Abstract
The influence of ionization states of hydroxamates and retrohydroxamates and the presence of zinc ions in the active site were investigated using the wild-type and E402Q mutant of MMP-9. The deprotonated hydroxamates showed a significantly enhanced enrichment factor in the presence of zinc ions. A pharmacophore model was developed based on the deprotonated compounds and was used to identify four structurally diverse compounds with antiproliferative activities.
Keywords: Protonation state, curcumin, chalcone, and zinc binding group
The matrix metalloproteinases (MMPs) are members of a large family of zinc-containing, calcium-dependent enzymes involved in a variety of diseases, including cancers and arthritis. Endothelial cells secrete MMPs, which break down almost any component of the extracellular matrix (ECM) surrounding cancer cells, allowing the endothelial cells to move to the cancer site and form new blood vessels for the growth of cancerous cells. In addition, the MMPs promote the mobility and invasiveness of cancerous cells.1 The breakdown of the ECM also helps to release neutrophils and monocytes/macrophages from inflammatory cells. Under normal physiological conditions, the activities of MMPs are balanced by the endogenous tissue inhibitors of matrix metalloproteinases (TIMPs), which block the active site of MMPs. An imbalanced MMP/TIMP has been found in phenotypes such as breast, colon, and lung tumors.2
Among the 23 identified MMPs, gelatinase B (MMP-9) has been observed in a variety of pathological processes, such as inflammation, autoimmune diseases, and the metastasis of cancer cells.3,4 MMP-9 efficiently and rapidly cleaves the type IV collagen (gelatin),5 releasing the soluble Kit ligand6 and/or bioactive vascular endothelial growth factor (VEGF).7 The newly released VEGF turns on the angiogenic switch, leading to tumor angiogenesis, that is, the formation of new blood vessels from preexisting vessels for tumor growth. A direct link between MMP-9 and tumor progression was revealed in an experiment that upon the activation of MMP-9 zymogen, the neutrophil MMP-9 induced angiogenesis.8 Significant increases in MMP-9 activities were observed in the acute phase after myocardial infarction. The cardiovascular protection via inhibition of MMP-9 was confirmed by the observation that the MMP-9 activity after myocardial infarction was suppressed by imidapril, an angiotensin-converting enzyme (ACE) inhibitor.9
Given the important role of MMP-9 in the progression of cancer, osteoarthritis, and rheumatoid arthritis,1 researchers around the world have synthesized a wide array of MMP-9 inhibitors. The vast majority of MMP inhibitors are hydroxamates (marimastat and prinomastat),10−12 reverse hydroxamates (ABT-518),13 or carboxylic acid derivatives (BAY-129566)14 (Figure 1).
Figure 1.
Chemical structures of MMP inhibitors.
The crystal structure of the MMP-9 and a trifluoromethyl hydroxamic acid (PDB ID: 2OW1) shows a penta-coordination with a zinc ion through His401, His405, and His411 and two oxygen atoms on the hydroxamate group.15 Obviously, the oxygen atom of the hydroxyl group in the hydroxamic acid should adopt a deprotonated state (i.e., ionized) to form such a penta-coordination with Zn2+. The deprotonated model of hydroxamic acid inhibitors coordinated with Zn2+ is illustrated in Figure 2. So far, all computational studies of binding affinity and quantitative structure−activity relationship of hydroxamate-based MMP inhibitors have been carried out with the hydrogen atom on the oxime hydroxyl group (i.e., in a protonated state).16−18 Such a treatment, however, may be potentially problematic due to the ionizing nature of the oxime hydroxyl group of the hydroxamic acid, similar to the ionization of the carboxylic acid-based MMP inhibitors. The limited success in modeling-based prediction for hydroxamate-like MMP inhibitors was attributed to the failure in modeling the van der Waals interactions.16 However, we hypothesized that it might as well be related to how the hydroxamates are treated, deprotonated or protonated. Therefore, it is of fundamental importance to determine the impact of the ionization states of hydroxamates and/or retrohydroxamates on MMP binding.
Figure 2.
Deprotonated model of hydroxamates in the MMP-9 active site. Three histidines important for coordination with a zinc ion were shown.
