagues reported that the RNA level of MUC13 was decreased in colon cancer; however, this was a small study with only 23 samples of colon cancer and 6 normal colon tissue samples (15). In a recently published report MUC13 mRNA was detected in the blood of colorectal cancer patients; however, MUC13 mRNA was also detected in the blood of healthy individuals (31), thus providing additional support for the importance of identifying glycosylation differences between MUC13 expressed under normal and cancer conditions. In our own studies, we have observed the overexpression of MUC13 in colon cancer compared with normal colon (unpublished data), and further studies are required to clarify the relationships between MUC13 expression and colon cancer stages and prognosis.
Gastric cancer
Shimamura and colleagues detected increased expression of MUC13 at both mRNA and protein levels in gastric cancer (64.9% of cases; ref. 17). MUC13 was also detected in 9 of 10 cases of intestinal metaplasia (precancerous lesions of intestinal type gastric cancer). In this study, MUC13 expression did not correlate with clinicopathologic factors (depth of invasion or lymph node metastasis), but MUC13 expression was associated with intestinal type of gastric cancer. MUC13 expression did not correlate with the expression of other mucins (MUC2, MUC5AC, MUC6, and CD10), suggesting that MUC13 expression may be regulated in a manner different from that of other mucin markers for gastric cancer (17). Recently, Lee and colleagues reported a 115-fold increase in MUC13 mRNA in intestinal metaplasia (compared with chief cells; ref. 32). They also reported that MUC13 protein was detected in 50% of all gastric cancers and 91% of intestinal type gastric cancer (32). In addition, although not statistically significant, the expression of cytoplasmic MUC13 (as opposed to membranous MUC13) was possibly associated with decreased survival rates (32).
Ovarian cancer: expression and oncogenic functions of MUC13
The aberrant expression of mucins has important roles in ovarian cancer pathogenesis and diagnosis (recently reviewed by Chauhan and colleagues, ref. 33). We analyzed the expression profile and functions of MUC13 to elucidate its potential role in ovarian cancer diagnosis and pathogenesis (14). We determined the expression profile of MUC13 by immunohistochemistry and found that the expression of MUC13 was significantly (P < 0.005) higher in epithelial ovarian cancer samples than in the normal ovary/benign tissues (66% stained positive). Among epithelial ovarian cancer types, MUC13 expression was highest in mucinous epithelial ovarian cancer (100% stained positive).
Exogenous expression of full-length MUC13 in ovarian cancer cells increased tumorigenesis in a xenograft mouse model system (14). In addition, we observed morphologic changes in ovarian cancer cells, including scattering of cells, marked reduction in cell–cell adhesion, and significant (p < 0.05) increases in cell motility and proliferation. These cellular changes correlated with upregulation of HER2, p21-activated kinase1 (PAK1), and p38 mitogen-activated protein kinase (MAPK) protein expression, all of which support increased tumor progression due to MUC13 expression. In fact, inhibiting the JNK, JNK2, and MAPK pathways abrogated the oncogenic effects of MUC13, suggesting that MUC13 expression alters cell signaling pathways. Our findings show the aberrant expression of MUC13 in ovarian cancer and also that its expression alters the cellular characteristics of ovarian cancer cells, implying a significant role of MUC13, potentially through modulation of cell signaling pathways, in ovarian cancer.
Regulation of MUC13 expression
MUC13 is overexpressed in a variety of cancers; however, the regulation of MUC13 expression has not been studied. Other secreted (MUC2, MUC5AC, MUC5B, MUC6) or membrane-bound mucins (MUC1, MUC3, and MUC4) have been well studied and seem to be regulated by a diverse set of transcription factors relating to various stimuli, such as cytokines, bacterial products, growth factors, and differentiation agents (18, 34). On the basis of an in silico analysis, the MUC13 promoter region contains binding sites for a number of transcription factors (35), including CREB (cAMP response element-binding protein), COUPTF (chicken ovalbumin upstream promoter transcription factor), Spz1 (spermatogenic leucine zipper 1), STAT5A/B (signal transducer and activator of transcription), and HNF-4 α1 and 2 (hepatocyte nuclear factor 4 α). A number of these transcription factors are known to be aberrantly expressed in various cancers. For example, aberrant CREB signaling is frequently reported in leukemia cells and endocrine tumors (36). COUP-TF may induce oncogenic cell signaling through multiple pathways including its positive regulation on the transcription of vascular endothelial factors C and D (37, 38). STAT5A/B are known to play a role in pathogenesis of both prostate cancer and breast cancer (39), and HNF-4 has been shown to be dysregulated in a variety of cancers, including gastric, hepatocellular, and colorectal carcinomas (40). Additional research on the regulation of MUC13 expression will enhance our understanding of MUC13 in cancer pathogenesis.
