Abstract
Microorganisms are rarely found in isolation. Frequently, they live as complex consortia or communities known as biofilms. The microbes within these complex structures are typically enmeshed in a matrix of macromolecules collectively known as the extracellular polymeric substances (EPS). The last decade has seen enormous growth in the breadth and depth of biofilm-related research. An important area of focus has been the study of pure culture biofilms of different model species. This work has informed us about the different genetic determinants involved in biofilm formation and the environmental conditions that influence the process. These studies have also highlighted both species-specific aspects of biofilm development and common trends observed across many different organisms. This report highlights some exciting findings in recent biofilm-related research.
Introduction
Recent work in biofilm research has brought certain topics to the fore. Several labs have made key contributions to our understanding of how surfaces are sensed by bacteria (highlighted in Figure 1). These reports relate the signal transduction events involved in linking adherence to a specific physiological response. Scientists are very interested in discerning the molecular mechanisms involved in responding to surfaces as a way to combat biofilm formation for pathogenic species.
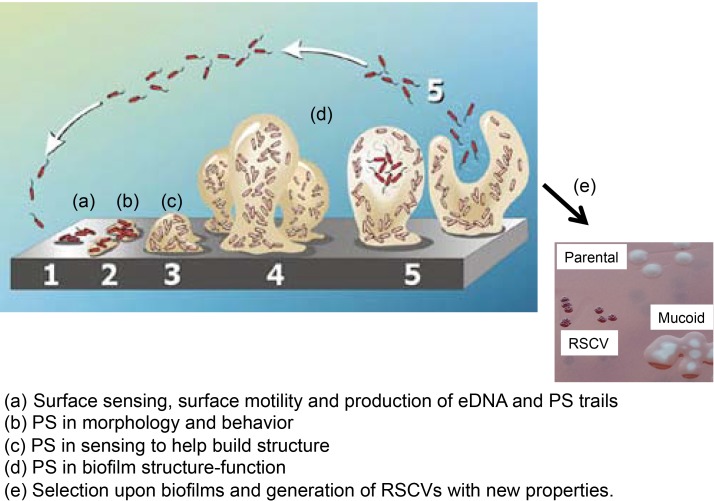
Figure 1. New events involved in biofilm development.
A traditional figure depicting different stages of biofilm development. Detailed in letters below are recent discoveries important for this process discussed in the text of the report. This includes the appearance of colony morphology variants that have been isolated from mature biofilms of Pseudomonas aeruginosa. Abbreviations: eDNA, extracellular DNA; PS, polysaccharide; RSCV, rugose small-colony variant.
The composition and function of biofilm EPS vary significantly among organisms and this has been the topic of several recent reviews [1-6]. Here, we will summarize recently described, exciting properties of one biofilm matrix component, exopolysaccharides (PSs). Three interrelated topics will be discussed: (a) linking PS structure with biological function; (b) PS signaling properties and (c) the roles of PSs in promoting biofilm morphology and behavior.
In addition, the importance of the secondary messenger, cyclic dimeric guanosine monophosphate (cyclic-di-GMP), in controlling the transition between biofilm and planktonic lifestyles is becoming increasingly apparent for many Gram-negative species [7-9]. The appearance of genetic variants with altered colony morphologies has been reported for a number of bacterial species and these variants have been linked to disease, cyclic-di-GMP signaling, and altered PS expression profiles. This short report will also address these themes with a focus on the model species Pseudomonas aeruginosa.
Recent insight into surface sensing
It is becoming increasingly clear that a wide variety of bacterial species are capable of sensing surfaces and adjusting their physiology accordingly. This is obviously important in that it allows the bacterium to transition rapidly to a biofilm lifestyle. Surface sensing presents an interesting challenge to bacteria. A surface represents a physical stimuli to a bacterium that encounters it; thus, the cell must in some way perceive such surface contact and transduce a signal.