There are seven reported MMP-9/ligand complexes. Among them, only 1GKC contains a native Glu402 residue.19 All other reported MMP-9/ligand complexes such as 2OVX, 2OVZ, 2OW0, 2OW1, 2OW2,15 and 1GKD(19) contain an E402Q mutant, in which Glu402 was mutated to the inactive Gln402 to prevent MMP-9 autodegradation during the crystallization process. It would be interesting to find out whether E402Q mutant has any effect on the binding.
With the aim of determining the effect of ionization of hydroxamates and the effect of the E402Q mutant on ligand binding, we carried out dockings studies of 47 MMP-9 inhibitors and 117 druglike molecules against MMP-9 proteins using the Glide program in Schrödinger software suite.20 Figure 3 shows the scaffolds of all 47 MMP-9 inhibitors, which were extracted from refs (12) and (13). The detailed structures of 47 MMP-9 inhibitors are listed in Tables S1−S4 in the Supporting Information.
Figure 3.
Template structures of MMP inhibitors (1−47) used in the docking study.
All 47 MMP-9 ligands were built based on the hydroxamate in 2OW1.15 Two lists of libraries were prepared as follows: one with hydrogen atom on the hydroxamic acid group (protonated model) and another characterized with a deprotonated oxygen atom (deprotonated model, Figure 2). One hundred seventeen druglike molecules were also docked to the model proteins to determine the enrichment factor (EF). To study the effect of Zn2+ and the E402Q mutation on the ligand binding, we built four model proteins for ligand docking: E402 with Zn2+ in the active site (E402_Zn), E402 without Zn2+ (E402_woZn), Q402 with Zn2+ (Q402_Zn), and Q402 without Zn2+ (Q402_woZn).
The effectiveness of Glide docking can be quantified by the EF. The EF is defined as the ratio of the percentage of active inhibitors in a subset (top 10% hits) divided by the percentage of active inhibitors in the entire database. EF has been used as a measurement of the efficiency of a docking program: the higher the EF is, the more accurate the docking program.
The number of active molecules in the top 10% best-fit docked poses of 164 molecules (ranked by the docking scores) and the EFs of the deprotonated and protonated ligands against four model proteins are listed in Table 1. The high EFs observed in model proteins E402_Zn with deprotonated ligands and E402_woZn with protonated ligands suggest that in the presence of Zn2+, the docking results from the deprotonated hydroxamates/retrohydroxamate models are in better agreement with experimental data than the protonated models, whereas in the absence of Zn2+ the latter yielded results closer to what is observed in experiments. The mutant E402Q yielded lower EFs than the wild-type MMP-9, indicating that this mutation may reduce binding affinity of MMP-9 inhibitors. Therefore, when modeling MMP-9 inhibition, it is very critical to keep in mind that whether MMP-9 is a wild-type or an E402Q mutant and that whether Zn2+ is present in the active site or not. All of these factors, along with the ionization state of the oxime hydroxyl group, determine the predictability of a docking program when it is used to evaluate the binding affinity on MMP-9/ligand interactions.
Table 1. Number of Active Molecules in the Top 10% (the Top 17 Hits) of Ligands Ranked by the Docking Scores and the EFs of 47 MMP-9 Inhibitors (Deprotonated and Protonated) against Four Model Proteinsa.
| protonated ligands |
deprotonated ligands |
|||||||
|---|---|---|---|---|---|---|---|---|
| MMP-9 | E402_Zn | E402_woZn | Q402_Zn | Q402_woZn | E402_Zn | E402_woZn | Q402_Zn | Q402_woZn |
| no. of active molecules | 2 | 11 | 2 | 8 | 13 | 5 | 11 | 2 |
| EF | 0.45 | 2.47 | 0.45 | 1.79 | 2.92 | 1.12 | 2.69 | 0.45 |
Note that the ideal EF for this study would be (17/17)/(43/164) = 3.81.
The docking scores of 47 MMP-9 inhibitors are listed in Table 2, and those of the 117 druglike molecules are listed in Table S5 in the Supporting Information. The docking scores of most potent MMP-9 inhibitors generally are more negative for the deprotonated ligands, indicating that the deprotonated ligand model in the presence of zinc ions is more in line with what might be observed experimentally.
