Abstract
Background
Prefrontal behavior and activity in humans are heritable. Studies in animals demonstrate an interaction between dopamine D2 receptors and nicotinic acetylcholine receptors on prefrontal behavior but evidence in humans is weak. Therefore, we hypothesize that genetic variation regulating dopamine D2 and nicotinic acetylcholine receptor signaling impact prefrontal cortex activity and related cognition. To test this hypothesis in humans, we explored the interaction between functional genetic variants in the D2 receptor gene (DRD2, rs1076560) and in the nicotinic receptor α5 gene (CHRNA5, rs16969968) on both dorsolateral prefrontal cortex mediated behavior and physiology during working memory and on prefrontal gray matter volume.
Methods
A large sample of healthy subjects was compared for genotypic differences for DRD2 rs1076560 (G>T) and CHNRA5 rs16969968 (G>A) on prefrontal phenotypes, including cognitive performance at the N-Back task, prefrontal physiology with BOLD fMRI during performance of the 2-Back working memory task, and prefrontal morphometry with structural MRI.
Results
We found that DRD2 rs1076560 and CHNRA5 rs16969968 interact to modulate cognitive function, prefrontal physiology during working memory, and prefrontal gray matter volume. More specifically, CHRNA5-AA/DRD2-GT subjects had greater behavioral performance, more efficient prefrontal cortex activity at 2Back working memory task, and greater prefrontal gray matter volume than the other genotype groups.
Conclusions
The present data extend previous studies in animals and enhance our understanding of dopamine and acetylcholine signaling in the human prefrontal cortex, demonstrating interactions elicited by working memory that are modulated by genetic variants in DRD2 and CHRNA5.
Introduction
Working memory is a highly heritable complex cognitive trait [1]–[3], defined as the ability to keep information immediately available for a short period of time to solve a task that may be delayed; it is, therefore, a fundamental component of higher-level functions [4]. The prefrontal cortex has been identified as a key neocortical region supporting working memory [4], [5]. Previous functional imaging studies in humans have demonstrated that working memory prefrontal activity is also heritable [6], [7], suggesting the importance of prefrontal cortex function in generating and testing various neuroimaging intermediate phenotypes for complex genetic brain disorders.
Several neurotransmitters [8] and related genetic variation [9]–[11] modulate the physiology of prefrontal cortex and interact in determining neuronal response to cognitive stimuli. It is well known that dopamine critically modulates prefrontal neuronal signal-to-noise during working memory processes [12]. By differentially acting on dopamine D1 and D2 receptors, dopamine directly regulates firing of pyramidal neurons and of their GABA inhibitory surround within prefrontal cortex to focus prefrontal cortical resources to the task at hand [13], [14]. Recent studies in animal models also implicate a specific role for prefrontal acetylcholine [15]–[17]. For example, rhesus monkeys with selective lesions of cholinergic input from the basal forebrain to the lateral and orbital prefrontal cortex are unimpaired in tests of decision making and episodic memory that also require intact prefrontal cortex, but are severely impaired on a spatial working memory task [16]. Pharmacological studies in animals and in humans complement cholinergic lesion studies, specifically implicating nicotinic acetylcholine receptors (nAChRs) in working memory performance [18], [19]. Systemic administration of high doses of mecamylamine, a nicotinic receptor antagonist, as well as intracranial infusion of the antagonist dihydro-β-erythroidine in the frontal cortex of rats lead to significant working memory performance deficits in the radial arm maze [20], [21]. Conversely, agonist-mediated activation of AchRs improves working memory performance in rats [20], [22], rabbits [23], non-human primates [24], and abstinent smokers [25]. Moreover, transdermally-administered nicotine in humans also improves performance in a variety of recall tasks through non-selective stimulationof nAChRs [26].
Neuronal nAChRs are pentameric ligand-gated channels, distinguished on the basis of subunit stoichiometry (α2- α10, β2–β4) [27]. α4β2-containing receptors are present on multiple cell types in multiple layers (L) of the human prefrontal cortex [28], where they modulate layer-specific activity of pyramidal neurons [29]. Specifically, LII/III pyramidal neurons are inhibited by nAChR stimulation, while LV and LVI pyramidal neurons are prominently activated. α4β2 nAChRs incorporating the α5 accessory subunit (α4β2α5) are important players in the regulation of prefrontal neuronal plasticity [30], [31]. The α5 subunit is more densely expressed on soma and axons of pyramidal neurons LVI of the murine medial prefrontal cortex [29], [32]–[35]. This subunit substantially increases the conductance [31] and currents [36] of α4β2-containing nAChRs, and drives developmental changes in the morphology and activation of medial prefrontal cortex LVI pyramidal neurons [37].
Because both dopaminergic and cholinergic systems modulate pyramidal neuron firing in the prefrontal cortex, they likely interact to shape prefrontal neuronal plasticity critical for information processing. Pharmacological studies in rodents support a potential interaction between these two neuromodulators. Radial maze performance, a behavioral measure of working memory in rats, is improved after application of a nAChR agonist [38] but impaired by an antagonist [39]. Interestingly, the detrimental effect of the nAChR antagonist can be reversed by a dopamine D2 receptor agonist [40], while co-administration of a D2 receptor and of a nAChR antagonist leads to an even stronger impairment compared with the effect of each pharmacological challenge alone [41]. The interaction is specific for dopamine D2 receptors as dopamine D1 agonists do not neutralize the detrimental effect of nAChR antagonists [42]. However, studies have yet to be performed in humans to evaluate this potential interaction in terms of prefrontal physiology during executive and cognitive control processes.
In the present study, we evaluated this interaction on prefrontal cortical activity in humans, by exploiting known functional genetic variants that have demonstrable effects on cortical dopamine and acetylcholine signaling in vivo. Specifically, we investigated dorsolateral prefrontal cortex (DLPFC) activity during working memory in healthy subjects for interactions between single nucleotide polymorphisms (SNPs) in genes encoding the D2 receptor (DRD2, rs1076560) and the nicotinic receptor α5 (CHRNA5, rs16969968). DRD2 is located on chromosome 11 and encodes two D2 isoforms, D2S (short) and D2L (long). D2L receptors mainly mediate post-synaptic signaling, while D2S receptors mainly serve as auto-receptors on pre-synaptic neurons [43], even though they are also found on post-synaptic neurons [44]. The minor allele (T) of DRD2 rs1076560 (G/T), located within intron 6 of DRD2, is associated with reduced expression of D2S in prefrontal cortex and striatum, and with altered activity of the striato-thalamic-prefrontal pathway during working memory in healthy subjects [45] and patients with schizophrenia [46]. DRD2 rs1076560 genotype also predicts putative steady-state striatal dopamine as assessed with SPECT and its correlation with prefrontal activity during performance of working memory, in that subjects carrying the T allele have reduced striatal D2 signaling and increased prefrontal activity during the 2-Back working memory task [47]. Recently, the T allele has been associated with risk for substance abuse related disorders including alcohol dependence [48], cocaine abuse [49] and opioid addiction [50]. Moreover, other DRD2 variants have been associated with nicotine dependence [51], [52] and alcoholism [53], [54]. CHRNA5, encoding the α5 nicotinic accessory subunit, is located on chromosome 15. The rs16969968 SNP within CHRNA5 changes the encoded amino acid sequence from aspartic acid (G allele) to asparagine (A allele) at position 398 (Asp398Asn) [55], [56]. The A allele resides almost exclusively on a haplotype associated with reduced CHRNA5 mRNA expression in the brain [57], [58]. Furthermore, it has been associated in vitro with lower agonist-evoked intracellular calcium response of α4β2α5 nAChRs, lower Ca2+ permeability and greater short-term desensitization compared to the α5 ancestral allele (G) [55], [59]. Moreover, the A allele has also been associated with increased risk for lung cancer [55], nicotine dependence and smoking behavior [60], [61], as well as with lower cognitive performance in healthy subjects [62]. More recently, two studies have demonstrated that this allele is also associated with increased susceptibility to schizophrenia and bipolar disorders [63], [64].