There have been a number of exciting studies investigating the mechanisms involving surface sensing at early stages of biofilm development. These studies have revealed that different species employ similar mechanisms for surface sensing. A recent study by the Brun lab demonstrated that surface interactions result in pili-mediated impairment of flagellar rotation in the well-studied alpha Proteobacteria, Caulobacter crescentus, and Agrobacterium tumefaciens [10]. This results in the rapid production of polar PSs, which is important for the initial steps promoting tight, irreversible binding of bacteria to a surface. Further experiments by Fuqua et al. have shown that this response to a surface involves an increase in cyclic-di-GMP signaling [11].
In Bacillus subtilis, surface attachment also impairs the rotation of the flagellum. This in turn stimulates the DegS/U two-component system, which is a key regulator for biofilm formation in this organism [12]. An important mechanism for this species controls the initiation of biofilm formation and has been described by the Kearns lab, which found that a glycosyl transferase, EpsE, plays a dual role in controlling flagellar activity and biofilm EPS production [13,14]. In this system, EpsE can act as a flagellar “clutch” by glycosylating the flagellum and inhibiting motility. This is in addition to playing a role in biofilm PS production. Thus, B. subtilis has at least two systems governing the transition from the free swimming to the biofilm mode of growth.
Collectively, these studies reinforce the importance of coordinating the expression of swimming motility functions and the production of biofilms. They also call to mind the seminal studies in Vibrio parahaemolyticus [15,16]. These studies, like some of those mentioned above, related inhibition of polar flagellar rotation as a key signal perceived on a surface. In this instance, rotation of the flagellum promoted the expression of lateral flagella. Lateral flagella expression was shown to be a key feature of surface motility for this species. Recent work from the McCarter lab has demonstrated the role of c-di-GMP sensing and PS production as a part of the surface-sensing process [17].
It is important to note that studies by other labs have identified surface-sensing mechanisms that appear to be distinct from those involving flagellar motility. In P. aeruginosa, the Harwood lab has identified a chemotaxis-like sensing system important for abiotic surface sensing [18,19]. The Wsp system involves a distinct methyl-accepting chemotaxis proteins (MCP)/chemotaxis-like system that is involved in surface sensing. Once activated, the output of this system is a cyclic-di-GMP synthase called WspR. Thus, the Wsp system becomes activated upon surface contact, and this in turn stimulates cyclic-di-GMP signaling and biofilm formation. Another motility-independent surface-sensing mechanism involves the Cpx two-component system in Escherichia coli [20]. This system responds to different types of membrane stress and it appears that surface contact induces its activity. These studies suggest that surface sensing may involve different mechanisms implicated in detecting different aspects of surface association. Thus, there is potential that multiple features of surface interaction are capable of inducing a physiological response of a given cell to a surface.
Roles of exopolysaccharides in promoting biofilm morphology and behavior
While the roles of PSs in promoting adhesion and biofilm structure are well accepted, less is known of the roles PSs play in other surface-related processes. Biofilm formation in most organisms is believed to be a developmental process whereby individual cells migrate both to and on a surface and initiate a complex, yet coordinated, behavioral process ultimately resulting in a mature structure [4,21]. Our understanding of biofilm behavior has been inspired by parallel developmental and morphological systems studied in Myxococcus, Bacillus, and the Streptomyces. Recent parallel work in two microbial biofilm systems has provided mechanistic insights into how cooperative motility and PSs intersect.
B. subtilis has long been known to produce the molecule surfactin, which aids the bacterial colonies to spread on a surface in the absence of flow [22]. This spreading results from the hydration of secreted EPS [23]. Until recently, whether surfactin-mediated effects impact B. subtilis biofilm development was not known. Studies by Angelini et al. showed that surfactin is released in “waves” and this generates surface tension gradients that promote cooperative spreading [24]. The surface tension gradients developed were dependent on the bacterial biofilm geometry, which in turn required specific PS production. This results in a concentration gradient of surfactin distribution that results in surface tension forces that pull the biofilm cells outward, away from the colony center.
