PI3K is the main downstream effector of PDGFRα signaling during murine skeletal development. Fantauzzo et al. discovered skeletal defects in embryos in which PDGFRα is unable to bind PI3K. They identified 56 proteins that are phosphorylated by Akt downstream from PI3K-mediated PDGFRα signaling. Several of these proteins, including Ybox1, mediate cell survival through regulation of p53. These findings identify p53 as a novel effector downstream from PI3K-engaged PDGFRα signaling that regulates survival and proliferation during skeletal development in vivo.
Keywords: PDGFRα, PI3K, skeleton, survival, proliferation, p53
Abstract
Previous studies have identified phosphatidylinositol 3-kinase (PI3K) as the main downstream effector of PDGFRα signaling during murine skeletal development. Autophosphorylation mutant knock-in embryos in which PDGFRα is unable to bind PI3K (PdgfraPI3K/PI3K) exhibit skeletal defects affecting the palatal shelves, shoulder girdle, vertebrae, and sternum. To identify proteins phosphorylated by Akt downstream from PI3K-mediated PDGFRα signaling, we immunoprecipitated Akt phosphorylation substrates from PDGF-AA-treated primary mouse embryonic palatal mesenchyme (MEPM) lysates and analyzed the peptides by nanoliquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS). Our analysis generated a list of 56 proteins, including 10 that regulate cell survival and proliferation. We demonstrate that MEPM cell survival is impaired in the presence of a PI3K inhibitor and that PdgfraPI3K/PI3K-derived MEPMs do not proliferate in response to PDGF-AA treatment. Several of the identified Akt phosphorylation targets, including Ybox1, mediate cell survival through regulation of p53. We show that Ybox1 binds both the Trp53 promoter and the p53 protein and that expression of Trp53 is significantly decreased upon PDGF-AA treatment in MEPMs. Finally, we demonstrate that introduction of a Trp53-null allele attenuates the vertebral defects found in PdgfraPI3K/PI3K neonates. Our findings identify p53 as a novel effector downstream from PI3K-engaged PDGFRα signaling that regulates survival and proliferation during skeletal development in vivo.
There are four PDGF ligands in mammals, PDGF-A–D, which signal through two receptor tyrosine kinases, PDGFRα and PDGFRβ (Andrae et al. 2008). Targeted disruption of Pdgfra in mice (Pdgfra−/−) results in embryonic lethality during midgestation, with homozygous mutant embryos surviving as late as embryonic day 16 (E16). Null embryos exhibit a cleft face, subepidermal blebbing, edema, hemorrhaging, abnormalities in neural tube development, abnormally patterned somites, and extensive skeletal defects affecting the craniofacial bones, shoulder girdle, vertebrae, ribs, and sternum (Soriano 1997). The homodimers PDGF-AA and PDGF-CC have been shown to exclusively activate PDGFRα signaling in vivo during mammalian development, and loss of both ligands in mice (Pdgfa−/−;Pdgfc−/−) phenocopies Pdgfra-null embryos (Ding et al. 2004). The developmental roles of PDGFRα appear to be conserved between mice and humans, as missense mutations in the human PDGFRA coding region as well as single-base-pair substitutions in the 3′ untranslated region (UTR) are associated with nonsyndromic cleft palate (Rattanasopha et al. 2012). Furthermore, PDGFRA promoter haplotypes with high transcriptional activity are overrepresented in patients with sporadic spina bifida (Joosten et al. 2001). Consistent with a critical role for PDGFRα in cranial and axial skeletal development, H2B-GFP expressed from the endogenous Pdgfra promoter is detected in the facial processes and somites at E10.5, among other sites (Hamilton et al. 2003).
Upon ligand binding, PDGFRs dimerize and activate cytoplasmic tyrosine kinase domains, which in turn autophosphorylate intracellular tyrosine residues. Signaling molecules containing phosphotyrosine recognition motifs, including Src family tyrosine kinases, the tyrosine phosphatase SHP-2, phosphatidylinositol 3-kinase (PI3K), Crk, and phospholipase Cγ (PLCγ), bind to specific phosphorylated residues in the cytoplasmic domains of the receptors and mediate downstream cellular responses (Heldin and Westermark 1999). To determine the roles of various signaling pathways downstream from PDGFRα signaling in vertebrate development, our laboratory previously generated an allelic series of autophosphorylation mutant knock-in mice at the Pdgfra locus (Klinghoffer et al. 2002). One allele (PdgfraPI3K) contained two tyrosine-to-phenylalanine mutations at residues (Y731 and Y742) that mediate the ability of PDGFRα to bind PI3K (Yu et al. 1991), and a second allele (PdgfraF7) included five additional mutations at residues (Y572, Y574, Y720, Y988, and Y1018) required for association with Src family kinases, SHP-2, and PLCγ (Rosenkranz et al. 1999). Homozygous PdgfraPI3K/PI3K embryos die perinatally and exhibit skeletal defects affecting the palatal shelves, shoulder girdle, vertebrae, and sternum, among other abnormalities (Klinghoffer et al. 2002). However, these embryos do not display a subset of skeletal defects found in Pdgfra−/−-null embryos, such as incomplete fusion of the anterior facial bones and rib fusions (Soriano 1997). Whereas homozygous PdgfraF7/F7 and hemizygous PdgfraF7/- embryos do not exhibit more severe abnormalities than those detected in homozygous PdgfraPI3K/PI3K embryos (Klinghoffer et al. 2002), the complete range of Pdgfra−/− skeletal defects is observed in PdgfraPI3K/PI3K;PdgfrbPI3K/PI3K double-homozygous mutant embryos in which PI3K signaling cannot be engaged through PDGFRα/β heterodimers (Klinghoffer et al. 2002). These results identify PI3K signaling as the main intracellular pathway downstream from PDGFRα signaling during embryogenesis in mice (Klinghoffer et al. 2002). Intriguingly, loss of all class Ia PI3K p85 regulatory subunit isoforms in mice (p85a−/−;p55a−/−;p50a−/−;p85b−/−) results in embryonic lethality at E12.5, with mutant embryos exhibiting phenotypes strikingly similar to those observed in Pdgfra−/−-null embryos, including facial clefting, subepidermal blebbing, hemorrhaging, and a wavy neural tube (Brachmann et al. 2005).
Activated PI3K signaling increases phosphatidylinositol-3,4,5-trisphosphate (PIP3) levels at the cell membrane, resulting in the recruitment of several pleckstrin homology domain-containing molecules, including Akt. Upon phosphorylation by PDK1 (T308) and mTORC2 (S473), active Akt phosphorylates an array of target proteins with roles in diverse cellular processes such as survival, proliferation, growth, and metabolism (Manning and Cantley 2007). Despite recent advances in the field, it is unclear which intracellular signaling pathways and corresponding cellular processes are governed by PI3K/Akt-mediated PDGFRα signaling during in vivo development.
Here we used a mass spectrometry-based phosphoproteomic approach to identify Akt phosphorylation targets downstream from PDGFRα signaling in primary mouse embryonic palatal mesenchyme (MEPM) cells. Our analysis identified both previously validated and novel Akt phosphorylation target proteins, a subset of which regulates cell survival and proliferation in part through modulation of p53 activity. We demonstrate that the functional interactions between these Akt phosphorylation targets and p53 are PDGF-AA-dependent and further show that introduction of a Trp53-null allele attenuates a subset of axial skeleton defects found in PdgfraPI3K/PI3K embryos. Taken together, our findings provide significant insight into the mechanisms of PDGFRα signaling in vivo, uncovering novel intracellular pathways governing cell survival and proliferation during skeletal development.
Results
Midgestation morphological abnormalities in PdgfraPI3K/PI3K mutant embryos
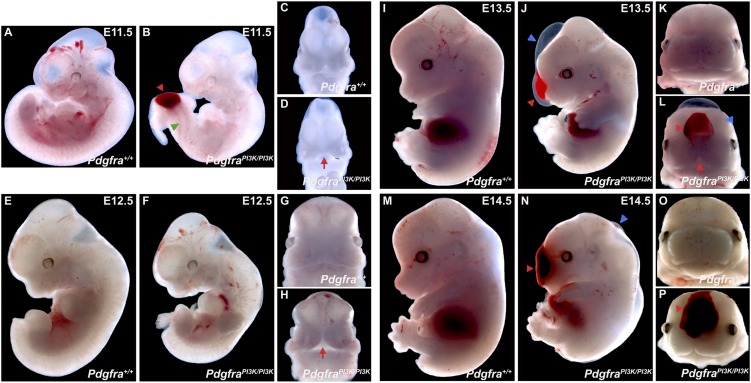
Gross morphological examination revealed that midgestation PdgfraPI3K/PI3K embryos were smaller than their wild-type littermates from E11.5 to E14.5 (Fig. 1). Superficially, fusion of the medial nasal processes was delayed in PdgfraPI3K/PI3K embryos at E11.5–E12.5 (Fig. 1C,D,G,H), and fusion of the maxillary region was variably delayed at E13.5 (Fig. 1K,L). Additionally, subepidermal blebbing was common in the mutant embryos (Fig. 1J,N), where it often spanned the facial midline and was accompanied by hemorrhaging (Fig. 1L,P). Mutant embryos also exhibited a wavy neural tube (Fig. 1B).
