Abstract
Goat is an important agricultural animal for meat production. Functional studies have demonstrated that microRNAs (miRNAs) regulate gene expression at the post-transcriptional level and play an important role in various biological processes. Although studies on miRNAs expression profiles have been performed in various animals, relatively limited information about goat muscle miRNAs has been reported. To investigate the miRNAs involved in regulating different periods of skeletal muscle development, we herein performed a comprehensive research for expression profiles of caprine miRNAs during two developmental stages of skeletal muscles: fetal stage and six month-old stage. As a result, 15,627,457 and 15,593,721 clean reads were obtained from the fetal goat library (FC) and the six month old goat library (SMC), respectively. 464 known miRNAs and 83 novel miRNA candidates were identified. Furthermore, by comparing the miRNA profile, 336 differentially expressed miRNAs were identified and then the potential targets of the differentially expressed miRNAs were predicted. To understand the regulatory network of miRNAs during muscle development, the mRNA expression profiles for the two development stages were characterized and 7322 differentially expressed genes (DEGs) were identified. Then the potential targets of miRNAs were compared to the DEGs, the intersection of the two gene sets were screened out and called differentially expressed targets (DE-targets), which were involved in 231 pathways. Ten of the 231 pathways that have smallest P-value were shown as network figures. Based on the analysis of pathways and networks, we found that miR-424-5p and miR-29a might have important regulatory effect on muscle development, which needed to be further studied. This study provided the first global view of the miRNAs in caprine muscle tissues. Our results help elucidation of complex regulatory networks between miRNAs and mRNAs and for the study of muscle development.
Introduction
MicroRNAs (miRNAs) are small (∼22 nucleotides) noncoding RNAs which are highly conserved among mammals [1]. They bind primarily to the 3′-UTR of target mRNAs to regulate their translation and accelerate their decay [1], [2]. Hundreds of miRNAs have been discovered in recent years and several have been functionally characterized. miRNAs play critical fine-tuning roles in diverse biological processes, such as cell proliferation and differentiation [3], [4], tumorigenesis [5], the morphogenesis of special organs [6] and viral defense [7].
Skeletal muscle occupies approximately 40% of the body weight [8], and its development is a complex process involving multiple factors which regulate the proliferation of myoblasts, their exit from the cell cycle and subsequent differentiation into multinucleated myotubes [9]. The discovery of miRNAs has open up a new field of factors controlling skeletal muscle development. Previous studies showed that miRNAs had important effect on skeletal muscle development. They could regulate myogenesis or hypertrophy under physiological and patho-physiological conditions. Skeletal muscle related miRNAs influence multiple facets of muscle development and function through the regulation on key genes controlling myogenesis [10]–[12]. Recently, several miRNAs that highly-enriched in skeletal muscle have been identified. The muscle specific miR-1, miR-133 and miR-206 are perhaps the most studied and best-characterized miRNAs. They play important roles in myoblast proliferation, differentiation, and hypertrophy [10]–[11], [13]–[14]. In addition to muscle specific miRNAs, non-muscle-specific miRNAs also regulate skeletal muscle differentiation in multiple ways. Muscle differentiation-related genes such as MyoD, MEF2, Pax3 and YY1, are regulated by these non-muscle-specific miRNAs [15]–[18]. For example, miR-699a is able to inhibit skeletal muscle differentiation,and shows down regulated expression during skeletal muscle differentiation [15]. Thus, globally identifying and characterizing skeletal muscle miRNAs during various phases of muscle development will significantly enhance our understanding of skeletal muscle biology and function.
Previous studies indicated that many miRNAs have multiple target genes and most of the genes are targeted by multiple miRNAs [19]. For example, YY1 is targeted by miRNA-1 [20] and miRNA-29a [21]. YY1 can inhibit skeletal muscle cell differentiation by inhibiting the synthesis of late-stage differentiation genes including muscle creatine kinase and myosin heavy chain [22]. Pax3 was reported to be targeted by miRNA-1 [11], miRNA-27b [23] and miRNA-206 [24]. High levels of Pax3 interfere with muscle cell differentiation, in both the embryo and adult [23]. Skeletal muscle stem cells are regulated by Pax3/7. Pax7 was targeted by miRNA-1 [25], miRNA-206 [26] and miRNA-486 [26]. On the other hand, the experimentally proved target genes for miRNA-1 include YY1, HDAC4, Cx43, Pax3 and Pax7. Thus, muscle miRNAs and their target genes may integrate into complex regulatory networks, an integrated analysis of expression of miRNAs and their targets can be helpful to identify miRNA/mRNA modules which may be involved in muscle development regulation.
Goat is one of the most important meat-producing livestock animals grown worldwide. Many studies have focused on miRNA, and a suite of miRNAs highly-enriched in skeletal muscle has been identified [14], [27]–[30]. However, few works have been done on the identification of goat muscle miRNAs. Therefore, global identification of the known and novel miRNAs involved in goat skeletal muscle development is essential. In addition, the mRNA profiles were analyzed in this study. Based on these, we briefly analyzed the regulation network of miRNA-mRNA, which will significantly enhance our understanding the effect of miRNAs on muscle development for goat.
Materials and Methods
Ethics statement
Huanghuai Goats in present study were bought from HeQiao caprine Co., Ltd. Huanghuai goat is famous for its meat-producing in China. All animals were raised and fed under the same standard management conditions according to the No. 5 proclamation of the Ministry of Agriculture, P. R. China. Sample collection was approved by the Animal Care Commission of the College of Animal Science and Technology, Northwest A&F University.
Tissue collection and high-throughput sequencing
Ninety day embryos were collected from a flock of ewes, which were estrus synchronized, at 90 days of pregnancy (gestation period 150 days). Embryos were collected into sterile physiological saline immediately after removal from the reproductive tract of slaughtered ewes at an abattoir. Longissimus thoracis muscle tissues were collected from fetal and six month old Huanghuai goat, and immediately snap-frozen in liquid nitrogen, then stored at −80°C until use.
Total RNAs were extracted from 8 fetal and 8 six month old Huanghuai goat longissimus thoracis. The total RNA was isolated from each sample using the Trizol reagent (Takara, Dalian, China). RNA was quantified using the Nano-Drop ND-2000 spectrophotometer (NanoDrop products, Wilmington, USA) and checked for purity and integrity in a Bioanalyzer-2100 (Agilent Technologies, Inc., Sant a Clara CA, USA). Then the RNAs from fetal and six month old goats were pooled, respectively. Subsequently, low molecular weight RNAs were separated by 15% polyacrylamide gel electrophoresis (PAGE), and RNA molecules in the range of 18–30 nt were enriched and ligated with proprietary adapters to the 5′and 3′termini. A reverse transcription reaction followed by low cycle PCR was performed to obtain sufficient product for Solexa technology (Beijing Genomics Institute, China). In the present study, two miRNA libraries were constructed.
Small RNA sequence analysis
After getting rid of reads with 5' primer contaminants and without 3' primer, removal of redundancy and reads smaller than 18 nt, the clean reads were mapped to the latest caprine genome assembly [http://goat.kiz.ac.cn/GGD/download.htm] using the program SOAP [31]. In order to determine the compositions of small RNA sample, the length distribution analysis was conducted using caprine mRNA [http://goat.kiz.ac.cn/GGD/download.htm], and CDS [http://goat.kiz.ac.cn/GGD/download3.htm], RepeatMasker [http://www.re peatmasker.org] and Sanger Rfam data (version 10.1), according to the length of the small RNA. For example, miRNA is normally 21 nt or 22 nt, siRNA is 24 nt. Subsequently, the remaining reads were searched against the Sanger miRBase (version 19.0) to identify the known miRNAs. Only those small RNAs whose mature and precursor sequences perfectly matched known bovine miRNAs and ovine miRNAs in miRBase were considered to be known caprine miRNAs. To discover potential novel miRNA precursor sequences, unique sequences that have more than 10 hits to the genome or match to known non-coding RNAs were removed. Then the flanking sequences (150 nt upstream and downstream) of each unique sequence were extracted for secondary structure analysis with M fold [http://www.bioinfo.rpi.edu/applications/mfold] and then evaluated by Mireap [http://source forge.net/project s/mireap/]. After prediction, the resulting potential miRNA loci were examined carefully based on the distribution and numbers of small RNAs on the entire precursor regions. Those sequences residing in the stem region of the stem-loop structure and ranging between 16–30 nt with free energy hybridization lower than -18 kcal/mol were considered. Then, all novel miRNA candidates were further subjected to MiPred (http://www.bioinf.seu.edu.cn/miRNA/) to filter out pseudo-pre-miRNAs [32].
MicroRNA expression analysis
To find out the differentially expressed miRNAs, we compared the known and novel miRNA expression profile between two samples. The expression of miRNA was shown in two samples by plotting a Log2-ratio figure and a Scatter Plot. The procedures were shown as below: (1) Normalize the expression of miRNA in two samples (fetal and six month longissimus thoracis) to get the expression of transcript permillion. Normalized expression (NE) = actual miRNA count/total count of clean reads. (2) Calculate fold-change and P-value from the normalized expression. Then generate the Log2-ratio plot and scatter plot. Fold-change formula: fold change = log2 (fetal NE/six month NE). P-value formula:
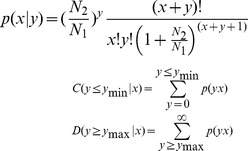 |
The x and y represented normalized expression levels, and the N1 and N2 represented total count of clean reads of a given miRNA in small RNA libraries of the fetal and six month stage, respectively.
