Abstract
Epigenetic mechanisms involving DNA methylation, histone modifications and noncoding RNAs regulate and maintain gene-expression states. Similar to genetic mutations, alterations in epigenetic regulation can lead to uncontrolled cell division, tumor initiation and growth, invasiveness and metastasis. Research in brain cancer, particularly gliomas, has uncovered global and gene-specific DNA hypomethylation, local DNA hypermethylation of gene promoters and the de-regulation of microRNA expression. Understanding epigenetic dysregulation in brain cancers has provided new tools for prognostication, as well as suggesting new approaches to therapy. There is significant interest in new sequencing-based technologies that map genetic and epigenetic alterations comprehensively and at high resolution. These methods are being applied to brain tumors, and will better define the contribution of epigenetic defects to tumorigenesis.
Keywords: DNA methylation, epigenetics, glioma, histone deacetylase, histone modification, microRNA, SAHA/vorinostat
Epigenetics is defined as mitotically heritable changes in gene expression that are not due to changes in the primary DNA sequence. Epigenetic mechanisms include enzymatic modification of DNA or associated histone proteins that together regulate and maintain gene-expression states, and have important roles in chromosome structure and stability. Epigenetic mechanisms are essential for mammalian development. The discovery of altered epigenetic profiles in human neoplasia has led to new integrative models in which both genetic and epigenetic mechanisms contribute to and interact in cancer, and perhaps many other common human diseases. Owing to their reversible nature, epigenetic alterations are being targeted for reversal by therapeutic agents in cancer clinical trials. Here, we discuss the contribution of epigenetic mechanisms to brain development and tumorigenesis, how these epigenetic modifications are used for diagnosis and prognosis, and how their reversal may have therapeutical value.
Epigenetics
DNA methylation involves the covalent, enzymatic addition of a methyl group to cytosine in DNA. In adult tissues, methylation occurs almost exclusively at a cytosine followed by guanine, or CpG dinucleotide. Presumably as a result of deamination of methylated CpGs over evolutionary time, CpG dinucleotides are fivefold under-represented in the human genome. CpG islands are an exception, as they are regions of several hundred base pairs to a few kilobases in length that contain a relative abundance of CpGs and higher GC content [1]. DNA methylation in promoter regions blocks transcription and suppresses activity of transposable elements, regulates or maintains gene imprinting and is critical for chromosome stability [2,3].
DNA methyltransferase proteins, the DNMTs, catalyze the addition of the methyl group to DNA and maintain DNA methylation patterns. DNMT3A and DNMT3B are de novo methyltransferases, which establish new methylation marks at unmethylated sites, while DNMT1 is the maintenance methyltransferase and maintains the pattern of DNA methylation after DNA replication [4,5]. DNMT3L, another member of the DNMT family, shares homo logy to DNMT3A and DNMT3B through a cysteine-rich region, but lacks a catalytic domain [4]. Recent structural analysis in mice has demonstrated that DNMT3A and DNMT3L can form a complex that has two active sites and can bind DNA [6]. Ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1) colocalizes with DNMT1 throughout S phase and may facilitate DNMT1 interaction with chromatin [7]. Furthermore, UHRF1 contains an SRA domain that has a strong preference for hemimethylated DNA, suggesting that it might help recruit DNMT1 during DNA replication [7].
The regulation of de novo and maintenance methylation has been a focus for many years; however, passive and active demethylation also play a vital role in cellular function and development of an organism [8,9]. Active DNA demethylation also takes place during postnatal stages at specific gene loci, in post-mitotic neurons upon induction of synaptic activity [10], and in T cells upon anti-CD3 mediated activation [11]. Thus, DNA demethylation appears to be a more dynamic and regulated process than previously appreciated. To date, an enzyme capable of directly removing a methyl group from cytosine has not been identified; by contrast, indirect mechanisms of active demethylation via DNA repair mechanisms involve deaminases, glycosylases such as MBD4, the DNA repair exonuclease XPG and the GADD45 family of proteins [12–14].
In addition to 5-methycytosine, 5-hydroxy-methylcytosine (HMC) is found in genomic DNA from T2, T4 and T6 bacteriophages [15], but also in mouse brain and human embryonic stem cells, whilst being curiously absent from a human cancer cell line [16,17]. In mammals, the enzyme TET1 can convert 5-methylcytosine to HMC [17]. It may be that HMC can lead to genomic demethylation, either through a passive mechanism via the inability of DNMT1 to bind [18] or maintain methylation during mitosis, or through an active mechanism acting as an intermediate for a glycosylase [13]. Loss of TET1 and its family member TET2 are associated with malignancies, and a TET1 fusion to the histone methyltransferase MLL has been described in acute myeloid leukemia [19–21].
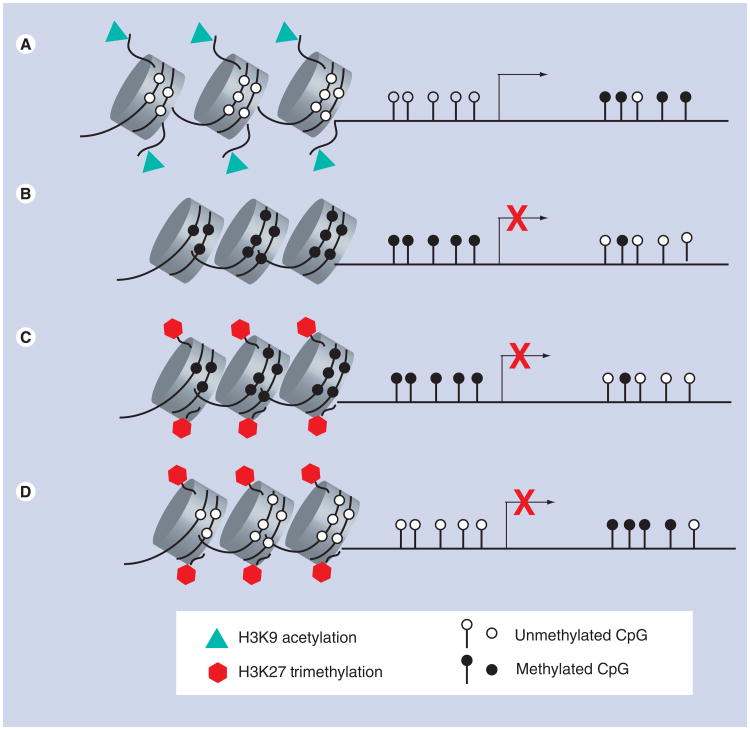
While modification of DNA by CpG methylation generally leads to gene silencing, post-translational modifications of histone proteins including acetylation, methylation, phosphorylation, ubiquitination or sumoylation can lead to either gene activation or repression, depending on which specific amino acid residue is modified. One of the best studied is the acetylation status of lysine residues, a reversible process catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs). The HAT-mediated addition of an acetyl group conveys structural changes to chromatin by decreasing the interaction between the negatively charged DNA backbone and the positively charged histone tail. This decrease in interaction can lead to a less compacted nucleosome, which permits greater access of transcription-factor complexes to the DNA (Figure 1A). Therefore, histone acetylation is a mark of actively transcribed genes and genes primed for induction [22,23]. Conversely, HDACs remove the acetyl group, potentially leading to repression of gene transcription.
Figure 1. DNA methylation and histone modifications at promoters of normal and cancer cells.
(A) At promoters of genes that are actively transcribed, the tails of histone proteins are marked with acetylation and the CpG dinucleotides are unmethylated. (B) Silenced genes can be marked by aberrant methylation of CpG dinucleotides alone, or (C) by histone H3 lysine 27 trimethylation and methylation of CpG dinucleotides or (D) By histone H3 lysine 27 trimethylation in the absence of CpG methylation. Initial reports in cancer cell lines suggest this pattern (D) is found in far fewer genes compared with aberrant DNA methylation. It is quite possible that other histone modifications change coordinately or inversely with DNA methylation, but have not yet been studied in gliomas.