Figure 1.
Midgestation morphological abnormalities in PdgfraPI3K/PI3K mutant embryos. (A–P) Gross morphology of Pdgfra+/+ versus PdgfraPI3K/PI3K embryos at E11.5–E14.5. Note the reduced size of mutant embryos at all time points. Fusion of the medial nasal processes and maxillary region is delayed (red arrows) at E11.5–E12.5 (D,H) and E13.5 (L), respectively, in PdgfraPI3K/PI3K embryos. Subepidermal blebbing (blue arrowheads) is common in the mutant embryos (J,N), where it often spans the facial midline and is accompanied by hemorrhaging (L,P; red arrowheads). Wavy neural tubes (green arrowhead) are often observed in earlier embryos and are occasionally accompanied by hemorrhaging (shown in B).
We next performed a detailed histological analysis of palate development in PdgfraPI3K/PI3K embryos from E12.5 to E15.5. The anterior palatal shelves of the mutant embryos were smaller than those of their wild-type counterparts, with consistently abnormal morphologies ranging from absence of the characteristic finger-like projection at E13.5 to decreased vertical outgrowth at E14.5 (Supplemental Fig. 1A–R). By E15.5, the PdgfraPI3K/PI3K palatal shelves had failed to elevate and/or reach the midline, resulting in a cleft palate (Supplemental Fig. 1S–X).
Akt signaling dynamics in the embryo
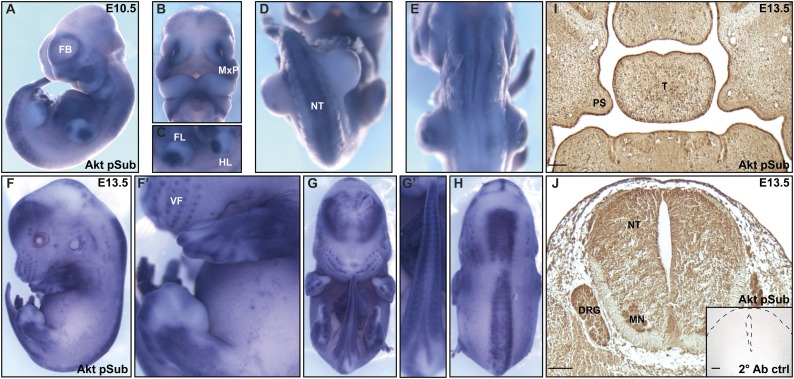
Multiple lines of evidence support a role for Akt as the main effector of PI3K signaling (Vasudevan and Garraway 2010). To identify sites of active Akt signaling in the embryo, we performed whole-mount immunohistochemistry with an anti-Akt phosphosubstrate antibody generated against the phosphorylated Akt consensus recognition motif (RxRxxpS/T) (Alessi et al. 1996; Zhang et al. 2002). While the antibody generated a faint signal throughout the entire embryo at E10.5, significantly increased staining was detected in the forebrain, in the maxillary processes from which the palatal shelves will extend, in the limb buds, and along the neural tube into the tail (Fig. 2A–E). This pattern was largely maintained at E13.5, with additional staining in the newly formed vibrissae and supra- and suborbital tactile follicles (Fig. 2F–H). Immunohistochemistry with this same antibody on paraffin sections confirmed staining in the palatal shelves, the neuroepithelium of the neural tube, and the dorsal root ganglia at E13.5 (Fig. 2I,J).
Figure 2.
Akt phosphosubstrate expression in the embryo. (A–H) Whole-mount immunohistochemistry of E10.5 (A–E) and E13.5 (F–H) embryos with an anti-Akt phosphosubstrate antibody generated against the phosphorylated Akt consensus recognition motif. Noticeably increased expression was detected in the forebrain (A,F), in the maxillary processes (B), in the tactile follicles (F,F′), in the limbs (C,F′), and along the neural tube into the tail (D,E,G–H). (I,J) Immunohistochemistry performed on paraffin sections with the anti-Akt phosphosubstrate antibody. Note the increased expression in the palatal shelves (I) and the neuroepithelium of the neural tube and the dorsal root ganglia (J) at E13.5. (J, inset) No signal was detected in a secondary antibody control section of the neural tube and surrounding tissue (outlined with dashed lines) at E13.5. (FB) Forebrain; (MxP) maxillary process; (FL) forelimb bud; (HL) hindlimb bud; (NT) neural tube; (VF) vibrissae follicles; (PS) palatal shelves; (T) tongue; (DRG) dorsal root ganglia; (MN) motor neurons. Bars, 100 μm.
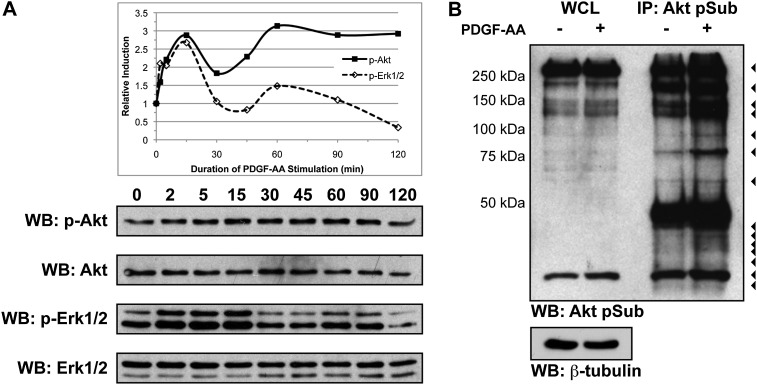
We subsequently examined the time course of PDGF-AA-dependent Akt phosphorylation in E13.5 primary MEPM cells. These cells uniformly express PDGFRα as well as several additional markers of palatal mesenchyme cells in vivo and are responsive to stimulation with numerous growth factors (Bush and Soriano 2010; data not shown), indicating that they serve as a faithful surrogate for embryonic palatal mesenchyme. Following serum starvation, low-passage cells were stimulated with 10 ng/mL PDGF-AA ligand for up to 2 h. Western blot analysis of whole-cell lysates revealed a peak of Akt induction 15 min after ligand stimulation (Fig. 3A). Immunoprecipitation of Akt targets from untreated and PDGF-AA-treated primary MEPM lysates using the anti-Akt phosphosubstrate antibody followed by Western blotting with the same antibody revealed at least 14 protein bands increased in intensity upon ligand stimulation (Fig. 3B).
Figure 3.
PDGF-AA treatment results in the differential phosphorylation of several Akt targets in primary MEPM cells. (A) PDGF-AA-dependent Akt and Erk1/2 phosphorylation time course in E13.5 primary MEPM lysates. Following serum starvation, low-passage MEPM cells were stimulated with 10 ng/mL PDGF-AA ligand for 2 min to 2 h. Western blot analysis of whole-cell lysates revealed peaks of both Akt and Erk1/2 phosphorylation 15 min after ligand treatment. (B) PDGF-AA-dependent Akt target phosphorylation in E13.5 primary MEPM lysates. Immunoprecipitation of Akt targets from lysates of primary MEPM cells that were untreated or treated for 15 min with 10 ng/mL PDGF-AA ligand with an anti-Akt phosphosubstrate antibody followed by Western blotting with the same antibody revealed 14 protein bands (arrowheads) that increased in intensity upon ligand stimulation. Large bands at ∼50 kDa correspond to the heavy chain of IgG. (WB) Western blot; (WCL) whole-cell lysate; (IP) immunoprecipitation.
Mass spectrometry-based identification of PDGF-AA-dependent Akt phosphorylation targets in primary MEPM cells
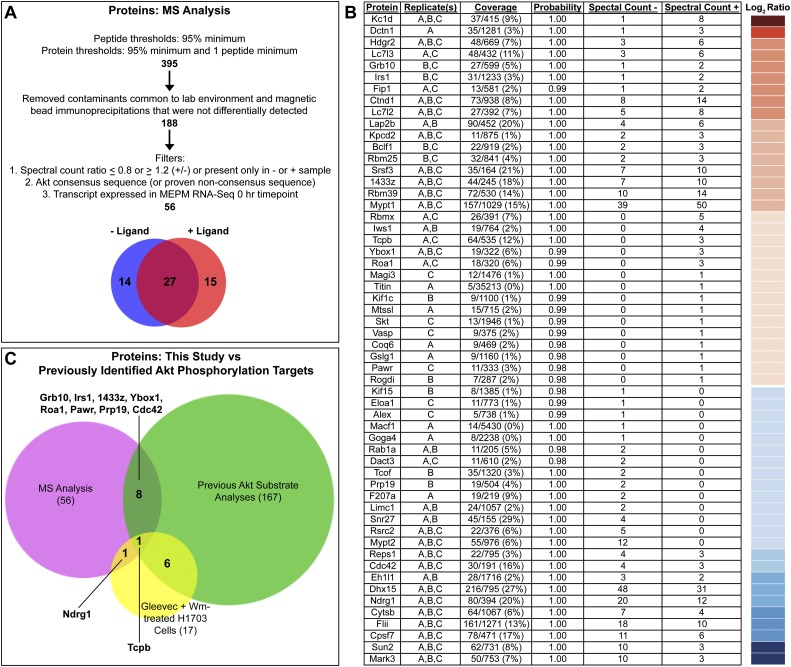
We next used a phosphoproteomic approach to identify proteins phosphorylated by Akt downstream from PI3K-mediated PDGFRα signaling in MEPM cells. We immunoprecipitated in parallel Akt targets from primary MEPM lysates that were untreated or stimulated with PDGF-AA for 15 min using the anti-Akt phosphosubstrate antibody and separated the proteins by SDS-PAGE. Whole-gel lanes were excised for comparative analysis to capture the full range of immunoprecipitated proteins and subjected to in-gel trypsin digestion, and the tryptic peptides were analyzed by nanoliquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS). Peptides and proteins were subsequently identified with the Mascot search engine, and statistical analyses were performed with Scaffold 3 software. After removing common contaminants that were not differentially detected, proteins were filtered based on three criteria: (1) quantitative spectral counts that differed by at least 20% between untreated and PDGF-AA-treated samples, (2) the presence of an Akt consensus sequence (or proven nonconsensus sequence), and (3) expression of the transcript encoding the protein in unstimulated MEPM cells, as determined by an RNA sequencing experiment performed in our laboratory (Fig. 4A; F He, H Vasudevan, and P Soriano, unpubl.). The full spectrum count report is in Supplemental Table 1.