Altogether, these findings suggest the crucial functional relevance of D2 and nAChR receptors as well as of genetic variation in DRD2 and CHRNA5 for prefrontal physiology. Furthermore, they implicate a complex and tight relationship between D2 and nAChR signaling, and call for further investigation of the impact of related genetic interaction on brain function [55]. Indeed, understanding the effect of genetic interactions on brain function has immediate clinical potential in elucidating the pathophysiology of complex neuropsychiatric disorders (i.e. Alzheimer Disease, Parkinson Disease, Schizophrenia) and in predicting therapeutic drug response [65].
Guided by the hypothesis that the DLPFC is especially vulnerable to the combined effect of suboptimal dopaminergic and cholinergic signaling, the aim of the present study was to investigate in healthy subjects the effect of CHRNA5 rs16969968 and its interaction with DRD2 rs1076560 on prefrontal physiology (as assessed with blood oxygenation level-dependent functional magnetic resonance imaging, BOLD fMRI), and mediated behavior during working memory. Furthermore, given compelling evidence in animals that both dopamine and acetylcholine signaling are involved in brain development and in ongoing local synaptic plasticity [37], [66], we also explored the potential effect of these two polymorphisms and their interaction on prefrontal gray matter volume.
Materials and Methods
Participants
Healthy Caucasian subjects from the region of Puglia, Italy, were recruited for the study and were evaluated with the Structured Clinical Interview for DSM-IV [67] to exclude any psychiatric disorder. Further exclusion criteria were: history of drug or alcohol abuse, active drug use in the past year, head trauma with loss of consciousness, and any significant medical condition revealed by clinical and magnetic resonance imaging. Handedness (Edinburgh Inventory)[68], and total IQ (WAIS-R) were also measured. The present study was approved by the local Institutional Review Board (Comitato Etico Locale Indipendente Azienda Ospedaliera “Ospedale Policlinico Consorziale” Bari). After complete description of the protocol and procedures, written informed consent was obtained by all participants, in accordance with the Helsinki Declaration. All subjects were genotyped for CHRNA5 rs16969968 and DRD2 rs1076560 and underwent one or more of the procedures described below.
The study involved a total number of 460 healthy subjects, with overlapping groups undergoing behavioral assessments, functional MRI (fMRI), and structural MRI (sMRI). A sample of 387 subjects (age, mean ± SD: 26.6±7.8; 194 males) underwent working memory behavioral assessment. A sample of 329 individuals (age: 27.1±7.8; 161 males) underwent fMRI during the N-Back working memory task, and a group of 211 individuals (age: 26.5±7.4; 114 males) underwent sMRI for Voxel Based Morphometry analysis. 166 subjects performed both fMRI and sMRI, 274 subjects performed both fMRI and WM behavioral assessment, while 173 subjects performed both sMRI and WM behavioral assessment.
In order to exclude that nicotine consumption may have been a confounding factor for our results, we also evaluated smoking status. Smokers were defined as those who smoked for at least 1 year and were currently smoking [69]. Chronic exposure was estimated in packs-year. All smokers were not allowed to tobacco use at least for 2 hours before scanning. Non smokers were defined by lifetime smoking of less than 20 cigarettes. Smoking status was available for a total of N = 221 subjects. More specifically, neuropsychological analyses were performed in a sample of N = 205 subjects, 114 Non-Smokers and 91 Smokers (age, mean ± SD: 26.42±6.90; 98 males). fMRI analyses were performed in a sample of N = 204 subjects, 137 Non-Smokers and 67 Smokers (age, mean ± SD: 26.42±6.90; 93 males).
Genotype determination
DNA was extracted from whole blood samples using standard procedures. CHRNA5 rs16969968 genotypes were determined by restriction fragment length polymorphism methods, using primers tagged with a fluorophore (forward 5′-TAGAAACACATTGGAAGCTGCG-3′ and reverse 5′- AATTCTGGCCCTCAATCTATGCT-3′). Taqα1 (from New England Biolabs, Ipswich, MA, USA) was used to cut the amplified gDNA ancestral allele, and the resultant fragment length was resolved and analyzed on an ABI 3730 DNA analyzer (Life Technologies). DRD2 rs1076560 genotypes were determined by direct sequencing. Amplification of the 213 bp DNA fragment containing the DRD2 rs1076560 polymorphism (G>T) was performed using forward 5′-GGCAGAACAGAAGTGGGGTA-3′ and reverse 5′-GACAAGTTCCCAGGCATCAG-3′ primers. PCR was performed on 100 ng genomic DNA in a standard 25 µL volume, containing 0.2 µM primers, 100 µM dNTPs, 2.5 µl reaction Gold buffer (Applied Biosystems, Foster City, CA), 2 mM MgCl2 and 2.5 U Ampli Taq Gold Polymerase (Applied Biosystems, Foster City, CA). Thermal cycler conditions were as follows: initial denaturation step at 94°C for 12 min; 94°C for 30 sec, 62°C for 30 sec, 72°C for 45 sec for 35 cycles; final elongation step at 72°C for 7 minutes. DRD2 rs1076560 PCR products were sequenced in both directions using BigDye Terminator chemistry and run on an ABI Prism 3130 DNA sequencer (Applied Biosystems, Foster City, CA, USA). Sequences were analyzed with SeqMan from Lasergene-DNASTAR package (DNASTAR Inc., Madison, Wis.).
All alleles displayed Hardy-Weinberg equilibrium. Given the low number of subjects homozygous for the DRD2-T minor allele, we combined these individuals (when present) with heterozygous subjects (GT) for further analyses, consistent with earlier studies evaluating polymorphisms with low minor allele frequencies [70]. In each of the study cohorts included in the experiments, the χ2 analysis demonstrated equal distribution of DRD2 genotypes in CHRNA5 groups and vice versa (all χ2<4.06; all p>0.13), indicating that the genotype groups were not differentially distributed in subpopulations.
N-Back Working Memory paradigm for behavioral study
Briefly, ‘N-back’ refers to how far back in the sequence of stimuli the subject had to recall. The stimuli consisted of numbers (1–4) shown in random sequence and displayed at the points of a diamond-shaped box. There was a visually paced motor task which also served as a non-memory guided control condition (0-Back) that simply required subjects to identify the stimulus currently seen. In the working memory conditions, the task required recollection of a stimulus seen one (1-Back) or two stimuli (2-Back) previously while continuing to encode additionally incoming stimuli. Performance data were recorded as the percentage (%) of correct responses (accuracy) and as reaction time (ms).
Statistical Analysis for demographics and behavioral performance
One-way ANOVAs and χ2 analyses were used to compare demographic data across genotype groups. General linear models with repeated measures for task conditions (1-Back and 2-Back) and with predictors CHRNA5 rs16969968 and DRD2 rs1076560 were used to evaluate behavioral differences across genotype groups. Fisher's Least Significant Difference Test and t-tests for dependent samples as appropriate were used for all post-hoc analyses.
Imaging Data Acquisition and Processing
Functional and structural MRI were performed on a General Electric (Milwaukee, WI) 3 Tesla scanner.
fMRI acquisition parameters
Each subject was scanned using a gradient-echo echo planar imaging sequence (repetition time, 2000 ms; echo time, 28 ms; 20 interleaved axial slices; thickness, 4 mm; gap, 1 mm; voxel size, 3.75×3.75×3.75; flip angle, 90°; field of view, 24 cm; matrix, 64×64). We used a simple block design in which each block consisted of eight alternating 0-Back and 2-Back conditions (each lasting 30 s), obtained in 4 min and 8 s, 120 whole-brain fMRI volumes. The first four scans at the beginning of each time series were acquired to allow the signal to reach a steady state and were not included in the final analysis.
fMRI image analysis. Preprocessing and statistical analyses
Data processing and analysis were performed with freely available Statistical Parametric Mapping software (SPM8; Wellcome Trust Centre for Neuroimaging, London, UK, http://www.fil.ion.ucl.ac.uk/spm). Images, for each subject, were realigned to the first volume in the time series and movement parameters were extracted to exclude subjects with excessive head motion (>2 mm of translation, >2° rotation). Images were then re-sampled to a 2 mm isotropic voxel size, spatially normalized into a standard stereotactic space (Montreal Institute on Neurology, MNI template) and smoothed using a 10 mm full-width half-maximum isotropic Gaussian kernel to minimize noise and to account for residual inter-subject differences. A box car model convolved with the hemodynamic response function at each voxel was modeled. In the first-level analysis, linear contrasts were computed producing a t statistical map at each voxel for the 2-Back condition, assuming the 0-Back condition as a baseline.