Methylation of histones is another form of epigenetic regulation that is reversible. Mono-, di- and tri-methylation on histone H3 at lysine 4 (H3K4) is associated with transcriptional activation, while the di- and tri-methylation on histone H3 at lysine 9 (H3K9me2 and H3K9me3) is indicative of transcriptional inhibition [24]. Another repressive mark, histone H3 lysine 27 trimethylation (H3K27me3), is placed by the polycomb group complex (PcG). PcG proteins are responsible for the initiation of heritable long-term silencing of gene expression [25]. H3K27me3 in cancer cells can recruit DNA methyltransferases to gene promoters, potentially leading to de novo methylation and gene silencing (Figure 1C) [26–28]. However, a subset of inactive genes marked by H3K27me3 in cancer cells do not undergo de novo methylation (Figure 1D). Genes with H3K27me3 but without DNA methylation have also been observed in normal cells [29]. Histone methylation is also reversible by histone demethylases. Lysine-specific histone demethylase 1 (LSD1/KDM1) can remove mono- and dimethylated lysines, while the JmjC-domain histone methylase family (JHDMs) can remove mono-, di- and tri-methyl groups from different lysines on histone H3 [30,31].
Another mechanism that is potentially epigenetic and modulates gene expression involves small RNAs, in particular microRNAs (miRNAs). MiRNAs are short RNAs (∼21–22 nucleotides) that bind to the 3′ untranslated region of target mRNAs and prevent translation of the mRNA [32]. MiRNAs can have multiple mRNA targets, and individual mRNA can be targeted by multiple miRNAs. MiRNAs have the capacity to fine-tune gene-expression programs and cell function through cell-type-specific and temporal regulation of their expression, which itself can be deregulated through epigenetic mechanisms in brain tumors.
Epigenetics in the CNS
DNA methylation plays an integral part in the development and function of the mammalian CNS, and research in this area may shed light on the causes and consequences of aberrant epigenetic mechanisms in brain tumors. Early studies reported that in mice Dnmt1 mRNA is expressed at an unexpectedly high level in postmitotic neurons, suggesting a role for DNMT1 in postnatal brain function [33]. Coupled with a later finding that DNMT3A is also expressed in the postnatal brain, it appears that DNA methylation might be important for neuronal maturation in the adult CNS [34]. Conditional removal of DNMT1 in mouse CNS precursor cells leads to global demethylation and perinatal death from a defect in neural control of respiration [35]. Thus, in addition to the unanticipated function in postmitotic cells of the adult brain, DNA methyltransferases are important for the normal development of the CNS.
The de novo DNMTs, DNMT3A and DNMT3B, are expressed in the developing murine CNS, with DNMT3B found during embryonic day E11 to E15, while DNMT3A is expressed from early neurogenesis into the postnatal CNS [34]. Interestingly, DNA methylation can also influence the timing of CNS development through regulation of the JAK–STAT pathway [36,37]. STAT3, an important member of this pathway, also plays an important role in gliomagenesis [38–40].
Proteins that bind to methylated DNA are also involved in epigenetic regulation of CNS development and function. Mutations in methyl CpG binding protein 2 (MECP2) are known to cause Rett syndrome, a developmental disease associated with mental retardation [41]. Mice with generalized or CNS-specific deletions of Mecp2 have virtually indistinguishable phenotypes, suggesting the function of MeCP2 is primarily related to development of the CNS [42–44]. In one of these mouse models, point mutations in Mecp2 lead to increased histone H3 acetylation in the cerebellum and cortex [44]. Methyl CpG binding domain protein 2 (MBD2) and MeCP2 also interact with HDACs to induce heterochromatin formation and gene silencing [45,46]. Taken together, these data provide further evidence that DNA methylation and its effectors are vital for CNS development and function.
Similar to methylation of DNA, histone modifications are also important in CNS functions, such as learning and memory [47]. The CREB-binding protein (CBP) exhibits HAT activity, and mutations of CBP underlie at least 50% of cases of Rubinstein–Taybi syndrome, a mental retardation disorder that also carries a risk of cancer, including medulloblastoma and glioma [48–50]. Histone methylation is also an important epigenetic mechanism for CNS function. Postmortem brain from individuals that had Huntington's disease and Friereich's ataxia exhibit misregulation of H3K9 methylation [51,52]. Mll1/KMT2A, a histone methyl-transferase and component of the trithorax chromatin remodeling complex, is essential for postnatal neurogenesis [53]. Similar to histone methylation, the demethylation of histones has a role in CNS function, as an X-linked mental retardation and autism gene, SMCX/KDM5C, is a Jumonji-containing protein that encodes an H3K4 demethylase [54,55]. These data suggest that specific post-translational histone modifications are crucial for the normal development and function of the CNS.
Gliomas & epigenetics
While the role of epigenetics in the development and progression of gliomas, particularly of glioblastoma multiforme (GBM), has been studied for many years, many new findings are still emerging. Alterations in DNA methylation, histone modifications and miRNA expression are found in GBMs, although how they arise and what effects they have on tumorigenesis remain under active investigation.
DNA hypomethylation in gliomas
Hypomethylation of genomic DNA has been found in many different cancers [56], including GBMs, where up to 80% exhibit demethylation compared with normal brain [57]. The amount of global DNA hypomethylation in GBMs can vary, from very little change to as much as 50% relative to normal brain [57], which is equivalent to approximately 10 million CpG sites in a single tumor. Loss of methylation is found at repetitive elements, as well as at single copy loci [57]. One particular gene, encoding the cancer-testis antigen MAGEA1, was demethylated and expressed in a subset of primary GBMs and in glioma cell lines [57]. GBMs with severe loss of global methylation are also among the most proliferative of these aggressive tumors [57,58]. While these studies demonstrate a role for the maintenance of global methylation levels, the exact cause of hypomethylation in GBMs and other tumors is not well understood. So far, loss of DNMT proteins has not been observed in tumor cells. However, the potential for counterbalancing of methyltransferase activity through active DNA demethylation, or involvement of histone modifications in potential downstream DNA hypomethylation, have not been ruled out.
DNA hypermethylation in gliomas
In addition to local and global hypomethylation, locus-specific hypermethylation is also found in GBMs. CpG islands in 5′-promoter regions are predominantly unmethylated in normal cells, but undergo hypermethylation in cancer cells (Figure 1B) [59], although the causative mechanism(s) is not fully understood. Many different pathways are affected by hypermethylation-related gene silencing in gliomas, including cell-cycle regulation, apoptosis, DNA repair, angiogenesis, drug resistance and invasion. For example, the tumor suppressor genes PTEN and RB are known to undergo DNA hypermethylation-induced silencing in GBM cell lines and hypermethylation in primary tumors [60,61]. PDCD4, a tumor suppressor that can suppress expression of target genes, is itself silenced in many cancers including gliomas through methylation of the 5′ CpG island [62]. Another candidate tumor suppressor, LRRC4, is silenced by CpG island promoter hypermethylation in both primary gliomas and glioma cell lines [63]. The imprinted gene PEG3, which has tumor-suppressor activity in glioma cell lines, exhibits both promoter hypo- and hyper-methylation in primary gliomas [64]. The changes in levels of PEG3 methylation are inversely correlated with gene expression, suggesting that the methylation status of the exonic CpG island can influence gene expression [64]. Hypermethylation of the promoter of the DNA repair protein ERCC1 may have a role in the development of resistance to cisplatin in gliomas, since the levels of ERCC1 mRNA are associated with sensitivity to cisplatin in glioma samples [65] and hypermethylation of ERCC1 in glioma cell lines and primary gliomas may confer resistance through suppression of ERCC1 mRNA and protein. CD133, a cell-surface marker that is expressed on neural progenitor cells and putative cancer stem cells, is silenced in both colon cancer and glioma cell lines by promoter CpG island methylation [66]. However, the data from primary gliomas and subsequent xenograft models are not as consistent as the cell lines, suggesting that the role of CD133 promoter methylation and expression in gliomas is not yet fully understood.
