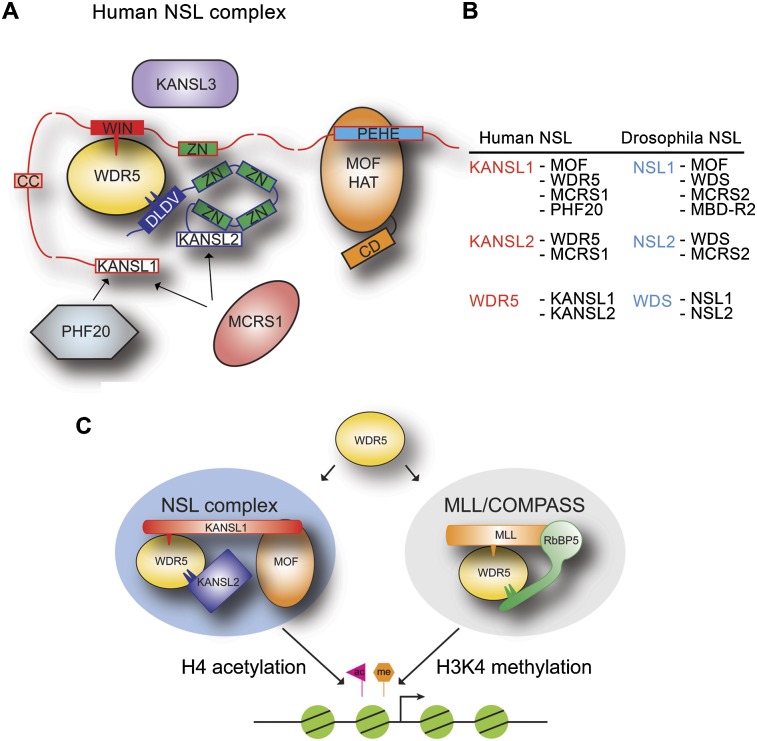
The subunits of the nonspecific lethal (NSL) complex, which include the histone acetyltransferase MOF, play important roles in transcription regulation and stem cell maintenance. Dias et al. provide the first biochemical and structural insights into the molecular architecture of this complex. The data show that the interactions of WDR5 with the MOF-containing NSL complex and the MLL/COMPASS histone methyltransferase complexes are mutually exclusive. The authors propose that WDR5 plays a key role in assembling distinct histone-modifying complexes with different epigenetic regulatory roles.
Keywords: chromatin, epigenetics, histone acetylation
Abstract
The subunits of the nonspecific lethal (NSL) complex, which include the histone acetyltransferase MOF (males absent on the first), play important roles in various cellular functions, including transcription regulation and stem cell identity maintenance and reprogramming, and are frequently misregulated in disease. Here, we provide the first biochemical and structural insights into the molecular architecture of this large multiprotein assembly. We identified several direct interactions within the complex and show that KANSL1 acts as a scaffold protein interacting with four other subunits, including WDR5, which in turn binds KANSL2. Structural analysis of the KANSL1/WDR5/KANSL2 subcomplex reveals how WDR5 is recruited into the NSL complex via conserved linear motifs of KANSL1 and KANSL2. Using structure-based KANSL1 mutants in transgenic flies, we show that the KANSL1–WDR5 interaction is required for proper assembly, efficient recruitment of the NSL complex to target promoters, and fly viability. Our data clearly show that the interactions of WDR5 with the MOF-containing NSL complex and MLL/COMPASS histone methyltransferase complexes are mutually exclusive. We propose that rather than being a shared subunit, WDR5 plays an important role in assembling distinct histone-modifying complexes with different epigenetic regulatory roles.
Eukaryotic transcription is controlled by a complex interplay between transcription regulators and local chromatin organization. Various post-translational modifications of histones, such as acetylation or methylation, help modify chromatin structure and are important for recruitment of effector proteins such as transcription factors or chromatin remodelers. MOF (males absent on the first), a member of the MYST family of histone acetyltransferases (HATs), is responsible for a large fraction of histone H4 Lys16 acetylation (H4K16ac) in mammalian cells (Taipale et al. 2005). Similar to other chromatin regulators, MOF does not exert its function alone but within multiprotein complexes that regulate its activity and specificity. For a long time, MOF has been known to be a component of the dosage compensation complex (DCC; or male-specific lethal [MSL] complex), which plays a key role in dosage compensation in Drosophila males (Conrad and Akhtar 2011). Recently, MOF was also shown to exist within the NSL (nonspecific lethal) complex involved in global transcription regulation (Raja et al. 2010).