All the individual contrast images were entered in a second level random effects analysis. A Factorial Analysis of Variance (ANOVA) was then performed, with CHRNA5 rs16969968 and DRD2 rs1076560 genotype as the between-subjects factors. Because of our strong hypothesis about α4β2α5 nAChRs and dopamine-D2 mediated modulation of dorsolateral prefrontal neuronal plasticity, we used a statistical threshold of p<0.05, with family-wise error (FWE) small-volume correction within a Region of Interest (ROI) comprehensive of Brodmann's areas 46 (BA46) as defined by the Wake Forest University PickAtlas 1.04 (WFU_PickAtlas) (http://www.fmri.wfubmc.edu/cms/software#PickAtlas). Because we did not have a priori hypotheses regarding the activity of brain regions outside of the ROI we used a statistical threshold of p<0.05, FWE-corrected for these whole-brain comparisons. Because no effects were detected with this threshold and for the sake of completeness, we also report exploratory analyses at p = 0.001 uncorrected, k = 10. Moreover, to further explore differences between genotype groups, post-hoc analysis outside of SPM8 was also performed on BOLD responses extracted from the cluster showing the interaction using MarsBar (http://marsbar.sourceforge.net/).
Finally, to evaluate the behavioral relevance of the interaction between CHRNA5 rs16969968 and DRD2 rs1076560 genotypes on DLPFC activity, we performed separate linear regression analyses within SPM8 using as predictor behavioral accuracy (%) at 2-Back working memory task both in the whole sample and within each genotype group. Again, a statistical threshold of p<0.05, with FWE small-volume correction within a ROI comprehensive of BA46 as defined by the WFU_PickAtlas was applied. All fMRI data are reported with reference to the MNI standard space within SPM8.
sMRI acquisition parameters
Three-dimensional images were acquired using a T1-weighted SPGR sequence (TE = min full; flip angle, 6°; field of view, 250 mm; bandwidth, 31.25; matrix, 256×256) with 124 1.3-mm-axial slices.
sMRI image analysis. Preprocessing and statistical analyses
Voxel Brain Morphometry Analysis (VBM) of the sMRI data was also performed using SPM8. The T1-weighted scans were partitioned into different tissue classes- gray matter (GM), white matter and non-brain voxels (cerebrospinal fluid, skull) - based on separate tissue probability maps for each tissue class using the “new segmentation” approach in SPM8 [71]. In order to compare brains of different subjects, the resulting segments were normalized to a population template generated from the complete dataset using a diffeomorphic registration algorithm [72]. This high-dimensional non-linear warping algorithm selects conserved features, which are informative for registration, thus minimizing structural variation among subjects and providing optimal inter-subject registration. Subsequently, all images were “modulated” by the Jacobian determinants from the normalisation steps to preserve initial volumes. Thus, images were smoothed by convolution with an isotropic Gaussian kernel of 8 mm full-width at half maximum.
We examined the SNP main effects and their interaction by creating voxel-based, whole-brain, statistical parametric maps using Gaussian random fields theory and the general linear model. More specifically, we used a full factorial Analisys of Covariance (ANCOVA) design with two level factors, DRD2 rs1076560 and CHRNA5 rs16969968. The statistical model also included orthogonalized first- and second-order polynomials of age, gender and total GM volume as “nuisance” variables, in order to control for any independent effects on our findings and to ensure that the analysis identified regionally specific “non-global” effects [73]. Because of our strong a priori hypothesis based on the effects of CHRNA5 and DRD2 variants on mRNA levels in prefrontal cortex [45], [57] and consistent with the fMRI analyses, the ANCOVA was masked with an ROI identified in BA46 using the WFU_PickAtlas. Statistical non-stationary inference [74] was performed at the cluster level at p<0.05 corrected within the ROI by using the ns toolbox (http://fmri.wfubmc.edu/cms/NS-General) implemented in SPM8, to avoid increased false-positive rate due to the non-stationary structural images. Exploratory whole-brain statistics outside the ROI was set at p = 0.001, uncorrected.
VBM results are reported with reference to the MNI standard space within SPM8. To further examine differences between genotype groups, post-hoc analysis outside of SPM8 was also performed on gray matter volumes extracted from the cluster showing a CHRNA5 rs16969968 by DRD2 rs1076560 interaction using MarsBar.
Results
Demographics (±SD) and genetics of the samples included in the experiments are reported in Table 1.
Table 1. Demographics (±SD) and genetics of the samples included in the experiments performed.
| Cognitive Behavior | fMRI | sMRI | |
| N | 387 | 329 | 211 |
| Gender (M/F) | 194/193 | 161/168 | 114/97 |
| Age | 26.61±7.76 | 27.06±7.76 | 26.47±7.42 |
| Handedness | 0.73±0.42 | 0.77±0.36 | 0.62±0.51 |
| IQ | 107.57±12.48 | 109.81±12.38 | 107.77±12.62 |
| N | |||
| CHRNA5 GG/DRD2 GG | 122 | 99 | 72 |
| CHRNA5 GG/DRD2 Tcarriers | 28 | 21 | 15 |
| CHRNA5 GA/DRD2 GG | 143 | 117 | 76 |
| CHRNA5 GA/DRD2 Tcarriers | 42 | 45 | 23 |
| CHRNA5 AA/DRD2 GG | 42 | 36 | 28 |
| CHRNA5 AA/DRD2 Tcarriers | 10 | 11 | 7 |
Association with Working Memory behavioral performance
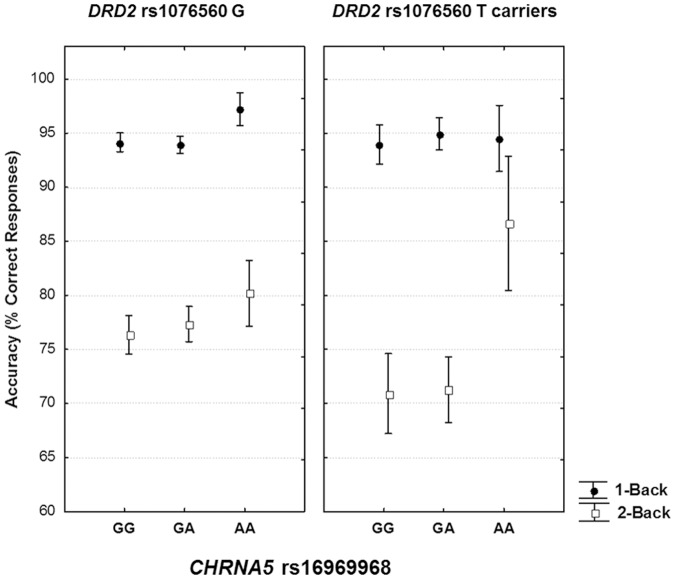
In the cognitive behavior sample (N = 387), genotype groups were matched in terms of gender, age, handedness, and IQ (all p>0.1). Repeated measures ANOVA on working memory load accuracy indicated no significant effect of DRD2 rs1076560 (F1,381 = 0.18, p = 0.66); a main effect of CHRNA5 rs16969968 (F2,381 = 3.10, p = 0.046) and an interaction between DRD2 rs1076560 and CHRNA5 rs16969968 (F2,381 = 3.16, p = 0.044) [Mean Squared Error (MSE): 1-Back = 94, 2-Back = 384] (Fig. 1). More specifically, post hoc analysis with t-test for dependent samples demonstrated a statistically significant drop in performance from 1-Back to 2-Back for all genotype groups (all p<0.001) with the exception of CHRNA5 AA/DRD2 GT subjects (p = 0.09) (Fig. 1). In other words, the interaction between the minor T allele of rs1076560 and the minor A allele of rs16969968 was associated with attenuated drop in performance which was instead observed from 1-Back to 2-Back for all other genotypes. Repeated measures ANOVA on working memory load reaction time indicated no significant effect of DRD2 rs1076560 (F1,381 = 3.66, p = 0.07); no significant effect of CHRNA5 rs16969968 (F2,381 = 1.04, p = 0.35), and no interaction between DRD2 rs1076560 and CHRNA5 rs16969968 (F2,381 = 1.18, p = 0.307).