Using Stem-loop RT-qPCR method to validation of differentially-expressed miRNAs
In order to evaluate the repeatability and reproducibility of miRNA expression data obtained by Small RNA-Seq, a Stem-loop real-time reverse transcription polymerase chain reaction (RT-PCR) with SYBR Green was used for the analysis of miRNA expression [33]. The isolated RNA of individual samples was converted to cDNA with a RT primer mixture (250 nM) using PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) in a total volume of 20µl containing 1µg of total RNA. The cDNA was then used for real-time PCR quantification of miRNA using the miRNA specific forward primer and the universal reverse primer. 18 S rRNA was used as the reference gene in the qRT-PCR detection of goat miRNAs. Real-time quantitative PCR was performed using an ABI Stepone plus Real Time Detection System (Applied Biosystems, Inc., Foster City, CA) and SYBR Green PCR Master Mix (TaKaRa, Dalian, China) in a 20µl reaction volume. Each sample was analyzed by triplicate. The PCR mix included 100 ng cDNA for each miRNA, 0.4µM forward and reverse primers, and 10µl 2× SYBR Green PCR Master Mix, thermal cycle was: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s. The threshold cycle (Ct) was defined as the cycle number at which the fluorescence intensity passed a predetermined threshold. The quantification of each miRNA relative to 18 S rRNA gene was calculated.
RNA-Seq (Transcriptome)
The mRNA was isolated from total RNA (described as above) to construct cDNA libraries. The cDNA was synthesized using the mRNA fragments as templates. Agilent 2100 Bioanaylzer and ABI Stepone Plus Real-Time PCR System were used in quantification and qualification of the sample library. The library was sequenced using Illumina HiSeq TM 2000. Then, the primary sequencing data was subjected to quality control (QC). After remove reads with adapters, reads in which unknown bases are more than 5% and low quality reads (we define the low quality base to be the base whose sequencing quality is no more than 10), raw reads are filtered into clean reads, which were aligned to the latest goat genome assembly [http://www.ncbi.nlm.nih.gov/bioproject/PRJNA158393] using SOAP aligner/SOAP2 [34]. No more than 5 mismatches are allowed in the alignment. Mapping was performed to the entire genome and gene.
Screening of DEGs and validated by RT-qPCR
Referring to Audic and Claverie (1997) [35], we have developed a strict algorithm to identify differentially expressed genes between two samples, and the method used is described as follow: denote the number of unambiguous clean tags (which means reads in RNA_Seq) from gene A as x, given every gene's expression occupies only a small part of the library, x yields to the Poisson distribution:
The total clean tag number of the FC is N1, and total clean tag number of SMC is N2; gene A holds x tags in FC and y tags in SMC. The probability of gene A expressed equally between two samples can be calculated with:
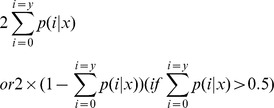 |
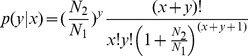 |
P-value corresponds to differential gene expression test. Since DEG analysis generate a large multiplicity problems in which thousands of hypothesis (is gene x differentially expressed between the two groups) are tested simultaneously, correction for false positive (type I errors) and false negative (type II) errors are performed using [36] FDR method. This is a standard approach for multiple hypothesis correction for most of the common used software for differential expression analysis. We used ‘FDR≤0.001 and the absolute value of Log2-Ratio≥1’ as the threshold to judge the significance of gene expression difference.
To validate the sequence data, the isolated RNA of individual samples was converted to cDNA with a RT primer mixture (250 nM) using Prime Script RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) in a total volume of 20µl containing 1µg of total RNA. The cDNA was then used for real-time PCR quantification of candidate DEGs. GAPDH was used as the reference gene in the qRT-PCR detection.
microRNA-mRNA integrated analysis
After small RNA sequencing and RNA-Seq (Transcriptome) analysis of the paired samples, the integrated analysis of differentially-expressed miRNAs and their predicted target genes were performed. Firstly, we predicted the potential targets of known miRNAs. The putative target genes for known miRNAs were searched by aligning the miRNA sequences with 3′UTR sequences of goat mRNA sequence. The target prediction tool RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) [37] was employed and the rules used for target prediction are based on those suggested by Allen et al. (2005) and Schwab et al. (2005) [38], [39], details were in accordance with Ji et al. (2012) [40]. Secondly, the target genes of differentially expressed miRNAs were sorted out. Then we compared these target genes with DEGs, and sorted out the intersections of the two gene sets, called them DE targets. Then we analyzed the regulating networks between differentially-expressed miRNAs and DE targets.
Pathway analysis of differential expression targets
To comprehensively describe the properties of the integrated analysis results, database for Annotation, Visualization, and Integrated Discovery (DAVID) [41]–[42] was used to perform pathway enrichment analysis of DE targets. The results of this integrated analysis of different functional databases (e.g. GO, KEGG pathways, SP-PIR keywords). FDR was used to correct the P-value. KEGG analysis has satisfaction values of P<0.05 and FDR≤0.05. Genes with FDR≤0.05 are considered as significantly enriched target gene candidates.
Results
Small RNA libraries from goat longissimus thoracis
To identify the small RNAs involved in goat muscle development, total RNAs from goat longissimus thoracis at fetal and six month-old stages were used to construct small RNA libraries for deep sequencing. After deleting some contaminant reads, we obtained 15,627,457 clean reads from the fetal caprine (FC) library and 15,593,721 clean reads from the six month old caprine (SMC) library (Table 1). Length distribution analysis showed that most reads ranged from 21 to 23 nt. The percentage of the 22 nt reads in the total reads accounted for 69.42% of the FC library and 79.75% of the SMC library (Figure 1). All Solexa reads were aligned against caprine genome assembly using the program SOAP, thesoap-v0-r2-M0-aclean.fa-Dref_genome.fa.index-omatch_genome.soap. miRNA is the most abundant RNA species in both libraries and represents 81.52% and 82.94 reads in the FC library and the SMC library (Table 2), respectively.
Table 1. Summary of small RNA sequencing date.
| Type | Fetal caprine muscle tissue | Six month-old caprine muscle tissue | ||
| Count | % | Count | % | |
| total_reads | 15731062 | 15676284 | ||
| high_quality | 15685279 | 100% | 15630959 | 100% |
| 3'adapter_null | 2683 | 0.02% | 3257 | 0.02% |
| insert_null | 2119 | 0.01% | 636 | 0.00% |
| 5'adapter_contaminants | 19517 | 0.12% | 17252 | 0.11% |
| smaller_than_18 nt | 33482 | 0.21% | 16062 | 0.10% |
| polyA | 21 | 0.00% | 31 | 0.00% |
| clean_reads | 15627457 | 99.63% | 15593721 | 99.76% |
Figure 1. Length distribution of small RNAs in SMC and FC libraries.
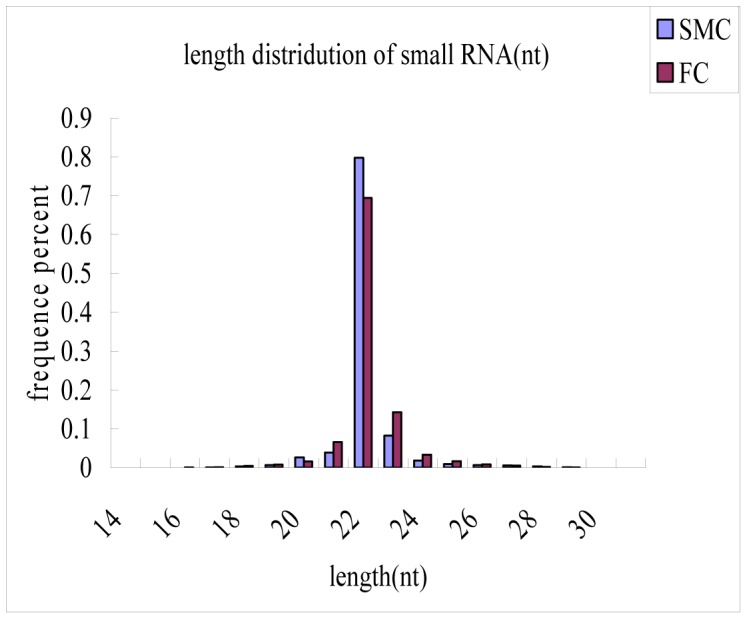
SMC: six month old caprine muscle tissue; FC: fetal caprine muscle tissue.