The NSL complex is an evolutionarily conserved multiprotein assembly consisting of at least MYST1 (KAT8)/MOF, KANSL1/NSL1, KANSL2/NSL2, KANSL3/NSL3, WDR5/WDS, MCRS1/MCRS2, and PHF20/MBD-R2 proteins in mammals and Drosophila, respectively (Mendjan et al. 2006; Cai et al. 2010; Raja et al. 2010). NSL1, NSL2, NSL3, and MCRS2 were shown to be essential for Drosophila viability (Mendjan et al. 2006). In Drosophila, the complex associates with promoters of >4000 genes, and loss of the NSL complex binding severely affects their expression levels (Raja et al. 2010). The majority of the NSL complex-bound targets were shown to belong to housekeeping genes, and the NSL complex was shown to be required for efficient recruitment of RNA polymerase II at their target promoters (Feller et al. 2012; Lam et al. 2012).
In mammals, loss of MOF leads to early embryonic lethality (Thomas et al. 2008), while PHF20 knockout mice die just after birth (Badeaux et al. 2012). MOF and H4K16ac have been shown to regulate autophagy (Fullgrabe et al. 2013). In humans, haploinsufficiency of KANSL1 causes a multisystem disorder called the 17q21.31 microdeletion syndrome, characterized by intellectual disability (Koolen et al. 2012; Zollino et al. 2012). These observations point to an essential but also a more complex function of these proteins in higher eukaryotes. Consistently, MOF was shown to catalyze p53 Lys120 acetylation, which is required for optimal transcription activation of p53 target genes (Sykes et al. 2006; Li et al. 2009), while the tudor domain of PHF20 associates with methylated Lys370 and Lys382 of p53 (Cui et al. 2012). Both MOF and WDR5 are important regulators of the embryonic stem cell core transcription network (Ang et al. 2011; Li et al. 2012; Taylor et al. 2013), and PHF20 is required for somatic cell reprogramming (Zhao et al. 2013a). Despite the emerging global importance of the NSL complex in transcription regulation, the details and specificity of its recruitment to chromatin as well as its mode of action are currently poorly understood. Similarly, the molecular architecture of the complex and individual protein–protein interactions among the subunits remain unknown.
The human KANSL1 subunit consists of 1105 amino acid residues. It is predicted to be mostly unstructured; however, its C terminus contains the PEHE domain, which interacts with the HAT domain of MOF (Kadlec et al. 2011). KANSL2 is not well characterized. In humans, it consists of 492 residues and contains four putative Zn-coordinating motifs possibly involved in DNA binding. WDR5 (WDS in Drosophila) is also an established subunit of the human MLL/COMPASS histone H3 Lys4 (H3K4) methyltransferase complexes (Shilatifard 2012). It contains a seven-bladed β-propeller domain that was first shown to interact with an H3R2 peptide (Couture et al. 2006; Han et al. 2006; Ruthenburg et al. 2006; Schuetz et al. 2006). Within the MLL complexes, WDR5 recognizes both a WIN (WDR5-interacting) motif of MLL proteins and a short motif of RbBP5. These interactions stimulate the methyltransferase activity of MLL proteins (Patel et al. 2008b; Odho et al. 2010; Avdic et al. 2011). The NSL and MLL/COMPASS complexes were proposed to form a larger assembly that would possess both H3K4 methylation and H4 acetylation activities (Dou et al. 2005; Li et al. 2009).
In this study, we focus on the role of KANSL1/NSL1 and WDR5/WDS within the NSL complex. We show that KANSL1/NSL1 is a scaffold of the complex. We identified two previously unknown interactions that the WDR5/WDS subunit makes with KANSL1 and KANSL2, mapped the interacting regions, and determined crystal structures of these subcomplexes. We show that the NSL1–WDS interaction is important for efficient targeting of the NSL complex to promoters of target genes in Drosophila and that loss of this interaction results in male and female lethality. Our study provides the first insights into the molecular interactions within the NSL complex and reveals a mutually exclusive division of labor for WDR5/WDS between the NSL and MLL/COMPASS complexes.