Figure 4.
Mass spectrometry-based identification of PDGF-AA-dependent Akt phosphorylation targets in primary MEPM cells. (A) Filters applied to the nano-LC-MS/MS data set. (B) Proteins identified are listed based on the ratio of the number of quantitative spectra detected in PDGF-AA-treated (+) versus untreated (−) samples. The log2 of this ratio for each protein is represented colorimetrically in a heat map at right. Additional information listed includes protein name, detection in biological replicates, coverage, protein identification probability, and quantitative spectral counts for untreated (−) and PDGF-AA-treated (+) samples as determined using Scaffold 3 software. Replicate(s), coverage, and protein identification probability pertain to the sample (untreated or PDGF-AA-treated) in which each protein had the highest number of quantitative spectra. (C) Comparison of the proteins identified in this study (56) versus previously identified Akt phosphorylation targets in humans, mice, and/or rats (167) versus proteins for which phosphorylation at an Akt consensus site is sensitive to independent treatment with the PDGFRα inhibitor Gleevec and the PI3K inhibitor Wortmannin (Wm) in H1703 cells, a non-small-cell lung cancer cell line driven by PDGFRα (17) (Moritz et al. 2010). Of the proteins identified in this study, 46 are potentially novel Akt phosphorylation targets.
Our parameters generated a list of 56 proteins (Fig. 4B), 27 of which were identified in both samples, while 14 and 15 were identified exclusively in the untreated and PDGF-AA-treated samples, respectively (Fig. 4A). The finding that the majority of target proteins are common to both samples in the nano-LC-MS/MS analysis is consistent with our immunoprecipitation experiments with the anti-Akt phosphosubstrate antibody, in which the majority of bands observed increased in intensity upon ligand stimulation (Fig. 3B). Treatment of unstimulated lysates with λ protein phosphatase prior to immunoprecipitation of Akt targets using the anti-Akt phosphosubstrate antibody followed by Western blotting with the same antibody revealed a significant decrease in band intensities upon phosphatase treatment (Supplemental Fig. 2A), indicating that most Akt phosphorylation targets in primary MEPM cells are phosphorylated at baseline levels before PDGF-AA ligand stimulation. Ten of the proteins identified in our analysis had previously been confirmed as Akt phosphorylation targets in mammals, and/or phosphorylation of an Akt consensus site within the protein was shown to be sensitive to independent treatment with the PDGFRα inhibitor Gleevec and the PI3K inhibitor Wortmannin in a PDGFRα-driven cancer cell line (Fig. 4C; Moritz et al. 2010). Importantly, our strategy made use of a primary cell line that is highly relevant to a morphogenetic process active during development, marking a fundamental departure from the vast majority of previous studies in the field that have sought to identify Akt signaling events in transformed or cancer cell lines. As such, 46 out of 56 proteins (82%) identified in our analysis are potentially novel Akt phosphorylation targets.
PI3K-engaged PDGFRα signaling mediates cell survival and proliferation in primary MEPM cells
After validating the PI3K-mediated phosphorylation of select target proteins upon PDGF-AA treatment in primary MEPM cells (Supplemental Fig. 2B), we grouped our targets into various categories based on their cellular functions. As expected, the target proteins identified have diverse roles in the cell, such as regulation of the cytoskeleton (14), RNA processing (13), regulation of Wnt signaling (nine), 14-3-3 binding (five), transcriptional regulation (five), cell adhesion (five), and migration (four). Notably, the most significant ontology term detected in a mammalian phenotype enrichment analysis of our list of 56 identified proteins using the Enrichr tool (Chen et al. 2013) was abnormal cell proliferation (MP:0000350), with an adjusted P-value of 0.00865. As such, we chose to focus our studies on a group of 10 proteins that regulate cell survival and proliferation: Grb10, Rbm25, 14-3-3ζ, Tcpb, Ybox1, Pawr, Tcof, Rsrc2, Cdc42, and Ndrg1. None of these proteins have previously been demonstrated to operate downstream from activated PDGFRα signaling or interact in a physical complex or network. Several lines of evidence point to a role for PDGFRα signaling, particularly mediated through PI3K activity, in regulating survival and proliferation. First, Pdgfra−/−-null embryos display increased apoptosis in the cephalic region, branchial arches, and somites by E10.5 (Soriano 1997). Second, in vitro studies in a mouse fibroblast cell line have demonstrated that PDGF-AA-induced proliferation and anti-apoptotic signaling downstream from PDGFRα are solely mediated by PI3K (Rosenkranz et al. 1999; Vantler et al. 2006). We performed a whole-mount in situ hybridization analysis to examine the expression of the transcripts encoding several of these target proteins (Supplemental Fig. 3), confirming their localization at sites of Pdgfra expression (Hamilton et al. 2003).
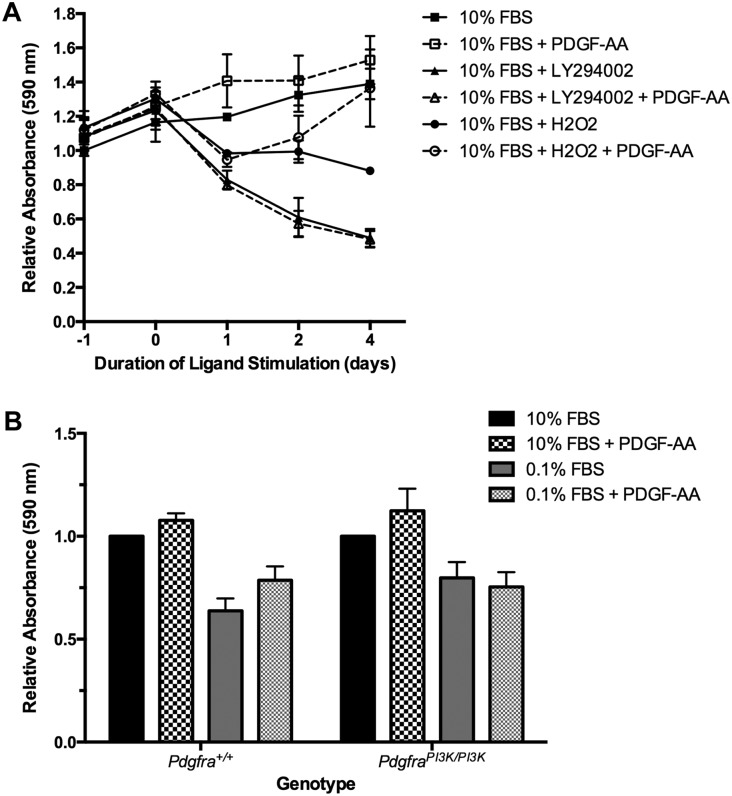
To extend the above in vitro results into the palatal mesenchyme, we next performed a series of cell growth assays to assess the role of PI3K-engaged PDGFRα signaling in mediating survival and proliferation in our primary MEPM culture system. We demonstrated that MEPM cell survival is impaired in the presence of the PI3K inhibitor LY294002 (64.88% ± 1.538% decrease in relative absorbance; P = 0.0130) regardless of PDGF-AA ligand stimulation (68.48% ± 0.2966% decrease in relative absorbance; P = 0.0197) and that PDGF-AA treatment provides protection from hydrogen peroxide-induced apoptosis (54.88% ± 26.34% increase in relative absorbance; P = 0.1653) after 4 d in culture (Fig. 5A). Furthermore, we showed that PdgfraPI3K/PI3K-derived MEPM cells under serum starvation conditions do not proliferate in response to PDGF-AA treatment, unlike their wild-type counterparts (Fig. 5B).
Figure 5.