Table 2. Distribution of the genome-mapped sequence reads in small RNA libraries.
| Locus | Fetal caprine muscle tissue | Six month-old caprine muscle tissue | ||
| Unique sequences | Reads | Unique sequences | Reads | |
| Total | 317200 (100%) | 15627457 (100%) | 249940(100%) | 15593721 (100%) |
| miRNA | 4229 (1.33%) | 12740265 (81.52%) | 2679 (1.07%) | 12934004 (82.94%) |
| exon_antisense | 892 (0.28%) | 965 (0.01%) | 396 (0.16%) | 430 (0.00%) |
| exon_sense | 28465 (8.97%) | 29875 (0.19%) | 35305 (14.13%) | 38078 (0.24%) |
| intron_antisense | 2087 (0.66%) | 5792 (0.04%) | 1045 (0.42%) | 2431 (0.02%) |
| intron_sense | 12410 (3.91%) | 54645 (0.35%) | 6261 (2.51%) | 34197 (0.22%) |
| rRNA | 42174 (13.30%) | 435237 (2.79%) | 38504 (15.41%) | 355165 (2.28%) |
| scRNA | 15 (0.00%) | 17 (0.00%) | 10 (0.00%) | 10 (0.00%) |
| snRNA | 2057 (0.65%) | 6055 (0.04%) | 1542 (0.62%) | 4218 (0.03%) |
| snoRNA | 1649 (0.52%) | 7294 (0.05%) | 1001 (0.40%) | 4476 (0.03%) |
| tRNA | 8800 (2.77%) | 54890 (0.35%) | 8510 (3.40%) | 65835 (0.42%) |
| unann | 214422 (67.60%) | 2292422 (14.67%) | 154687 (61.89%) | 2154877 (13.82%) |
Identification and profiling of known caprine miRNAs
Because caprine miRNAs database is not available in miRbase version 19.0 (http://www.mirbase.org/index.shtml), we compared tentative caprine miRNA sequences with the known bovine and ovine sequences. There are currently 821 miRNA precursors and 858 mature miRNAs containing 605 miRNAs, 127 miRNA-5p, 126 miRNA-3p in currently miRBbase. By Blastn searched against the miRBbase19.0, 4,283 unique sequences (12,740,411 reads) were annotated as miRNA candidates in the FC library as well as 2720 unique sequences (12,934,111 reads) in the SMC library, the rest were unannotated. The caprine miRNA candidates were then clustered into 457 and 384 categories corresponding to 437 and 383 independent genomic loci in the two libraries (Table 3 ).
Table 3. Summary of known miRNAs in each sample.
| miR | miR-5p | miR-3p | pre-miRs | Unique matched to pre-miRs | Read matched to pre-miRs | |
| Known miRs | 605 | 127 | 126 | 821 | ||
| Fetal caprine muscle tissue | 291 | 82 | 84 | 437 | 4283 | 12740411 |
| Six month caprine muscle tissue | 245 | 64 | 75 | 383 | 2720 | 12934111 |
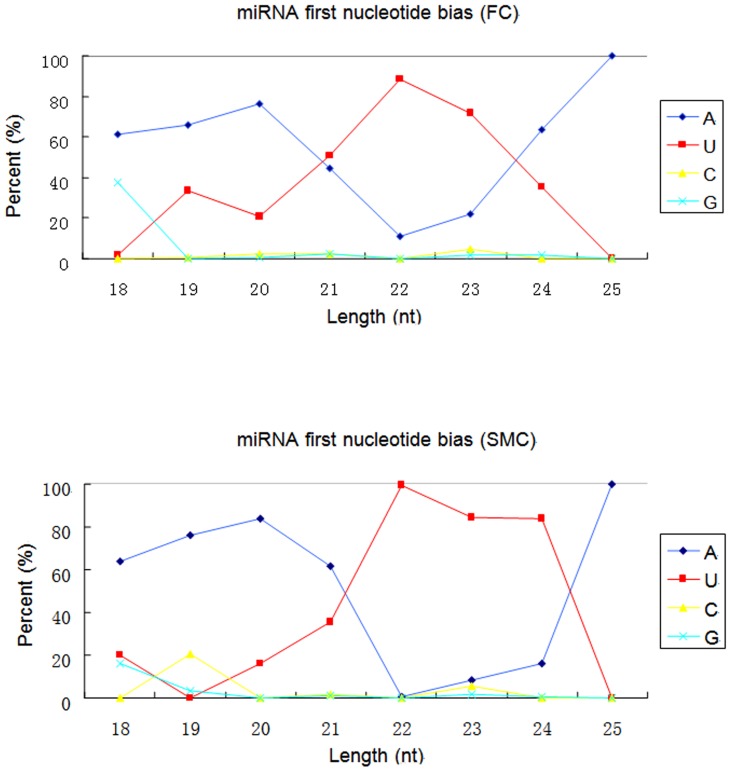
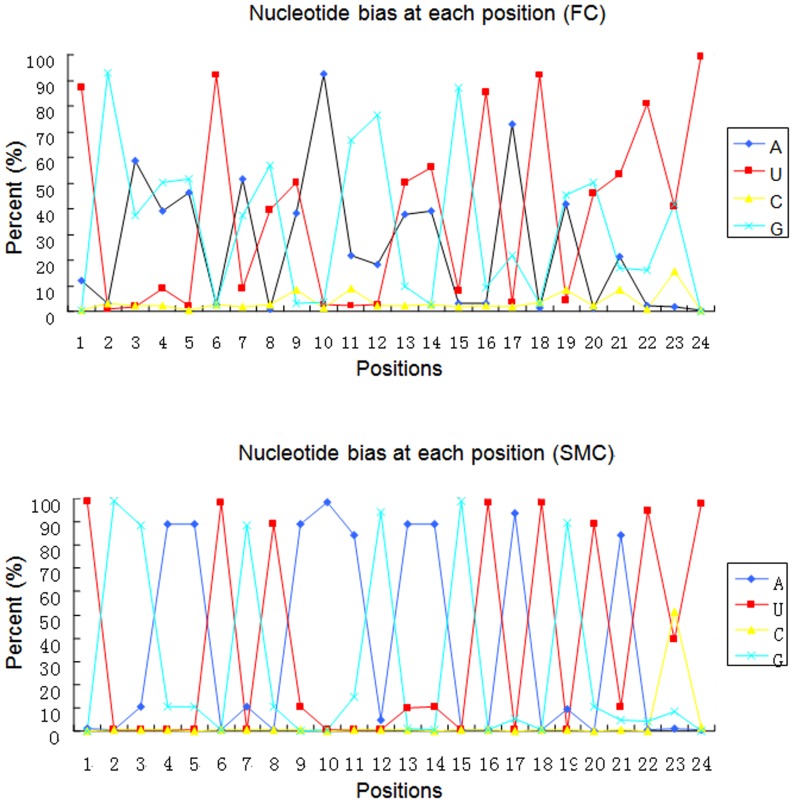
We identified 464 known miRNAs in the two libraries (Table S1). 364 known miRNAs are found in both FC and SMC libraries, and the 20 most abundant miRNAs of them were listed in Table 4. 103 miRNAs are displayed in the lowest sequencing frequencies, with no more than 10 reads in FC or SMC library (Table S1). To better characterize the identified known miRNAs, we conducted nucleotide bias analysis. Nucleotide bias shows that first nucleotide bias was different with various sequence lengths in the two libraries. As shown in Figure 2, U was the most frequent first nucleotide in the miRNAs in both the two library at a proportion of 86.88% in FC and 99.0% in SMC. The (A+U) content was found most abundantly, with an average percentage of 99.9% in SMC and 99.29% in FC, while C or G was seldom used as the first nucleotide (Figure 2 and Table S2a). In addition, an analysis of the four nucleotides at each position along the length revealed that there are different nucleotide biases at each position (Figure 3 and Table S2b). For example, U was predominated at positions 1th, 6th, 16th, 18th, 22th and 24th in both the two library. The G base had a high frequency in the 2th, 3th, 7th, 12th, 15th, 19th positions in SMC library. Notable skewing in the distribution of nucleotides at specific positions included significant low G+C content at position 1 (0.71% in FC, 0.09% in SMC) and position 9 (11.63%in FC, 11.58% in SMC).
Table 4. Twenty most abundance co-expressed miRNAs.
| miR_Name | miRNA abundance | Length | miRNA sequence | |
| Total reads-FC | Total reads-SMC | |||
| chi-miR-206 | 3477840 | 1172192 | 22 | AATGTAAGGAAGTGTGTGGTTT |
| chi-miR-1 | 1929430 | 9986601 | 22 | TGGAATGTAAAGAAGTATGTAT |
| chi-let-7f | 1529359 | 521706 | 22 | TGAGGTAGTAGATTGTATAGTT |
| chi-let-7a-5p | 1521553 | 413551 | 22 | TGAGGTAGTAGGTTGTATAGTT |
| chi-let-7b | 821526 | 225830 | 22 | TGAGGTAGTAGGTTGTGTGGTT |
| chi-let-7c | 632168 | 93292 | 22 | TGAGGTAGTAGGTTGTATGGTT |
| chi-miR-199a-3p | 180808 | 24392 | 22 | ACAGTAGTCTGCACATTGGTTA |
| chi-miR-432 | 177423 | 1786 | 23 | TCTTGGAGTAGGTCATTGGGTGG |
| chi-let-7e | 173110 | 9774 | 22 | TGAGGTAGGAGGTTGTATAGTT |
| chi-miR-3958-3p | 671096 | 22323 | 22 | AGATATTGCACGGTTGATCTCT |
| chi-let-7g | 100560 | 42530 | 22 | TGAGGTAGTAGTTTGTACAGTT |
| chi-miR-499 | 88088 | 30477 | 21 | TTAAGACTTGCAGTGATGTTT |
| chi-let-7i | 79377 | 8212 | 22 | TGAGGTAGTAGTTTGTGCTGTT |
| chi-let-7d | 77925 | 12377 | 22 | AGAGGTAGTAGGTTGCATAGTT |
| chi-miR-103 | 77519 | 15021 | 23 | AGCAGCATTGTACAGGGCTATGA |
| chi-miR-495 | 75454 | 727 | 22 | AAACAAACATGGTGCACTTCTT |
| chi-miR-107 | 71806 | 12072 | 23 | AGCAGCATTGTACAGGGCTATCA |
| chi-miR-140 | 67024 | 49671 | 23 | TACCACAGGGTAGAACCACGGAC |
| chi-miR-128 | 64515 | 17542 | 21 | TCACAGTGAACCGGTCTCTTT |
| chi-miR-543 | 62573 | 489 | 22 | AAACATTCGCGGTGCACTTCTT |
Figure 2. Nucleotide bias of the first base in different sequence lengths.