Results
NSL1 interacts with multiple subunits of the NSL complex
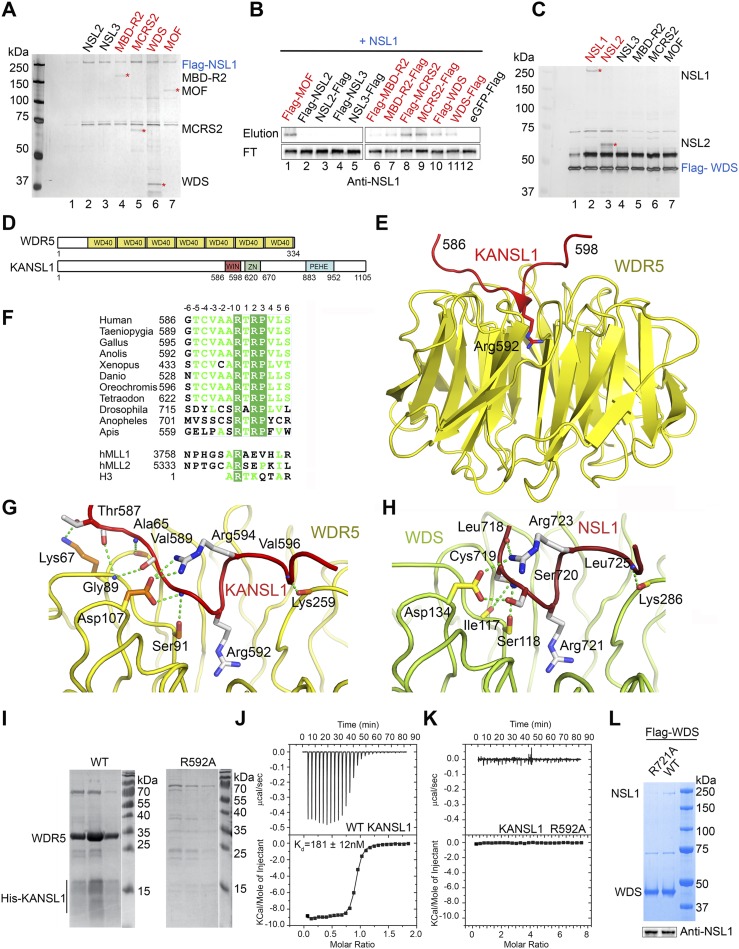
The acetyltransferase MOF/KAT8/MYST1 is recruited into the NSL complex via its interaction with the C-terminal PEHE domain of NSL1, analogous to the interaction between MOF and MSL1 in the DCC or the MSL complex (Kadlec et al. 2011). Both NSL1 and MSL1 are predicted to be mostly intrinsically disordered, and we demonstrated previously that within the MSL complex, MSL1 functions as a scaffold protein interacting independently with MSL2, MSL3, and MOF (Kadlec et al. 2011; Hallacli et al. 2012). To test whether NSL1 has an analogous function within the NSL complex, we expressed individual subunits of the Drosophila NSL complex in insect cells and performed pull-down experiments using 3xFlag-tagged NSL1 as the bait. While MOF, WDS, MCRS2, and MBD-R2 copurified with NSL1 (Fig. 1A, lanes 4–7; Supplemental Fig. S1), no binding was observed for NSL2 and NSL3. None of the NSL subunits copurified with a 3xFlag-tagged GFP control, confirming specificity of the interaction (data not shown). In the inverse experiment, N-terminal or C-terminal 3xFlag fusions of MOF, MBD-R2, MCRS2, and WDS copurified untagged NSL1 (Fig. 1B, lanes 1,6–11; Supplemental Fig. S1B). While the interactions with MOF and MCRS2 have already been described (Raja et al. 2010; Kadlec et al. 2011), the additional direct binding of NSL1 to WDS and MBD-R2 was not known and indicates that NSL1 might function as the scaffold subunit of the NSL complex. Since NSL1 is predicted to contain no known globular domains and to be predominantly intrinsically disordered, it is likely to interact with its binding partners via short linear motifs, similar to MSL1. Indeed, in humans, the interaction with MOF requires only a short motif spanning residues 883–952 (Kadlec et al. 2011).
