In this perspective, Clarke and Graves examine Esnault et al.’s genome-wide analysis of the role of actin-responsive MRTF transcriptional coactivators in the serum response factor network and the coordination of the cytoskeletal response to growth factor signaling.
Keywords: SRF, MRTF, TCF, Rho, signal transduction, chromatin
Abstract
Esnault and colleagues (pp. 943–958) take a genomics approach to investigate the role of SRF (serum response factor) in the serum response of fibroblasts. The well-established dual role of SRF with alternative cofactors and responsiveness to two signaling pathways is illustrated at the genome-wide level, yet new insight comes from this global picture.
Cell signaling into the nucleus directs changes in gene expression that mediate a cell’s responsiveness to environmental changes during development and homeostasis. Surprisingly, a single mammalian transcription factor, SRF (serum response factor), responds to two distinct signaling pathways and mediates expression of distinct target genes (Fig. 1A). The SRF story begins in the early 1980s with the study of a simple environmental change: activation of quiescent mouse fibroblasts by adding serum to the culture medium. The changes in gene expression, which became known as the serum response, served as a mammalian model system for gene activation and were exemplified by the rapid activation of c-fos expression (Linzer and Nathans 1983; Greenberg and Ziff 1984). A decade of study of the serum responsiveness of this gene uncovered SRF as a transcription factor that binds DNA near the transcription start site of c-fos (Treisman 1986). At that site, SRF functions in a ternary complex with one of several related proteins, so-called ternary complex factors (TCFs), belonging to the ETS family of transcription factors. Ras–MAPK signaling was identified as the route from extracellular growth factors in serum to transcription factor phosphorylation and altered activity (Treisman 1994). A surprising connection to actin dynamics was also noted (Hill et al. 1995), and this alternative role of SRF was confirmed by independent investigation of gene expression changes during muscle development and function (Wang et al. 2001). Genes of interest appeared to have common regulatory control regions and DNA-binding activities, which were identified to be SRF. SRF in this context requires myocardin-related transcription factors (MRTFs), which are regulated by binding to free actin (G-actin) that drives their nuclear compartmentalization. Rho signaling responds to serum treatment to activate this route in the fibroblast system (Olson and Nordheim 2010). These classic studies helped define the role of DNA-binding transcription factors and their responsiveness to signals; however, the full picture of the broad and diverse use of a single factor, such as SRF, awaited the power of today’s genomic approaches.
Figure 1.
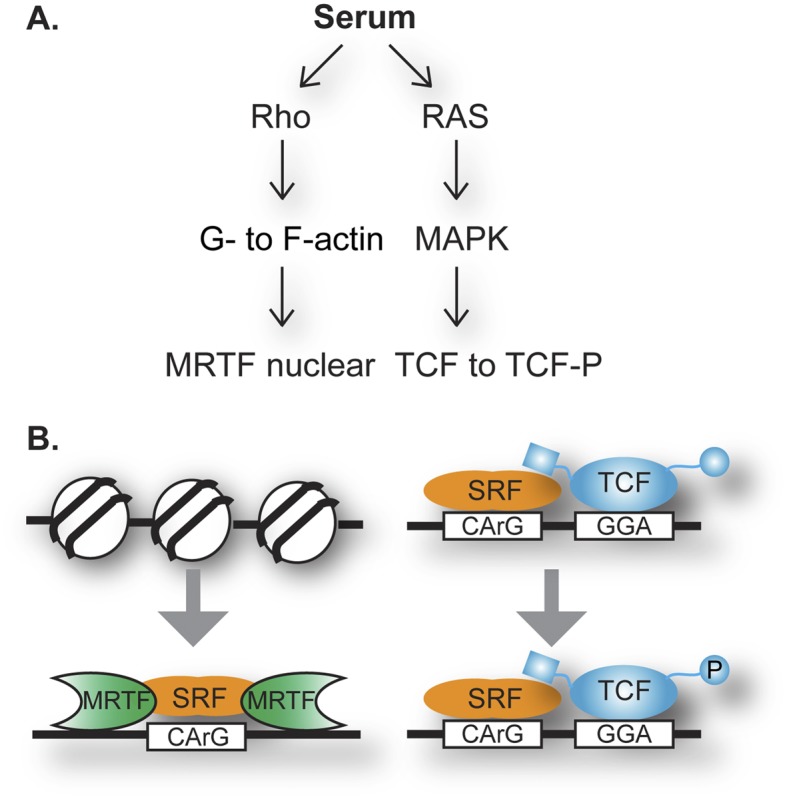
SRF directs the nuclear response to two signaling pathways via TCFs or MRTFs. (A) Serum activates Rho signaling (which changes actin dynamics) and Ras/MAPK kinase cascade (which phosphorylates TCFs). G-actin binds MRTF directly and modulates its nucleo–cytoplasmic trafficking (Mouilleron et al. 2011). (B) In response to Rho-actin signaling, SRF binding to DNA is stimulated, which involves nucleosome displacement. MRTF’s DNA binding is interdependent with SRF DNA binding (Zaromytidou et al. 2006). In response to Ras/MAPK signaling, prebound SRF functions with TCF, which binds to DNA independently and binds SRF via a dedicated B-box (Mo et al. 2001).