Figure 1. Interaction between Working Memory behavioral performance, DRD2 rs1076560 and CHNRA5 rs16969968 genotypes.
Mean ± Standard Errors correct responses (DRD2-GG, left panel; DRD2-Tcarriers, right panel) showing the interaction between the two genotypes. See text for statistics.
To test whether nicotine consumption may have confounded these results, we also performed ANCOVA covarying for smoking status in N = 205 subjects, including 114 Non-Smokers and 91 Smokers. Similar to the above analysis, ANCOVA demonstrated an interaction between DRD2 rs1076560 and CHRNA5 rs16969968 on working memory load accuracy (F2,198 = 5.10; p = 0.007; MSE 1-Back = 77.1, 2-Back = 327.4). More specifically, post hoc analysis with t-test for dependent samples demonstrated a statistically significant drop in performance from 1-Back to 2-Back for all genotype groups (all p<0.0001) with the exception of CHRNA5 AA/DRD2 GT subjects (p = 0.25). No significant genotype effects or interactions on working memory load reaction time were detected in this sample (all p>0.07).
Association with Working Memory DLPFC activity measured with fMRI
In the fMRI sample (N = 329), genotype groups were also matched in terms of gender, age, handedness and IQ (all p>0.1). No genotype effects or interaction were present on accuracy and reaction time at the N-Back task in this sample (all p>0.1), thus allowing us to compare brain responses in the absence of behavioral differences. For behavioral performance see Table S1.
Effect of the Working Memory task
As expected from previous studies with the N-Back task (Callicott et al.1999, 2000; Bertolino et al. 2004, 2006), performance of the 2-Back working memory condition was associated with activity in a distributed network of brain regions including the prefrontal cortex, the parietal cortex, the anterior cingulate, and the striatum bilaterally.
Genotype main effects and interaction during working memory
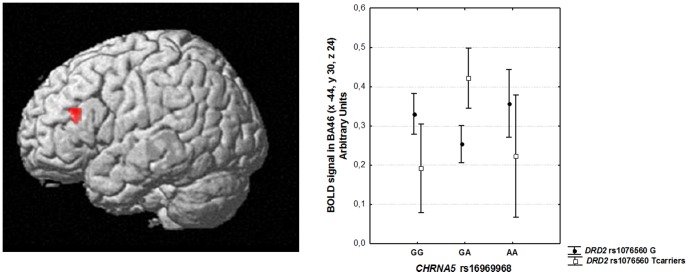
No statistically significant main effect of CHRNA5 rs16969968 or DRD2 rs1076560 genotype in the DLPFC ROI was found. On the other hand, ANOVA revealed a DRD2 by CHRNA5 interaction in left DLPFC (BA 46: x -44 y 30 z 24; K = 28; corrected pFWE = 0.036; Fig. 2). Post hoc analysis of BOLD response from this cluster indicated that within DRD2-GT genotype, CHRNA5-GA subjects have greater prefrontal activity compared with DRD2-GT CHRNA5- GG (p = 0.02) or -AA subjects (p = 0.02) (Fig. 2b). No significant differences emerged in the context of DRD2-GG genotype. Results of the uncorrected exploratory whole-brain analyses are reported in Table 2.
Figure 2. Interaction between DRD2 rs1076560 and CHNRA5 rs16969968 on prefrontal physiology at 2-Back.
3Dimensional rendering (left, image thresholded at p<0.005, uncorrected) and mean ±0.95 CIs of BOLD response (right) of the interaction between DRD2 rs1076560 and CHNRA5 rs16969968 on working memory DLPFC activity.
Table 2. Results of exploratory uncorrected whole brain statistics (p<0.001, k = 10) showing the main effect of DRD2 and its interaction with CHRNA5 on brain physiology at 2-Back WM Task.
| Main effect of DRD2 genotype | |||||||
| MNI | |||||||
| Region | Brodmann Area | k | Z-score | p | x | y | z |
| Middle Temporal Gyrus | BA 39 | 19 | 3.59 | 0.0001 | 40 | −54 | 22 |
| Middle Frontal Gyrus | BA 10 | 15 | 3.35 | 0.0001 | 32 | 56 | 6 |
Again, to test whether nicotine consumption may have confounded these results, we also performed ANCOVA covarying for smoking status on BOLD responses identified in the above analysis. This analysis included N = 204 subjects, of whom 137 were Non-Smokers and 67 Smokers. These analysis indeed demonstrated an interaction between DRD2 by CHRNA5 (p = 0.008). Similar to the analysis in the whole sample, post hoc analysis indicated within DRD2-GT genotype, CHRNA5-GA subjects have greater prefrontal activity compared with DRD2-GT CHRNA5- GG (p = 0.01) or -AA subjects (p = 0.007). No significant differences emerged in the context of DRD2-GG genotype. These results suggest that smoking status did not significantly confound the identified interaction.
Relationship between DLPFC activity and behavioral performance at 2-Back
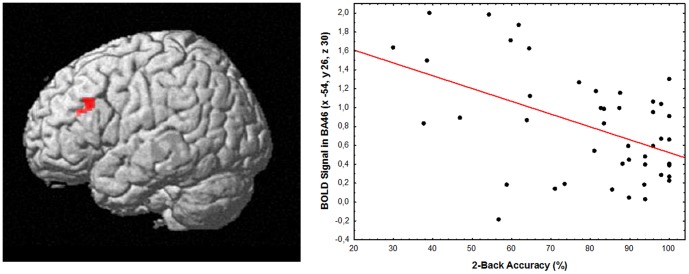
Regression analysis in SPM8 demonstrated a negative correlation between activity in DLPFC and accuracy (%) at 2-Back in the CHRNA5-GA/DRD2-GT group (BA 46: x -56 y 26 z 30; K = 39; corrected pFWE = 0.02; Fig. 3). Also, exploratory analyses which did not survive correction for multiple comparisons suggested a negative correlation in the CHRNA5-AA/DRD2-GT group (x -42 y 30 z 22; K = 15; p = 0.002 uncorrected), and a positive correlation in the CHRNA5-GG/DRD2-GT group (x -36 y 56 z 30; K = 20; p = 0.002 uncorrected) (See Fig. S1).
Figure 3. Correlation between BOLD fMRI in prefrontal cortex and Working Memory accuracy in CHRNA5-GA/DRD2-GT subjects.
3Dimensional rendering (left, image thresholded at p<0.005, uncorrected) of the correlation between BOLD response in DLPFC and percentage of correct responses at 2-Back task in CHRNA5-GA/DRD2-GT subjects. On the right, relative scatterplot is shown indicating individual data points.
Association with DLPFC gray matter volume measured with sMRI
In the sMRI sample (211 subjects), genotype groups were also matched in terms of gender, age, handedness and IQ (all p>0.1).
Genotype main effects and interaction
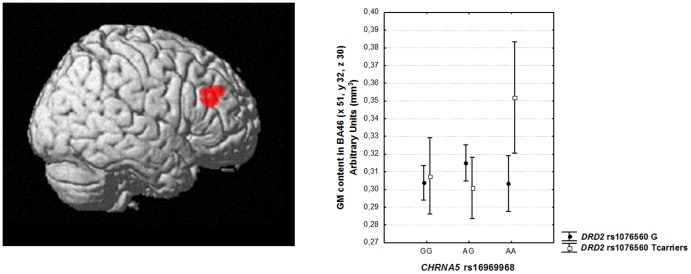
There was no statistically significant main effect of CHRNA5 rs16969968 or of DRD2 rs1076560 genotype in the DLPFC ROI. However, an interaction between CHRNA5 and DRD2 genotypes was found in right DLPFC (BA 46: x 51, y 32, z 30, k = 321, Z = 3.96, p = 0.006 cluster-level corrected; Fig. 4). Post hoc analysis of gray matter volume extracted from the interaction cluster indicated that CHRNA5-AA/DRD2-GT subjects have greater DLPFC gray matter volume compared to all other genotype groups (all p<0.02; Fig. 4).