Recent studies from three groups have revealed similar behavior in P. aeruginosa involving the coordination of type IV pilin-dependent motility with extracellular DNA (eDNA) or PSs [25-27]. Two of these studies utilized individual cell tracking algorithms to monitor the behavior of each cell during the migration of newly colonized surfaces. Collectively, the reports show that coordinated groups of P. aeruginosa migrate along surfaces and create “furrows” or “trails” along which following cells preferentially migrate, providing positive feedback to the community. Cells actively released eDNA [25] or the PS Psl [26,27], which was necessary for the formation of furrows or trails, respectively. This behavior appears to be stimulated by nutritional deprivation that occurs within the microcolony [26]. Psl trails have been shown to impact the surface behavior of cells that subsequently encounter these trails. Future work is required to elucidate which specific signal coupled Psl/eDNA release and type IV pilin-dependent motility and how the signals regulate these two phenomena. This may in some ways be similar to the PS trails deposited by Myxococcus xanthus as it swarms on an agar surface.
Exopolysaccharide signaling properties
It has long been recognized that mammals are capable of specifically responding to microbial-derived PSs. Conserved molecular patterns present in the cell wall PSs of bacteria and fungi bind to germ line-encoded receptors on host cells. For example, bacterial peptidoglycan and fungal mannans or beta-glucans are recognized by Toll-like receptor-2 and -4 or C-type lectins, respectively. Recognition of these PS molecules leads to activation of host signaling pathways and the production of antimicrobial compounds [28,29]. Similar recognition and signaling occurs in plant-microbe interactions [30]. A recent discovery that certain plant-derived PSs trigger B. subtilis biofilm formation on plant roots provides another example of PS-derived signaling [31]. These cases illustrate the interplay of PS signals on the dynamic nature of microbe-host interactions. A new twist on PS-mediated signaling involving only prokaryotes has recently emerged in P. aeruginosa [32]. Depending upon the conditions, biofilms produced by this organism require one or more of three distinct PSs synthesized by the proteins encoded by the alg, pel, or psl gene clusters [33]. Irie et al. discovered that, in addition to playing a major structural role in biofilms, the self-produced matrix PS Psl acts as an extracellular signal to activate further matrix PS production [34]. Although the details regarding the pathway of Psl recognition and signaling remain to be discovered, Psl stimulated SadC and SiaD, two diguanylate cyclases that produced elevated levels of cyclic-di-GMP. Because elevated cyclic-di-GMP further stimulates matrix polysaccharide production [35], this regulatory circuit constitutes a unique feed-forward loop. These findings indicate that this biofilm matrix PS does not simply play a passive structural or protective role for biofilm cells. Instead, Psl appears to be a dynamic structure with signaling properties in some ways similar to those of the extracellular matrix of eukaryotic tissues.
Linking exopolysaccharide structure with biological function
PSs provide critical functions within biofilms. These include adhesion, cohesion, and aggregation, a barrier that protects cells against antimicrobials or biocides, retention of fluids and nutrients, and a role as a structural scaffold for the community. Several recent studies that defined interactions of biofilm matrix proteins with PSs or eDNA have provided mechanistic insights into how specific the biofilm matrix can modulate these disparate activities.
One such seminal study employed an in vivo labeling strategy coupled with super-resolution imaging microscopy to visualize interactions of four V. cholerae biofilm matrix components in living and developing biofilms [36]. Vibrio sp. are naturally aquatic organisms but can establish symbiotic or pathogenic interactions with eukaryotes. For V. cholerae, biofilm formation is a critical component of the transmission cycle as surface water in cholera-endemic areas harbor large aggregates of these bacteria bound to chitin on a variety of seawater fauna, including crabs, shrimp, and zooplankton [37]. These biofilms consist of at least three matrix proteins, RbmA, Bap1, and RbmC, along with the Vps PS. Berk and colleagues [36] showed that these components promoted three levels of spatial structure: cells, clusters of cells, and clusters of clusters. Cells were organized into clusters with boundaries defined by three-dimensional envelopes of Vps, RbmC, and Bap1. However, RbmA was found throughout the biofilm structure, presumably promoting clustering of cells and aggregates of cells. The amounts and timing of production of each component were critical as mutations in genes encoding these molecules greatly impacted biofilm structure.