PI3K-engaged PDGFRα signaling mediates cell survival and proliferation in primary MEPM cells. (A) Cell growth as assessed by crystal violet staining in E13.5 primary MEPM cells grown in medium containing 10% FBS and treated once with LY294002 or H2O2 and/or daily with PDGF-AA for up to 4 d. MEPM cell survival is impaired in the presence of the PI3K inhibitor LY294002 (64.88% ± 1.538% decrease in relative absorbance; P = 0.0130) regardless of PDGF-AA ligand stimulation (68.48% ± 0.2966% decrease in relative absorbance; P = 0.0197). PDGF-AA treatment provides protection from hydrogen peroxide-induced apoptosis (54.88% ± 26.34% increase in relative absorbance; P = 0.1653). (B) Cell growth as assessed by crystal violet staining in Pdgfra+/+- and PdgfraPI3K/PI3K-derived E13.5 primary MEPM cells grown in medium containing 10% or 0.1% FBS and treated with PDGF-AA for 1 d. PdgfraPI3K/PI3K-derived MEPM cells under serum starvation conditions do not proliferate in response to PDGF-AA treatment. Data are presented as mean ± SEM.
p53 activity is inhibited downstream from PDGFRα signaling
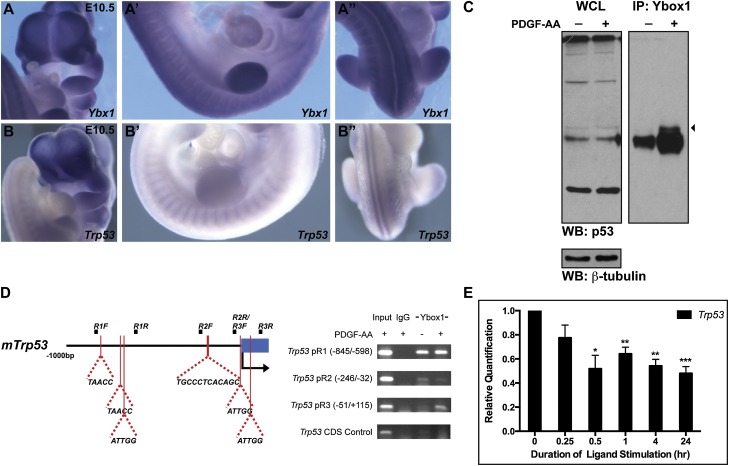
Several of the Akt phosphorylation targets that we identified, including 14-3-3ζ, Ybox1, Tcof, and Ndrg1, are known to mediate cell survival in part through regulation of p53 (Kurdistani et al. 1998; Lasham et al. 2003; Stein et al. 2004; Homer et al. 2005; Gu et al. 2006; Jones et al. 2008). Ybox1 is a particularly appealing target protein, as its Akt consensus sequence (RKYLRS) is fully conserved among vertebrates ranging from humans to zebrafish. Ybx1 transcripts are robustly expressed in the facial processes, somites, and the neural tube at E10.5, among other sites (Fig. 6A), where they overlap with Pdgfra (Hamilton et al. 2003) and Trp53 expression (Fig. 6B). Furthermore, Ybx1−/−-null embryos die during embryogenesis, surviving as late as E18.5, and exhibit phenotypes common to both Pdgfra−/− and Trp53−/− embryos, including abnormalities in neural tube development and craniofacial bone hypoplasia, and several additional phenotypes characteristic of Pdgfra−/− embryos, such as retarded growth, edema, and hemorrhaging (Soriano 1997; Sah et al. 1995; Uchiumi et al. 2006; Rinon et al. 2011).
Figure 6.
p53 transcription is inhibited downstream from PDGFRα signaling. (A–A″) Ybx1 transcripts are robustly expressed in the facial processes, somites, and neural tube at E10.5, as determined by whole-mount in situ hybridization. (B–B″) Trp53 transcripts also localize to the facial processes, somites, and neural tube at E10.5. (C) Ybox1 bound the p53 protein (arrowhead) following 30 min of PDGF-AA treatment in coimmunoprecipitation experiments using primary MEPM lysates. (WB) Western blot; (WCL) whole-cell lysate; (IP) immunoprecipitation. (D) Ybox1 bound up to two sites in the Trp53 promoter (−8/−4 and +50/+54) within the promoter region 3 (pR3) amplicon (−51/+115) exclusively following 30 min of PDGF-AA treatment in endogenous ChIP experiments in primary MEPM cells. No binding was observed in this region of the promoter in an IgG control sample or in the absence of ligand. Furthermore, no binding was observed in a coding sequence (CDS) negative control region. (E) Bar graph depicting qRT-PCR values revealing significantly reduced expression of Trp53 transcripts 30 min (48.15% ± 11.14%; P = 0.0124) to 24 h (51.99% ± 5.61%; P = 0.0008) after PDGF-AA treatment of primary MEPM cells. Data are presented as mean ± SEM. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Phosphorylation of Ybox1 by Akt has been shown to promote its translocation to the nucleus (Sutherland et al. 2005) and result in the repression of p53 activity through two mechanisms: (1) binding to the p53 protein and thereby inhibiting the ability of p53 to transactivate cell death genes (Homer et al. 2005) and (2) binding to the Trp53 promoter and repressing Trp53 transcription (Lasham et al. 2003). To assess whether these interactions are relevant downstream from PDGFRα signaling, we first performed coimmunoprecipitation experiments using primary MEPM lysates, demonstrating that Ybox1 binds p53 30 min after PDGF-AA stimulation (Fig. 6C). As Ybox1 is known to bind inverse CCAAT motifs in DNA (Didier et al. 1988), we next scanned the Trp53 promoter within 1 kb of the transcriptional start site, identifying six potential binding sites through exon 1 (Fig. 6D), including one (−197/−186) that has been experimentally confirmed (Lasham et al. 2003). Using endogenous chromatin immunoprecipitation (ChIP) assays, we revealed that Ybox1 binds up to two of these sites (−8/−4 and +50/+54) (Fig. 6D pR3 gel) exclusively following 30 min of PDGF-AA treatment, while no binding was observed in this region of the promoter in an IgG control sample or in the absence of ligand (Fig. 6D).
To examine whether the signaling events downstream from PDGFRα activation result in a decrease in p53 activity, we first treated primary MEPM cells with PDGF-AA ligand for up to 24 h, harvested RNA at several timepoints, and performed quantitative real-time PCR (qRT-PCR) analysis examining the expression of Trp53. We found that Trp53 expression was significantly reduced 30 min (48.15% ± 11.14%; P = 0.0124) to 24 h (51.99% ± 5.61%; P = 0.0008) after PDGF-AA ligand treatment (Fig. 6E). Moreover, similar expression analyses of p53 transcriptional targets regulating cell cycle arrest and cell death demonstrated that several (Ccng1, Trp53inp1, Bax, Gadd45a, Perp, and Zmat3) were also significantly down-regulated upon PDGF-AA treatment (Supplemental Fig. 4A,B). Finally, additional qRT-PCR analyses comparing expression of these same transcripts in RNA derived from Pdgfra+/+ versus PdgfraPI3K/PI3K E13.5 head tissue revealed modest increases in the expression of Trp53, Trp53inp1, Bax, Perp, and Zmat3 in the mutant samples (Supplemental Fig. 4C). Collectively, these results demonstrate that PDGFRα signaling inhibits the transcription of Trp53 and a subset of p53 targets regulating cell cycle arrest and cell death, at least in part, through modulation of Ybox1 activity.
Introduction of a Trp53-null allele attenuates a subset of axial skeleton defects found in PdgfraPI3K/PI3K neonates
We next performed genetic epistasis experiments, examining whether introduction of a Trp53-null allele could rescue the skeletal phenotypes observed in PdgfraPI3K/PI3K mice. Pdgfra+/PI3K;Trp53+/− mice were intercrossed, and the perinatal progeny were harvested. Genotyping revealed that PdgfraPI3K/PI3K;Trp53+/+ pups were recovered slightly below Mendelian frequency at birth (seven pups vs. 12 expected pups out of 190 total; P = 0.16), while PdgfraPI3K/PI3K;Trp53+/− pups were recovered significantly below Mendelian frequency at this time point (seven pups vs. 24 expected pups out of 190 total; P = 5.9 × 10−4). Importantly, however, PdgfraPI3K/PI3K;Trp53+/− pups that survived through late gestation were more likely to be alive at birth (five of seven live pups, 71%) than their PdgfraPI3K/PI3K;Trp53+/+ littermates (two of seven live pups, 29%) (Supplemental Table 2).