Figure 1.
Structural analysis of the KANSL1–WDR5 interaction. (A) SDS-PAGE and silver staining analysis of Flag pull-down assays using N-terminally 3xFlag-NSL1 as the bait and other untagged NSLs as prey. All proteins are from baculovirus-expressed protein extracts. Interacting proteins are indicated by asterisks and highlighted in red. (See Supplemental Fig. S1A for the input sample). (B) Western blot analysis of the reverse Flag pull-down assays of A using untagged NSL1 as the prey and either N-terminal or C-terminal 3xFlag-tagged NSLs as bait. (FT) Flowthrough sample. Interacting proteins are highlighted in red. The eluted prey proteins are shown in Supplemental Figure S1B. (C) SDS-PAGE and silver staining analysis of Flag pull-down assays using 3xFlag-WDS as the bait and other untagged NSLs as prey. All proteins are from baculovirus-expressed protein extracts. Interacting proteins are indicated by asterisks and highlighted in red. (See Supplemental Fig. S1C for the input sample). (D) Schematic representation of the domain structure of human WDR5 and KANSL1. KANSL1 contains a PEHE domain involved in the binding of MOF (Kadlec et al. 2011) and a WIN motif identified in this study. (E) Ribbon diagram of the minimal human WDR5–KANSL1 complex structure. WDR523–334 is shown in yellow, and KANSL1 is in red. The key interacting KANSL1 Arg592 is shown as sticks. (F) Sequence alignment of NSL1 proteins. Only the sequence of the fragment involved in the interaction with WDR5 is shown. Identical residues are in green boxes. Numbering of the motif residues centered on Arg592 (in humans) is shown above the alignment. Representative sequences of corresponding MLL and H3 motifs are aligned with NSL1. (G) Details of the interaction between the β-propeller domain of WDR5 and the WIN motif of KANSL1. Interacting WDR5 residues are shown in orange, and KANSL1 residues are in gray. Residues without side chains represent main chain interactions. Contacts with Arg592 of KANSL1 are similar to the ones of H3 and MLL proteins and are not shown for clarity (see Supplemental Fig. S3B). (H) Interaction interface of the complex between the β-propeller domain of WDS (green) and the WIN motif of NSL1 (red). (I) SDS-PAGE analysis of the binding of His-tagged KANSL1584–690 (wild type [WT] and the R592A mutant) to coexpressed untagged WDR523–334 after purification using Ni2+ resin. KANSL1584–690 is only detectable (but still degrading) when bound to WDR5. (J) ITC measurement of the interaction between WDR523–334 and the KANSL1 WIN motif-containing peptide (585-DGTCVAARTRPVLS-598-Y). The bottom panel represents a fit of the calorimetric data to single-site-binding model. Dissociation constant (Kd) derived from the fit is indicated. (K) ITC measurement of the interaction between WDR523–334 and the mutated KANSL1 WIN motif-containing peptide (585-DGTCVAAATRPVLS-598-Y). (L, top panel) 3xFlag-WDS was coexpressed with either wild-type NSL1 (WT) or mutant NSL1 (R721A) in a single baculovirus and purified using Flag-M2 resin. (Bottom panel) The Western blot shows that equal amounts of wild-type and mutant NSL1 (R721A) were present in the input extracts, yet only the wild-type NSL1 interaction is detected with WDS in the Coomassie blue gel.