Figure 4. Interaction between DRD2 rs1076560 and CHNRA5 rs16969968 on prefrontal gray matter volume.
3Dimensional rendering (left, image thresholded at p<0.005, uncorrected) and mean ±0.95 CIs of gray matter content (right) of the interaction between DRD2 rs1076560 and CHNRA5 rs16969968 on DLPFC gray matter.
Results of the uncorrected exploratory whole-brain analyses are reported in Table 3.
Table 3. Results of exploratory uncorrected whole brain statistics (p<0.001, k = 10) showing the main effect of genotypes and their interaction on gray matter volume.
| Main effect of CHRNA5 genotype | |||||||
| MNI | |||||||
| Region | Brodmann Area | K | Z-score | p | x | y | z |
| Inferior Frontal Gyrus | BA 47 | 169 | 4.01 | 0.0001 | −28 | 30 | −5 |
| Occipital Lobe, Cuneus | BA 19 | 137 | 3.82 | 0.0001 | −10 | −95 | 30 |
| Cerebellum, Posterior Lobe | 466 | 3.68 | 0.0001 | 28 | −82 | −24 | |
Discussion
The results of the present study demonstrate that variants in two genes implicated in dopamine and acetylcholine signaling interact to modulate the biology and physiology of the prefrontal cortex during working memory. More specifically, the interaction between CHNRA5 rs16969968 and DRD2 rs1076560 genotypes differentially predicted cognitive behavior with increasing working memory load, in that CHRNA5-AA/DRD2-GT subjects have better behavioral performance. In addition, we found that the effect of CHRNA5 rs16969968 in dorsolateral prefrontal activity at 2-Back is only evident in the context of DRD2 rs1076560 genotype, such that CHRNA5 demonstrates an inverted U shaped prefrontal response in DRD2-GT subjects (see below).
As a further demonstration of the functional effects of these polymorphisms, CHRNA5 rs16969968 and DRD2 rs1076560 also interacted on gray matter volume of the dorsolateral prefrontal cortex (DLPFC). Once again, the effect of CHNRA5 was mostly evident in the context of DRD2-GT genotype.
Our behavioral findings during working memory as elicited by the N-Back task are consistent with a previous report by Markett et al. (2010) in healthy subjects (N = 101), showing an interaction between a functional SNP in CHRNA4 (rs1044396) and a haplotype block covering three SNPs in DRD2 (rs1800497, rs6277, rs2283265) on working memory capacity [75]. As in our sample, this effect only became apparent at greater working memory load, suggesting that the CHRNA5 by DRD2 interaction affects the efficiency by which relevant information is encoded during the trial-wise updating of working memory items. Unlike all other genotype groups, CHRNA5-AA/DRD2-GT subjects showed no statistically significant reduction in behavioral performance with increasing working memory load, leading to the speculation that the prefrontal neuronal signal-to-noise affected by the genetically determined balance of cholinergic and dopaminergic signaling is increased in CHRNA5-AA/DRD2-GT subjects allowing greater performance with increasing working memory load. Moreover, this interaction is similar to the reciprocal rescue of minor allele risk found in association with other biological phenotypes [76]. Of note, our results suggest that the pattern of genotype effect on working memory load accuracy was not moderated by reaction time.
Our functional imaging data also indicate that the interaction between CHRNA5 rs16969968 and DRD2 rs1076560 genotypes differentially predicted the efficiency of the prefrontal cortex at 2- Back condition. Earlier fMRI studies found that genetic variation in dopamine signaling in the prefrontal cortex affects the efficiency or signal-to-noise ratio of the physiological response during the N-Back following an inverted U-shaped response function [77]–[79]. In the present study, we found that only within DRD2- GT genotype, CHRNA5 demonstrates an inverted U shaped prefrontal response at 2-Back working memory task, suggesting that CHRNA5 rs16969968 further affects the signal-to-noise ratio of prefrontal cortex in subjects with greater dopamine signaling (i.e. GT subjects) [45]–[47]. More specifically, within the context of DRD2-GT genotype: CHRNA5-AA subjects are more efficient, because the combination of behavioral data and imaging results suggests that they have reduced activity for greater behavioral performance; CHRNA5-AG subjects are less efficient, because they show greater activity for reduced behavioral performance, which is also consistent with the negative relationship between BOLD response and behavioral accuracy; and CHRNA5-GG have reduced engagement of prefrontal resources to the task at hand for reduced behavioral performance, as also suggested by the positive relationship between prefrontal activity and behavioral performance.
Thus, the DRD2 GT subjects show an overdominance effect for the CHRNA5 genotype with heterozygous revealing greater/more inefficient prefrontal activity compared with individuals homozygous for either allele. While this effect in genetics would be regarded as a “heterozygous advantage”, at the brain imaging level it may actually reflect an inefficient prefrontal activity during information processing. This finding is of particular interest since central dopamine-acetylcholine imbalance in synaptic plasticity is responsible for cognitive deficits in Parkinson disease [80] and likely in psychosis, as suggested by heavy smoking in patients with schizophrenia [81].
There may be several and complex molecular/neuronal mechanisms in human DLPFC which are responsible for the interaction we have measured in vivo with BOLD fMRI, and further work on molecular and cellular models is warranted. Still, previous work examining cortical anatomy and physiology allows us to speculate on the biology underlying our observations. D2 receptors in prefrontal cortex are mainly found pre-synaptically on dopamine terminals [82], modulating dopamine release and D2S autoreceptors are relatively more abundant in the prefrontal cortex compared to D2Ls [45]. Thus, DRD2-GT genotype associated with reduced D2S may increase dopamine levels in the prefrontal cortex, and in turn increase its activity. nAchRs containing the α5 subunit in prefrontal cortex are mainly expressed on soma and axon of LVI pyramidal neurons [29], [83], where they are responsible for strong activation of the neuronal population of this layer [29]. Similarly to DRD2 genotype, the CHRNA5-A allele which is associated with reduced total CHRNA5 mRNA expression in prefrontal cortex tissue and signaling in vitro, may alter the neuronal activation of LVI pyramidal neurons. However, as mentioned above, the effect of CHRNA5 rs16969968 is only manifest in the context of DRD2 rs1076560 GT genotype, suggesting that the physiological relevance of this SNP, in terms of overall prefrontal cortex activity during working memory, occurs only in the context of genetic variation modulating dopamine signaling.
Alternatively, the interaction observed in prefrontal cortex could also be influenced by activity within the cortico-striato-thalamic pathway. Dopamine is an important modulator of this circuit. Specifically, greater release of dopamine in the striatum increases activity of the whole network. We have previously demonstrated in healthy subjects that DRD2-GT genotype is associated with reduced pre-synaptic DAT and post-synaptic D2 receptor density, reduced striatal dopamine signaling [47], greater caudate activity and greater prefrontal activity during working memory performance [46]. The α4β2α5 nAChRs in striatum are expressed on dopaminergic terminals [84], where they dominantly regulate dopamine release in the dorsal caudate-putamen [85], overriding the release mediated by ascending dopaminergic somata firing [86]. Thus, it is possible that the DRD2 by CHRNA5 genotype interaction might be associated with modulation of dopamine release in the striatum, which would increase activity in the whole circuit. However, we did not detect any CHRNA5 rs16969968 effect or interaction with DRD2 rs1076560 on striatal activity, and further studies are necessary to test this hypothesis.
The behavioral and functional imaging findings complement our VBM results, which provide in vivo evidence that DRD2 and CHRNA5 interact to affect gray matter volume in human DLPFC. This is consistent with previous data in animal models demonstrating an effect of dopamine and acetylcholine in brain development and in ongoing local neural plasticity [66], [87]. Increasing synaptic dopamine in developing brains through prenatal cocaine exposure leads to specific neurodevelopmental alterations including abnormal dendritic growth and abnormal arborization of pyramidal cells that persist postnatally [88]. Conversely, neonatal dopamine denervation in rat produces permanent differential changes in prefrontal cortex dendritic morphology, i.e. atrophy of proximal apical and basilar dendrites [89]. Cholinergic inputs to the cortex also appear early during brain development and are widespread in rat by the third week of post-natal life [87], [90], likely influencing the normal morphological development of pyramidal neurons. Two elegant studies have indicated that direct nicotinic stimulation can modulate growth or retraction of neurites in cultured neurons [91], [92]. More recently, Bailey et al (2012) have demonstrated that α5 nAChRs underlie the neurodevelopmental peak in the nicotinic excitation of murine medial prefrontal cortex LVI neurons that occurs during the third week of postnatal life, and that it is likely to influence a specific ontogenetic retraction of apical dendrites in LVI pyramidal neurons. However, it is possible that α5 nAChRs on dopaminergic neurons [33], [34] may also influence the morphology of prefrontal cortex neurons.