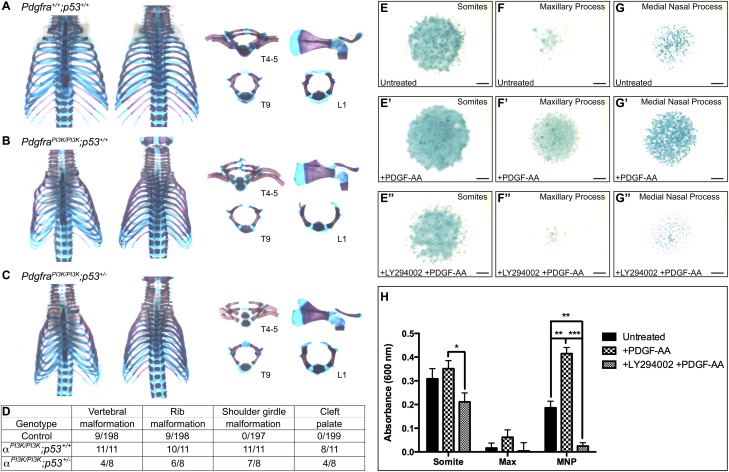
Skeletal preparations were then generated from the perinatal Pdgfra+/PI3K;Trp53+/− intercross progeny. PdgfraPI3K/PI3K;Trp53+/+ skeletons exhibited rib malformations (10 of 11, 91%) (Fig. 7B), shoulder girdle malformations (11 of 11, 100%) (Fig. 7B), and cleft palate (eight of 11, 73%) (Supplemental Fig. 5B) at incidences (Fig. 7D) comparable with those previously observed (Klinghoffer et al. 2002) and with those detected in PdgfraPI3K/PI3K;Trp53+/− skeletons (six of eight rib malformations, 75%; seven of eight shoulder girdle malformations, 88%; four of eight cleft palate, 50%) (Fig. 7C,D; Supplemental Fig. 5C). Morphometric analyses of PdgfraPI3K/PI3K;Trp53+/+ and PdgfraPI3K/PI3K;Trp53+/− skeletal preparations confirmed that there were no significant differences in the dimensions of the craniofacial structures derived from either the paraxial mesoderm or neural crest between the two genotypes (Supplemental Fig. 5D). Notably, however, PdgfraPI3K/PI3K;Trp53+/− skeletons exhibited significantly reduced incidences (P = 0.0181) (Fig. 7D) of vertebral malformations (four of eight, 50%) compared with their PdgfraPI3K/PI3K;Trp53+/+ counterparts (11 of 11, 100%). Vertebral defects that were rescued in PdgfraPI3K/PI3K;Trp53+/− skeletons included bifurcated cervical vertebrae, asymmetric thoracic vertebral fusions, and incomplete closure of the thoracic and lumbar vertebral arches resulting in spina bifida (Fig. 7B,C). Shoulder girdle malformations and cleft palate were never detected in at least 197 control skeletons, and vertebral and rib malformations, when rarely observed (Fig. 7D), were never as severe as those found in experimental skeletons (Fig. 7A; Supplemental Fig. 5A). Analysis of transverse sections at E12.5 revealed that the cartilage primordia of the ventral neural arches of PdgfraPI3K/PI3K;Trp53+/+ embryos (Supplemental Fig. 6B) had not advanced as far dorsally as those of wild-type littermates (Supplemental Fig. 6A), while those of PdgfraPI3K/PI3K;Trp53+/− embryos had (Supplemental Fig. 6C). Moreover, Ki67 immunofluorescence analyses demonstrated that proliferation of the mesenchyme precartilage primordia of the dorsal neural arches, which was detected below wild-type levels (Supplemental Fig. 6A) in PdgfraPI3K/PI3K;Trp53+/+ embryos (Supplemental Fig. 6B), was recovered in PdgfraPI3K/PI3K;Trp53+/− embryos (Supplemental Fig. 6C).
Figure 7.
A subset of axial skeleton defects in PdgfraPI3K/PI3K;Trp53+/+ neonates are rescued in PdgfraPI3K/PI3K;Trp53+/− littermates. (A–C) Skeletal preparations of the ribs, vertebrae, and scapular blades with acromions generated from Pdgfra+/+;Trp53+/+ (A), PdgfraPI3K/PI3K;Trp53+/+ (B), and PdgfraPI3K/PI3K;Trp53+/− (C) littermate pups. Note the asymmetric vertebral fusions of thoracic vertebrae 4–5 and the incomplete closure of the vertebral arches of thoracic vertebra 9 and lumbar vertebra 1 in a PdgfraPI3K/PI3K;Trp53+/+ skeleton (B) that are rescued in a PdgfraPI3K/PI3K;Trp53+/− skeleton (C). (T) Thoracic vertebra; (L) lumbar vertebra. (D) Pdgfra+/PI3K;Trp53+/− intercross skeletal phenotype scoring. (E–G″) Alcian blue-stained micromass cultures generated from E11.5 somites (E–E″), maxillary processes (F–F″), and medial nasal processes (G–G″). Bars, 1 mm. (H) Bar graph depicting absorbance readings from Alcian blue-stained micromass cultures. While all three cultures showed an increase in chondrogenesis upon PDGF-AA ligand treatment and a decrease in the presence of the PI3K inhibitor LY294002, these responses were more significant in the cultures derived from the facial processes (122.8% ± 3.790% increase upon PDGF-AA treatment in medial nasal processes, P = 0.0038; 94.00% ± 2.954% decrease upon LY294002 treatment in medial nasal processes, P = 0.0002) than those derived from the somites (13.74% ± 5.457% increase upon PDGF-AA treatment, P = 0.4467; 39.88% ± 5.108% decrease upon LY294002 treatment, P = 0.0336). Data are presented as mean ± SEM. (Max) Maxillary process; (MNP) medial nasal process. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
The finding that the vertebral but not the craniofacial defects were attenuated in the above in vivo interaction experiments led us to examine any potential differences in the chondrogenic response of somite versus facial process mesenchyme to PDGF-AA ligand treatment. We established high-density micromass cultures from E11.5 somites, maxillary processes, and medial nasal processes and measured the extent of chondrogenesis in each by Alcian blue staining after 6 d in culture. While all three cultures showed an increase in chondrogenesis upon PDGF-AA ligand treatment (Fig. 7E,E′,F,F′,G,G′,H) and a decrease in the presence of the PI3K inhibitor LY294002 (Fig. 7E″,F″,G″,H), these responses were more robust in the cultures derived from the facial processes (122.8% ± 3.790% increase upon PDGF-AA treatment in medial nasal processes, P = 0.0038; 94.00% ± 2.954% decrease upon LY294002 treatment in medial nasal processes, P = 0.0002) than those derived from the somites (13.74% ± 5.457% increase upon PDGF-AA treatment, P = 0.4467; 39.88% ± 5.108% decrease upon LY294002 treatment, P = 0.0336). Taken together, these results highlight different responses of various embryonic skeletal populations to PI3K-mediated PDGFRα signaling and indicate that craniofacial-derived structures may be especially sensitive to loss of activity downstream from the receptor, consistent with the findings of our in vivo interaction experiments.
Discussion
An extensive number of studies have sought to identify phosphorylation targets of Akt and link these post-translational modification events to changes in cell behavior (Manning and Cantley 2007; Vasudevan and Garraway 2010). Most of this biochemical work, similar to early studies of PDGF signaling, has been conducted in transformed or cancer cell lines and functionally validated in vitro. Despite the critical advances gained from these studies, the targets of Akt signaling that operate downstream from distinct receptor tyrosine kinases and their role in mediating receptor function during development remain largely unknown. Here, we identified a novel intracellular pathway downstream from PI3K/Akt-mediated PDGFRα signaling that regulates survival and proliferation in skeletal development in vivo. We demonstrate that a subset of Akt target proteins regulate p53 activity in a PDGF-AA-dependent manner and further show that introduction of a Trp53-null allele attenuates the vertebral skeleton defects found in Pdgfra mutant embryos. These findings provide unique insights into the mechanisms of PDGFRα signaling during development.
Interestingly, we found that Akt phosphosubstrate expression, while ubiquitous, is not uniform within the midgestation mouse embryo. Several explanations could account for this finding. First, Akt itself could be differentially phosphorylated across various tissues due to changes in the expression or activation of upstream regulators at those sites. Second, the expression level of individual substrate proteins and/or number of substrate proteins present at unique regions could vary within the embryo. Based on our whole-mount in situ hybridization results, we know that at least several of the Akt phosphorylation targets identified in this study are expressed in discrete domains during development. The composite Akt phosphosubstrate expression observed likely reflects both of these phenomena, however, resulting in localized up-regulation of Akt phosphosubstrate expression in the forebrain, maxillary processes, limb buds, and neural tube.
Our results collectively highlight the fact that PDGFRα signaling leads to subtle differences in the quantitative phosphorylation of Akt and its targets, which in turn have a substantial impact on embryonic development. In fact, we demonstrate that Akt phosphorylation peaks at approximately threefold baseline levels upon PDGF-AA treatment by Western blot analysis of cultured MEPM cells and further show in our mass spectrometry screen that the most significant spectral count ratio between untreated and PDGF-AA-treated samples is eight (for the protein Kc1d), while the ratios for the other identified proteins were often considerably lower. Notably, a previous study from our laboratory found that signaling through another receptor tyrosine kinase, EphB3, led to subtle changes in the phosphorylation of Erk1/2 (also peaking at approximately threefold baseline levels upon ligand treatment) and additional proteins that had significant effects on palatal shelf proliferation (Bush and Soriano 2010).
Of the proteins identified in our mass spectrometry screen, several are known to be enriched in the branchial arches, neural tube, and/or somites during development and display defects in the structures derived from these cell populations in null mouse models. For example, Csnk1d (encoding Kc1d), Grb10, Ybx1, and Hnrnpa1 (encoding Roa1) were all identified in a screen for genes involved in mouse craniofacial development that employed suppression subtractive hybridization to enrich for transcripts expressed in E10.5 branchial arches 1 and 2 (Fowles et al. 2003), lending support to our findings. While we chose to focus here on a group of proteins that regulates cell survival and proliferation, proteins in other functional categories point to additional, novel roles for PDGFRα signaling in vivo, such as regulation of RNA processing, and will be the subject of future studies.