Diagnostic and therapeutic potential of MUC13
The development of screening or diagnostic blood tests is an important area of cancer research. Because of their abundant expression and potential for altered glycosylation patterns in cancer, mucins are promising targets as early screening/diagnostic markers. An important attribute of a molecular marker for cancer is the ability to detect the marker in the blood or other accessible body fluid. Membrane proteins can release their extracellular domains into circulation by metalloproteinase-dependent cleavage near the cell membrane (shedding; ref. 41). MUC13 has been detected within goblet cells thecae and in secreted material in the large intestine (9). In addition, biochemical characterization showing cleavage of MUC13 increases the potential of MUC13 to be an effective screening/diagnostic marker due to the expectation that a substantial amount of soluble MUC13 will be present in patients with tumors expressing high levels of MUC13. However, further research is needed to determine whether MUC13 can be detected in body fluids. With a high level of expression in gastric, colon, and ovarian cancers (ranging between 65% and 100% positive expression), we expect that MUC13 holds significant potential in the screening, diagnosis, and treatment of cancer.
The altered glycosylation patterns of mucins, such as MUC13, expressed by cancer cells create potential targets for antibodies that bind to cancer-specific epitopes. Targeted cancer therapy is a rapidly advancing field. One of the most promising techniques is the development of immunonanoparticles capable of both drug loading and antibody-mediated delivery.
Summary
The recently generated antibodies have brought significant progress in characterizing the role of MUC13 in cancer. Further studies that expand particular research areas of interest include (i) defining the role of the cytoplasmic tail of MUC13 in cell signaling, (ii) determining the effect of MUC13 on cell–cell and cell–ECM interactions, (iii) exploring the regulation of MUC13 expression, and (iv) determining the potential of MUC13 as an early marker for cancer and a target for antibody-guided therapy. These studies will increase understanding about the role of MUC13 in pathologic conditions and will eventually provide additional therapies to reduce cancer burden.
Acknowledgments
The authors thank Cathy Christopherson for editorial assistance and Mara Ebeling for her valuable work on MUC13.
Grant Support
This work was supported by grants from Sanford Research/USD, Department of Defense (DOD; PC073887), Governor's Cancer 2010, and NIH RO1 (CA142736) awarded to S.C. Chauhan and DOD (PC073643) and Governor's Cancer 2010 grants awarded to M. Jaggi.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 2.Gendler SJ, Spicer AP. Epithelial mucin genes. Annu Rev Physiol. 1995;57:607–34. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- 3.Moniaux N, Escande F, Porchet N, Aubert JP, Batra SK. Structural organization and classification of the human mucin genes. Front Biosci. 2001;6:D1192–206. doi: 10.2741/moniaux. [DOI] [PubMed] [Google Scholar]
- 4.Andrianifahanana M, Moniaux N, Schmied BM, Ringel J, Friess H, Hollingsworth MA, et al. Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin Cancer Res. 2001;7:4033–40. [PubMed] [Google Scholar]
- 5.Balague C, Audie JP, Porchet N, Real FX. In situ hybridization shows distinct patterns of mucin gene expression in normal, benign, and malignant pancreas tissues. Gastroenterology. 1995;109:953–64. doi: 10.1016/0016-5085(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 6.Dong Y, Walsh MD, Cummings MC, Wright RG, Khoo SK, Parsons PG, et al. Expression of MUC1 and MUC2 mucins in epithelial ovarian tumours. J Pathol. 1997;183:311–7. doi: 10.1002/(SICI)1096-9896(199711)183:3<311::AID-PATH917>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Giuntoli RL, 2nd, Rodriguez GC, Whitaker RS, Dodge R, Voynow JA. Mucin gene expression in ovarian cancers. Cancer Res. 1998;58:5546–50. [PubMed] [Google Scholar]
- 8.Chauhan SC, Singh AP, Ruiz F, Johansson SL, Jain M, Smith LM, et al. Aberrant expression of MUC4 in ovarian carcinoma: diagnostic significance alone and in combination with MUC1 and MUC16 (CA125). Mod Pathol. 2006;19:1386–94. doi: 10.1038/modpathol.3800646. [DOI] [PubMed] [Google Scholar]
- 9.Williams SJ, Wreschner DH, Tran M, Eyre HJ, Sutherland GR, McGuckin MA. MUC13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J Biol Chem. 2001;276:18327–36. doi: 10.1074/jbc.M008850200. [DOI] [PubMed] [Google Scholar]
- 10.Lang T, Hansson GC, Samuelsson T. An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics. 2006;7:197. doi: 10.1186/1471-2164-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NCBI [2010 Mar 2];Entrez Gene MUC13. Available from: http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=retrieve&dopt=-ull_report&list_uids=56667#geneGenomic%20regions.