In this issue of Genes & Development, Esnault et al. (2014) identified SRF genomic binding sites by chromatin immunoprecipitation (ChIP) and massively parallel DNA sequencing (ChIP-seq). ChIP-seq identified a large number of sites (>3000); thus, a goal of this study was to determine which of these sites are functional. In support of the relevance, a subset of SRF sites was co-occupied with TCF factors, and another nonoverlapping set was co-occupied by MRTFs. The observed mutual exclusivity is reminiscent of biochemical experiments that showed competition of the cofactors for the same interface on SRF (Wang et al. 2004). This competition contributes to the binary choice for SRF that is specific to each binding site. A notable contrast in the use of cofactors is the almost complete overlap of MRTF sites with a subset of SRF sites, while the TCFs have many SRF-independent binding events.
The original serum induction regimen detected many changes in expression of genes, but early techniques could not address how many might be direct targets of SRF. The genomics approach, with its ability to link transcription factor binding to proximity of nearby genes, strongly implicates genes as direct transcriptional targets. With the caveat of defining “near,” this is an enormous benefit of genome-wide occupancy studies. Esnault et al. (2014) assayed for changes in gene expression—in this case, following serum stimulation—to build an argument for functional relevance of the nearby binding. One-third of genes near SRF-binding sites were induced in a short 30-min window, and thus these 960 genes were proposed to be direct targets of SRF’s serum responsiveness. Surprisingly, the vast majority of SRF’s direct targets were bound in complex with MRTFs rather than TCFs. Thus, in sharp contrast to a bias created by early studies on c-fos, the dominant role of SRF as a serum response factor appears to be in signaling through Rho-actin dynamics, at least in the fibroblast serum response model. The remaining genes with nearby SRF-binding sites were not induced but rather displayed constitutive expression and may not even need SRF binding. This finding may be an interesting direction to pursue, as many of these genes fall into housekeeping classes, such as metabolism and DNA synthesis, and may require a distinct regulatory route for maintenance of constitutive expression.
Genomic occupancy by transcription factors pinpoints potential functional elements within a genome’s rich supply of evolutionarily conserved DNA sequences. Thus, a critical follow-up of ChIP-seq is the identification of enriched sequence motifs underlying peaks of sequencing reads. These data, which are represented by position weight matrices, can be compared with previously derived “consensus” sites of DNA-binding proteins, most often derived from biochemical selection experiments, such as SELEX. As expected, SRF binding is associated with a dyad sequence motif, matching the CArG box and consistent with its homodimeric configuration. Divergence from the perfect consensus, which might be anticipated to be useful in biological regulation, can be measured in a genome-wide approach. Indeed, MRTF–SRF peaks had CArG boxes with a closer match to the consensus than the SRF–TCF peaks. In deeper bioinformatics analyses, motifs for ETS factors were found near SRF–TCF peaks, as expected, as well as SP-1- and NFY-binding motifs. In contrast, the MRTF–SRF sites displayed significant enrichment for AP-1 and TEAD motifs.