Many of these genes were chosen for investigation using a candidate gene approach, but it remains possible that a subset of glioma genes might be inactivated primarily by methylation, and thus, only through methylation screening could such genes be identified. Such methylation screens have uncovered new potential tumor suppressors in RUNX3 and TESTIN [67,68]. Another screen found that BEX1 and BEX2, novel X-linked genes, are hypermethylated and silenced in gliomas [69]. When re-expressed in cells, these genes led to increased sensitivity to chemotherapy-induced apoptosis [69]. While these screens identified candidate genes in glioma cell lines, complementary approaches have investigated aberrant DNA methylation in primary glioma samples. A study looking at 87 primary GBMs using a bead array system found 32 genes that exhibit changes in their methylation profile compared with a normal brain sample [70]. In this study, HOXA11, CD81, PR KCDBP, TES, MEST, TNFRSF10A and FZD9 were hypermethylated in more than 50% of the samples [70].
An integrated genomic and epigenomic approach using copy-number analysis and DNA-methylation scanning identified both SLC5A8 and WNK2 methylation in primary human gliomas [71,72]. Methylation of SLC5A8 is correlated with decreased expression, while re-expression of exogenous SLC5A8 led to inhibition of colony formation in vitro [71]. Mouse models of oligodendroglioma also exhibited silencing of mSLC5A8, suggesting a possible evolutionary conserved pathway for glioma formation or progression [71].
WNK2 is a member of the WNK family (WNK1–4), which is known to have genetic point mutations in many types of solid tumors [73–75]. In gliomas, WNK2 exhibits aberrant methylation of its promoter and concurrent silencing of gene expression. Reintroduction of WNK2 into glioma cell lines leads to decreased colony formation, which along with many examples of point mutations in other common cancer types, supports a role for WNK2 as a suppressor of cell growth [72]. On a pathway level, WNK2 epigenetic silencing may synergize with genetic amplification of EGFR, a potent oncogene in gliomas, as WNK2 suppresses the activity of MEK1 through the Rho–GTPase pathway mechanism, and reduces EGFR signaling [76]. These studies underscore the importance of integrative genetic and epigenetic screens to discover novel mutations or alterations, and how they give rise to brain cancers.
Hypermethylation of the O6-methylguanine-DNA-methyltransferase (MGMT) promoter is associated with increased sensitivity to combined chemo- and radio-therapy. MGMT is a cellular DNA repair protein that removes alkylation damage at the O6 position of guanine. Glioma patients are treated with chemotherapeutic drugs such as temozolomide (Temodar®) that induce alkylation damage at the O6 position of guanine, among other sites on DNA. If left unrepaired, the lesions lead to DNA crosslinks and induce apoptosis. While high levels of MGMT are correlated with resistance to alkylating agents in glioma cell lines and xenografts [77–81], promoter hypermethylation of MGMT is associated with increased patient survival when coupled with radiation and treatment with temozolomide [82,83]. The Cancer Genome Atlas Project analyzed MGMT methylation status in 91 GBMs, and discovered a link between MGMT promoter hypermethylation and the mismatch repair pathway [84]. Recurrent GBMs with methylated MGMT exhibited a high incidence of G–C to A–T transition mutations at non-CpG sites (81%), also seen in mismatch repair genes (MMR), similar to those reported for p53 [85]. This is in contrast to tumors with unmethylated MGMT, which do not have a high incidence of G–C to A–T transitions at non-CpG sites (23%), suggesting that MGMT methylation and MMR deficiency in the context of treatment influence the mutational spectrum [84].
Gliomas & histone modification changes
To date, there have only been a few studies investigating the role of histone modifications in tumorigenesis and progression, particularly for gliomas. Changes in some post-translational histone modifications are common in cancers, including the loss of H4K16 acetylation (H4K16ac) and H4K20 trimethylation (H4K20me3) [86], and genes encoding histone modifying enzymes can be mutated genetically. In addition, low levels of H3K18 acetylation (H3K18ac) are correlated with prostate tumor recurrence [87]. These results suggest an important role for maintaining normal levels of histone acetylation. There are 18 known mammalian HDACs, which can be classified into four main groups based on structure, cellular localization and homology. In GBMs, class II and IV HDAC mRNAs are downregulated relative to lower grade astrocytomas and normal brain tissue [88]. One study also observed increased H3 acetylation in GBM compared with normal brain [88]. However, this is in opposition to the results reported for other types of cancer [86,87], which exhibit a loss of histone H3 acetylation. Even though the role of histone acetylation in GBMs is not fully understood, new therapies (discussed in more detail below) involving HDAC inhibitors (HDACi) are currently in clinical trials and others are in development [89–96].
Histone modifications and DNA methylation are interdependent at the level of the marks, but also at the level of protein–protein interaction between the histone- or DNA-modifying enzymes. For example, suppression of DNMT1 or DNMT3b in glioma cell lines led to re-expression of three genes, DUSP5, SDC2 and TMTC1 [97], but surprisingly, DNA hypermethylation at these promoters was unchanged. Instead, a reduction of H3K9me2, H3K9me3 and H3K27me3 levels was observed for the three genes [97]. Although it is not clear how altered DNA methyltransferase levels impacted these specific histone modifications, a mechanism involving local decreased DNA methylation is unlikely.
Deregulation of miRNAs in gliomas
Studies investigating the roles of miRNAs in gliomas are just emerging. While not strictly defined as an epigenetic mechanism, in model organisms miRNAs regulate heterochromatin formation, and in mammals miRNA can influence expression of many genes without altering the genetic sequence. MiRNA profiling has shown that miR-21 is overexpressed in primary GBMs, early-passage GBM cultures, and GBM cell lines [98]. miR-21 is overexpressed in a variety of cancers, including breast, lung, colon, prostate, pancreas, ovarian and stomach cancer [99,100]. miR-21 targets the p53, TGF-β, and mitochondrial apoptosis tumor-suppressive pathways [101]. Knockdown of miR-21 leads to repression of cell growth, increased apoptosis, and cell-cycle arrest, as well as decreased ability to migrate and invade [101,102]. Expression of another miRNA, miR-7, is downregulated in GBMs, and reintroducing miR-7 into GBM cell lines results in decreased viability and invasiveness [103]. As a potential tumor suppressor, miR-7 inhibits the Akt pathway and suppresses EGF receptor expression [103]. However, others, miR-124a and miR-137 for example, exhibit decreased expression in both anaplastic astrocytomas and glioblastomas compared with normal brain tissue [104]. Treatment of glioblastoma cell lines with the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-aza) resulted in increased expression of miR-137, suggesting that its expression might be regulated by DNA methylation [104].
miR-128 is associated with epigenetic alternations by way of its interaction with a histone-modifying enzyme, and is downregulated in GBMs [105]. Re-expression of miR-128 in glioma cell lines leads to a decrease in Bmi-1 expression [105]. Bmi-1 is a member of the PcG family and also plays a role in stem-cell renewal [106,107]. The silencing of Bmi-1 via miR-128 results in a global reduction of H3K27me3 [105]. While these studies focus on specific targets for individual miRNAs, in general, multiple targets exist for individual miRNA. These studies underscore the importance of investigating miRNAs in brain cancers, both in terms of how they are dysregulated and the mRNAs they target.
Pediatric brain cancers & epigenetics
Medulloblastoma is a malignant tumor in the cerebellum that occurs primarily in children and carries 5-year survival rates of 70–80% for standard risk patients [108]. Sporadic forms of medullo blastoma are associated with loss of chromosomal arm 17p where p53 resides, although p53 mutations are not commonly associated with medulloblastoma [109]. Methylation screens of the 17p region led to the discovery of hypermethylated in cancer 1 (HIC1), a zinc finger transcriptional repressor that itself is activated by p53 and is hypermethylated in a variety of pediatric and adult tumors [110,111]. HIC1 targets at least two different genes, the class III HDAC SIRT1 and a proneural transcription factor, ATOH1 [112,113]. Hypermethylation and gene silencing of HIC1 is commonly found in medulloblastomas of all stages and pathology [114–116], and treatment with 5-aza can reactivate HIC1 [114,116].