Some potential limitations of the present study should be addressed. First, the study includes assumptions that we expect protein expression to correspond to mRNA levels, as we did not directly measure dopamine and acetylcholine receptor levels in our cohort, we can only speculate about the molecular mechanisms relying on previous studies demonstrating functional consequences of the chosen SNPs. Hence, it remains to be determined whether and how genetic variation in dopaminergic and cholinergic signaling to cortical signal-to-noise may directly affect differential engagement of DLPFC at a cellular level, especially in humans. Second, our neuropsychological findings are based on a relatively small group size for the CHRNA5 AA/DRD2 GT genotype (N = 10). Although computation of the maximal effect size d and achieved power (respectively, 1.1 and 0.82) support their statistical robustness, replication of our results is necessary. Furthermore, Levene's test indicates homogeneity of variance of behavioral performance (difference between 2-Back and 1-Back accuracy) across DRD2/CHRNA5 genotype groups (MS Effect = 106.33; MS Error = 85.30; F = 1.24; p = 0.28) supporting that our results were not influenced by unequal population variances. Another caveat of our study is that our neuropsychological and fMRI findings could be affected by tobacco use of subjects. Rs16969968 has been associated with nicotine dependence [55], although it does not alter per se sensitivity of α4β2α5 nAChRs to nicotine [59]. However, the additional analyses we performed including smoking status as covariate allowed us to exclude it as confounding factor.
The present study advances our understanding of the in vivo interactions between dopamine and acetylcholine signaling in the prefrontal cortex, specifically through the DRD2 and CHRNA5 receptors. Our observations of these gene-gene interactions on neurophysiology and cognition begin to build a more solid foundation for explaining the neurobiology underlying complex human behaviors and lend insight into disease susceptibility. Furthermore, our results have relevant potential implications for the therapeutic approach of various neurological and psychiatric disorders in which altered cholinergic transmission potentially contributes to cognitive deficits, such as those observed in schizophrenia.
Supporting Information
Correlation between BOLD fMRI in prefrontal cortex and Working Memory accuracy in CHRNA5-AA/DRD2-GT subjects (S1a) and in CHRNA5-GG/DRD2-GT subjects (S1b).
(DOCX)
Behavioral data (mean ± SD) at the 2-Back task for each genotype group.
(DOCX)
Acknowledgments
This study was in part supported by a grant from the US National Institutes of Health, General Medical Sciences, U01 GM092655.
Funding Statement
This study was in part supported by a grant from the US National Institutes of Health, General Medical Sciences, U01 GM092655. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1. Ando J, Ono Y, Wright MJ (2001) Genetic structure of spatial and verbal working memory. Behav Genet 31: 615–624. [DOI] [PubMed] [Google Scholar]
- 2. Luciano M, Wright M, Smith GA, Geffen GM, Geffen LB, et al. (2001) Genetic covariance among measures of information processing speed, working memory, and IQ. Behav Genet 31: 581–592. [DOI] [PubMed] [Google Scholar]
- 3. Polderman TJ, Gosso MF, Posthuma D, Van Beijsterveldt TC, Heutink P, et al. (2006) A longitudinal twin study on IQ, executive functioning, and attention problems during childhood and early adolescence. Acta Neurol Belg 106: 191–207. [PubMed] [Google Scholar]
- 4. Goldman-Rakic PS (1995) Architecture of the prefrontal cortex and the central executive. Ann N Y Acad Sci 769: 71–83. [DOI] [PubMed] [Google Scholar]
- 5. Wang Y, Markram H, Goodman PH, Berger TK, Ma J, et al. (2006) Heterogeneity in the pyramidal network of the medial prefrontal cortex. Nat Neurosci 9: 534–542. [DOI] [PubMed] [Google Scholar]
- 6. Blokland GA, McMahon KL, Thompson PM, Martin NG, de Zubicaray GI, et al. (2011) Heritability of working memory brain activation. J Neurosci 31: 10882–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Callicott JH, Egan MF, Mattay VS, Bertolino A, Bone AD, et al. (2003) Abnormal fMRI response of the dorsolateral prefrontal cortex in cognitively intact siblings of patients with schizophrenia. Am J Psychiatry 160: 709–719. [DOI] [PubMed] [Google Scholar]
- 8. Robbins TW, Roberts AC (2007) Differential regulation of fronto-executive function by the monoamines and acetylcholine. Cereb Cortex 17 Suppl 1i151–160. [DOI] [PubMed] [Google Scholar]
- 9. Blasi G, Napolitano F, Ursini G, Taurisano P, Romano R, et al. (2011) DRD2/AKT1 interaction on D2 c-AMP independent signaling, attentional processing, and response to olanzapine treatment in schizophrenia. Proc Natl Acad Sci U S A 108: 1158–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papaleo F, Burdick MC, Callicott JH, Weinberger DR (2013) Epistatic interaction between COMT and DTNBP1 modulates prefrontal function in mice and in humans. Mol Psychiatry. [DOI] [PMC free article] [PubMed]
- 11. Tan HY, Chen Q, Sust S, Buckholtz JW, Meyers JD, et al. (2007) Epistasis between catechol-O-methyltransferase and type II metabotropic glutamate receptor 3 genes on working memory brain function. Proc Natl Acad Sci U S A 104: 12536–12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tritsch NX, Sabatini BL (2012) Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron 76: 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang M, Vijayraghavan S, Goldman-Rakic PS (2004) Selective D2 receptor actions on the functional circuitry of working memory. Science 303: 853–856. [DOI] [PubMed] [Google Scholar]
- 14. Seamans JK, Yang CR (2004) The principal features and mechanisms of dopamine modulation in the prefrontal cortex. Prog Neurobiol 74: 1–58. [DOI] [PubMed] [Google Scholar]
- 15. Zhou X, Qi XL, Douglas K, Palaninathan K, Kang HS, et al. (2011) Cholinergic modulation of working memory activity in primate prefrontal cortex. J Neurophysiol 106: 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Croxson PL, Kyriazis DA, Baxter MG (2011) Cholinergic modulation of a specific memory function of prefrontal cortex. Nat Neurosci 14: 1510–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chudasama Y, Dalley JW, Nathwani F, Bouger P, Robbins TW (2004) Cholinergic modulation of visual attention and working memory: dissociable effects of basal forebrain 192-IgG-saporin lesions and intraprefrontal infusions of scopolamine. Learn Mem 11: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Granon S, Poucet B, Thinus-Blanc C, Changeux JP, Vidal C (1995) Nicotinic and muscarinic receptors in the rat prefrontal cortex: differential roles in working memory, response selection and effortful processing. Psychopharmacology (Berl) 119: 139–144. [DOI] [PubMed] [Google Scholar]
- 19. Levin ED, McClernon FJ, Rezvani AH (2006) Nicotinic effects on cognitive function: behavioral characterization, pharmacological specification, and anatomic localization. Psychopharmacology (Berl) 184: 523–539. [DOI] [PubMed] [Google Scholar]
- 20. Chan WK, Wong PT, Sheu FS (2007) Frontal cortical alpha7 and alpha4beta2 nicotinic acetylcholine receptors in working and reference memory. Neuropharmacology 52: 1641–1649. [DOI] [PubMed] [Google Scholar]
- 21. Levin ED, Simon BB (1998) Nicotinic acetylcholine involvement in cognitive function in animals. Psychopharmacology (Berl) 138: 217–230. [DOI] [PubMed] [Google Scholar]
- 22. Levin ED, Rose JE, Abood L (1995) Effects of nicotinic dimethylaminoethyl esters on working memory performance of rats in the radial-arm maze. Pharmacol Biochem Behav 51: 369–373. [DOI] [PubMed] [Google Scholar]