Another elegant example linking biofilm structure with function involves the B. subtilis EPS matrix protein, BslA. B. subtilis biofilms are involved in a mutually beneficial association with plant surfaces. The matrix consists of PSs, amyloid-like proteins containing the structural protein TasA and an accessory protein BslA [38]. The surface of B. subtilis biofilms is extremely hydrophobic and displays persistent resistance to liquid, gas, and antimicrobial infiltration. However, until recently, it was unclear how the assembly of these matrix components resulted in biofilm structures with such resiliency. Two independent reports revealed that BslA is a major contributor to the hydrophobic surface properties of B. subtilis biofilms [39,40]. BslA was localized to the biofilm matrix via PS interactions and formed a shell around the periphery of biofilms [39,40]. The structure of BslA revealed it to be a member of the immunoglobulin (Ig) superfamily with numerous solvent-exposed hydrophobic amino acids that were critical for BslA-mediated biofilm structure and repulsion activities [39].
Recent studies with several bacteria have revealed eDNA as a critical component of the biofilm matrix (reviewed in [41]). Biofilm eDNA can be derived either from lysed bacterial cells in the community or from neutrophils that release it during a process known as NETosis [42]. While biofilms formed by some bacteria contain eDNA arranged in unique patterns [25,43], it is not clear how eDNA interacts with cells and other matrix components to promote biofilm structure. New studies have linked the DNABII family of DNA-binding proteins (e.g. integration host factor, or IHF) in promoting structural integrity of biofilms through its interaction with eDNA [44-47]. Moreover, treatment of biofilms formed by numerous bacteria with anti-IHF resulted in significant decreases in biofilm biomass and integrity, and active IHF immunization of animals experimentally infected with nontypeable Haemophilus influenzae biofilms enhanced disease resolution [44].
Colony morphology variants linking biofilms to disease
For several bacterial species, colony morphology variants have been isolated that exhibit elevated PS production. For example, colony morphology variants of Staphylococcus aureus and Staphylococcus epidermidis, Enterococcus faecium, Burkholderia sp., E. coli, and V. cholerae have been isolated from infections caused by these species. In several cases, the appearance of these colony variants has been correlated to disease and several of these variants exhibit biofilm-linked traits and heightened tolerance to antibiotic treatment and host defenses. Collectively, these studies indicate that the appearance of colony variants may represent a general adaptive strategy to the disease environment.
P. aeruginosa commonly undergoes a phenotypic conversion from a nonmucoid to mucoid colony morphotype [48-51] during cystic fibrosis (CF) airway infections. Mucoid colonies result from the overproduction of alginate, a PS that confers a selective advantage to the CF environment. Another colony morphology variant observed in CF is the rugose small-colony variant (RSCV) phenotype. RSCVs are actively selected for in CF and result from mutations that promote the overexpression of the Pel and Psl PS. RSCVs display increased biofilm formation and heightened resistance to antibiotics, suggesting that they represent a persistent subpopulation in CF [52-55]. RSCVs are also associated with increased tolerance of aminoglycoside, an important class of antibiotics used in the prophylactic therapy of patients with CF [53].
Recent findings have shed some light on the genetic basis for the RSCV phenotype. Mutations resulting in elevated levels of cyclic-di-GMP can cause the RSCV phenotype. Four sets of genetic elements have been linked to the RSCV phenotype in P. aeruginosa: the wsp operon, the tpb (also denoted aws or yfi) loci, dsbA, and genes of the rsm signaling pathway [19,56-60]. Both wsp and tpb loci contain a gene encoding a diguanylate cyclase, a type of enzyme that synthesizes cyclic-di-GMP. Mutations derepressing the activity of these diguanylate cyclases cause the increase in cyclic-di-GMP that results in the RSCV phenotype. DsbA is thought to modulate cyclic-di-GMP signaling by mediating the correct folding of a periplasmic repressor of the diguanylate cyclase TpbB. It is not yet clear how mutations in the rsm pathway are linked to cyclic-di-GMP signaling. RSCVs harboring mutations in the wsp or yfi loci have also been observed among CF isolates [61].
The adaptive advantage of the RSCV phenotype appears to be multifactorial. Besides promoting biofilm formation, these variants have other advantages, which should promote chronic infection. The RSCV phenotype has been shown to promote both oxygen utilization and maintenance of redox balance in cells growing under microaerobic conditions (likely similar to those encountered in the CF airways) [62,63]. In addition, experiments have shown that RSCVs reduce the oxidative burst produced by neutrophils compared with parental non-mucoid strains [64]. These experiments suggest that these variants are particularly well adapted to evading the host immune system.