Epigenetic screens have been used to identify numerous other genes that are misexpressed in medulloblastoma, such as SPINT2, S100 and MCJ. SPINT2, a serine protease inhibitor and putative tumor suppressor, exhibits promoter hypermethylation in 34.3% of medulloblastomas [117]. The hypermethylation leads to reduced SPINT2 expression, which is reversible by treatment with 5-aza [117]. Re-expression of SPINT2 leads to reduced cell motility, proliferation and anchorage-independent growth with an increase of overall survival time in xenograft models [117]. The S100 family of proteins is associated with malignant progression. In medulloblastoma cell lines, two members of this family (S100A6 and S100A10) exhibit promoter hypermethylation [118]. The hypermethylation of S100A6 is associated with the aggressive large cell/anaplastic morphophenotype [118]. MCJ, a member of the DNAJ protein family, is silenced via promoter methylation in medulloblastomas [119]. Interestingly, the methylation patterns of the MCJ promoter fall into two categories – complete methylation of the promoter CpG island or limited methylation concentrated at the 5′ end of the of CpG island [119]. Furthermore, epigenetic loss of MCJ expression can result from biallelic promoter methylation or methylation on one allele combined with genetic loss of the other allele [119]. Other genes such as DIKKOPF-1 and COL1A2 are also silenced via DNA methylation in medulloblastomas [120,121].
DNA methylation is not the only widespread epigenetic alteration found in medulloblastoma. A genome-wide SNP screen identified genomic regions that are altered in copy number, affecting genes that add, remove or ‘read’ histone lysine methylation [122]. Restoration of expression of one of these genes, L3MBTL, led to a decrease in proliferation of medulloblastoma cell lines [122]. The genetic alteration of such epigenetic-modifying genes leads to a global loss of H3K9me2, and might also result in a genome-wide redistribution or global loss of other histone methylation at specific lysine residues. Thus, alteration of multiple epigenetic mechanisms, either directly through epigenetic ‘mutations’ or indirectly through genetic alterations of genes encoding epigenetic modifying enzymes, contributes to the formation of medulloblastomas.
Prognosis & epigenetics
Despite the identification of a large number of epigenetic alterations in brain tumors, very little progress has been made in finding marks with prognostic value. The most well studied epigenetic mark for prognosis of treatment for gliomas is the methylation status of MGMT. Hypermethylation of the MGMT promoter region, when sufficiently dense, leads to MGMT silencing in gliomas, lymphomas, breast cancer, prostate cancer and retinoblastoma [123]. Hypermethylation of the MGMT promoter region along with radiation and treatment with temozolomide, is associated with longer survival of patients with GBMs and those low-grade gliomas treated with temozolomide only [82,124]. A subset of GBM patients receiving this therapeutic regimen can survive up to, and perhaps beyond 5 years [83]. It should be noted that the typical assay used to assess MGMT methylation, methylation-sensitive PCR (MSP), does not measure precisely within the core promoter, nor is it quantitative. These issues undoubtedly contribute to the observed lack of correlation between methylation, assessed by MSP, and MGMT expression, assessed by reverse transcription (RT)-PCR, immunohistochemistry or enzymatic activity. In contrast to GBM, hypermethylation of MGMT occurs later in progressive oligodendrogliomas, suggesting that its prognostic effect in oligodendrogliomas might be different than for GBMs and low-grade gliomas [125].
A second DNA methylation-related prognostic marker of GBM and astrocytoma patient survival is the level of p-SMAD2, an indicator of TGFβ activity, as high levels are associated with poor survival [126]. TGFβ is an inhibitor of proliferation in various cell types including astrocytes and is a putative tumor suppressor [127]. However, some tumors such as gliomas lose TGFβ-mediated suppression of proliferation and instead TGFβ then promotes proliferation, angiogenesis, invasion and metastasis [127]. The investigation into a potential ‘switch’ that causes TGFβ to undergo transition from tumor suppressor to oncogene, uncovered a correlation between PDGF-β induction and high p-SMAD2 levels. This induction is dependent on the methylation status of the PDGF-β promoter, as a hypermethylated promoter does not lead to induction by TGFβ and these tumors are typically less aggressive [126]. Thus, the methylation status of PDGF-β may influence the TGFβ signaling pathway in gliomas, and could be useful as a prognostic marker.
A third potential prognostic epigenetic mark is the methylation status of the target of methylation induced silencing 1 (TMS1/ASC) gene, a gene that can activate the NF-κB pathway [128]. In typical GBM patients compared to long-term survivors, TMS1/ASC promoter hypermethylation was found significantly more often in long-term survivors [128]. A mechanism by which promoter hypermethylation of TMS1/ASC associates with long-term survival requires further investigation.
A fourth epigenetic mark with a potential prognostic significance was recently identified in a retrospective study of 27 GBMs, which found methylation of the tumor suppressor p15 promoter in 37% of the samples. p15 is found on chromosome 9p, a region often deleted in GBM patients, and a region significantly associated with risk of developing GBM [129]. Patients with methylation of p15 had a significant shorter overall survival (16.9 vs 23.8 months) [130]. This study provides another possible prognostic marker for GBM, pending replication on a larger sample set.
Therapeutics & epigenetics
With the knowledge that many loci exhibit epigenetic alterations in brain cancers, it is no surprise that therapies that can reverse these changes are being pursued. In particular, the DNA methylation inhibitor decitabine (5-aza-2′-deoxycytidine; Dacogen®, MGI Pharma Inc., MN, USA) and the HDACi suberoylanilide hydroxamic acid (SAHA; vorinostat, Patheon, Inc., Ontario, Canada) are being used for cancer patients, although only vorinostat is currently in clinical trials for GBM.
Theoretically, modulation of DNA methylation could provide therapeutic benefit by reversing hypermethylation and restoring expression of silenced genes in gliomas. Treatment of gliomas in culture with decitabine decreased cell proliferation and increased apoptosis [131]. Treatment of glioma cell lines with decitabine and Taxol® (paclitaxel; Bristol-Myers-Squibb, NY, USA) can lead to silencing of telomerase and decreased cell viability [132]. However, decitabine treatment can also activate MGMT, which might increase resistance to alkylating agents such as temozolomide, depending on the relative balance of local and global effects. An alternative to decitabine involves reduction of DNMT1 levels through double-stranded (ds) RNA knockdown experiments [133]. This approach, coupled with either paclitaxel or temozolomide, results in decreased glioma cell viability [133]. Thus, in cell lines, the anti-tumor effect appears to predominate over potential pro-resistance effects when DNA methyltransferase is inhibited. Nevertheless, more appropriate in vitro and in vivo models are needed to confirm these results, as the epigenetic profile of glioma cell lines is substantially different from that of primary gliomas.
Inhibition of HDACs could also provide a therapeutic benefit alone, or in combination with temozolomide. HDACs catalyze the removal of acetyl groups from histone lysine residues, as well as nonhistone proteins. The HDAC family consists of 18 members in humans that are divided into five different classes. HDACs of class I (HDACs 1, 2, 3 and 8), class IIA (HDACs 4, 5, 7 and 9), class IIB (HDACs 6 and 10) and class IV (HDAC 11) all contain zinc in their active sites, while the class III HDACs (sirtuins) do not contain zinc. The removal of acetyl groups on core histone proteins can lead to more condensed chromatin that is not as accessible to transcription factors, which can result in gene silencing.
HDACis have been used to treat many different malignancies, though it is not yet clear exactly how HDACi functions on a cellular level, nor which HDAC or other protein is their most important target for anti-tumor activity [134]. Similarly, it is not yet known how extensively HDACi changes the global landscape of acetylation specifically, or how it may alter the entire epigenome secondary to modifying acetylation. One speculation is that HDAC inhibition results in increased acetylation in promoters and activates critical growth regulating or apoptotic genes [135]. Another speculation is that HDAC inhibition generically opens chromatin genome-wide to increase DNA damage by chemo therapeutic agents [135]. In any case, it is known that HDACi treatment synergizes with DNA damaging agents such as temozolomide in arresting glioma cells in vitro [91].