- 23. Woodruff-Pak DS (2003) Mecamylamine reversal by nicotine and by a partial alpha7 nicotinic acetylcholine receptor agonist (GTS-21) in rabbits tested with delay eyeblink classical conditioning. Behav Brain Res 143: 159–167. [DOI] [PubMed] [Google Scholar]
- 24. Spinelli S, Ballard T, Feldon J, Higgins GA, Pryce CR (2006) Enhancing effects of nicotine and impairing effects of scopolamine on distinct aspects of performance in computerized attention and working memory tasks in marmoset monkeys. Neuropharmacology 51: 238–250. [DOI] [PubMed] [Google Scholar]
- 25. Loughead J, Ray R, Wileyto EP, Ruparel K, Sanborn P, et al. (2010) Effects of the alpha4beta2 partial agonist varenicline on brain activity and working memory in abstinent smokers. Biol Psychiatry 67: 715–721. [DOI] [PubMed] [Google Scholar]
- 26. Howe MN, Price IR (2001) Effects of transdermal nicotine on learning, memory, verbal fluency, concentration, and general health in a healthy sample at risk for dementia. Int Psychogeriatr 13: 465–475. [DOI] [PubMed] [Google Scholar]
- 27. Gotti C, Zoli M, Clementi F (2006) Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci 27: 482–491. [DOI] [PubMed] [Google Scholar]
- 28. Sihver W, Gillberg PG, Nordberg A (1998) Laminar distribution of nicotinic receptor subtypes in human cerebral cortex as determined by [3H](-)nicotine, [3H]cytisine and [3H]epibatidine in vitro autoradiography. Neuroscience 85: 1121–1133. [DOI] [PubMed] [Google Scholar]
- 29. Poorthuis RB, Bloem B, Schak B, Wester J, de Kock CP, et al. (2013) Layer-specific modulation of the prefrontal cortex by nicotinic acetylcholine receptors. Cereb Cortex 23: 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gotti C, Clementi F, Fornari A, Gaimarri A, Guiducci S, et al. (2009) Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem Pharmacol 78: 703–711. [DOI] [PubMed] [Google Scholar]
- 31. Ramirez-Latorre J, Yu CR, Qu X, Perin F, Karlin A, et al. (1996) Functional contributions of alpha5 subunit to neuronal acetylcholine receptor channels. Nature 380: 347–351. [DOI] [PubMed] [Google Scholar]
- 32. Marks MJ, Pauly JR, Gross SD, Deneris ES, Hermans-Borgmeyer I, et al. (1992) Nicotine binding and nicotinic receptor subunit RNA after chronic nicotine treatment. J Neurosci 12: 2765–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salas R, Orr-Urtreger A, Broide RS, Beaudet A, Paylor R, et al. (2003) The nicotinic acetylcholine receptor subunit alpha 5 mediates short-term effects of nicotine in vivo. Mol Pharmacol 63: 1059–1066. [DOI] [PubMed] [Google Scholar]
- 34. Wada E, McKinnon D, Heinemann S, Patrick J, Swanson LW (1990) The distribution of mRNA encoded by a new member of the neuronal nicotinic acetylcholine receptor gene family (alpha 5) in the rat central nervous system. Brain Res 526: 45–53. [DOI] [PubMed] [Google Scholar]
- 35. Winzer-Serhan UH, Leslie FM (2005) Expression of alpha5 nicotinic acetylcholine receptor subunit mRNA during hippocampal and cortical development. J Comp Neurol 481: 19–30. [DOI] [PubMed] [Google Scholar]
- 36. Bailey CD, De Biasi M, Fletcher PJ, Lambe EK (2010) The nicotinic acetylcholine receptor alpha5 subunit plays a key role in attention circuitry and accuracy. J Neurosci 30: 9241–9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bailey CD, Alves NC, Nashmi R, De Biasi M, Lambe EK (2012) Nicotinic alpha5 subunits drive developmental changes in the activation and morphology of prefrontal cortex layer VI neurons. Biol Psychiatry 71: 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levin ED, Rose JE (1991) Nicotinic and muscarinic interactions and choice accuracy in the radial-arm maze. Brain Res Bull 27: 125–128. [DOI] [PubMed] [Google Scholar]
- 39. Levin ED, Castonguay M, Ellison GD (1987) Effects of the nicotinic receptor blocker mecamylamine on radial-arm maze performance in rats. Behav Neural Biol 48: 206–212. [DOI] [PubMed] [Google Scholar]
- 40. Levin ED, McGurk SR, South D, Butcher LL (1989) Effects of combined muscarinic and nicotinic blockade on choice accuracy in the radial-arm maze. Behav Neural Biol 51: 270–277. [DOI] [PubMed] [Google Scholar]
- 41. McGurk SR, Levin ED, Butcher LL (1989) Radial-arm maze performance in rats is impaired by a combination of nicotinic-cholinergic and D2 dopaminergic antagonist drugs. Psychopharmacology (Berl) 99: 371–373. [DOI] [PubMed] [Google Scholar]
- 42. Levin ED, McGurk SR, Rose JE, Butcher LL (1989) Reversal of a mecamylamine-induced cognitive deficit with the D2 agonist, LY 171555. Pharmacol Biochem Behav 33: 919–922. [DOI] [PubMed] [Google Scholar]
- 43. Usiello A, Baik JH, Rouge-Pont F, Picetti R, Dierich A, et al. (2000) Distinct functions of the two isoforms of dopamine D2 receptors. Nature 408: 199–203. [DOI] [PubMed] [Google Scholar]
- 44. Centonze D, Gubellini P, Usiello A, Rossi S, Tscherter A, et al. (2004) Differential contribution of dopamine D2S and D2L receptors in the modulation of glutamate and GABA transmission in the striatum. Neuroscience 129: 157–166. [DOI] [PubMed] [Google Scholar]
- 45. Zhang Y, Bertolino A, Fazio L, Blasi G, Rampino A, et al. (2007) Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci U S A 104: 20552–20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bertolino A, Fazio L, Caforio G, Blasi G, Rampino A, et al. (2009) Functional variants of the dopamine receptor D2 gene modulate prefronto-striatal phenotypes in schizophrenia. Brain 132: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bertolino A, Taurisano P, Pisciotta NM, Blasi G, Fazio L, et al. (2010) Genetically determined measures of striatal D2 signaling predict prefrontal activity during working memory performance. PLoS One 5: e9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sasabe T, Furukawa A, Matsusita S, Higuchi S, Ishiura S (2007) Association analysis of the dopamine receptor D2 (DRD2) SNP rs1076560 in alcoholic patients. Neurosci Lett 412: 139–142. [DOI] [PubMed] [Google Scholar]
- 49. Moyer RA, Wang D, Papp AC, Smith RM, Duque L, et al. (2011) Intronic polymorphisms affecting alternative splicing of human dopamine D2 receptor are associated with cocaine abuse. Neuropsychopharmacology 36: 753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Clarke TK, Weiss AR, Ferarro TN, Kampman KM, Dackis CA, et al. (2014) The Dopamine Receptor D2 (DRD2) SNP rs1076560 is Associated with Opioid Addiction. Ann Hum Genet 78: 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Voisey J, Swagell CD, Hughes IP, van Daal A, Noble EP, et al. (2012) A DRD2 and ANKK1 haplotype is associated with nicotine dependence. Psychiatry Res 196: 285–289. [DOI] [PubMed] [Google Scholar]
- 52. Wei J, Chu C, Wang Y, Yang Y, Wang Q, et al. (2012) Association study of 45 candidate genes in nicotine dependence in Han Chinese. Addict Behav 37: 622–626. [DOI] [PubMed] [Google Scholar]
- 53. Connor JP, Young RM, Lawford BR, Saunders JB, Ritchie TL, et al. (2007) Heavy nicotine and alcohol use in alcohol dependence is associated with D2 dopamine receptor (DRD2) polymorphism. Addict Behav 32: 310–319. [DOI] [PubMed] [Google Scholar]
- 54. Wang TY, Lee SY, Chen SL, Huang SY, Chang YH, et al. (2013) Association between DRD2, 5-HTTLPR, and ALDH2 genes and specific personality traits in alcohol- and opiate-dependent patients. Behav Brain Res 250: 285–292. [DOI] [PubMed] [Google Scholar]