First nucleotide bias in different sequence lengths from 18–25 nt for known miRNAs; SMC: six month-old caprine muscle tissue; FC: fetal caprine muscle tissue.
Figure 3. Nucleotide bias at each position.
Nucleotide bias at each position (1–24) for known miRNAs. SMC: six month-old caprine muscle tissue; FC: fetal caprine muscle tissue.
Family analysis showed that the 464 known miRNAs belonged to 270 miRNA families in the two libraries. Most of the identified miRNA families have been shown to be conserved in a variety of species. For example, mir-9, mir-34, mir-124, mir-29 and let-7 families have been found in 72, 61, 73, 56, 68 species, respectively. However, some miRNAs have been shown to less conserved, such as miRNA-1246, miRNA-1260a, miRNA-1248, miRNA-1343 and miRNA-1307 only been found in 3, 2, 5, 5, 6 species, respectively (Table S3). In the present study, the largest miRNA family size identified was let-7 and miRNA-376, which consisted of 9 and 8 members, respectively. miRNA-2284, miRNA-2285 and miRNA-30 possessed 7, 6 and 7 members, respectively. The other miRNA families, such as miRNA-1, miRNA-221 and miRNA-206, had only one member.
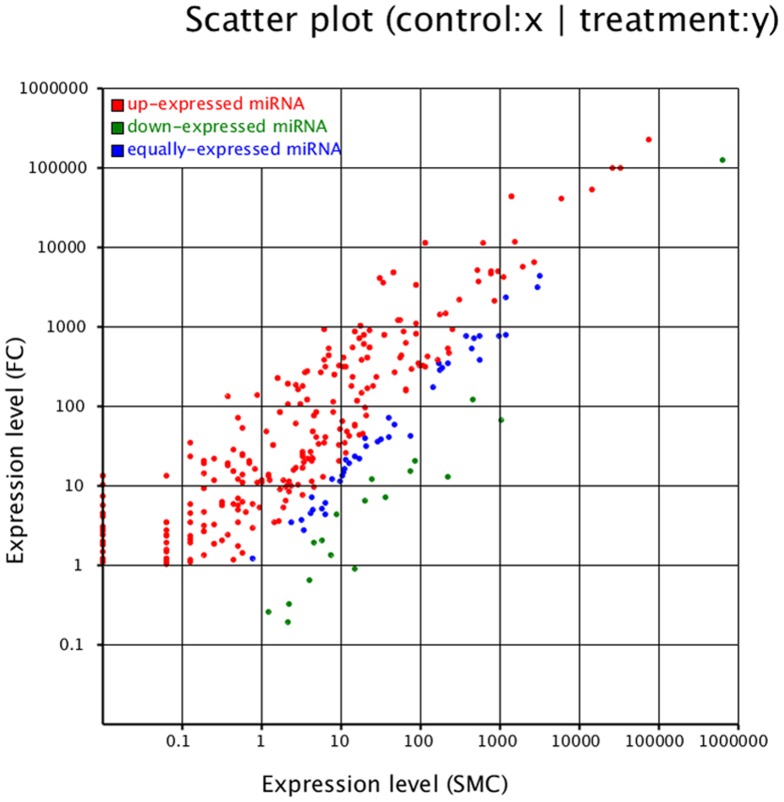
To gain insight into possible stage-dependent roles of miRNAs during the development, we also investigated the differential expression profiles of the goat muscle tissue miRNAs. Expression of known miRNAs in the two libraries was demonstrated by plotting Log2-ratio and scatter plot (Figure 4). The expression profiles between the two libraries are shown in Table S4. According to the changes in relative miRNA abundance between the two libraries, 336 miRNAs are significantly differently expressed between FC and SMC (Table S4). Compared with miRNA expression in FC library, 317 miRNAs in SMC library were significantly down-regulated (Table S4). 26 miRNAs were significantly up-regulated in SMC library (Table S4). Among the up-regulated miRNAs, miR-487a has the highest fold-change (10 fold), while miR-29c has the highest fold-change (4 fold) in the down-regulated miRNAs.
Figure 4. The differential expression of caprine known miRNA between fetal and six month old caprine muscle tissues.
Expression level (SMC): Expression level of six month-old caprine musscle tissue; Expression level (FC): Expression level of fetal caprine muscle tissue. Red points represent miRNAs with a fold change>2, blue points represent miRNAs with 1/2<fold change≤2, green points represent miRNA with a fold change≤1/2.
Identification and profiling of novel miRNAs candidates
The characteristic hairpin structure of miRNA precursors have been used to predict novel miRNAs. We predicted novel miRNAs by exploring the secondary structure, the Dicer cleavage site and the minimum free energy of the unannotated small RNA reads. A total of 4,447,299 unannotated sequences in the two libraries were used to predict novel miRNAs. We identified 83 potential novel miRNAs, which corresponded to 95 genomic loci. The length of the novel miRNA sequences ranged from 19 to 24 nt, with a distribution peak at 22 nt (Table S5). Among the 83 candidates, 74 novel miRNAs were identified in the fetal goat library and 36 were identified in the six month-old library, 27 miRNAs were expressed in both the two libraries (Table S5). 40 miRNAs were differentially expressed between the two development stages (Table S6). Compared with miRNAs expression in SMC library, 33 miRNAs were up-regulated in FC library, only 7 miRNAs were down-regulated.
The predicted hairpin structures of these novel miRNAs ranged from 65 to 99 nt in length and the folding energies ranged from −64.4 to −23.3 kcal/mol (Table S5). The read number for each novel miRNA was lower than that for the majority of known miRNAs. For example, the total reads of chi-novel-miRNA-30, chi-novel-miRNA-84 and chi-novel-miRNA-70 that had the highest reads number, were only 18540, 3436 and 3436, respectively. Moreover, the MFE index (MFEI) can be used for assaying miRNAs and distinguishing miRNAs from other coding and non-coding RNAs. We calculated the MFEI value for each individual miRNA precursor sequence. The values of MFEI for majority of miRNA precursors were greater than 0.85, a value that serves as a key discriminator to distinguish pre-miRNAs from other small RNAs.
mRNA transcriptome profiling
miRNA regulates mRNA stability and translation. To further analyze the role of caprine miRNAs during muscle development, we characterized the mRNAs expression to do a miRNA-mRNA integrated analysis. After deep sequencing, we obtained 27,512,850 clean reads from the FC library and 27,582,908 clean reads from the SMC library (Table S7). Clean reads were mapped to a reference sequences using SOAPaligner/SOAP2. The ratio of reads mapped to reference genome was 67.92% for FC and 74.52% for SMC. 12,493,828 and 13,985,770 reads perfectly match the reference genome in the FC library and the SMC library (Table S7), respectively. 62.41% were unique match reads in the FC library as well as 66.89% in the SMC library. When mapped to the reference gene, the ratio of reads mapped to the reference gene was 68.18% for FC and 51.91% for SMC. 13,979,498 and 10,902,671 reads perfectly match the reference gene in the FC library and in the SMC library, respectively. 64.77% and 50.76% of mapped reads are unique matched reads in the FC library and SMC library (Table S7), respectively.
To reveal the molecular events during different development stages, genes that differentially expressed between the two libraries were identified. There are 7322 mRNAs with at least a two-fold difference in expression between FC and SMC libraries, of which 1329 genes are up-regulated, 5993 are down-regulated in the six month old caprine muscle tissue compared to that of the fetal, and 6907 are expressed in both libraries. The details of the gene-expression, including gene length, RPKM, log2 Ratio, P value and FDR were shown in Table S8.
miRNA-mRNA integrated analysis
We firstly predicted the potential target genes of all the known miRNAs. Larger numbers of annotated mRNA transcripts were selected as potential targets for each library. Then, we screened out the potential targets of differentially expressed miRNAs which have a −3≥log2 ratio≥3. Then, these target genes were compared to the DEGs (listed in Table S8), and selected the intersection of the two gene sets and called them DE targets. The regulative relationship between these differentially expressed miRNAs and the DE targets were shown in Table S9. The result showed that most individual miRNAs had multiple gene targets and each target was regulated by multiple miRNAs. All DE-targets were then processed by KEGG pathway analysis. In total, 2719 targets significantly matched in the database, and were assigned to 231 KEGG pathways (Table S10). The pathways were listed according to the P-value in the table. It was shown that these DE-targets have a wide range of diverse functions.