Clinical trials for glioma patients have included four different HDACi therapies (TABLE 1). One of these, vorinostat, is an inhibitor of class I and class II HDACs and binds to the zinc ion in the active site of HDACs [96]. Vorinostat has been approved by the US FDA for the treatment of cutaneous T-cell lymphoma [136]. Preclinical studies found that vorinostat induces apoptosis and inhibition of cell growth in glioma cells in vitro, ex vivo and in vivo [94,137,138]. These and other studies led to the inclusion of vorinostat in seven clinical trials for primary GBM patients, with promising results reported from a Phase II study [139]. In recurrent GBMs, treatment with vorinostat alone resulted in modest 6-month progression-free survival [139]. Histone acetylation and gene-expression analysis determined that vorinostat was affecting its intended target within the tumor [139], although the effect on nonhistone protein acetylation sites was not assessed.
Table 1.
Clinical trials using epigenetic therapy.
| Trial | HDAC class targeted | Disease | Clinical trial phase | Status | Protocol ID |
|---|---|---|---|---|---|
| Vorinostat, isotretinoin, carboplatin | I, II | Recurrent GBM | I, II | Active | NCT00555399 |
| Vorinostat, bevacizumab, temozolomide | I, II | Recurrent GBM | I, II | Active | NCT00939991 |
| Vorinostat, borezomib | I, II | Recurrent GBM | II | Active | NCT00641706 |
| Vorinostat, temozolomide | I, II | Malignant gliomas | I | Active | NCT00268385 |
| Vorinostat, temozolomide, radiation | I, II | GBM | I, II | Temporarily closed | NCT00731731 |
| Vorinostat | I, II | Progressive or recurrent GBM | II | Closed | NCT00238303 |
| Vorinostat, bevacizumab, irinotecan | I, II | Recurrent GBM | I | Closed | NCT00762255 |
| Panobinostat, bevacizumab | I, II | Recurrent high-grade glioma | I, II | Active | NCT00859222 |
| Panobinostat | I, II | Malignant brain tumor | II | Active | NCT00848523 |
| Valproic acid, temozolomide, radiation | I, IIa | GBM | II | Active | NCT00313664 |
| Valproic acid | I, IIa | High-grade glioma in children | II | Active | NCT00879437 |
| Valproic acid, etoposide | I, IIa | Neuronal tumors & brain metastases | I | Active | NCT00513162 |
| Valproic acid | I, IIa | Recurrent or refractory CNS tumors in children | I | Closed | NCT00107458 |
| Romidepsin | I, II, IV | Recurrent high-grade glioma | I, II | Closed | NCT00085540 |
GBM: Glioblastoma multiforme; HDAC: Histone deacetylases.
Trials are also currently underway for two additional HDACis, valproic acid (Depakene; Depakote, Abbott Laboratories, IL, USA) and panobinostat (LBH589; Novartis, Basel, Switzerland). Valproic acid is active against class I and IIA HDACs and is being tested against GBM in combination with temozolomide plus radiation, and in a broader second trial against neuronal tumors and brain metastases in combination with etoposide. The use of valproic acid and radiation is also being tested in children with high-grade gliomas. Panobinostat, a potent inhibitor of class I and II HDACs [140], is also in active clinical trials for gliomas. One trial is examining the effects of panobinostat in patients with recurrent malignant gliomas, while another trial is testing its effects in combination with bevacizumab in recurrent high-grade gliomas. A Phase I/II trial was conducted with depsipeptide (Romidepsin; FK-228, Gloucester Pharmaceuticals, MA, USA). Depsipeptide is active against class I HDACs, and depsipeptide monotherapy was tested against high-grade gliomas. Another HDACi not yet in clinical trials is pivaloyloxymethyl butyrate (AN-9), a derivative of butyrate. Butyrate is an aliphatic acid HDACis effective against class I and IIA HDACs. AN-9 demonstrates efficacy in GBM cell culture and animal models, with studies showing that AN-9 sensitized GBM mouse xenografts to radiation and showed decreased tumor growth and increased survival [91,141]. In addition, there are several HDACi that have shown efficacy against other forms of cancer in cell lines but have not yet been tested for gliomas, including belinostat (PXD101; Topotarget, Copenhagen, Denmark) [142,143] and OSU-HDAC42 [144].
An attractive idea for the therapeutic application is the combined treatment with a DNA methylation inhibitor and HDACi, as in vitro studies in gliomas and other tumor cells show they act synergistically [145–147]. Changes in gene expression or cell viability are observed when treating the cancer cell lines with either 5-aza or HDACi alone [145–147]. However, treatment with 5-aza and HDACi leads to substantially increased gene activation [145,147] or increased radiosensitivity [146]. Trials with valproic acid and decitabine are underway in acute myeloid leukemia and solid tumors. While the in vitro results are encouraging, there are no current clinical trials in gliomas for combined DNA methylation inhibitors and HDACi.
Epigenome analysis
As previously discussed, epigenome-wide screens have been employed to detect cancer-specific changes in DNA methylation, histone modifications and miRNA expression, typically by microarray approaches. However, 40–50% of the genome is composed of repetitive elements that cannot be studied by microarray, and thus nearly half of the genome has been ignored despite having important regulatory functions. However, recent advances in high-throughput sequencing present a new unbiased methodology to uncover epigenetic alterations in brain cancers. Original epigenetic screens in brain tumors were carried out with restriction landmark genome scanning, a technique that utilizes methylation-sensitive enzymes [148–153] that assessed approximately 1000 sites for each enzyme combination. More recent techniques for mapping genome-wide DNA methylation include methylated DNA immunoprecipitation or methylation-sensitive restriction enzymes, coupled with either DNA microarrays or massively parallel sequencing [154–157]. Alternatively, the gold-standard bisulfte conversion has been used in reduced representation bisulfte sequencing (RRBS) [158,159]. These array and sequencing technologies can also be applied to mapping histone modifications and small RNAs [160,161]. Thus, there are many possibilities for uncovering new epigenetic alterations with prognostic value in human brain tumors.
Future perspective
With the advances in high-throughput sequencing, it appears that a much more detailed and comprehensive analysis of glioma epigenetics is possible in the next few years. In particular, genome-wide maps of DNA methylation, histone modifications and miRNA expression can be generated from primary tumors and compared with normal brain tissue. The hope is that these genome-wide comparisons can uncover epigenetic marks that might offer better prognosis and/or targets for therapeutics, as well as advancing our understanding of the epigenetic and genetic underpinnings of drug response and gliomagenesis.
Epigenetic therapies for gliomas are currently limited to HDACi. However, it remains to be seen if other histone modifications are aberrant in gliomas and whether these could be reversed for therapeutic benefit. In addition, the lack of an effective DNA demethylating agent with low toxicity and ease of delivery to the brain has prevented more widespread use of DNA methylation-blocking therapies. However, as previously mentioned, clinical trials investigating the combined effects of decitabine and valproic acid in other tumor types are ongoing. This combined approach of altering DNA methylation and histone acetylation, which results in synergistic effects in vitro at lower doses, might reduce toxicity and increase efficacy sufficiently to promote clinical trials in brain tumors. Furthermore, the use of small RNAs as therapeutic agents is becoming more realistic with improved delivery modalities, and could be used to target the mRNA corresponding to epigenetic modifying enzymes. Currently, there are multiple clinical trials of miRNA- and siRNA-based therapies against malignancies, though none for gliomas. With more specific knowledge of critical pathways affected by epigenetic alterations, new small RNA-type therapies may be designed to be more target-specific. Recent work to design better HDACi drugs has led to OSU–HDAC42, an HDACi that may be more potent than SAHA [144].
Executive Summary.
Gliomas & epigenetics
Genome-wide scans and candidate approaches have identified genes that exhibit promoter hyper- and hypo-methylation in gliomas compared with normal brain.
Relatively little is known about changes in post-translational histone modifications in primary gliomas, although histone deacetylase (HDAC) inhibitors are already in clinical trails for glioblastoma multiforme (GBM) patients.
miRNA expression can be regulated by DNA methylation and microRNAs can also target proteins that modulate histone modifications in gliomas.
Understanding the epigenetic mechanisms present in brain development may shed light on their contribution to gliomagenesis.
Prognosis
Promoter region methylation of MGMT is a associated with a longer survival of patients with GBMs and low-grade gliomas treated with temozolomide and radiation.
New epigenetic marks of potential prognostic significance include the promoter methylation status of PDGF-β, TMS1/ASC and p15.