- 55. Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, et al. (2008) Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry 165: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, et al. (2007) Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 16: 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith RM, Alachkar H, Papp AC, Wang D, Mash DC, et al. (2011) Nicotinic alpha5 receptor subunit mRNA expression is associated with distant 5′ upstream polymorphisms. Eur J Hum Genet 19: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang JC, Spiegel N, Bertelsen S, Le N, McKenna N, et al. (2013) Cis-Regulatory Variants Affect CHRNA5 mRNA Expression in Populations of African and European Ancestry. PLoS One 8: e80204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuryatov A, Berrettini W, Lindstrom J (2011) Acetylcholine receptor (AChR) alpha5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (alpha4beta2)(2)alpha5 AChR function. Mol Pharmacol 79: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, et al. (2007) Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet 16: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, et al. (2009) Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet 150B: 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Winterer G, Mittelstrass K, Giegling I, Lamina C, Fehr C, et al. (2010) Risk gene variants for nicotine dependence in the CHRNA5-CHRNA3-CHRNB4 cluster are associated with cognitive performance. Am J Med Genet B Neuropsychiatr Genet 153B: 1448–1458. [DOI] [PubMed] [Google Scholar]
- 63. Hong LE, Yang X, Wonodi I, Hodgkinson CA, Goldman D, et al. (2011) A CHRNA5 allele related to nicotine addiction and schizophrenia. Genes Brain Behav 10: 530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jackson KJ, Fanous AH, Chen J, Kendler KS, Chen X (2013) Variants in the 15q25 gene cluster are associated with risk for schizophrenia and bipolar disorder. Psychiatr Genet 23: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Falcone M, Smith RM, Chenoweth MJ, Kumar Bhattacharjee A, Kelsoe JR, et al. (2013) Neuroimaging in psychiatric pharmacogenetics research: the promise and pitfalls. Neuropsychopharmacology 38: 2327–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lambe EK, Krimer LS, Goldman-Rakic PS (2000) Differential postnatal development of catecholamine and serotonin inputs to identified neurons in prefrontal cortex of rhesus monkey. J Neurosci 20: 8780–8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.First M, Gibbon M, Spitzer R, Williams J (1996) Guide for the structured clinical interview for DSM-IV axis I disorders-Research version. New York: Biometrics Research. [Google Scholar]
- 68. Oldfield RC (1971) The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia 9: 97–113. [DOI] [PubMed] [Google Scholar]
- 69. Hong LE, Hodgkinson CA, Yang Y, Sampath H, Ross TJ, et al. (2010) A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc Natl Acad Sci U S A 107: 13509–13514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tan HY, Nicodemus KK, Chen Q, Li Z, Brooke JK, et al. (2008) Genetic variation in AKT1 is linked to dopamine-associated prefrontal cortical structure and function in humans. J Clin Invest 118: 2200–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ashburner J (2007) A fast diffeomorphic image registration algorithm. Neuroimage 38: 95–113. [DOI] [PubMed] [Google Scholar]
- 72. Ashburner J, Friston KJ (2005) Unified segmentation. Neuroimage 26: 839–851. [DOI] [PubMed] [Google Scholar]
- 73. Ashburner J, Friston KJ (2000) Voxel-based morphometry—the methods. Neuroimage 11: 805–821. [DOI] [PubMed] [Google Scholar]
- 74. Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE (2004) Nonstationary cluster-size inference with random field and permutation methods. Neuroimage 22: 676–687. [DOI] [PubMed] [Google Scholar]
- 75. Markett SA, Montag C, Reuter M (2010) The association between dopamine DRD2 polymorphisms and working memory capacity is modulated by a functional polymorphism on the nicotinic receptor gene CHRNA4. J Cogn Neurosci 22: 1944–1954. [DOI] [PubMed] [Google Scholar]
- 76. Sullivan D, Pinsonneault JK, Papp AC, Zhu H, Lemeshow S, et al. (2013) Dopamine transporter DAT and receptor DRD2 variants affect risk of lethal cocaine abuse: a gene-gene-environment interaction. Transl Psychiatry 3: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, et al. (2001) Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A 98: 6917–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mattay VS, Goldberg TE, Fera F, Hariri AR, Tessitore A, et al. (2003) Catechol O-methyltransferase val158-met genotype and individual variation in the brain response to amphetamine. Proc Natl Acad Sci U S A 100: 6186–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mattay VS, Tessitore A, Callicott JH, Bertolino A, Goldberg TE, et al. (2002) Dopaminergic modulation of cortical function in patients with Parkinson's disease. Ann Neurol 51: 156–164. [DOI] [PubMed] [Google Scholar]
- 80. Calabresi P, Picconi B, Parnetti L, Di Filippo M (2006) A convergent model for cognitive dysfunctions in Parkinson's disease: the critical dopamine-acetylcholine synaptic balance. Lancet Neurol 5: 974–983. [DOI] [PubMed] [Google Scholar]
- 81. Kuehn BM (2006) Link between smoking and mental illness may lead to treatments. JAMA 295: 483–484. [DOI] [PubMed] [Google Scholar]
- 82. Pickel VM, Chan J, Nirenberg MJ (2002) Region-specific targeting of dopamine D2-receptors and somatodendritic vesicular monoamine transporter 2 (VMAT2) within ventral tegmental area subdivisions. Synapse 45: 113–124. [DOI] [PubMed] [Google Scholar]
- 83. Kassam SM, Herman PM, Goodfellow NM, Alves NC, Lambe EK (2008) Developmental excitation of corticothalamic neurons by nicotinic acetylcholine receptors. J Neurosci 28: 8756–8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zoli M, Moretti M, Zanardi A, McIntosh JM, Clementi F, et al. (2002) Identification of the nicotinic receptor subtypes expressed on dopaminergic terminals in the rat striatum. J Neurosci 22: 8785–8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Exley R, McIntosh JM, Marks MJ, Maskos U, Cragg SJ (2012) Striatal alpha5 nicotinic receptor subunit regulates dopamine transmission in dorsal striatum. J Neurosci 32: 2352–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Threlfell S, Lalic T, Platt NJ, Jennings KA, Deisseroth K, et al. (2012) Striatal dopamine release is triggered by synchronized activity in cholinergic interneurons. Neuron 75: 58–64. [DOI] [PubMed] [Google Scholar]
- 87. Mechawar N, Descarries L (2001) The cholinergic innervation develops early and rapidly in the rat cerebral cortex: a quantitative immunocytochemical study. Neuroscience 108: 555–567. [DOI] [PubMed] [Google Scholar]
- 88. Levitt P, Harvey JA, Friedman E, Simansky K, Murphy EH (1997) New evidence for neurotransmitter influences on brain development. Trends Neurosci 20: 269–274. [DOI] [PubMed] [Google Scholar]
- 89. Sherren N, Pappas BA (2005) Selective acetylcholine and dopamine lesions in neonatal rats produce distinct patterns of cortical dendritic atrophy in adulthood. Neuroscience 136: 445–456. [DOI] [PubMed] [Google Scholar]
- 90. Kristt DA (1979) Acetylcholinesterase-containing neurons of layer VIb in immature neocortex: possible component of an early formed intrinsic cortical circuit. Anat Embryol (Berl) 157: 217–226. [DOI] [PubMed] [Google Scholar]
- 91. Lipton SA, Frosch MP, Phillips MD, Tauck DL, Aizenman E (1988) Nicotinic antagonists enhance process outgrowth by rat retinal ganglion cells in culture. Science 239: 1293–1296. [DOI] [PubMed] [Google Scholar]
- 92. Pugh PC, Berg DK (1994) Neuronal acetylcholine receptors that bind alpha-bungarotoxin mediate neurite retraction in a calcium-dependent manner. J Neurosci 14: 889–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between BOLD fMRI in prefrontal cortex and Working Memory accuracy in CHRNA5-AA/DRD2-GT subjects (S1a) and in CHRNA5-GG/DRD2-GT subjects (S1b).
(DOCX)
Behavioral data (mean ± SD) at the 2-Back task for each genotype group.
(DOCX)