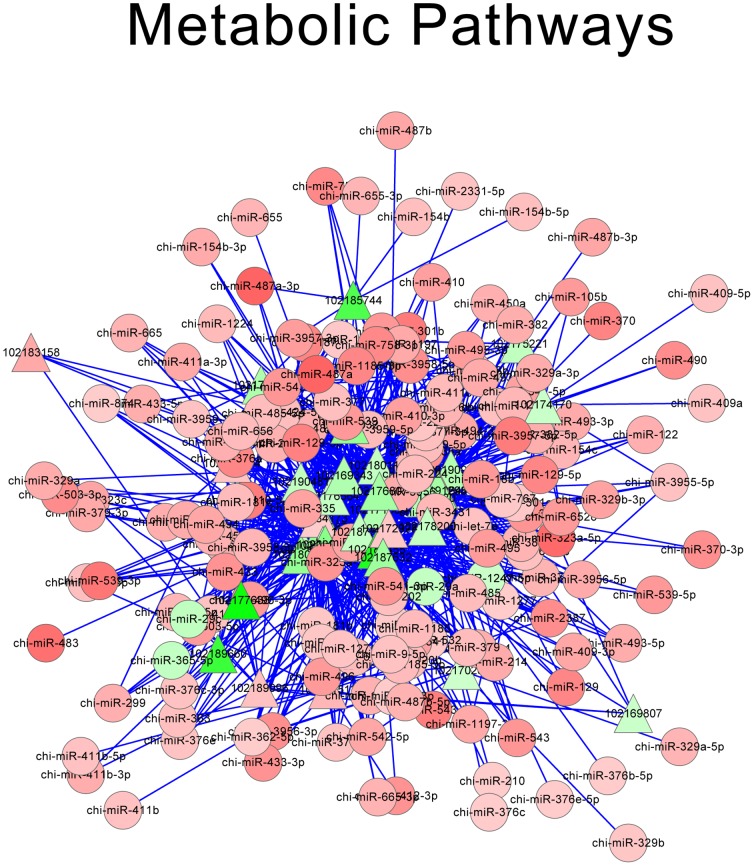
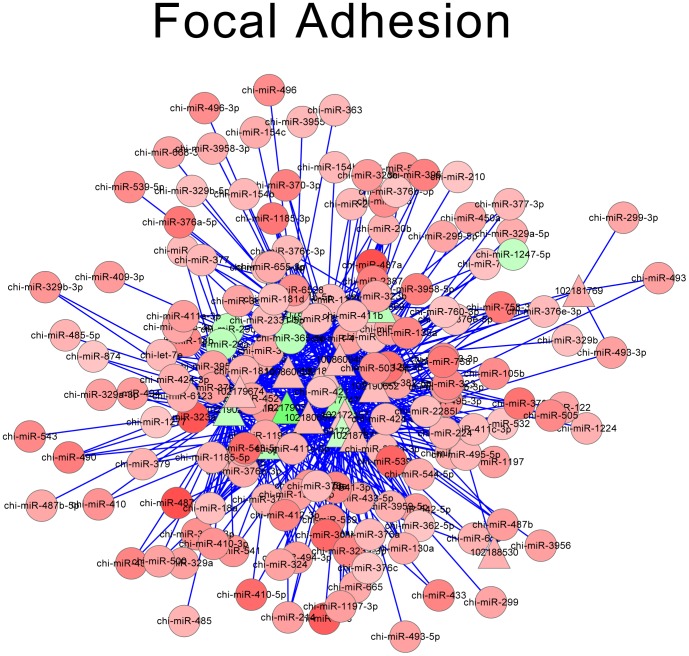
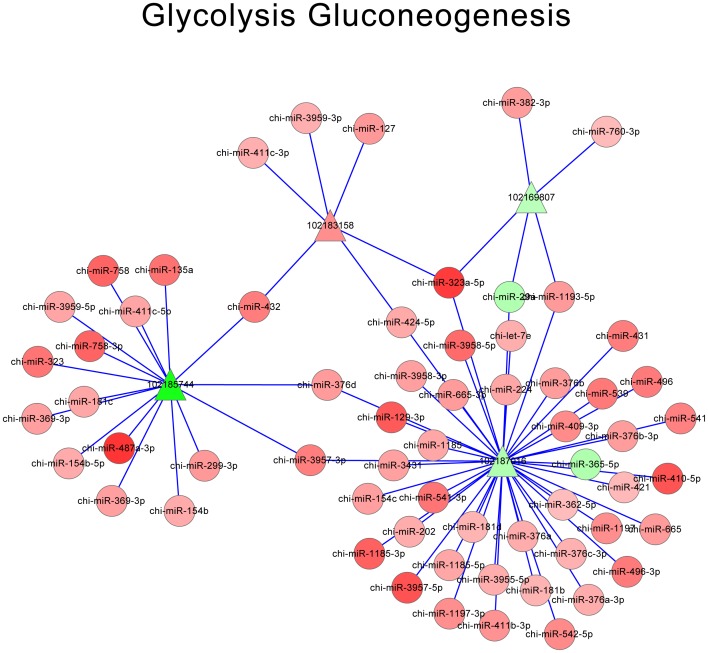
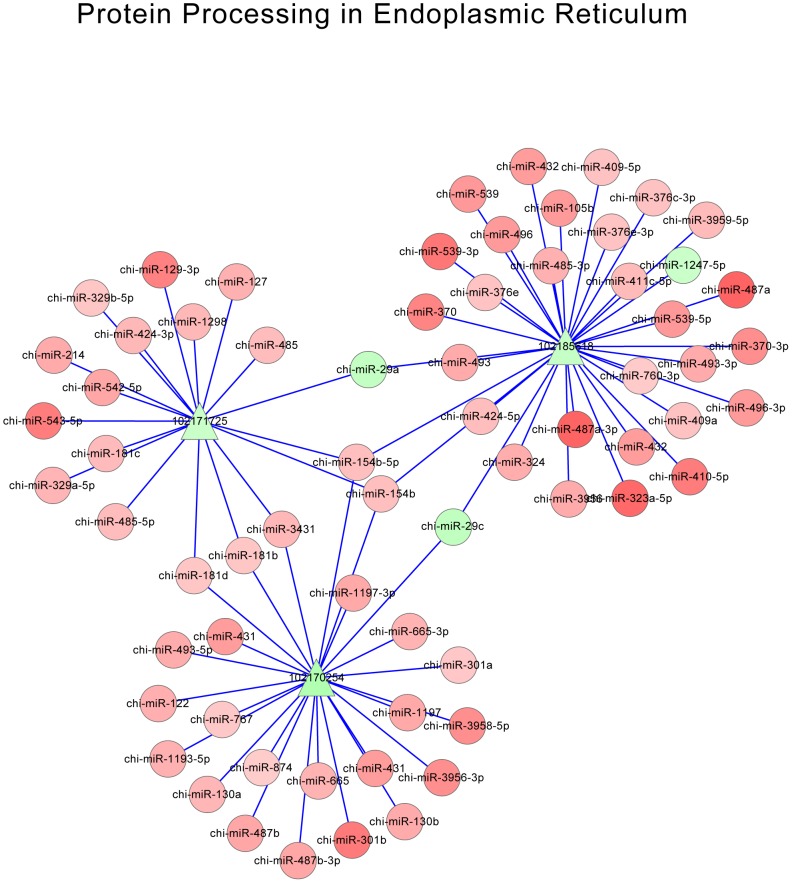
According to miRNA-mRNA-KEGG annotation, 10 pathways with the smallest P-value were screened out, which were shown in Table 5. Each pathway comprised of multiple miRNAs and DE-targets. Then the interactions of miRNAs and DE-targets for these 10 pathways were integrated to construct possible regulatory networks, which were shown as network pictures. Net work of metabolic pathways, focal adhesion, glycolysis gluconeogenesis and protein processing in endoplasmic reticulum were shown in Figure 5, 6, 7 and 8, respectively. Oxidative phosphorylation, pentose phosphate pathway, ribosome, alzheimers disease, huntingtons disease, parkinsons disease were shown in Figure S1. The network of metabolic pathways included 169 miRNAs and 35 DE-targets (Table 5). In this pathway, miR-424-5p regulated most numbers (16) of DE targets, which suggested its important role in this pathway. When it came to focal adhesion (Figure 6), 163 miRNAs and 17 genes were involved in, and miR-424-5p regulated 14 DE-targets, which was the largest number of DE targets. It was very interesting that among the 7 networks, miR-424-5p were very active in that it regulates largest numbers of DE-targets and at the center of the networks. We also find miRNA-29a is involved in most of the networks. Except miR-424-5p and miRNA-29a, miR-129-3p, miR-181b and miR-181d were also involved in multiple pathways. This indicated that these miRNA might have very important biological function on myoblast proliferation or differentiation.
Table 5. Network of ten smallest P-value pathways.
| Pathways | miRNA number | Targets genes number involved in network | P-value |
| Metabolic_pathways | 169 | 35 | 3.25E-13 |
| Focal adhesion | 163 | 17 | 3.50E-07 |
| Ribosome | 38 | 5 | 8.44E-14 |
| Oxidative phosphorylation | 21 | 1 | 5.74E-08 |
| Glycolysis Gluconeogenesis | 65 | 4 | 2.51E-06 |
| Pentose phosphate pathway | 66 | 3 | 6.72E-06 |
| Protein processing in endoplasmic reticulum | 68 | 3 | 9.39E-06 |
| Parkinson's disease | 56 | 4 | 1.53E-11 |
| Alzheimer's disease | 54 | 3 | 1.53E-09 |
| Huntington's disease | 66 | 5 | 3.82E-09 |
Figure 5. Network of metabolic pathways.
Red triangle: up-regulated DE-targets; Green triangle: down-regulated DE-targets; Red roundness: up-regulated miRNAs; Green roundness: down-regulated miRNAs. The deeper the color, the stronger the trend.
Figure 6. Network of focal adhesion.
Red triangle: up-regulated DE-targets; Green triangle: down-regulated DE-targets; Red roundness: up-regulated miRNAs; Green roundness: down-regulated miRNAs. The deeper the color, the stronger the trend.