Therapeutics
HDAC inhibitors are promising therapeutics for treatment of gliomas.
Vorinostat is in clinical trials for GBM patients.
Combined DNA methylation inhibitors and HDAC inhibitors are synergistic in gene activation in glioma and other cancer cell lines, but have not yet been tested in glioma clinical trials.
Epigenome analysis
Recent advances in high-throughput sequencing allow high-resolution comprehensive mapping of DNA methylation and histone modifications in normal brain and glioma genomes. As these methods are sequencing based, genetic mutations can be discovered simultaneously.
Footnotes
Financial & competing interests disclosure: The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Shaun D Fouse, Brain Tumor Research Center, Department of Neurosurgery, Helen Diller Family Comprehensive Cancer Center, University of California San Francisco, San Francisco, CA 94158, USA.
Joseph F Costello, Brain Tumor Research Center, Department of Neurosurgery, Helen Diller Family Comprehensive Cancer Center, University of California San Francisco, San Francisco, CA 94158, USA, Tel.: +1 415 514 1183, Fax: +1 415 502 6779.
Bibliography
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.McClelland M, Ivarie R. Asymmetrical distribution of CpG in an ‘average’ mammalian gene. Nucleic Acids Res. 1982;10:7865–7877. doi: 10.1093/nar/10.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 3.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 5.Bestor TH. Cloning of a mammalian DNA methyltransferase. Gene. 1988;74:9–12. doi: 10.1016/0378-1119(88)90238-7. [DOI] [PubMed] [Google Scholar]
- 6.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 8.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 10.Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 11.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 12.Barreto G, Schafer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 13.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Wyatt GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953;55:774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. Describes a novel base in eukaryotic DNA that is found in the adult CNS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. Describes a novel base whose formation is catalyzed by a protein with assoications with malignancies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 19.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 20.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 21.Viguie F, Aboura A, Bouscary D, et al. Common 4q24 deletion in four cases of hematopoietic malignancy: early stem cell involvement? Leukemia. 2005;19:1411–1415. doi: 10.1038/sj.leu.2403818. [DOI] [PubMed] [Google Scholar]
- 22.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 23.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 24.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 26.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 27.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 28.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 32.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 33.Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 34.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 35•.Fan G, Beard C, Chen RZ, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. First paper to demonstrate a crucial role for DNA methylation in CNS development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan G, Martinowich K, Chin MH, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK–STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 37.Takizawa T, Nakashima K, Namihira M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 38.Lo HW, Cao X, Zhu H, Ali-Osman F. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res. 2008;14:6042–6054. doi: 10.1158/1078-0432.CCR-07-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren W, Duan Y, Yang Y, Ji Y, Chen F. Down-regulation of Stat3 induces apoptosis of human glioma cell: a potential method to treat brain cancer. Neurol Res. 2008;30:297–301. doi: 10.1179/016164107X230784. [DOI] [PubMed] [Google Scholar]
- 40.Brantley EC, Nabors LB, Gillespie GY, et al. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res. 2008;14:4694–4704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 42.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 43.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 44.Shahbazian M, Young J, Yuva-Paylor L, et al. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 45.Ng HH, Zhang Y, Hendrich B, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 46.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 47.Jiang Y, Langley B, Lubin FD, et al. Epigenetics in the nervous system. J Neurosci. 2008;28:11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrij F, Giles RH, Dauwerse HG, et al. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 49.Miller RW, Rubinstein JH. Tumors in Rubinstein–Taybi syndrome. Am J Med Genet. 1995;56:112–115. doi: 10.1002/ajmg.1320560125. [DOI] [PubMed] [Google Scholar]
- 50.Roelfsema JH, Peters DJ. Rubinstein–Taybi syndrome: clinical and molecular overview. Expert Rev Mol Med. 2007;9:1–16. doi: 10.1017/S1462399407000415. [DOI] [PubMed] [Google Scholar]
- 51.Ryu H, Lee J, Hagerty SW, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc Natl Acad Sci USA. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Mahdawi S, Pinto RM, Ismail O, et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 53.Lim DA, Huang YC, Swigut T, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwase S, Lan F, Bayliss P, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 55.Adegbola A, Gao H, Sommer S, Browning M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) Am J Med Genet A. 2008;146A:505–511. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- 56.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 58.Yu J, Zhang H, Gu J, et al. Methylation profiles of thirty four promoter-CpG islands and concordant methylation behaviours of sixteen genes that may contribute to carcinogenesis of astrocytoma. BMC Cancer. 2004;4:65. doi: 10.1186/1471-2407-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 60.Baeza N, Weller M, Yonekawa Y, Kleihues P, Ohgaki H. PTEN methylation and expression in glioblastomas. Acta Neuropathol. 2003;106:479–485. doi: 10.1007/s00401-003-0748-4. [DOI] [PubMed] [Google Scholar]
- 61.Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest. 2001;81:77–82. doi: 10.1038/labinvest.3780213. [DOI] [PubMed] [Google Scholar]
- 62.Gao F, Wang X, Zhu F, et al. PDCD4 gene silencing in gliomas is associated with 5′CpG island methylation and unfavorable prognosis. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00497.x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Z, Li D, Wu M, et al. Promoter hypermethylation-mediated inactivation of LRRC4 in gliomas. BMC Mol Biol. 2008;9:99. doi: 10.1186/1471-2199-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Otsuka S, Maegawa S, Takamura A, et al. Aberrant promoter methylation and expression of the imprinted PEG3 gene in glioma. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:157–165. doi: 10.2183/pjab.85.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H, Shao C, Shi H, Mu Y, Sai K, Chen Z. Single nucleotide polymorphisms and expression of ERCC1 and ERCC2 vis-a-vis chemotherapy drug cytotoxicity in human glioma. J Neurooncol. 2007;82:257–262. doi: 10.1007/s11060-006-9290-2. [DOI] [PubMed] [Google Scholar]
- 66.Yi JM, Tsai HC, Glockner SC, et al. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer Res. 2008;68:8094–8103. doi: 10.1158/0008-5472.CAN-07-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mueller W, Nutt CL, Ehrich M, et al. Downregulation of RUNX3 and TES by hypermethylation in glioblastoma. Oncogene. 2007;26:583–593. doi: 10.1038/sj.onc.1209805. [DOI] [PubMed] [Google Scholar]
- 68.Kunitz A, Wolter M, van den Boom J, et al. DNA hypermethylation and aberrant expression of the EMP3 gene at 19q13.3 in human gliomas. Brain Pathol. 2007;17:363–370. doi: 10.1111/j.1750-3639.2007.00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Foltz G, Ryu GY, Yoon JG, et al. Genome-wide analysis of epigenetic silencing identifies BEX1 and BEX2 as candidate tumor suppressor genes in malignant glioma. Cancer Res. 2006;66:6665–6674. doi: 10.1158/0008-5472.CAN-05-4453. [DOI] [PubMed] [Google Scholar]
- 70.Martinez R, Martin-Subero JI, Rohde V, et al. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–264. doi: 10.4161/epi.9130. [DOI] [PubMed] [Google Scholar]