Whereas the role of WDR5 within the MLL/COMPASS methyltransferase complex is well characterized (Patel et al. 2008a; Song and Kingston 2008; Odho et al. 2010; Avdic et al. 2011), nothing is known about the function of WDR5/WDS within the NSL or other chromatin-modifying complexes. Interestingly, it has been proposed that the WDR5 subunit might serve as a physical link between MOF and MLL/COMPASS complexes, forming a complex that would possess both H3K4 methylation and H4 acetylation activities to promote transcription initiation at target genes (Dou et al. 2005; Li et al. 2009). To better understand the details of the function and the recruitment of WDR5/WDS into the NSL complex, we first assessed how WDS interacts with other subunits. Using pull-down assays with 3xFlag-tagged WDS and untagged NSL subunits, we could show that in addition to NSL1, WDS also directly interacts with NSL2, while it does not interact with MOF (Fig. 1C, lanes 2,3,7; Supplemental Fig. S1C). These results indicate that the interaction between MOF and WDS is mediated by NSL1.
WDR5/WDS–KANSL1 structure
The mapping of the WDS- and NSL1-interacting domains was performed using their human orthologs, as they could be more easily produced in bacteria. To identify the region involved in the interaction with WDR5, KANSL1, predicted to be mostly intrinsically disordered, was divided into four ∼25-kDa segments, and their interactions with His-tagged WDR5 (residues 23–334) were tested in a coexpression experiment. When expressed alone, all four fragments were unstable/degraded (data not shown). However, when coexpressed with WDR5, the KANSL1 fragment spanning residues 537–773 could be efficiently copurified (Supplemental Fig. S2A, lane 3), indicating that it possesses a WDR5-binding region. This fragment contains several stretches of well-conserved residues and four highly conserved cysteine and histidine residues, possibly forming a Zn-coordinating structure. When loaded onto an S75 gel filtration columns, the WDR5 and KANSL1537–773 coeluted as a complex in the same elution volume, distinct from that of WDR5 alone (Supplemental Fig. S2B,C). KANSL1, however, suffered from significant degradation, consistent with its predicted unstructured nature (Supplemental Fig. S2C). Using mass spectrometry analysis, we mapped the minimal WDR5-binding fragment to a region encompassing a conserved sequence around Arg592. We then determined by X-ray crystallography the structure of the WDR5 β-propeller domain (residues 23–334) in complex with this KANSL1 fragment spanning residues 585–598 at a resolution of 1.5Å (Fig. 1D–F). Interestingly, the structure revealed that the KANSL1 motif centered on Arg592 interacts with WDR5 in a manner similar to MLL proteins and histone H3. Arg592 inserts into the central pore of the β propeller of WDR5, forming hydrogen bonds and stacking interactions with Phe133, Phe263, Ser91, and Cys261 (Fig. 1E; Supplemental Fig. S3A,B). Compared with the known WDR5 structures, Thr587 and Val589 make additional hydrogen bonds with Lys67, Ala65, and Gly89, and the highly conserved Arg594 makes a salt bridge with Asp107 (Fig. 1F,G; Supplemental Fig. S3A). Downstream from Arg592, Val596 interacts with a shallow hydrophobic surface lined by Tyr191, Pro216, and Leu234 in a way similar to MLL2 or MLL3 (Supplemental Fig. S3C).
We also identified a corresponding WDS-interacting motif in Drosophila NSL1 that spans residues 714–729 and solved its crystal structure in complex with WDS. Compared with the human structure, the residues in position −4 to −7 relative to the central arginine are not ordered (Fig. 1F,H; Supplemental Fig. S3D). The unusual feature of the NSL1 WIN motif is a serine residue (Ser720) in the −1 position that is invariantly an alanine in all known WIN motif structures (Fig. 1H). This serine, however, fits well into the interaction interface and makes an additional hydrogen bond with Asp134 (Fig. 1H). While Arg592, Arg594, and Pro595 of KANSL1 are well conserved among species, residues mediating additional contacts in these two structures are conserved only among vertebrates or insects, respectively (Fig. 1F).
His-tagged KANSL1584–690 could be copurified and stabilized with untagged WDR5 in a coexpression experiment (Fig. 1I). However, when expressed on its own, the fragment became unstable and degraded. To assess the importance of Arg592 for the interaction with WDR5, we mutated this residue to alanine. The R592A mutant could not engage in a stabilizing interaction with WDR5 and was therefore degraded (Fig. 1I). To confirm the role of Arg592 in the interaction, we used isothermal titration calorimetry (ITC). We observed that WDR5 binds the KANSL1 peptide (585–598) with a dissociation constant (Kd) of 180 nM (Fig. 1J). As expected, no binding was observed for the KANSL1 peptide containing the R592A mutation (Fig. 1K). The NSL1–WDR5 dissociation constant is in the same range as the reported values for MLL WIN motifs (Dharmarajan et al. 2012). In addition, we also tested the corresponding R721A mutation within Drosophila NSL1, which significantly reduced the interaction between full-length WDS and NSL1 (Fig. 1L).