- 12.Linden SK, Florin TH, McGuckin MA. Mucin dynamics in intestinal bacterial infection. PLoS One. 2008;3:e3952. doi: 10.1371/journal.pone.0003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moehle C, Ackermann N, Langmann T, Aslanidis C, Kel A, Kel-Margoulis O, et al. Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med. 2006;84:1055–66. doi: 10.1007/s00109-006-0100-2. [DOI] [PubMed] [Google Scholar]
- 14.Chauhan SC, Vannatta K, Ebeling MC, Vinayek N, Watanabe A, Pandey KK, et al. Expression and functions of transmembrane mucin MUC13 in ovarian cancer. Cancer Res. 2009;69:765–74. doi: 10.1158/0008-5472.CAN-08-0587. [DOI] [PubMed] [Google Scholar]
- 15.Packer LM, Williams SJ, Callaghan S, Gotley DC, McGuckin MA. Expression of the cell surface mucin gene family in adenocarcinomas. Int J Oncol. 2004;25:1119–26. [PubMed] [Google Scholar]
- 16.Walsh MD, Young JP, Leggett BA, Williams SH, Jass JR, McGuckin MA. The MUC13 cell surface mucin is highly expressed by human colorectal carcinomas. Hum Pathol. 2007;38:883–92. doi: 10.1016/j.humpath.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 17.Shimamura T, Ito H, Shibahara J, Watanabe A, Hippo Y, Taniguchi H, et al. Overexpression of MUC13 is associated with intestinal-type gastric cancer. Cancer Sci. 2005;96:265–73. doi: 10.1111/j.1349-7006.2005.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrianifahanana M, Moniaux N, Batra SK. Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochim Biophys Acta. 2006;1765:189–222. doi: 10.1016/j.bbcan.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Jonckheere N, Van Seuningen I. The membrane-bound mucins: how large O-glycoproteins play key roles in epithelial cancers and hold promise as biological tools for gene-based and immunotherapies. Crit Rev Oncog. 2008;14:177–96. doi: 10.1615/critrevoncog.v14.i2-3.30. [DOI] [PubMed] [Google Scholar]
- 20.Patsos G, Corfield A. Management of the human mucosal defensive barrier: evidence for glycan legislation. Biol Chem. 2009;390:581–90. doi: 10.1515/BC.2009.052. [DOI] [PubMed] [Google Scholar]
- 21.Peracaula R, Barrabes S, Sarrats A, Rudd PM, de Llorens R. Altered glycosylation in tumours focused to cancer diagnosis. Dis Markers. 2008;25:207–18. doi: 10.1155/2008/797629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaturvedi P, Singh AP, Batra SK. Structure, evolution, and biology of the MUC4 mucin. FASEB J. 2008;22:966–81. doi: 10.1096/fj.07-9673rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prosite. [2010 Nov 1];EGF-like domain signatures and profile. [Apr 2006; pattern revised]. Available from: http://www.expasy.ch/prosite/PDOC00021.