Factor binding, although essential for the readout of the inherited programming of gene expression, is not the full story of gene activation. Nevertheless, further analysis of factor assembly at a genome-wide scale can help develop mechanistic hypotheses. Esnault et al. (2014) again found a divergence from previous single-gene analysis and a contrast between the dual roles of SRF (Fig. 1B). TCFs and SRF are prebound to gene control elements of c-fos, with phosphorylation of the TCFs being the serum response (Herrera et al. 1989). SRF and TCF co-occupied sites displayed this trend; however, SRF with MRTF displayed stronger occupancy following serum activation, suggesting an induction of DNA binding. Differences were also observed in nucleosome coverage, with preclearing appearing at SRF–TCF sites, while nucleosomes were displaced during serum stimulation near MTRF–SRF-bound sites. Another intriguing trend was the broader sequence conservation around the SRF–TCF peaks than in the MRTF–SRF context, suggesting stronger roles for additional transcription factors that would recognize motifs within that DNA. An alternative that was not considered by the investigators would be a role for this sequence conservation in nucleosome dynamics.
With a rigorous list of putative transcriptional targets that are inducible by serum and occupied by SRF and one of the two cofactors, Esnault et al. (2014) next analyzed the potential biological roles of such genes by ontology programs. The SRF–MRTF complex regulates a constellation of genes important to the actin and microtubule cytoskeleton, cell matrix, cell–cell junctions, extracellular matrix (ECM) components, and vesicle trafficking, consistent with its relevance to muscle development and roles in adhesion, motility, and wound healing. The picture for the SRF–TCF side, as also observed by genomic studies focused on TCFs (Boros et al. 2009), was consistent with roles in cell proliferation and responsiveness to growth factors, including targets known to be important in T-cell activation. Nevertheless, the genomics approach yielded previously unsuspected transcriptional targets. The gene lists from SRF–MRTF targets overlapped with those of the YAP–TAZ pathway and Hippo signaling. The finding of TEAD motifs, which bind transcription factors for this pathway, near the SRF–MRTF peaks supports this possible connection. The gene lists also implicated a role for SRF and MRTF in circadian cycling that was further explored by the investigators.
SRF research over almost three decades provided a robust system for investigating signaling into the nucleus. This study expanded our perspective from single-gene analyses to genome-wide studies, deepened our understanding of the contrasting roles of SRF, and demonstrated how diversity of biological regulation can be built from common elements. The research demonstrated that genomics, done carefully and thoughtfully, complements reductionist approaches. Nevertheless, the new insight must lead back to biochemistry and cellular physiology studies for mechanistic discoveries and biological significance. Future such experimental directions should explore new interesting observations, such as the contrasting transcriptional activation mechanisms and chromatin dynamics in the two contexts and the biological role of SRF in Hippo signaling and circadian cycling.
Acknowledgments
The work was supported by National Institutes of Health R01GM38663 to B.J.G.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.242420.114.
Freely available online through the Genes & Development Open Access option.
References
- Boros J, Donaldson IJ, O’Donnell A, Odrowaz ZA, Zeef L, Lupien M, Meyer CA, Liu XS, Brown M, Sharrocks AD 2009. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res 19: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R 2014. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev (this issue). doi: 10.1101/gad.239327.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB 1984. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 311: 433–438 [DOI] [PubMed] [Google Scholar]
- Herrera RE, Shaw PE, Nordheim A 1989. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature 340: 68–70 [DOI] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R 1995. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Linzer DI, Nathans D 1983. Growth-related changes in specific mRNAs of cultured mouse cells. Proc Natl Acad Sci 80: 4271–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Ho W, Johnston K, Marmorstein R 2001. Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex. J Mol Biol 314: 495–506 [DOI] [PubMed] [Google Scholar]
- Mouilleron S, Langer CA, Guettler S, McDonald NQ, Treisman R 2011. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal 4: ra40. [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 11: 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R 1986. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell 46: 567–574 [DOI] [PubMed] [Google Scholar]
- Treisman R 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev 4: 96–101 [DOI] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105: 851–862 [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN 2004. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428: 185–189 [DOI] [PubMed] [Google Scholar]
- Zaromytidou AI, Miralles F, Treisman R 2006. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol Cell Biol 26: 4134–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]