Figure 7. Network of glycolysis gluconeogenesis.
Red triangle: up-regulated DE-targets; Green triangle: down-regulated DE-targets; Red roundness: up-regulated miRNAs; Green roundness: down-regulated miRNAs. The deeper the color, the stronger the trend.
Figure 8. Network of protein processing in endoplasmic reticulum.
Red triangle: up-regulated DE-targets; Green triangle: down-regulated DE-targets; Red roundness: up-regulated miRNAs; Green roundness: down-regulated miRNAs. The deeper the color, the stronger the trend.
Validate the sequence data
Stem-loop RT-PCR for selected miRNAs
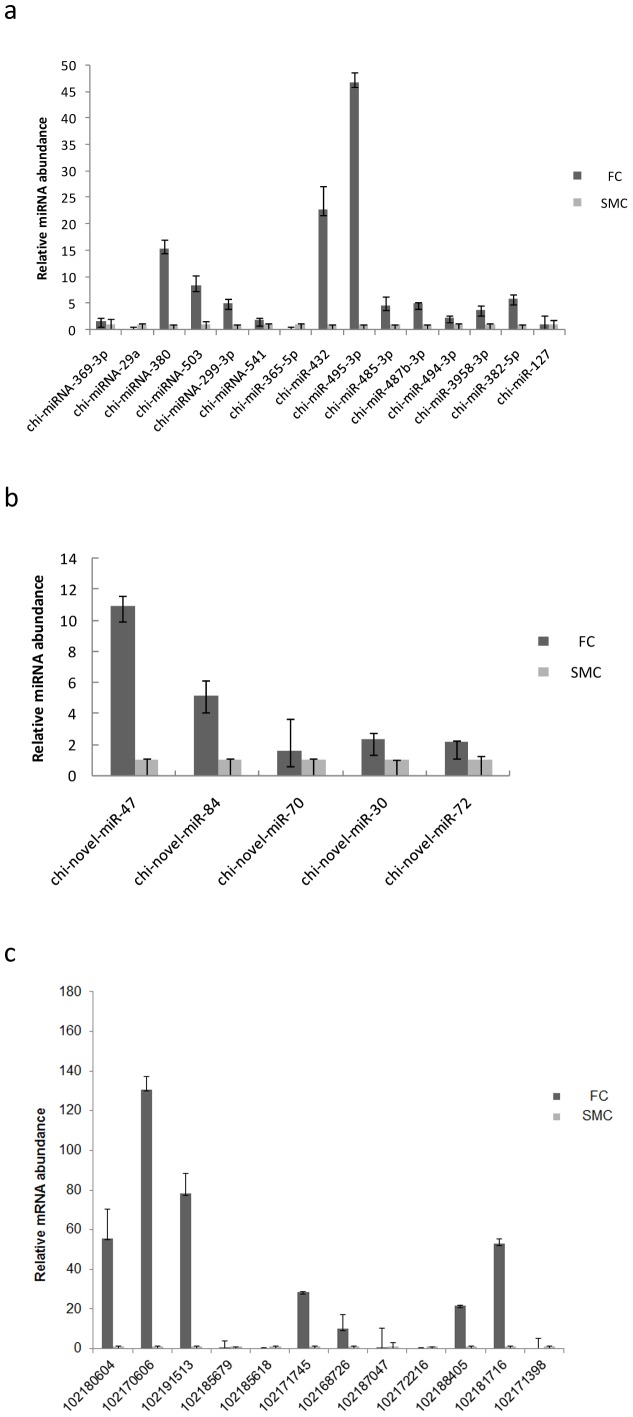
Stem-loop Real-time RT-PCR was performed on 15 differential expressed known miRNAs including chi-miR-485, chi-miR-487b, chi-miR-494, chi-miR-382-5p, chi-miR-432, chi-miR-495, chi-miR-365-5p, chi-miR-369-3p, chi-miR-29a, chi-miR-380, chi-miR-503, chi-miR-541, chi-miR-127, chi-miR-299-3p, chi-miR-3958, and 5 novel miRNAs, including chi-novel-miR-47, chi-novel-miR-84, chi-novel-miR-70, chi-novel-miR-30, chi-novel-miR-72. MiRNAs were chosen randomly. 18SrRNA was used as the internal reference RNA as it shows negligible expression differences along the sequence experiment. The sequences of primers were listed in the Table S11. As shown in Figure 9a, for the known miRNAs, the relative expression abundant of chi-miR-485, chi-miR-487b, chi-miR-494, chi-miR-382-5p, chi-miR-432, chi-miR-495, chi-miR-369-3p, chi-miR-380, chi-miR-503, chi-miR-541, chi-miR-127, chi-miR-299-3p and chi-miR-3958 were significantly higher in FC library than that of SMC library except chi-miR-29a and chi-miR-365-5p, which were highly correlated with sequence data. In addition, five novel miRNAs were chosen to detect the relative abundance. All of the five chosen miRNAs were shown to have higher expression level in FC library than in SMC library (Figure 9b).
Figure 9. The expression of miRNAs and mRNAs in caprine muscle tissue were validated by RT-PCR.
FC: fetal caprine; SMC: six month old caprine.
Quantitative real-time PCR for selected DEGs
To measure gene expression levels, 12 genes were chosen to validate the sequence data using quantitative real-time PCR (Figure 9c). GAPDH gene was used as the internal reference gene as it shows negligible expression differences along the sequence experiment. The primer sequences are listed in Table S12. The result showed a highly correlation between sequence data and qPCR data (Figure 9c).
Discussion
The development of the muscle results from the increase of muscle fibers and the expansion of cell volume, which entirely depend on the muscle cell proliferation and differentiation. Great deals of miRNAs have been reported to be enriched in muscle tissues and play an important role in the regulation of myoblast proliferation and differentiation. In recent years, the genomics of growth and development has gradually been elucidated through gene structure and function analyses to determine the gene expression regulation mechanism. miRNAs play critical roles in various biological and metabolic processes, by regulating gene expression at the post-transcriptional level. To date, many miRNA expression profiles have been reported in sheep, cow, chicken and pig, and other domestic animals in various tissues [43]–[46], but there is limited information about goat muscle miRNAs. The fetal stage is crucial for skeletal muscle development [47]. In livestock, all muscle fibers are formed during the prenatal stage, and 90-day around fetal life is the crucial stage of caprine myogenesis [48]. In contrast, postnatal skeletal muscle development is mainly due to the increase in muscle fiber size [49], and the period between birth and about eight months is the important stage of muscle fiber hypertrophy. Hence, in this study, 90 day fetal and six month old Huanghuai goat longissimus thoracis were collected for Solexa SBS technology sequencing.
The developmental skeletal muscle miRNA profile of several meat producing animal, including broiler, duck, pig, cattle and sheep, have been reported [28], [43], [50]–[52]. In the present study, 457 and 384 known miRNAs were detected in the library generated from fetal goat muscle tissue and six month old goat muscle tissue, respectively. Most of the sequences are 22nt in length, and majority of the small RNAs are 21–23nt in length, which is the typical size range of Dicer-derived products. The results are consistent with the small RNA distribution of Boer goat [53], sheep [54] and cattle [52], [55]. The most abundant miRNAs identified in our study were highly consisted with study on Boer goat, which reported that miR-133, miR-1, miR-378 and miR-206 families were the top 20 miRNAs in 6 month old Boer goat longissimus tissue [53], indicating the importance of these miRNAs on skeletal muscle development and growth. Interestingly, the most abundant miRNAs in our libraries were also highly consisted with that in bovine muscle libraries [52]. For instance, miR-1, miR-206, let-7f, let-7a-5p, let-7b, let-7c, let-7g, miR-378 etc, indicating that miRNA was highly conserved between cattle and goat. Thus, bovine miRbase could act as a credible reference for caprine miRNAs. The muscle specific miR-1, miR-133 and miR-206 are three of the most studied miRNAs up to now. Several studies demonstrate that these miRNAs are necessary for proper skeletal muscle development and function. MiR-1 or miR-206 promotes myogenic differentiation, while miR-133 over-expression enhances myoblast proliferation, but represses differentiation [10]–[11], [56]. Moreover, miR-1 and miR-133 have been implicated in skeletal muscle hypertrophy [14]. In the present study, the expression pattern of miR-206 and miR-133a are consistent with previous study on fetal bovine and adult bovine muscle tissue [52]. In addition, the expression level of miR-1 is significant higher (log2 ratio:−2.37) in six month old goat muscle. As six month-old is the important phase of muscle hypertrophy, we suppose that miR-1 may play an important role in caprine skeletal muscle hypertrophy. Moreover, many studies have reported that let-7 gene family is highly expressed and conserved across animal species, including mammals, flies, worms and plants [52], [57]–[59]. In the present study, let-7 gene family is sequenced at a high frequency in the caprine muscle tissue, these data are similar to those from other studies on miRNAs in different tissues, which indicated that the let-7 family is very important for some fundamental biological processes [58]. The family analysis of the identified miRNAs in our study showed that most members of conserved miRNAs families were expressed in caprine skeletal muscle, supporting the idea that regulatory or functional diversification has occurred [60]–[61].