- 24.Funes M, Miller JK, Lai C, Carraway KL, 3rd, Sweeney C. The mucin Muc4 potentiates neuregulin signaling by increasing the cell-surface populations of ErbB2 and ErbB3. J Biol Chem. 2006;281:19310–9. doi: 10.1074/jbc.M603225200. [DOI] [PubMed] [Google Scholar]
- 25.Bork P, Patthy L. The SEA module: a new extracellular domain associated with O-glycosylation. Protein Sci. 1995;4:1421–5. doi: 10.1002/pro.5560040716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wreschner DH, McGuckin MA, Williams SJ, Baruch A, Yoeli M, Ziv R, et al. Generation of ligand-receptor alliances by “SEA” module-mediated cleavage of membrane-associated mucin proteins. Protein Sci. 2002;11:698–706. doi: 10.1110/ps.16502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda T, Inoue M, Koshiba S, Yabuki T, Aoki M, Nunokawa E, et al. Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16). J Biol Chem. 2004;279:13174–82. doi: 10.1074/jbc.M309417200. [DOI] [PubMed] [Google Scholar]
- 28.Palmai-Pallag T, Khodabukus N, Kinarsky L, Leir SH, Sherman S, Hollingsworth MA, et al. The role of the SEA (sea urchin sperm protein, enterokinase and agrin) module in cleavage of membrane-tethered mucins. FEBS J. 2005;272:2901–11. doi: 10.1111/j.1742-4658.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 29.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, et al. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283:26985–95. doi: 10.1074/jbc.M805036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen Y, Caffrey TC, Wheelock MJ, Johnson KR, Hollingsworth MA. Nuclear association of the cytoplasmic tail of MUC1 and beta-catenin. J Biol Chem. 2003;278:38029–39. doi: 10.1074/jbc.M304333200. [DOI] [PubMed] [Google Scholar]
- 31.Lauriola M, Ugolini G, Rosati G, Zanotti S, Montroni I, Manaresi A, et al. Identification by a Digital Gene Expression Displayer (DGED) and test by RT-PCR analysis of new mRNA candidate markers for colorectal cancer in peripheral blood. Int J Oncol. 2010;37:519–25. doi: 10.3892/ijo_00000701. [DOI] [PubMed] [Google Scholar]
- 32.Lee HJ, Nam KT, Park HS, Kim MA, Lafleur BJ, Aburatani H, et al. Gene expression profiling of metaplastic lineages identifies CDH17 as a prognostic marker in early stage gastric cancer. Gastroenterology. 2010;139:213–25. e3. doi: 10.1053/j.gastro.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauhan SC, Kumar D, Jaggi M. Mucins in ovarian cancer diagnosis and therapy. J Ovarian Res. 2009;2:21. doi: 10.1186/1757-2215-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonckheere N, Van Seuningen I. The membrane-bound mucins: from cell signalling to transcriptional regulation and expression in epithelial cancers. Biochimie. 2010;92:1–11. doi: 10.1016/j.biochi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 35.SABioscience [2010 Sep 14];DECODE (Decipherment of DNA Elements) Available from: http://www.sabiosciences.com/chipqpcr-search.php?species_id=0&factor=Over+200+TF&gene=MUC13&nfactor=n&ninfo=n&ngene=n&B2=Search.
- 36.Siu YT, Jin DY. CREB–a real culprit in oncogenesis. FEBS J. 2007;274:3224–32. doi: 10.1111/j.1742-4658.2007.05884.x. [DOI] [PubMed] [Google Scholar]
- 37.Schafer G, Wissmann C, Hertel J, Lunyak V, Hocker M. Regulation of vascular endothelial growth factor D by orphan receptors hepatocyte nuclear factor-4 alpha and chicken ovalbumin upstream promoter transcription factors 1 and 2. Cancer Res. 2008;68:457–66. doi: 10.1158/0008-5472.CAN-07-5136. [DOI] [PubMed] [Google Scholar]
- 38.Nagasaki S, Suzuki T, Miki Y, Akahira J, Shibata H, Ishida T, et al. Chicken ovalbumin upstream promoter transcription factor II in human breast carcinoma: possible regulator of lymphangiogenesis via vascular endothelial growth factor-C expression. Cancer Sci. 2009;100:639–45. doi: 10.1111/j.1349-7006.2008.01078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan SH, Nevalainen MT. Signal transducer and activator of transcription 5A/B in prostate and breast cancers. Endocr Relat Cancer. 2008;15:367–90. doi: 10.1677/ERC-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka T, Jiang S, Hotta H, Takano K, Iwanari H, Sumi K, et al. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol. 2006;208:662–72. doi: 10.1002/path.1928. [DOI] [PubMed] [Google Scholar]
- 41.Hooper NM, Karran EH, Turner AJ. Membrane protein secretases. Biochem J. 1997;321:265–79. doi: 10.1042/bj3210265. [DOI] [PMC free article] [PubMed] [Google Scholar]