Table 2. Docking Scores and in Vitro IC50 (nM) of 47 MMP-9 Inhibitors (Protonated and Deprotonated) against Four Model Proteins.
| protonated ligand models |
deprotonated ligand models |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| entry ID | MMP9 | E402_Zn | E402_woZn | Q402_Zn | Q402_woZn | E402_Zn | E402_woZn | Q402_Zn | Q402_woZn |
| 1 | 58 | −9.66 | −7.95 | −4.73 | −9.8 | −9.57 | −4.87 | −12.42 | −2.67 |
| 2 | 11 | −9.39 | −9.02 | −5.94 | −9.46 | −10.29 | −8.71 | −10.74 | −8.68 |
| 3 | 13 | −8.99 | −9.94 | −9.31 | −10.59 | −10.76 | −9.4 | −6.21 | −9.12 |
| 4 | 107 | −7.92 | −7.11 | −7.83 | −8.24 | −9.31 | −8.18 | −9.79 | −8.1 |
| 5 | 11 | −7.21 | −3.06 | −3.28 | −3.18 | −9.95 | −3.35 | −8.36 | −7.79 |
| 6 | 83 | −5.59 | −7.16 | −7.34 | −6.86 | −9.3 | −3.18 | −8.79 | −1.37 |
| 7 | 120 | −5.37 | −5.19 | −6.6 | −2.19 | −10.23 | −5.17 | −7.41 | −5.88 |
| 8 | 63 | −5.48 | −9.1 | −3.99 | −3.42 | −3.41 | −3.03 | −8.16 | −6.4 |
| 9 | 9 | −4.99 | −8.8 | −6.72 | −7.29 | −3.31 | −2.2 | −5.61 | −4.94 |
| 10 | 38 | −6.95 | −5.68 | −5.44 | −7.32 | −8.49 | −2.96 | −7.46 | −8.06 |
| 11 | 141 | −5.21 | −6.95 | −7.63 | −5.41 | −7.83 | −5.54 | −9.69 | −7.9 |
| 12 | 9 | −6.96 | −6.15 | −7.66 | −8.37 | −8.77 | −7.74 | −5.77 | −6.16 |
| 13 | 16 | −8.68 | −8.34 | −4.05 | −6.34 | −9.52 | −8.39 | −9.85 | −3.17 |
| 14 | 35 | −5.08 | −7.51 | −5.56 | −7.75 | −8.25 | −5.74 | −10.31 | −7.48 |
| 15 | 0.5 | −7.98 | −7.2 | −3.5 | −5.8 | −9.2 | −5.04 | −8.31 | −6.96 |
| 16 | 11 | −3.97 | −5.04 | −5.91 | −8.15 | −10.04 | −5.81 | −9.75 | −7.46 |
| 17 | 23 | −7.32 | −7.25 | −6.58 | −5.64 | −6.72 | −7.69 | −9.4 | −6.36 |
| 18 | 8 | −2.57 | −5.79 | −7.89 | −7.2 | −10.39 | −3.12 | −10.87 | −7.17 |
| 19 | 197 | −8.56 | −7.85 | −5.17 | −7.68 | −9.25 | −4.56 | −12.7 | −9.51 |
| 20 | 1.1 | −7.55 | −7.03 | −6.98 | −7.22 | −9.91 | 1.03 | −8.91 | −5.09 |
| 21 | 1.1 | −7.43 | −9.44 | −5.67 | −2.76 | −9.62 | −6.41 | −7.96 | −8.63 |
| 22 | 2 | −8.81 | −9.87 | −7.68 | −8.09 | −11.2 | −8.01 | −9.07 | −7.61 |
| 23 | 0.79 | −8.17 | −9.84 | −7.71 | −5.14 | −10.58 | −2.15 | −11.09 | −5.59 |
| 24 | 2 | −7.37 | −2.42 | −5.91 | −7.69 | −6.42 | −1.14 | −6.73 | −3.12 |