Further analyzation of known miRNAs was conducted for nucleotide bias analysis. The base composition is an important feature of a nucleotide sequence for it can influence the physiochemical and biological properties of nucleotide sequences through effects on base pairing and related features, including thermodynamic folding of secondary structures. These characteristics, in turn, influence the biochemistry of nucleotide sequences as evidenced by the influence of secondary structure of miRNA [62]–[64]. In this study, characterization of the frequency of the position-specific nucleotide composition along the length of miRNA revealed that (Fig. 3) nucleotide positions 1 and 9 are significantly enriched for U relative to other bases, with a concordant depletion of G in FC library. Positions 2 to 8 of a mature miRNA is called the seed region, which is highly conserved. This “seed region” has been shown to be critical for miRNA targeting of mRNAs [65]. Positions 1 and 9 in mature miRNAs are adjacent both upstream and downstream of the “seed region”. Our results point to a potential role of these seed region flanking sequences in miRNA biogenesis or target site recognition. These were consisted with a previous report, who reported the dominance of U in position 1 and 9 [66]. This result showed that U was under strong selection at the ‘seed region’, and this selection may have important functions on either miRNA biogenesis or mRNA target recognition.
The known miRNAs were used for studying the expression profiles between the two development stages. A total of 336 miRNAs were differentially expressed between the two libraries, of which 317 were up-regulated and 26 were down-regulated in the FC library compared to the SMC library. Previous study about bovine muscle tissue reported 251 differentially expressed miRNAs consisted of 230 up expressed miRNAs and 21 down-expressed miRNAs in fetal bovine muscle tissue compared to adult bovine muscle tissue [52]. The result suggested that the miRNA concentration in the muscle tissue decreased in postnatal stage compared to the fetal stage. This is also consistent with previous studies that many miRNAs are highly expressed in the embryo as compared with that in adulthood, and the roles of these miRNAs are generally involved in embryo development and tissue identity maintenance [50], [67]–[68]. We speculated that the miRNAs differentially expressed between the FC and SMC may participate in regulating muscle tissue development and maintaining their distinct function. Thus, identifying the role and regulation of skeletal muscle miRNAs during various phases of muscle development, will significantly enhance our understanding of skeletal muscle biology and function.
In our database, 74 and 36 novel miRNAs were detected in the library generated from fetal goat and six month old goat muscle tissue, respectively. In addition, we found that a majority of pre-miRNA sequences had an MFE index (MFEI) values higher than 0.85, which were significant higher than other RNAs. This would be a better criterion for assaying and distinguishing miRNAs from other RNAs. Similar results in bovine have been reported [52]. Previous studies have shown that in plant and animals, the MFEI is a credible criterion for assaying miRNAs and distinguishing miRNAs from other coding and non-coding RNAs [52], [69].
The mature miRNAs exert their functions by binding the 3′UTR of the target mRNAs to degrade or repress their translation. Large numbers of miRNAs and their target gene may form a complex network, thus, the regulation between various miRNAs and mRNAs was complicated [70]. Establishing the interactions between miRNA and mRNA will enhance our understanding of embryonic and postnatal muscle development. In the present study, we employed a novel and generalizable method to efficiently identify functional miRNA-target interactions in caprine muscle tissue, namely, an integrated analysis of the miRNA profile and mRNA profile. To do the integrated analysis, we characterized the repertoire of differentially expressed transcripts in the six month old muscle tissue as compared to the fetal muscle tissue, and detected 7322 genes with at least a one-fold difference in expression between FC and SMC libraries (Table S8), which indicated that there were great differences between the two development stages. Several of the DEGs identified in our analyses were previously reported in the muscle tissue and some of them have been shown to affect muscle development [71]. For instance, the expression level of MRF and MEF2 families were significantly different between fetal stage and six month old stage. Through the integrated analysis, the interactions of differentially expressed miRNAs and target genes were shown in Table S9. We found that each miRNA had multiple targets, and each target was regulated by multiple miRNAs.
In order to further understand the miRNA-mRNA interactions, function of DE targets were determined. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database is a knowledge base for systematic analysis of gene functions in terms of networks of genes and molecules in the cells and their variants specific to particular organisms [72]. Genes within the same pathway usually cooperate with each other to exercise their biological function, which indicate that pathway-based analysis is a useful tool to further understand the biological functions of genes [73]. In our study, the top ten pathways that have the lowest P-value were mainly involved in the basic physiological and biochemical process of cell. The result also indicated that, except muscle specific miRNA (for instance, miR-1, miR-206, miR-133 etc), many non-muscle special miRNAs also played important roles in myocyte proliferation or differentiation through target some myogenic transcription factors, this was consist with previous studies that reported non-muscle-specific miRNAs regulate skeletal muscle differentiation [74], [75]. In the integrated analysis, the related miRNAs or DE-targets in each network were differentially-expressed miRNAs or genes, most of the miRNAs and DE-targets were involved in multiple pathways, for instance, miR-424-5p, miR-29a, bta-miR-129-3p, miR-181b and miR-181d. miR-424-5p was involved in most of the 10 pathways, and regulated the largest number of DE-targets. This indicated that miR-424-5p might play an important role in regulation muscle development. However, the biological characteristics, molecular mechanisms, and targets of miR-424-5p were not well-understood. miR-29a was a down-regulated miRNA and involved in most of the network. Previous study reported that miRNA-29a could suppress cell proliferation [76]. In brief, we described the network of miRNAs and DE-targets from a macroscopic view. miR-424-5p was involved in most of the pathways, and significantly differentially expressed during different development stages, suggesting its important regulatory effect. Moreover, except miR-424-5p and miR-29a, there were many other miRNAs that have important regulate effect in the network, further studies on these miRNAs would enable us to determine the mechanism of miRNAs in muscle development and miRNA-mRNA interaction.
Conclusions
In conclusion, the data presented herein represent three advancements. First, using deep sequencing technologies, 464 known miRNAs were identified in skeletal muscle from fetal and six month old Huanghuai goat. The base composition analysis indicated that U is under strong selection in the position 1 and 9, which were the seed region flanking sequences, and this selection may have important function in either miRNA biogenesis or mRNA target recognition. Second, we have identified 83 novel miRNAs in longissimus thoracis from fetal and six month old goat. Most of the predicted novel miRNAs have an MEFI better than 0.85. Third, through intergrating the miRNA profile and the mRNA profile, the interaction of the miRNA and mRNA in this study were analyzed. Based on the networks, we found that miRNA-424-5p and miR-29a had important regulatory effect during muscle development.
Data deposition details
The data set supporting the results of this article is available in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) repository, with access number GSE49258 (RNA-seq) and GSE49260 (small RNA-seq).
Supporting Information
Network of miRNA and targets. Note: Red triangle: up-regulated DE-targets; Green triangle: down-regulated DE-targets; Red roundness: up-regulated miRNAs; Green roundness: down-regulated miRNAs. The deeper the color, the stronger the trend.
(PDF)
Known miRNAs in this study.
(XLS)
Nucleotide compositions of known miRNAs in this study.
(XLSX)
Family analysis of known miRNA.
(XLSX)
Known miRNAs expression profiles of the two libraries.
(XLS)
Novel miRNAs identified in this study.
(XLSX)
Novel miRNAs expression profiles.
(XLS)
Statistics of alignment (map to reference genome and gene).
(DOCX)
Differentially expressed genes that have FDR≤0.001 and Fold Change >2.
(XLS)
MiRNA-mRNA integrated analysis.
(XLSX)
Pathway of differential expression targets.
(XLSX)
The primer sequences of the stem-loop qPCR experiments.
(XLS)
The primer sequences of the qPCR experiments.