Among the subset of proteins identified in our screen that we categorized as regulating cell survival and proliferation, including Ybox1, several have corresponding mutant mouse models that exhibit phenotypic overlap with Pdgfra−/− and/or PdgfraPI3K/PI3K mutant embryos. Tcof1 is expressed at the edges of the neural fold and in the developing branchial arches (Supplemental Fig. 3F; Dixon et al. 1997), and Tcof1−/−-null embryos exhibit neonatal lethality, embryonic growth arrest, abnormalities in neural fold formation and neural tube closure, defective neural crest cell proliferation and migration, craniofacial bone hypoplasia, and cleft palate (Dixon et al. 2006). In humans, TCOF1 mutations underlie Treacher Collins syndrome, characterized by hypoplasia of the facial bones and cleft palate (The Treacher Collins Syndrome Collaborative Group 1996). Furthermore, conditional deletion of Cdc42 in various mesenchymal populations throughout the embryo using a Prx1-Cre driver results in decreased body size, cleft palate, abnormal cartilage development, and a split sternum (Aizawa et al. 2012), while deletion in neural crest cells using a Wnt1-Cre driver results in facial clefting and underdeveloped dorsal root ganglia due to reduced mitotic activity and increased cell cycle exit in neural crest-derived structures (Fuchs et al. 2009). Finally, Ndrg1−/−-null embryos display decreased body size and skeletal muscle atrophy in addition to peripheral nerve defects (Okuda et al. 2004).
Importantly, previous work by our laboratory demonstrated that cell proliferation is decreased in the medial nasal processes of PdgfraPI3K/PI3K embryos at E11.5 compared with their wild-type counterparts, as assessed by BrdU staining (He and Soriano 2013). Lineage tracing in PdgfraPI3K/PI3K;Wnt1-Cre+/Tg;R26R+/− embryos revealed no apparent defect in cranial neural crest cell migration, however (He and Soriano 2013), indicating a particular requirement for PI3K-mediated PDGFRα signaling in the regulation of cell proliferation in the facial processes. Conversely, an additional study examining the role of PDGFRα signaling in somite-derived structures revealed a cell migration defect in the somite-derived dorsal mesenchyme but no apoptotic or proliferation abnormalities in PdgfraPI3K/PI3K neural arches or sclerotome (Pickett et al. 2008), pointing to regional differences in the response of embryonic tissues to PI3K-mediated PDGFRα signaling.
In line with the above findings, our in vivo genetic interaction experiments and micromass culture results uncovered differences in the chondrogenic response of various embryonic skeletal tissues to PI3K-mediated PDGFRα signaling during development. More specifically, the vertebral defects of PdgfraPI3K/PI3K embryos were attenuated upon introduction of a Trp53-null allele, while the cleft palate, rib, and shoulder girdle malformations were not. One likely possibility to explain these findings is that the palate and other structures are particularly sensitive to loss of PI3K-mediated PDGFRα signaling such that loss of one copy of Trp53 cannot rescue the underlying survival and proliferation defects. In support of this model, micromass cultures derived from the facial processes have a much more robust response to PDGF-AA treatment and PI3K inhibition than those derived from the somites.
Despite limited investigation into the role of p53 during embryogenesis, several developmental abnormalities have been reported for Trp53−/− embryos. A variable percentage of homozygous mutant embryos exhibit exencephaly, characterized by failure of the anterior neural tube to close followed by overgrowth of neural tissue (Sah et al. 1995). A more recent study revealed a reduction in the mass of the frontal (neural crest cell-derived) and parietal (paraxial mesoderm-derived) bones in Trp53−/− embryos and showed that overexpression or stabilization of p53 in the chick cranial neural tube reduced cranial neural crest cell proliferation and delamination (Rinon et al. 2011). Rinon et al. (2011) further demonstrated that p53 coordinates these processes by regulating cell cycle gene expression, including Ccng1 and Mycn. Importantly, the abnormalities in neural tube development and craniofacial bone hypoplasia found in Trp53−/− embryos overlap with a subset of affected tissues in Pdgfra−/− embryos.
However, p53 is not the only effector protein of the Akt phosphorylation targets regulating cell survival and proliferation identified in our screen. For example, Grb10 is known to mediate cell survival through phosphorylation (and inactivation) of Bad (Kebache et al. 2007); Rbm25 mediates apoptosis through regulation of Bcl2l1 isoform expression (Zhou et al. 2008); Ybox1 has been shown to additionally promote cell survival through repression of Fas expression (Lasham et al. 2000) and promote cell proliferation through activation of Ccna1, Ccnb1 (Jurchott et al. 2003), and Mmp2 expression (Mertens et al. 1997); Pawr activates the Fas prodeath pathway, inhibits NFκB survival activity (Chakraborty et al. 2001), and transcriptionally represses Bcl2 under various conditions (El-Guendy and Rangnekar 2003); and Cdc42 up-regulates Ccnd1 expression (Bauerfeld et al. 2001). As such, the extent of rescue observed in PdgfraPI3K/PI3K;Trp53+/− neonates is remarkable and points to a new, critical role for PDGFRα signaling in modulating p53 activity to regulate cell survival during skeletal development.
In summary, we investigated Akt signaling dynamics in the embryo and identified p53 as a novel effector downstream from PDGFRα signaling that regulates cell survival and proliferation during development of the craniofacial and axial skeleton. Our findings reveal regional differences in the response of the embryonic skeleton to PI3K-mediated PDGFRα signaling and provide considerable insight into the mechanisms of PDGF signaling in vivo.
Materials and methods
Mouse strains
All animal experimentation was approved by the Institutional Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai. Pdgfra+/tm5Sor mice (Klinghoffer et al. 2002), referred to in the text as Pdgfra+/PI3K, were maintained on a 129S4 coisogenic genetic background. Trp53+/tm1Tyj mice (Jacks et al. 1994), referred to in the text as Trp53+/−, were a gift of Dr. James Manfredi, Icahn School of Medicine at Mount Sinai, and were maintained on a mixed 129S4/C57BL/6J genetic background. Statistics regarding number of progeny were performed using a χ2 test.
Immunohistochemistry
Whole-mount immunohistochemistry was performed based on a previously published protocol (Joyner and Wall 2008) using a phospho-(Ser/Thr) Akt substrate primary antibody (1:200; Cell Signaling Technology, Inc.) and goat anti-rabbit IgG peroxidase-conjugated secondary antibody (1:500; Jackson ImmunoResearch Laboratories, Inc.). Stained embryos were photographed using a Leica DFC290 digital camera (Leica Microsystems, Inc.) fitted onto a Nikon SMZ-U stereomicroscope (Nikon, Inc.). Paraffin sections (10 μm) were deparaffinized and rehydrated prior to antigen unmasking in subboiling 10 mM sodium citrate buffer (pH 6.0). Sections were then incubated in 3% H2O2 for 10 min, washed in 1× Tris-buffered saline with 0.1% Tween-20 (TBST), blocked in 5% normal goat serum in TBST for 1 h, and incubated in phospho-(Ser/Thr) Akt substrate primary antibody (1:250; Cell Signaling Technology, Inc.) diluted in SignalStain antibody diluent (Cell Signaling Technology, Inc.) overnight at 4°C. Sections were incubated in anti-rabbit IgG biotinylated secondary antibody (1:200; Vector Laboratories, Inc.) diluted in TBST for 30 min, followed by ABC reagent (Vector Laboratories, Inc.) for 30 min, and finally DAB substrate (Vector Laboratories, Inc.) for ∼2 min. Sections were dehydrated, permanently mounted with Permount (Thermo Fisher Scientific, Inc.), and photographed using a Leica DFC290 digital camera (Leica Microsystems, Inc.) fitted onto an Axioplan microscope (Carl Zeiss, Inc.).
Cell culture and growth assays
Primary MEPM cells were isolated from E13.5 (day of plug considered 0.5 d) embryo palatal shelves and cultured in medium (Dulbecco’s modified Eagle’s medium [Gibco, Invitrogen] supplemented with 50 U/mL penicillin [Gibco], 50 μg/mL streptomycin [Gibco], and 2 mM L-glutamine [Gibco]) containing 10% fetal bovine serum (FBS) (HyClone Laboratories, Inc.) as previously described (Bush and Soriano 2010). For cell growth assays, passage 2 MEPM cells were cultured in medium containing 10% FBS or 0.1% FBS and treated once with 50 μM LY294002 (Sigma-Aldrich) or 100 μM H2O2 and/or daily with 10 ng/mL PDGF-AA ligand (R&D Systems) for up to 4 d. Cells were subsequently fixed in 4% paraformaldehyde (PFA) in PBS, stained with 0.1% crystal violet in 10% ethanol, and extracted with 10% acetic acid, and the absorbance was measured at 590 nm. Micromass cultures were established from E11.5 embryo somites, maxillary processes, and medial nasal processes based on previously published protocols (Ralphs 1992; Tallquist et al. 2000). Single-cell suspensions were plated at a density of 2 × 107 cells per milliliter in 20-μL droplets, flooded with medium containing 0.1% FBS after 1.5 h, and treated with 10 μM LY294002 and/or 10 ng/mL PDGF-AA ligand. Medium, chemical inhibitor, and growth factor were replaced daily for 6 d. Cultures were subsequently fixed in 4% PFA in PBS, stained overnight with 1% Alcian blue in 0.1 N HCl, and extracted with 6 M guanidine hydrochloride, and the absorbance was measured at 600 nm. Data represent results from at least two independent trials. Statistical analyses were performed with Prism 6 (GraphPad Software, Inc.) using a two-tailed unpaired t-test.