| 25 | 2 | −8.04 | −7.92 | −6.7 | −9.07 | −11.03 | −3.46 | −10.29 | −7.09 |
| 26 | 1.7 | −6.95 | −9.11 | −7.17 | −10.67 | −10.79 | −4.76 | −8.94 | −4.98 |
| 27 | 0.092 | −8.39 | −6.37 | −7.12 | −7.3 | −9.75 | −8.21 | −9.02 | −4.71 |
| 28 | 0.49 | −6.32 | −7.24 | −7.1 | −10.09 | −12.07 | −4.02 | −8.66 | −4.48 |
| 29 | 0.3 | −6.74 | −2.92 | −7.94 | −4.2 | −9.55 | −7.71 | −10.52 | −4.58 |
| 30 | 0.9 | −6.98 | −9.69 | −7.04 | −7.75 | −9.3 | −6.2 | −8.6 | −8.4 |
| 31 | 0.26 | −6.98 | −2.93 | −6.83 | −7.12 | −7.82 | 0.4 | −8.69 | −3.86 |
| 32 | 1.7 | −7.03 | −7.96 | −6.81 | −4.24 | −9.53 | −1.19 | −8.76 | −4.82 |
| 33 | 0.9 | −7.18 | −9.48 | −7.05 | −8.05 | −9.42 | −3.87 | −8.68 | −4.38 |
| 34 | 0.53 | −8.42 | −6.997 | −8.59 | −9.48 | −11.1 | −2.29 | −9.41 | −5.43 |
| 35 | 0.98 | −8.48 | −9.47 | −9.01 | −10.75 | −11.28 | −9.31 | −12.69 | −6.81 |
| 36 | 37 | −8.45 | −11.61 | −9.11 | −9.94 | −9.96 | −5 | −9.51 | −5.85 |
| 37 | 2.2 | −7.86 | −3.42 | −7.73 | −7.31 | −11.36 | 2.06 | −10.83 | −4.19 |
| 38 | 4.1 | −7.98 | −4.83 | −7.17 | −4.66 | −10.49 | −1.5 | −9.51 | −10.49 |
| 39 | 0.5 | −7.55 | −5.09 | −6.93 | −8.38 | −9.54 | −2.88 | −8.87 | −4.47 |
| 40 | 1.4 | −7.34 | −8.65 | −6.95 | 0.51 | −6.22 | −4.96 | −5.65 | −0.68 |
| 41 | 0.34 | −8.76 | −8.39 | −9.01 | −9.77 | −10.24 | −4.85 | −8.09 | −8.82 |
| 42 | 1.4 | −7.97 | −5.37 | −7.51 | −7.64 | −10.06 | −0.28 | −9.63 | −4.29 |
| 43 | 1.8 | −7.64 | −8.3 | −7.43 | −8.52 | −9.24 | −6.72 | −8.46 | −6.34 |
| 44 | 37 | −7.73 | −6.68 | −7.06 | −8.02 | −8.79 | −5.83 | −9.9 | −6.69 |
| 45 | 3.9 | −7.08 | −7.19 | −7.44 | −7.52 | −6.82 | −5.83 | −8.14 | −7.05 |
| 46 | 2 | −7.17 | −5.58 | −7.35 | −4.67 | −8.73 | −6.84 | −7.75 | −4.8 |
| 47 | 4.1 | −7.95 | −10.75 | −8.93 | −8.8 | −10.59 | −8.62 | −11.07 | −4.12 |
We built a pharmacophore model based on the five potent inhibitors (compounds 22, 23, 28, 34, and 35) using the Pharmacophore Query protocol in MOE.21 Our pharmacophore model shows that two aromatic rings, two H-bond receptors, and one anion group/H-bond acceptor are essential for the MMP-9 binding activities (Figure 4). The distances between any two pharmacophore features are listed.
Figure 4.