(XLS)
Funding Statement
This study was supported by Research Fund for the Doctor Program of Higher Education of China (No. 20120204110007),A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the College Industrialization Project of Jiangsu Province(No. JHB2012-32), Agricultural Science and Technology Innovation Projects of Shaanxi Province (No. 2012NKC01-13),Science and Technology Planning Project of Xuzhou City(No. XF12C052). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 2. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- 3. Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, et al. (2010) MicroRNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett 476: 18–22. [DOI] [PubMed] [Google Scholar]
- 4. Song L, Tuan RS (2006) MicroRNAs and cell differentiation in mammalian development. Birth Defects Res C Embryo Today 78: 140–149. [DOI] [PubMed] [Google Scholar]
- 5. Sini RA, Trink B, Nissan A (2009) The role of microRNA in tumorigenesis: key players or innocent bystanders. J Surg Oncol 99: 135–136. [DOI] [PubMed] [Google Scholar]
- 6. Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, et al. (2007) Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129: 303–317. [DOI] [PubMed] [Google Scholar]
- 7. Pedersen IM, Cheng G, Wieland S, Croce CM, Chisari FV, et al. (2007) Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449: 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guller I, Russell AP (2010) MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. J Physiol 588: 4075–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buckingham M (2006) Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev 16: 525–532. [DOI] [PubMed] [Google Scholar]
- 10. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, et al. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim HK, Lee YS, Sivaprasad U, Malhotra A Dutta A (2006) Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rao PK, Kumar RM, Farkhondeh M, Baskerville S Lodish HF (2006) Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA 103: 8721–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao Y, Srivastava D (2007) A developmental view of microRNA function. Trends Biochem Sci 32: 189–197. [DOI] [PubMed] [Google Scholar]
- 14. McCarthy JJ, Esser KA (2007) MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol 102: 306–313. [DOI] [PubMed] [Google Scholar]
- 15. Crippa S, Cassano M, Messina G, Galli D, Galvez BG, et al. (2011) MiR669a and miR669q prevent skeletal muscle differentiation in postnatal cardiac progenitors. J Cell Biol 193: 1197–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, et al. (2006) A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 103: 18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun Q, Zhang Y, Yang G, Chen X, Cao G, et al. (2008) Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res 2008 36: 2690–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong CF, Tellam RL (2008) MicroRNA-26a targets the histone methyltransferase enhancer of Zeste homolog 2 during myogenesis. J Biol Chem 283: 9836–9843. [DOI] [PubMed] [Google Scholar]
- 19. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- 20. Anderson C, Catoe H, Werner R (2006) MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res 34: 5863–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang XH, Hu Z, Klein JD, Zhang L, Fang F, et al. (2011) Decreased miR-29 suppresses myogenesis in CKD. J. Am. Soc. Nephrol 22: 2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caretti G, Di PM, Micales B, Lyons GE, Sartorelli V (2004) The Poly-comb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, et al. (2009) Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA 106: 13383–13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen JF, Tao Y, Li J, Deng Z, Yan Z, et al. (2010) MicroRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol 190: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu L, Zhou L, Chen EZ, Sun K, Jiang P, et al. (2012) A novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. Plos One 7: e27596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dey BK, Gagan J, Dutta A (2011) MiR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol 31: 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Callis TE, Deng Z, Chen JF, Wang DZ (2008) Muscling through the microRNA world. Exp Biol Med 233: 131–138. [DOI] [PubMed] [Google Scholar]
- 28. van Rooij E, Liu N, Olson EN (2008) MicroRNAs flex their muscles. Trends Genet 24: 159–166. [DOI] [PubMed] [Google Scholar]
- 29. van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, et al. (2009) A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, et al. (2010) Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA 107: 4218–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24: 713–714. [DOI] [PubMed] [Google Scholar]
- 32. Jiang P, Wu H, Wang W, Ma W, Sun X, et al. (2007) MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res 35: W339–W344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. (2009) SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- 35. Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res. 7: 986–995. [DOI] [PubMed] [Google Scholar]
- 36. Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics 29: 1165–1188. [Google Scholar]
- 37. Rehmsmeier M, Steffen P, Ho chsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- 39. Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, et al. (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- 40. Ji ZB, Wang GZ, Xie ZJ, Wang JM, Zhang CL, et al. (2012) Identification of Novel and Differentially Expressed MicroRNAs of Dairy Goat Mammary Gland Tissues Using Solexa Sequencing and Bioinformatics. PLoS One. 7: e49463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 42. Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang S, Zhao F, Wei C, Sheng X, Ren H, et al. (2013) Identification and characterization of the miRNA transcriptome of Ovis aries. PLoS One 8(3): e58905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Muroya S, Taniguchi M, Shibata M, Oe M, Ojima K, et al. (2013) Profiling of differentially expressed microRNA and the bioinformatic target gene analyses in bovine fast- and slow-type muscles by massively parallel sequencing. J Anim Sci 91: 90–103. [DOI] [PubMed] [Google Scholar]
- 45. Huang P, Gong YZ, Peng XL, Li SJ (2010) The expression profile analysis of sexual dimorphism miRNAs expressed in chicken embryo at early stages of sex differentiation. J. Agricultura Biotechnol 18: 1149–1155. [Google Scholar]
- 46. Qin L, Chen Y, Liu X, Ye S, Yu K, et al. (2013) Integrative Analysis of Porcine microRNAome during Skeletal Muscle Development. Plos One 8(9): e72418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu MJ, Ford SP, Nathanielsz PW, Du M (2004) Effect of maternal nutrient restriction in sheep on the development of fetal skeletal muscle. Bio. Reprod 71: 1968–1973. [DOI] [PubMed] [Google Scholar]
- 48. Zhu MJ, Ford SP, Means WJ, Hess BW, Nathanielsz PW, et al. (2006) Maternal nutrient restriction affects properties of skeletal muscle in offspring. J Physiol 575: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hocquette JF (2010) Endocrine and metabolic regulation of muscle growth and body composition in cattle. Animal 4: 1797–1809. [DOI] [PubMed] [Google Scholar]
- 50. Li T, Wu R, Zhang Y, Zhu D (2011) A systematic analysis of the skeletal muscle miRNA transcriptome of chicken varieties with divergent skeletal muscle growth identifies novel miRNAs and diff1 erentially expressed miRNAs. BMC Genomics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gu L, Xu T, Huang W, Xie M, Sun S, et al. (2014) Identification and Profiling of MicroRNAs in the Embryonic Breast Muscle of Pekin Duck. PLoS One 9: e86150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun JJ, Li MJ, Li ZJ, Xue J, Lan XY, et al. (2013) Identification and profiling of conserved and novel microRNAs from Chinese Qinchuan bovine longissimus thoracis. BMC Genomics 14: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ling YH, Ding JP, Zhang XD, Wang LJ, Zhang YH, et al. (2013) Characterization of microRNAs from goat (Capra hircus) by Solexa deep-sequencing technology. Genet Mol Res 12: 1951–1961. [DOI] [PubMed] [Google Scholar]
- 54. Caiment F, Charlier C, Hadfield T, Cockett N, Georges M, et al. (2010) Assessing the effect of the CLPG mutation on the microRNA catalog of skeletal muscle using high-throughput sequencing. Genome Res 20: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen X, Gao C, Li H, Huang L, Sun Q, et al. (2010) Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res 20: 1128–1137. [DOI] [PubMed] [Google Scholar]
- 56. Boutz PL, Chawla G, Stoilov P, Black DL (2007) MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev 21: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roush S, Slack FJ (2008) The let-7 family of microRNAs. Trends Cell Biol 18: 505–516. [DOI] [PubMed] [Google Scholar]
- 58.Li ZJ, Lan XY, Guo WJ, Sun JJ, Huang YZ, et al. (2012) Comparative Transcriptome Profiling of Dairy Goat MicroRNAs from Dry Period and Peak Lactation Mammary Gland Tissues. Plos one 7: : 12, e52388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yan X, Ding L, Li Y, Zhang X, Liang Y, et al. (2012) Identification and profiling of microRNAs from skeletal muscle of the common carp. PloS One 7: e30925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, et al. (2005) Expression of Arabidopsis miRNA Genes. Plant Physiol 138: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, et al. (2007) Most caenorhabditis elegans microRNAs are individually not essential for development or viability. Plos Genet 3: e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arhondakis S, Auletta F, Torelli G, D'Onofrio G (2004) Base composition and expression level of human genes. Gene 325: 165–169. [DOI] [PubMed] [Google Scholar]
- 63. Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M (2006) High guanine and cytosine content increases mRNA levels in mam malian cells. PLoS Biol 4: 0933–0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. D'Onofrio G, Ghosh TC, Saccone S (2007) Different functional classes of genes are characterized by different compositional properties. FEBS Lett 581: 5819–5824. [DOI] [PubMed] [Google Scholar]
- 65. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, oftenflanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 66. Zhang B, Stellwag EJ, Pan X (2009) Large-scale genome analysis reveals unique features of microRNAs. Gene 443: 100–109. [DOI] [PubMed] [Google Scholar]
- 67. Huang TH, Zhu MJ, Li XY, Zhao SH (2008) Discovery of porcine microRNAs and profiling from skeletal muscle tissues during development. Plos One 3: e3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu Z, Jian Z, Shen SH, Purisima E, Wang E (2007) Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res 35: 152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA (2006) Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci 63: 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Okamura K, Ishizuka A, Siomi H, Siomi MC (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Davis E, Jensen CH, Schroder HD, Farnir F, Shay-Hadfield T, et al. (2004) Ectopic Expression of Dlkl Protein in Skeletal Muscle of Padumnal H eterozygotes Causes the Callipyge Phenotype. Curr Biol 14: 1858–1862. [DOI] [PubMed] [Google Scholar]
- 72. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, et al. (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wenping H, Yuan Z, Jie S, Lijun Z, Zhezhi W (2011) De vono transcriptome sequencing in Salvia miltiorrhiza to identify genes involved in the biosynthesis of active ingredients. Genomics 98: 272–279. [DOI] [PubMed] [Google Scholar]
- 74. Wang L, Chen X, Zheng YY, Fen L, Zheng L, et al. (2012) MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp cell res 318: 2324–2334. [DOI] [PubMed] [Google Scholar]
- 75. Feng Y, Cao JH, Li XY, Zhao SH (2011) Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts. Cell Biochem Funct 29: 378–383. [DOI] [PubMed] [Google Scholar]
- 76. Cui Y, Su WY, Xing J, Wang YC, Wang P, et al. (2011) MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. Plos one 0.1371/journal. pone.0025872 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Network of miRNA and targets. Note: Red triangle: up-regulated DE-targets; Green triangle: down-regulated DE-targets; Red roundness: up-regulated miRNAs; Green roundness: down-regulated miRNAs. The deeper the color, the stronger the trend.
(PDF)
Known miRNAs in this study.
(XLS)
Nucleotide compositions of known miRNAs in this study.
(XLSX)
Family analysis of known miRNA.
(XLSX)
Known miRNAs expression profiles of the two libraries.
(XLS)
Novel miRNAs identified in this study.
(XLSX)
Novel miRNAs expression profiles.
(XLS)
Statistics of alignment (map to reference genome and gene).
(DOCX)
Differentially expressed genes that have FDR≤0.001 and Fold Change >2.
(XLS)
MiRNA-mRNA integrated analysis.
(XLSX)
Pathway of differential expression targets.
(XLSX)
The primer sequences of the stem-loop qPCR experiments.
(XLS)
The primer sequences of the qPCR experiments.
(XLS)