- 71.Hong C, Maunakea A, Jun P, et al. Shared epigenetic mechanisms in human and mouse gliomas inactivate expression of the growth suppressor SLC5A8. Cancer Res. 2005;65:3617–3623. doi: 10.1158/0008-5472.CAN-05-0048. [DOI] [PubMed] [Google Scholar]
- 72.Hong C, Moorefeld KS, Jun P, et al. Epigenome scans and cancer genome sequencing converge on WNK2, a kinase-independent suppressor of cell growth. Proc Natl Acad Sci USA. 2007;104:10974–10979. doi: 10.1073/pnas.0700683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davies H, Hunter C, Smith R, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 74.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 75.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moniz S, Matos P, Jordan P. WNK2 modulates MEK1 activity through the Rho GTPase pathway. Cell Signal. 2008;20:1762–1768. doi: 10.1016/j.cellsig.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 77.Hotta T, Saito Y, Fujita H, et al. O6-alkylguanine-DNA alkyltransferase activity of human malignant glioma and its clinical implications. J Neurooncol. 1994;21:135–140. doi: 10.1007/BF01052897. [DOI] [PubMed] [Google Scholar]
- 78.Belanich M, Pastor M, Randall T, et al. Retrospective study of the correlation between the DNA repair protein alkyltransferase and survival of brain tumor patients treated with carmustine. Cancer Res. 1996;56:783–788. [PubMed] [Google Scholar]
- 79.Jaeckle KA, Eyre HJ, Townsend JJ, et al. Correlation of tumor O6-methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: a Southwest Oncology Group study. J Clin Oncol. 1998;16:3310–3315. doi: 10.1200/JCO.1998.16.10.3310. [DOI] [PubMed] [Google Scholar]
- 80.Friedman HS, McLendon RE, Kerby T, et al. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol. 1998;16:3851–3857. doi: 10.1200/JCO.1998.16.12.3851. [DOI] [PubMed] [Google Scholar]
- 81.Silber JR, Blank A, Bobola MS, Ghatan S, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase-defcient phenotype in human gliomas: frequency and time-to-tumor progression after alkylating agent-based chemotherapy. Clin Cancer Res. 1999;5:807–814. [PubMed] [Google Scholar]
- 82.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 83••.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised Phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. Demonstrates how promoter hypermethylation of O6-methylguanine-DNA methyltransferase is associated with a better response to temozolomide. [DOI] [PubMed] [Google Scholar]
- 84••.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. Large-scale comprehensive analysis of 206 cases of glioblastoma multiforme (GBM), including sequence and epigenetic analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Esteller M, Risques RA, Toyota M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- 86.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 87••.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. Study correlates levels of histone modifications to outcome of prostate cancer patients, independent of tumor stage, prostate-specific antigen level or capsule invasion. [DOI] [PubMed] [Google Scholar]
- 88.Lucio-Eterovic AK, Cortez MA, Valera ET, et al. Differential expression of 12 histone deacetylase (HDAC) genes in astrocytomas and normal brain tissue: class II and IV are hypoexpressed in glioblastomas. BMC Cancer. 2008;8:243. doi: 10.1186/1471-2407-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lopez CA, Feng FY, Herman JM, Nyati MK, Lawrence TS, Ljungman M. Phenylbutyrate sensitizes human glioblastoma cells lacking wild-type p53 function to ionizing radiation. Int J Radiat Oncol Biol Phys. 2007;69:214–220. doi: 10.1016/j.ijrobp.2007.04.069. [DOI] [PubMed] [Google Scholar]
- 90.Camphausen K, Cerna D, Scott T, et al. Enhancement of in vitro and in vivo tumor cell radio sensitivity by valproic acid. Int J Cancer. 2005;114:380–386. doi: 10.1002/ijc.20774. [DOI] [PubMed] [Google Scholar]
- 91.Entin-Meer M, Yang X, VandenBerg SR, et al. In vivo effcacy of a novel histone deacetylase inhibitor in combination with radiation for the treatment of gliomas. Neuro Oncol. 2007;9:82–88. doi: 10.1215/15228517-2006-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim EH, Kim SU, Shin DY, Choi KS. Roscovitine sensitizes glioma cells to TRAIL-mediated apoptosis by downregulation of survivin and XIAP. Oncogene. 2004;23:446–456. doi: 10.1038/sj.onc.1207025. [DOI] [PubMed] [Google Scholar]
- 93.Sawa H, Murakami H, Kumagai M, et al. Histone deacetylase inhibitor, FK228, induces apoptosis and suppresses cell proliferation of human glioblastoma cells in vitro and in vivo. Acta Neuropathol. 2004;107:523–531. doi: 10.1007/s00401-004-0841-3. [DOI] [PubMed] [Google Scholar]
- 94.Ugur HC, Ramakrishna N, Bello L, et al. Continuous intracranial administration of suberoylanilide hydroxamic acid (SAHA) inhibits tumor growth in an orthotopic glioma model. J Neurooncol. 2007;83:267–275. doi: 10.1007/s11060-007-9337-z. [DOI] [PubMed] [Google Scholar]
- 95.Wetzel M, Premkumar DR, Arnold B, Pollack IF. Effect of trichostatin A, a histone deacetylase inhibitor, on glioma proliferation in vitro by inducing cell cycle arrest and apoptosis. J Neurosurg. 2005;103:549–556. doi: 10.3171/ped.2005.103.6.0549. [DOI] [PubMed] [Google Scholar]
- 96.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 97•.Foltz G, Yoon JG, Lee H, et al. DNA methyltransferase-mediated transcriptional silencing in malignant glioma: a combined whole-genome microarray and promoter array analysis. Oncogene. 2009;28:2667–2677. doi: 10.1038/onc.2009.122. Knockdown of DNA methyltransferase (DNMT)-1 or -3B leads to changes in gene expression in glioma cell lines involving changes in histone acetylation, but not DNA methylation. [DOI] [PubMed] [Google Scholar]
- 98.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 99.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Papagiannakopoulos T, Shapiro A, Kosik KS. microRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 102.Gabriely G, Wurdinger T, Kesari S, et al. microRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kefas B, Godlewski J, Comeau L, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 104.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105•.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. Links miR-128 to the expression of stem cell renewal factor Bmi-1 in gliomas. Implicates microRNAs in helping to make gliomas more ‘stem-cell’ like. [DOI] [PubMed] [Google Scholar]
- 106.Zencak D, Lingbeek M, Kostic C, et al. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J Neurosci. 2005;25:5774–5783. doi: 10.1523/JNEUROSCI.3452-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 108.Gilbertson RJ. Medulloblastoma: signalling a change in treatment. Lancet Oncol. 2004;5:209–218. doi: 10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- 109.Eberhart CG. Medulloblastoma in mice lacking p53 and PARP: all roads lead to Gli. Am J Pathol. 2003;162:7–10. doi: 10.1016/S0002-9440(10)63792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wales MM, Biel MA, el Deiry W, et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1:570–577. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- 111.Fleuriel C, Touka M, Boulay G, Guerardel C, Rood BR, Leprince D. HIC1 (hypermethylated in cancer 1) epigenetic silencing in tumors. Int J Biochem Cell Biol. 2009;41:26–33. doi: 10.1016/j.biocel.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 113.Briggs KJ, Corcoran-Schwartz IM, Zhang W, et al. Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev. 2008;22:770–785. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lindsey JC, Lusher ME, Anderton JA, et al. Identification of tumour-specific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis. 2004;25:661–668. doi: 10.1093/carcin/bgh055. [DOI] [PubMed] [Google Scholar]
- 115.Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–3797. [PubMed] [Google Scholar]
- 116.Waha A, Koch A, Meyer-Puttlitz B, et al. Epigenetic silencing of the HIC-1 gene in human medulloblastomas. J Neuropathol Exp Neurol. 2003;62:1192–1201. doi: 10.1093/jnen/62.11.1192. [DOI] [PubMed] [Google Scholar]
- 117.Kongkham PN, Northcott PA, Ra YS, et al. An epigenetic genome-wide screen identifies SPINT2 as a novel tumor suppressor gene in pediatric medulloblastoma. Cancer Res. 2008;68:9945–9953. doi: 10.1158/0008-5472.CAN-08-2169. [DOI] [PubMed] [Google Scholar]