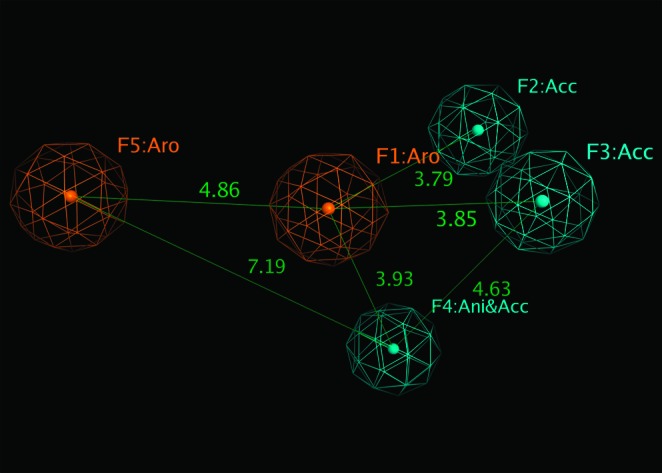
Pharmacophore model for MMP-9 inhibitors. Distance between pharmacophores (orange, aromatic rings; and cyan, H-bond acceptors) are listed (unit, Å).
We applied the pharmacophore model to search against a database of thousands of molecules. Nine molecules from commercially available sources as well as from our in-house synthesized compounds (Figure 5) were identified to satisfy this pharmacophore model. We carried out antiproliferative activities assay—the SVR proliferation assay, which was developed by Arbiser et al.22 and has been described in our previous papers.23 This assay is a direct measure of the candidate compound to serve as a potential antiangiogenic and antitumor agent.
Figure 5.
Chemical structures of compounds identified in the SVR proliferation assay.
The SVR proliferation assay shows that compounds A, B, E, and H had a significant effect on antiproliferative activities, that is, significantly lower cell counts than the control curcumin after 48 h of incubation of the individual test compound at 20 μg/mL concentration (Figure 6). Curcumin is a polyphenol derivative in the plant turmeric. Curcumin was able to suppress proliferation of tumor cells and to down-regulate MMP-9, epidermal growth factor receptor (EGFR), NF-κB, and other enzymes.24,25 Our SVR proliferation assay shows that chalcone derivative compound E, a structural homologue of curcumin, exhibited greater antiproliferative activity than curcumin. The three other structurally diverse lead compounds are A, B, and H. We docked compounds A−H and curcumin to MMP-9 model E402_Zn. Our docking results show that curcumin forms H-bonds with MMP-9 via Ala191 and Leu188, compound A forms H-bonds with Tyr423, and compound H forms H-bonds with Arg424. These interacting residues were also observed in the crystal structure 2OW1 (MMP-9/ligand complex).15 The identification of these compounds as potential antiproliferative agents is not surprising since they are potential bidentate ligands (via the lone pair electrons on the nitrogen or oxygen atoms) that may efficiently coordinate with the zinc ions.
Figure 6.
SVR proliferation assay for nine compounds with curcumin as the control.
In summary, the presence of a zinc ion in the MMP-9 active site is critical because the catalytic Zn2+ participates in the MMP-directed substrate cleavage. With the presence of Zn2+ in the active site, it is important for hydroxamates/retrohydroxamates to adopt deprotonated forms. This would be applicable to other zinc-binding groups such as carboxylic acid-based inhibitors. A pharmacophore model built based on five potent MMP-9 inhibitors was used to identify four structurally diverse compounds with antiproliferative activities greater than that of curcumin. Whether the anti-proliferative activity is due to the inhibition of MMP-9 or other regulatory enzymes such as EGFR remains to be determined.
This paper was published ASAP on the Web on April 7, 2011, as a Technology Note. After ASAP, the paper was changed to a Letter. The corrected version was posted on April 26, 2011.
Supporting Information Available
Experimental methods, structures of compounds 1−47, and docking scores of druglike molecules. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was partially supported by the Research Corporation for Science Advancement.