Conclusions
Biofilm research continues to reveal interesting aspects of bacterial life on a surface. In this commentary, we have highlighted some of the recent interesting work that indicates that biofilm formation is an intricate process involving the sensing of surface-related stimuli and the production of an extracellular matrix that is surprisingly complex in its structure and function. The early days of biofilm research questioned whether the process involved a developmental program. It is now clear that it does indeed and that there are several general trends emerging that are important for a number of different species. The role of biofilms in disease is becoming clearer, and several newly identified features of biofilms contribute to the process. The next few years should be illuminating as researchers continue to unravel important aspects of biofilm development.
Acknowledgments
The authors acknowledge David G. Davies for his permission to use an adapted version of the image shown in Figure 1.
Abbreviations
- Cyclic-di-GMP
cyclic dimeric guanosine monophosphate
- CF
cystic fibrosis
- eDNA
extracellular DNA
- EPS
extracellular polymeric substance
- IHF
integration host factor
- PS
polysaccharide
- RSCV
rugose small-colony variant
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/6/26
Contributor Information
Daniel J. Wozniak, Email: daniel.wozniak@osumc.edu.
Matthew R. Parsek, Email: parsem@u.washington.edu.
References
- 1.Flemming H, Neu TR, Wozniak DJ. The EPS matrix: the “house of biofilm cells”. J Bacteriol. 2007;189:7945–7. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flemming H, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–33. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 4.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310–47. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vu B, Chen M, Crawford RJ, Ivanova EP. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14:2535–54. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Römling U, Simm R. Prevailing concepts of c-di-GMP signaling. Contrib Microbiol. 2009;16:161–81. doi: 10.1159/000219379. [DOI] [PubMed] [Google Scholar]
- 9.Yildiz FH. Cyclic dimeric GMP signaling and regulation of surface-associated developmental programs. J Bacteriol. 2008;190:781–3. doi: 10.1128/JB.01852-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13395976
- 11.Xu J, Kim J, Koestler BJ, Choi J, Waters CM, Fuqua C. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol. 2013;89:929–48. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718027908
- 12.Cairns LS, Marlow VL, Bissett E, Ostrowski A, Stanley-Wall NR. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol Microbiol. 2013;90:6–21. doi: 10.1111/mmi.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718050650
- 13.Guttenplan SB, Blair KM, Kearns DB. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010;6:e1001243. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718352807
- 14.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–8. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1116605
- 15.McCarter L, Hilmen M, Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988;54:345–51. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 16.Belas R, Simon M, Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–8. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira RBR, Chodur DM, Antunes LCM, Trimble MJ, McCarter LL. Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J Bacteriol. 2012;194:914–24. doi: 10.1128/JB.05807-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Güvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–73. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1103883
- 19.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–7. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718352808
- 20.Otto K, Silhavy TJ. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc Natl Acad Sci USA. 2002;99:2287–92. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1004058
- 21.Monds RD, O'Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Kinsinger RF, Shirk MC, Fall R. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol. 2003;185:5627–31. doi: 10.1128/JB.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seminara A, Angelini TE, Wilking JN, Vlamakis H, Ebrahim S, Kolter R, Weitz DA, Brenner MP. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci USA. 2012;109:1116–21. doi: 10.1073/pnas.1109261108. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/14001968
- 24.Angelini TE, Roper M, Kolter R, Weitz DA, Brenner MP. Bacillus subtilis spreads by surfing on waves of surfactant. Proc Natl Acad Sci USA. 2009;106:18109–13. doi: 10.1073/pnas.0905890106. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718352809
- 25.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, Monahan LG, Cavaliere R, Charles IG, Wand MP, Gee ML, Prabhakar R, Whitchurch CB. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci USA. 2013;110:11541–6. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718042394
- 26.Wang S, Parsek MR, Wozniak DJ, Ma LZ. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2013;15:2238–53. doi: 10.1111/1462-2920.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luijten E, Parsek MR, Wong GCL. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–91. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718008684