Immunoprecipitations and Western blotting
To induce PDGFRα signaling, passage 2 MEPM cells at ∼70% confluence were serum-starved for 24 h in medium containing 0.1% FBS and stimulated with 10 ng/mL PDGF-AA ligand for the indicated length of time. When applicable, cells were pretreated with 50 μM LY294002 1 h before PDGF-AA ligand stimulation. Cells were harvested in ice-cold lysis buffer (20 mM Tris-HCl at pH 8, 150 mM NaCl, 10% glycerol, 1% Nonidet P-40, 2 mM EDTA, 1× complete Mini protease inhibitor cocktail [Roche Applied Science], 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4, 25 mM β-glycerophosphate), and total cell lysates were collected by centrifugation at 12,000 rpm for 20 min at 4°C. For immunoprecipitations, cell lysates were incubated with unconjugated (1:175 phospho-[Ser/Thr] Akt substrate [Cell Signaling Technology]; 1:433 Yb1 [C-terminal] [Sigma-Aldrich]) or magnetic bead-conjugated (1:30 phospho-Akt substrate [110B7E] rabbit mAb [Cell Signaling Technology]) primary antibodies overnight at 4°C. When unconjugated primary antibodies were used, cell lysates were incubated with 20 μg of protein A-coupled agarose beads (Santa Cruz Biotechnology) for 2 h at 4°C the following day. Beads were washed with lysis buffer five times, and the precipitated proteins were eluted with Laemmli buffer containing 10% β-mercaptoethanol (except in the case of samples blotted with Yb1 antibody), heated for 5 min at 95°C, and separated by SDS-PAGE. Western blot analysis was performed according to standard protocols using horseradish peroxidase-conjugated secondary antibodies. Quantifications of signal intensity were performed with ImageJ software (version 1.44o; National Institutes of Health). Antibody information and dilutions are provided in the Supplemental Material. The protein interactions from this study have been submitted to the International Molecular Exchange Consortium (IMEx; http://www.imexconsortium.org) through IntAct (Kerrien et al. 2012) and assigned the identifier IM-22193.
ChIP
Passage 2 MEPM cells derived from E13.5 embryos were serum-starved for 24 h in medium containing 0.1% FBS as above and stimulated with 10 ng/mL PDGF-AA ligand for 30 min. Cells were treated with 1% formaldehyde for 10 min at 37°C, washed twice with cold PBS containing 1× complete miniprotease inhibitor cocktail and 1 mM PMSF, and harvested. ChIP was carried out using the ChIP assay kit (EMD Millipore Corporation) according to the manufacturer’s instructions. Cell lysates were precipitated with 1 µg of either an anti-Yb1 rabbit polyclonal antibody (Sigma-Aldrich) or normal rabbit IgG (Santa Cruz Biotechnology, Inc.) as a negative control. After elution, DNA was recovered using the QIAquick PCR purification kit (Qiagen Inc.). PCR reactions were performed using input, IgG-precipitated, and Yb1-precipitated DNA samples, 1× ThermoPol buffer (0.2 M Tris at pH 8.8, 0.1 M KCl, 0.1 M [NH4]SO4, 20 mM MgSO4, 1% Triton X-100), 0.2 mM dNTPs, 0.4 μM primers (Integrated DNA Technologies, Inc.), and 1.2 U of Taq polymerase in a 25-μL reaction volume. The primers used for the various promoter regions as well as coding sequence negative controls are in Supplemental Table 3. The following PCR protocol was used: step 1, 5 min at 94°C; step 2, 45 sec at 94°C; step 3, 30 sec at 55°C; step 4, 1 min at 72°C; repeat steps 2–4 for 31–36 cycles; and step 5, 10 min at 72°C. PCR products were electrophoresed on a 1.5% agarose/TBE gel containing ethidium bromide and photographed on a Bio-Rad Molecular Imager Gel Doc system (Bio-Rad). Positive immunoprecipitation results were confirmed in two independent trials. The protein interactions from this study have been submitted to IMEx (http://www.imexconsortium.org) through IntAct (Kerrien et al. 2012) and assigned the identifier IM-22193.
qRT-PCR
For analysis of wild-type cells treated with PDGF-AA ligand, passage 2 MEPM cells derived from E13.5 embryos were serum-starved for 24 h in medium containing 0.1% FBS as above and stimulated with 10 ng/mL PDGF-AA for 15 min to 24 h. For analysis of Pdgfra+/+ and PdgfraPI3K/PI3K head samples, three independent sets of E13.5 littermates were harvested. Total RNA was isolated using the RNeasy minikit (Qiagen, Inc.) according to the manufacturer’s instructions. First strand cDNA was synthesized using a ratio of 2:1 random primers: oligo (dT) primer and SuperScript II RT (Invitrogen) according to the manufacturer’s instructions. qRT-PCR was performed on a Bio-Rad iQ5 multicolor RT-PCR detection system and analyzed with iQ5 optical system software (version 2.0; Bio-Rad). All reactions were performed with PerfeCTa SYBR Green FastMix for iQ (Quanta Biosciences, Inc.), 300 nM primers (Integrated DNA Technologies, Inc.), and 100 ng of cDNA in a 20-μL reaction volume. The following PCR protocol was used: step 1, 3 min at 95°C; step 2, 10 sec at 95°C; step 3, 30 sec at 60°C; repeat steps 2 and 3 for 40 cycles; step 4 (melting curve), 30 sec at 55°C; and repeat step 4 for 81 cycles. All samples were run in triplicate for three independent runs and normalized against an endogenous internal control, B2m. Statistical analyses were performed with Prism 6 (GraphPad Software, Inc.) using a two-tailed unpaired t-test. The qRT-PCR primers used are in Supplemental Table 4.
Skeletal preparations
E18.5 embryos and P0 (postnatal day 0) pups were skinned, eviscerated, fixed in 95% ethanol, and stained in 0.015% Alcian blue/0.005% alizarin red/5% glacial acetic acid in 70% ethanol at 37°C. Pups were then cleared in 1% KOH and transferred to solutions of decreasing KOH concentrations and increasing glycerol concentrations. Statistical analyses of skeletal phenotype scoring were performed with Prism 6 (GraphPad Software, Inc.) using a two-tailed Fisher’s exact test. Morphometric analyses were performed by simultaneously photographing littermate control and experimental craniofacial skeletons using a Leica DFC290 digital camera (Leica Microsystems, Inc.) fitted onto a Nikon SMZ-U stereomicroscope (Nikon, Inc.) and subsequently measuring pixel lengths in Preview (Apple, Inc.). Statistical analyses of morphometric measurements were performed with Prism 6 (GraphPad Software, Inc.) using a two-tailed unpaired t-test.
Acknowledgments
We are grateful to Dr. Hediye Erdjument-Bromage and Dr. Ronald C. Hendrickson of the Memorial Sloan-Kettering Cancer Center Proteomics and Microchemistry Core for their advice and assistance with mass spectrometry. We thank Aryel Heller, Tony Chen, and Anne Levine for genotyping and technical assistance. We are grateful to Dr. James Manfredi of the Icahn School of Medicine at Mount Sinai for the Trp53+/tm1Tyj mice. We thank members of the Soriano laboratory and Dr. Robert Krauss for their helpful discussions and critical comments on the manuscript, and Dr. Jeffrey Bush for his advice on the proteomics experiments. This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) grant R01DE022363 to P.S., and a NIH/NIDCR Ruth L. Kirschstein NRSA Individual Post-doctoral Fellowship F32DE022719 to K.A.F.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.238709.114.