Supplementary Material
References
- Whittaker M.; Floyd C. D.; Bowen P.; Gearing A. J. H. Design and therapeutic application of matrix metalloproteinase inhibitors. Chem. Rev. 1999, 99, 2735–2776. [DOI] [PubMed] [Google Scholar]
- Weinstat-Saslow D. L.; Zabrenetzky V. S.; vanHoutte K.; Frazier W. A.; Roberts D. D.; Steeg P. S. Transfection of thrombospondin 1 complementary DNA into a human breast carcinoma cell line reduces primary tumor growth, metastatic potential, and angiogenesis. Cancer Res. 1994, 54, 6504–6511. [PubMed] [Google Scholar]
- Johnson C.; Sung H. J.; Lessner S. M.; Fini M. E.; Galis Z. S. Matrix metalloproteinase-9 is required for adequate angiogenic revascularization of ischemic tissues: Potential role in capillary branching. Circ. Res. 2004, 94, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram M.; Sherer Y.; Shoefeld Y. Matrix metalloproteinase-9 and autoimmune diseases. J. Clin. Immunol. 2006, 26, 299–307. [DOI] [PubMed] [Google Scholar]
- Hamano Y.; Zeisberg M.; Sugimoto H.; Lively J. C.; Maeshima Y.; Yang C.; Hynes R. O.; Werb Z.; Sudhakar A.; Kalluri R. Physiological levels of tumstatin, a fragment of collagen IV α3 chain, are generated by MMP-9 proteolysis and suppress angiogenesis via αVβ3 integrin. Cancer Cell 2003, 3, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; DeBusk L. M.; Fukuda K.; Fingleton B.; Green-Jarvis B.; Shyr Y.; Matrisian L. M.; Carbone D. P.; Lin P. C. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell 2004, 6, 409–421. [DOI] [PubMed] [Google Scholar]
- Bergers G.; Brekken R.; McMahon G.; Vu T. H.; Itoh T.; Tamaki K.; Tanzawa K.; Thorpe P.; Itohara S.; Werb Z.; Hanahan D. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2000, 2, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardi V. C.; Kupriyanova T. A.; Deryugina E. I.; Quigley J. P. Human neutrophils uniquely release TIMP-free MMP-9 to provide a potent catalytic stimulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 20262–20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D.; Takai S.; Jin D.; Inagaki S.; Tanaka K.; Miyazaki M. Molecular mechanism of imidapril for cardiovascular protection via inhibition of MMP-9. J. Mol. Cell. Cardiol. 2007, 43, 670–676. [DOI] [PubMed] [Google Scholar]
- Becker D. P.; Villamil C. I.; Barta T. E.; Bedell L. J.; Boehm T. L.; DeCrescenzo G. A.; Freskos J. N.; Getman D. P.; Hockerman S.; Heintz R.; Howard S. C.; Li M. H.; McDonald J. J.; Carron C. P.; Funckes-Shippy C. L.; Mehta P. P.; Munie G. E.; Swearingen C. A. Synthesis and structure-activity relationships of β- and α-piperidine sulfone hydroxamic acid matrix metalloproteinase inhibitors with oral antitumor efficacy. J. Med. Chem. 2005, 48, 6713–6730. [DOI] [PubMed] [Google Scholar]
- Moroy G.; Denhez C.; Mourabit H. E.; Toribio A.; Dassonville A.; Decarme M.; Renault J.-H.; Mirand C.; Bellon G.; Sapi J.; Alix A. J. P.; Hornebeck W.; Bourguet E. Simultaneous presence of unsaturation and long alkyl chain at P1́ of Ilomastat confers selectivity for gelatinas A (MMP-2) over gelatinase B (MMP-9) inhibition as shown by molecular modeling studies. Bioorg. Med. Chem. 2007, 15, 4753–4766. [DOI] [PubMed] [Google Scholar]
- Aranapakam V.; Grosu G. T.; Davis J. M.; Hu B.; Ellingboe J.; Baker J. L.; Skotnicki J. S.; Zask A.; DiJoseph J. F.; Sung A.; Sharr M. A.; Killar L. M.; Walter T.; Jin G.; Cowling R. Synthesis and structure-activity relationship of α-sulfonylhydroxamic acids as novel, orally active matrix metalloproteinase inhibitors for the treatment of osteoarthritis. J. Med. Chem. 2003, 46, 2361–2375. [DOI] [PubMed] [Google Scholar]