- 28.Sorbara MT, Philpott DJ. Peptidoglycan: a critical activator of the mammalian immune system during infection and homeostasis. Immunol Rev. 2011;243:40–60. doi: 10.1111/j.1600-065X.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- 29.Sukhithasri V, Nisha N, Biswas L, Anil Kumar V, Biswas R. Innate immune recognition of microbial cell wall components and microbial strategies to evade such recognitions. Microbiol Res. 2013;168:396–406. doi: 10.1016/j.micres.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Soto MJ, Domínguez-Ferreras A, Pérez-Mendoza D, Sanjuán J, Olivares J. Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol. 2009;11:381–8. doi: 10.1111/j.1462-5822.2009.01282.x. [DOI] [PubMed] [Google Scholar]
- 31.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA. 2013;110:E1621–30. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717998613
- 32.Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013;41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irie Y, Borlee BR, O'Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109:20632–6. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717999927
- 35.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 36.Berk V, Fong JCN, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–9. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717951614
- 37.Pruzzo C, Vezzulli L, Colwell RR. Global impact of Vibrio cholerae interactions with chitin. Environ Microbiol. 2008;10:1400–10. doi: 10.1111/j.1462-2920.2007.01559.x. [DOI] [PubMed] [Google Scholar]
- 38.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11:157–68. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DMF, Stanley-Wall NR. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci USA. 2013;110:13600–5. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718056350
- 40.Kobayashi K, Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 41.Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol. 2013 doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 42.Yipp BG, Kubes P. NETosis: how vital is it? Blood. 2013;122:2784–94. doi: 10.1182/blood-2013-04-457671. [DOI] [PubMed] [Google Scholar]
- 43.Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–75. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–37. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718352810
- 45.Gustave JE, Jurcisek JA, McCoy KS, Goodman SD, Bakaletz LO. Targeting bacterial integration host factor to disrupt biofilms associated with cystic fibrosis. J Cyst Fibros. 2013;12:384–9. doi: 10.1016/j.jcf.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718024340
- 46.Justice SS, Li B, Downey JS, Dabdoub SM, Brockson ME, Probst GD, Ray WC, Goodman SD. Aberrant community architecture and attenuated persistence of uropathogenic Escherichia coli in the absence of individual IHF subunits. PLoS ONE. 2012;7:e48349. doi: 10.1371/journal.pone.0048349. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718352811
- 47.Novotny LA, Amer AO, Brockson ME, Goodman SD, Bakaletz LO. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS ONE. 2013;8:e67629. doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718033261
- 48.Darzins A, Chakrabarty AM. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984;159:9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fyfe JA, Govan JR. Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J Gen Microbiol. 1980;119:443–50. doi: 10.1099/00221287-119-2-443. [DOI] [PubMed] [Google Scholar]
- 50.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–74. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Govan JR, Fyfe JA. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid from to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J Antimicrob Chemother. 1978;4:233–40. doi: 10.1093/jac/4.3.233. [DOI] [PubMed] [Google Scholar]
- 52.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces "insurance effects" in biofilm communities. Proc Natl Acad Sci USA. 2004;101:16630–5. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1022613
- 53.Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740–3. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1005946
- 54.Kirisits MJ, Prost L, Starkey M, Parsek MR. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71:4809–21. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starkey M, Hickman JH, Ma L, Zhang N, Long S de, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492–503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78:158–72. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malone JG, Jaeger T, Manfredi P, Dötsch A, Blanka A, Bos R, Cornelis GR, Häussler S, Jenal U. The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 2012;8:e1002760. doi: 10.1371/journal.ppat.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718352815
- 58.Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C, Kaever V, Landmann R, Jenal U. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 2010;6:e1000804. doi: 10.1371/journal.ppat.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885) PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol. 2011;13:3128–38. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 61.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–92. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13892
- 62.Kempes CP, Okegbe C, Mears-Clarke Z, Follows MJ, Dietrich LEP. Morphological optimization for access to dual oxidants in biofilms. Proc Natl Acad Sci USA. 2014;111:208–13. doi: 10.1073/pnas.1315521110. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718206741
- 63.Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–80. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718352816
- 64.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 2012;14:95–106. doi: 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]