References
- Aizawa R, Yamada A, Suzuki D, Iimura T, Kassai H, Harada T, Tsukasaki M, Yamamoto G, Tachikawa T, Nakao K, et al. 2012. Cdc42 is required for chondrogenesis and interdigital programmed cell death during limb development. Mech Dev 129: 38–50 [DOI] [PubMed] [Google Scholar]
- Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P 1996. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett 399: 333–338 [DOI] [PubMed] [Google Scholar]
- Andrae J, Gallini R, Betsholtz C 2008. Role of platelet-derived growth factors in physiology and medicine. Genes Dev 14: 731–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerfeld CP, Hershenson MB, Page K 2001. Cdc42, but not RhoA, regulates cyclin D1 expression in bovine tracheal myocytes. Am J Physiol Lung Cell Mol Physiol 280: L974–L982 [DOI] [PubMed] [Google Scholar]
- Brachmann SM, Yballe CM, Innocenti M, Deane JA, Fruman DA, Thomas SM, Cantley LC 2005. Role of phosphoinositide 3-kindase regulatory isoforms in development and actin rearrangement. Mol Cell Biol 25: 2593–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Soriano P 2010. Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries. Genes Dev 24: 2068–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Qiu SG, Vasudevan KM, Rangnekar VM 2001. Par-4 drives trafficking and activation of Fas and Fasl to induce prostate cancer cell apoptosis and tumor regression. Cancer Res 61: 7255–7263 [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD 1988. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci 85: 7322–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Wu X, Boström H, Kim I, Wong N, Tsoi B, O’Rourke M, Koh GY, Soriano P, Betsholtz C, et al. 2004. A specific requirement for PDGF-C in palate formation and PDGFR-α signaling. Nat Genet 36: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Dixon J, Hovanes K, Shiang R, Dixon MJ 1997. Sequence analysis, identification of evolutionary conserved motifs and expression analysis of murine tcof1 provide further evidence for a potential function for the gene and its human homologue, TCOF1. Hum Mol Genet 6: 727–737 [DOI] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey J-P, Dixon MJ, Trainor PA 2006. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci 103: 13403–13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Guendy N, Rangnekar VM 2003. Apoptosis by Par-4 in cancer and neurodegenerative diseases. Exp Cell Res 283: 51–66 [DOI] [PubMed] [Google Scholar]
- Fowles LF, Bennetts JS, Berkman JL, Williams E, Koopman P, Teasdale RD, Wicking C 2003. Genomic screen for genes involved in mammalian craniofacial development. Genesis 35: 73–87 [DOI] [PubMed] [Google Scholar]
- Fuchs S, Herzog D, Sumara G, Büchmann-Moller S, Civenni G, Wu X, Chrostek-Grashoff A, Suter U, Ricci R, Relvas JB, et al. 2009. Stage-specific control of neural crest stem cell proliferation by the small Rho GTPases Cdc42 and Rac1. Cell Stem Cell 4: 236–247 [DOI] [PubMed] [Google Scholar]
- Gu Y-M, Jin Y-H, Choi J-K, Baek K-H, Yeo C-Y, Lee K-Y 2006. Protein kinase A phosphorylates and regulates dimerization of 14-3-3ζ. FEBS Lett 580: 305–310 [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P 2003. Evolutionary divergence of platelet-derived growth factor α receptor signaling mechanisms. Mol Cell Biol 23: 4013–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Soriano P 2013. A critical role for PDGFRα signaling in medial nasal process development. PLoS Genet 9: e1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH, Westermark B 1999. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev 79: 1283–1316 [DOI] [PubMed] [Google Scholar]
- Homer C, Knight DA, Hananeia L, Sheard P, Risk J, Lasham A, Royds JA, Braithwaite AW 2005. Y-box factor YB1 controls p53 apoptotic function. Oncogene 24: 8314–8325 [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey J-P, Glynn EF, Ellington L, Du C, Dixon J, et al. 2008. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med 14: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten PH, Toepoel M, Mariman EC, Van Zoelen EJ 2001. Promoter haplotype combinations of the platelet-derived growth factor α-receptor gene predispose to human neural tube defects. Nat Genet 27: 215–217 [DOI] [PubMed] [Google Scholar]
- Joyner A, Wall N 2008. Immunohistochemistry of whole-mount mouse embryos. Cold Spring Harb Protoc 2008: pdb.prot4820. [DOI] [PubMed] [Google Scholar]
- Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, Piaggio G, Fietze E, Dietel M, Royer HD 2003. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem 278: 27988–27996 [DOI] [PubMed] [Google Scholar]
- Kebache S, Ash J, Annis MG, Hagan J, Huber M, Hassard J, Stewart CL, Whiteway M, Nantel A 2007. Grb10 and active Raf-1 kinase promote Bad-dependent cell survival. J Biol Chem 282: 21873–21883 [DOI] [PubMed] [Google Scholar]
- Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. 2012. The IntAct molecular interaction database in 2012. Nucleic Acids Res 40: D841–D846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghoffer RA, Hamilton TG, Hoch R, Soriano P 2002. An allelic series at the PDGFαR locus indicates unequal contributions of distinct signaling pathways during development. Dev Cell 2: 103–113 [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Arizti P, Reimer CL, Sugrue MM, Aaronson SA, Lee SW 1998. Inhibition of tumor cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res 58: 4439–4444 [PubMed] [Google Scholar]
- Lasham A, Lindridge E, Rudert F, Onrust R, Watson J 2000. Regulation of the human fas promoter by YB-1, Purα and AP-1 transcription factors. Gene 252: 1–13 [DOI] [PubMed] [Google Scholar]
- Lasham A, Moloney S, Hale T, Homer C, Zhang YF, Murison JG, Braithwaite AW, Watson J 2003. The Y-box-binding protein, YB1, is a potential negative regulator of the p53 tumor suppressor. J Biol Chem 278: 35516–35523 [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC 2007. AKT/PKB signaling: navigating downstream. Cell 129: 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens PR, Harendza S, Pollock AS, Lovett DH 1997. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J Biol Chem 272: 22905–22912 [DOI] [PubMed] [Google Scholar]
- Moritz A, Li Y, Guo A, Villén J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, et al. 2010. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal 3: ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, Higashi Y, Kokame K, Tanaka C, Kondoh H, Miyata T 2004. Ndrg1-deficient mice exhibit a progressive demyelinating disorder of peripheral nerves. Mol Cell Biol 24: 3949–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett E, Olsen GS, Tallquist MD 2008. Disruption of PDGFRα-initiated PI3K activation and migration of somite derivatives leads to spina bifida. Development 135: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralphs JR 1992. Chondrogenesis and myogenesis in micromass cultures of mesenchyme from mouse facial primordia. In Vitro Cell Dev Biol 28A: 369–372 [DOI] [PubMed] [Google Scholar]
- Rattanasopha S, Tongkobpetch S, Srichomthong C, Siriwan P, Subhapeetiporn K, Shotelersuk V 2012. PDGFRa mutations in humans with isolated cleft palate. Eur J Hum Genet 20: 1058–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinon A, Molchadsky A, Nathan E, Yovel G, Rotter V, Sarig R, Tzahor E 2011. p53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/delamination processes. Development 138: 1827–1838 [DOI] [PubMed] [Google Scholar]
- Rosenkranz S, DeMali KA, Gelderloos JA, Bazenet C, Kazlauskas A 1999. Identification of the receptor-associated signaling enzymes that are required for platelet-derived growth factor-AA-dependent chemotaxis and DNA synthesis. J Biol Chem 274: 28335–28343 [DOI] [PubMed] [Google Scholar]
- Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T 1995. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet 10: 175–180 [DOI] [PubMed] [Google Scholar]
- Soriano P 1997. The PDGF α receptor is required for neural crest cell development and for normal patterning of the somites. Development 124: 2691–2700 [DOI] [PubMed] [Google Scholar]
- Stein S, Thomas EK, Herzog B, Westfall MD, Rocheleau JV, Jackson RS II, Wang M, Liang P 2004. NDRG1 is necessary for p53-dependent apoptosis. J Biol Chem 279: 48930–48940 [DOI] [PubMed] [Google Scholar]
- Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, Turbin D, Dedhar S, Nelson C, Pollak M, et al. 2005. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene 24: 4281–4292 [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellström M, Soriano P 2000. Early myotome specification regulates PDGFA expression and axial skeleton development. Development 127: 5059–5070 [DOI] [PubMed] [Google Scholar]
- The Treacher Collins Syndrome Collaborative Group. 1996. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet 12: 130–136 [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Fotovati A, Sasaguri T, Shibahara K, Shimada T, Fukuda T, Nakamura T, Izumi H, Tsuzuki T, Kuwano M, et al. 2006. YB-1 is important for an early stage embryonic development: neural tube formation and cell proliferation. J Biol Chem 281: 40440–40449 [DOI] [PubMed] [Google Scholar]
- Vantler M, Huntgeburth M, Caglayan E, ten Freyhaus H, Schnabel P, Rosenkranz S 2006. PI3-kinase/Akt-dependent antiapoptotic signaling by the PDGFα receptor is negatively regulated by Src family kinases. FEBS Lett 580: 6769–6776 [DOI] [PubMed] [Google Scholar]
- Vasudevan KM, Garraway LA 2010. AKT signaling in physiology and disease. Curr Top Microbiol Immunol 347: 105–133 [DOI] [PubMed] [Google Scholar]
- Yu JC, Heidaran MA, Pierce JH, Gutkind JS, Lombardi D, Ruggiero M, Aaronson SA 1991. Tyrosine mutations within the α platelet-derived growth factor receptor kinase insert domain abrogate receptor-associated phosphatidylinositol-3 kinase activity without affecting mitogenic or chemotactic signaling transduction. Mol Cell Biol 11: 3780–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ 2002. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem 277: 39379–39387 [DOI] [PubMed] [Google Scholar]
- Zhou A, Ou AC, Cho A, Benz EJ Jr, Huang SC 2008. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol Cell Biol 28: 5924–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]