- 118.Lindsey JC, Lusher ME, Anderton JA, Gilbertson RJ, Ellison DW, Clifford SC. Epigenetic deregulation of multiple S100 gene family members by differential hypomethylation and hypermethylation events in medulloblastoma. Br J Cancer. 2007;97:267–274. doi: 10.1038/sj.bjc.6603852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lindsey JC, Lusher ME, Strathdee G, et al. Epigenetic inactivation of MCJ (DNAJD1) in malignant paediatric brain tumours. Int J Cancer. 2006;118:346–352. doi: 10.1002/ijc.21353. [DOI] [PubMed] [Google Scholar]
- 120.Vibhakar R, Foltz G, Yoon JG, et al. Dickkopf-1 is an epigenetically silenced candidate tumor suppressor gene in medulloblastoma. Neuro Oncol. 2007;9:135–144. doi: 10.1215/15228517-2006-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Anderton JA, Lindsey JC, Lusher ME, et al. Global analysis of the medulloblastoma epigenome identifies disease-subgroup-specific inactivation of COL1A2. Neuro Oncol. 2008;10:981–994. doi: 10.1215/15228517-2008-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122•.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. Amplification or deletion of multiple genes can lead to dysregulation of histone lysine methylation in medulloblastoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 124.Everhard S, Kaloshi G, Criniere E, et al. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60:740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 125.Lavon I, Zrihan D, Zelikovitch B, et al. Longitudinal assessment of genetic and epigenetic markers in oligodendrogliomas. Clin Cancer Res. 2007;13:1429–1437. doi: 10.1158/1078-0432.CCR-06-2050. [DOI] [PubMed] [Google Scholar]
- 126••.Bruna A, Darken RS, Rojo F, et al. HighTGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. Detailed analysis of TGFβ signaling pathway identifies new prognostic mark for gliomas that is downstream of epigenetic control of PDGF-β expression. [DOI] [PubMed] [Google Scholar]
- 127.Seoane J. Escaping from the TGFβ anti-proliferative control. Carcinogenesis. 2006;27:2148–2156. doi: 10.1093/carcin/bgl068. [DOI] [PubMed] [Google Scholar]
- 128.Martinez R, Schackert G, Esteller M. Hypermethylation of the proapoptotic gene TMS1/ASC: prognostic importance in glioblastoma multiforme. J Neurooncol. 2007;82:133–139. doi: 10.1007/s11060-006-9264-4. [DOI] [PubMed] [Google Scholar]
- 129••.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. Genome-wide association study that found three loci that are associated with risk for glioblastoma multiforme (GBM) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wemmert S, Bettscheider M, Alt S, et al. p15 promoter methylation – a novel prognostic marker in glioblastoma patients. Int J Oncol. 2009;34:1743–1748. doi: 10.3892/ijo_00000305. [DOI] [PubMed] [Google Scholar]
- 131.Eramo A, Pallini R, Lotti F, et al. Inhibition of DNA methylation sensitizes glioblastoma for tumor necrosis factor-related apoptosis-inducing ligand-mediated destruction. Cancer Res. 2005;65:11469–11477. doi: 10.1158/0008-5472.CAN-05-1724. [DOI] [PubMed] [Google Scholar]
- 132.Patel R, Shervington L, Lea R, Shervington A. Epigenetic silencing of telomerase and a non-alkylating agent as a novel therapeutic approach for glioma. Brain Res. 2008;1188:173–181. doi: 10.1016/j.brainres.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 133.Shervington A, Patel R. Silencing DNA methyltransferase (DNMT) enhances glioma chemosensitivity. Oligonucleotides. 2008;18:365–374. doi: 10.1089/oli.2008.0128. [DOI] [PubMed] [Google Scholar]
- 134.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 135.Mottet D, Castronovo V. Histone deacetylases: target enzymes for cancer therapy. Clin Exp Metastasis. 2008;25:183–189. doi: 10.1007/s10585-007-9131-5. [DOI] [PubMed] [Google Scholar]
- 136.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 137.Eyupoglu IY, Hahnen E, Buslei R, et al. Suberoylanilide hydroxamic acid (SAHA) has potent anti-glioma properties in vitro, ex vivo and in vivo. J Neurochem. 2005;93:992–999. doi: 10.1111/j.1471-4159.2005.03098.x. [DOI] [PubMed] [Google Scholar]
- 138.Kumagai T, Wakimoto N, Yin D, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer. 2007;121:656–665. doi: 10.1002/ijc.22558. [DOI] [PubMed] [Google Scholar]
- 139•.Galanis E, Jaeckle KA, Maurer MJ, et al. Phase II trial of vorinostat in recurrent glioblastoma multiforme: a north central cancer treatment group study. J Clin Oncol. 2009;27:2052–2058. doi: 10.1200/JCO.2008.19.0694. First published Phase II study of histone deacetylase inhibitors in glioma patients demonstrates that vorinostat can have modest single-agent activity in patients with recurrent GBM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ellis L, Pan Y, Smyth GK, et al. Histone deacetylase inhibitor panobinostat induces clinical responses with associated alterations in gene expression profiles in cutaneous T-cell lymphoma. Clin Cancer Res. 2008;14:4500–4510. doi: 10.1158/1078-0432.CCR-07-4262. [DOI] [PubMed] [Google Scholar]
- 141.Entin-Meer M, Rephaeli A, Yang X, Nudelman A, VandenBerg SR, Haas-Kogan DA. Butyric acid prodrugs are histone deacetylase inhibitors that show antineoplastic activity and radiosensitizing capacity in the treatment of malignant gliomas. Mol Cancer Ther. 2005;4:1952–1961. doi: 10.1158/1535-7163.MCT-05-0087. [DOI] [PubMed] [Google Scholar]
- 142.Gimsing P, Hansen M, Knudsen LM, et al. A Phase I clinical trial of the histone deacetylase inhibitor belinostat in patients with advanced hematological neoplasia. Eur J Haematol. 2008;81:170–176. doi: 10.1111/j.1600-0609.2008.01102.x. [DOI] [PubMed] [Google Scholar]
- 143.Qian X, Ara G, Mills E, LaRochelle WJ, Lichenstein HS, Jeffers M. Activity of the histone deacetylase inhibitor belinostat (PXD101) in preclinical models of prostate cancer. Int J Cancer. 2008;122:1400–1410. doi: 10.1002/ijc.23243. [DOI] [PubMed] [Google Scholar]
- 144.Yang YT, Balch C, Kulp SK, Mand MR, Nephew KP, Chen CS. A rationally designed histone deacetylase inhibitor with distinct antitumor activity against ovarian cancer. Neoplasia. 2009;11:552–563. doi: 10.1593/neo.09204. 3 p following 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 146.Cho HJ, Kim SY, Kim KH, et al. The combination effect of sodium butyrate and 5-aza-2′-deoxycytidine on radiosensitivity in RKO colorectal cancer and MCF-7 breast cancer cell lines. World J Surg Oncol. 2009;7:49. doi: 10.1186/1477-7819-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Oi S, Natsume A, Ito M, et al. Synergistic induction of NY-ESO-1 antigen expression by a novel histone deacetylase inhibitor, valproic acid, with 5-aza-2′-deoxycytidine in glioma cells. J Neurooncol. 2009;92:15–22. doi: 10.1007/s11060-008-9732-0. [DOI] [PubMed] [Google Scholar]
- 148.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 149.Nakamura M, Konishi N, Tsunoda S, et al. Genomic alterations of human gliomas detected by restriction landmark genomic scanning. Brain Tumor Pathol. 1997;14:13–17. doi: 10.1007/BF02478863. [DOI] [PubMed] [Google Scholar]
- 150.Nakamura M, Konishi N, Tsunoda S, et al. Analyses of human gliomas by restriction landmark genomic scanning. J Neurooncol. 1997;35:113–120. doi: 10.1023/a:1005712308061. [DOI] [PubMed] [Google Scholar]
- 151.Nakamura M, Konishi N, Tsunoda S, et al. Genomic alterations in human glioma cell lines detected by restriction landmark genomic scanning. J Neurooncol. 1997;34:203–209. doi: 10.1023/a:1005714811327. [DOI] [PubMed] [Google Scholar]
- 152.Nakamura M, Konishi N, Inui T, et al. Genetic variations in recurrent astrocytic tumors detected by restriction landmark genomic scanning. Brain Tumor Pathol. 1998;15:1–6. doi: 10.1007/BF02482093. [DOI] [PubMed] [Google Scholar]
- 153.Costello JF, Plass C, Cavenee WK. Aberrant methylation of genes in low-grade astrocytomas. Brain Tumor Pathol. 2000;17:49–56. doi: 10.1007/BF02482735. [DOI] [PubMed] [Google Scholar]
- 154.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 155.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 156.Fouse SD, Shen Y, Pellegrini M, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Down TA, Rakyan VK, Turner DJ, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159•.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. Genome-wide quantitative mapping of site-specific DNA methylation in a variety of mouse cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Morin RD, O'Connor MD, Griffth M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]