- Wada C. K.; Holms J. H.; Curtin M. L.; Dai Y.; Florjancic A. S.; Garland R. B.; Guo Y.; Heyman H. R.; Stacey J. R.; Steinman D. H.; Albert D. H.; Bouska J. J.; Elmore I. N.; Goodfellow C. L.; Marcotte P. A.; Tapang P.; Morgan D. W.; Michaelides M. R.; Davidsen S. K. Phenoxyphenyl sulfone N-formylhydroxylamines (retrohydroxamates) as potent, selective, orally bioavailable matrix metalloproteinase inhibitors. J. Med. Chem. 2002, 45, 219–232. [DOI] [PubMed] [Google Scholar]
- Natchus M. G.; Bookland R. G.; Laufersweiler M. J.; Pikul S.; Almstead N. G.; De B.; Janusz M. J.; Hsieh L. C.; Gu F.; Pokross M. E.; Patel V. S.; Garver S. M.; Peng S. X.; Branch T. M.; King S. L.; Baker T. R.; Foltz D. J.; Mieling G. E. Development of new carboxylic acid-based MMP inhibitors derived from functionalized propargylglycines. J. Med. Chem. 2001, 44, 1060–1071. [DOI] [PubMed] [Google Scholar]
- Tochowicz A.; Maskos K.; Huber R.; Oltenfreiter R.; Dive V.; Yiotakis A.; Zanda M.; Bode W.; Goettig P. Crystal structures of MMP-9 complexes with five inhibitors: Contribution of the flexible Arg424 side-chain to selectivity. J. Mol. Biol. 2007, 371, 989–1006. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Hou T. J.; Qiao X. B.; Huai S.; Xu X. J. Binding affinity of hydroxamate inhibitors of matrix metalloproteinase-2. J. Mol. Model. 2004, 10, 112–120. [DOI] [PubMed] [Google Scholar]
- Gupta S. P.; Kumaran S. A quantitative structure-activity relationship study on some series of anthranilic acid-based matrix metalloproteinase inhibitors. Bioorg. Med. Chem. 2005, 13, 5454–5462. [DOI] [PubMed] [Google Scholar]
- Gupta S. P.; Kumar D.; Kumaran S. A Quantitative structure-activity relationship study of hydroxamate matrix metalloproteinase inhibitors derived from functionalized 4-aminoprolines. Bioorg. Med. Chem. 2003, 11, 1975–1981. [DOI] [PubMed] [Google Scholar]
- Roswell S.; Hawtin P.; Minshull C. A.; Jepson H.; Brockbank S. M. V.; Barratt D. G.; Slater A. M.; PcPheat W. L.; Waterson D.; Henney A. M.; Pauptit R. A. Crystal structure of human MMP9 in complex with a reverse hydroxamate inhibitor. J. Mol. Biol. 2002, 319, 173–181. [DOI] [PubMed] [Google Scholar]
- Glide, Schrödinger, LLC, Portland, OR. Web address: www.schrodinger.com, 2009. [Google Scholar]
- MOE software; Chemical Computing Group Inc.: Montreal, Canada, 2009. Web address: http://www.chemcomp.com. [Google Scholar]
- Arbiser J. L.; Moses M. A.; Fernandez C. A.; Ghiso N.; Cao Y.; Klauber N.; Frank D.; Brownlee M.; Flynn E.; Parangi S.; Byers H. R.; Folkman J. Oncogenic H-ras stimulates tumor angiogenesis by two distinct pathways. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T. R.; Hubbard R. B. IV; Ehlers T.; Arbiser J. L.; Goldsmith D. J.; Bowen J. P. Synthesis and biological evaluation of aromatic enones related to curcumin. Bioorg. Med. Chem. 2005, 13, 4007–4013. [DOI] [PubMed] [Google Scholar]
- Aggarwal B. B.; Kumar A.; Bharti A. C. Anticancer potential of curcumin: Preclinical and clinical studies. Anticancer Res. 2003, 23, 363–398. [PubMed] [Google Scholar]
- Mohan R.; Sivak J.; Ashton P.; Russo L. A.; Pham B. Q.; Kasahara N.; Raizman M. B.; Fini M. E. Curcuminoids inhibit the angiogenic response stimulated by fibroblast growth factor-2, including expression of matrix metalloproteinase gelatinase B. J. Biol. Chem. 2000, 275, 10405–10412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