WDR5/WDS–KANSL2 structure
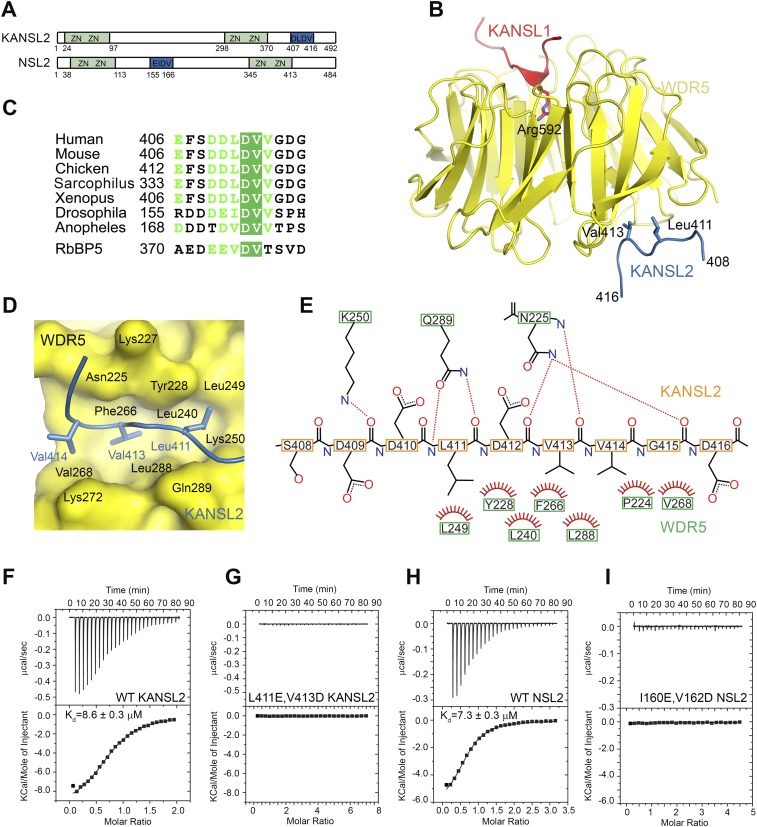
Within the MLL complexes, the β-propeller domain of WDR5 interacts with the MLL proteins and RbBP5 using two separate interaction surfaces (Odho et al. 2010; Avdic et al. 2011). Having shown that within the NSL complex, WDR5 interacts with both KANSL1 and KANSL2, we wondered whether a similar situation arose (Fig. 1C). To map the WDR5-binding region of KANSL2, we prepared several truncation constructs of KANSL2. While constructs covering the putative Zn-coordinating domains could not be expressed in bacteria (data not shown), a C-terminal construct spanning residues 381–492 yielded soluble protein. When loaded onto an S200 gel filtration column, KANSL2381–492 coeluted in complex with WDR5, while WDR5 alone eluted in a separate peak (Fig. 2A; Supplemental Fig. S4), indicating that KANSL2 interacts with WDR5 with its C terminus. This KANSL2 fragment as well as its truncated versions suffered from degradation and resisted crystallization. Eventually, a shorter KANSL2 fragment (residues 406–417) could be cocrystallized with the WDR5/KANSL1 subcomplex, and its structure could be solved at a resolution of 2 Å (Fig. 2B–E). Interestingly, the interaction between KANSL2 and WDR5 indeed resembles that of WDR5 with RbBP5 (Odho et al. 2010; Avdic et al. 2011). KANSL2 binds on the opposite side of the WDR5 β-propeller domain from KANSL1 (Fig. 2B). Three hydrophobic residues (Leu411, Val413, and Val414) of KANSL2 point into a large hydrophobic cavity in a cleft between blades 5 and 6 of the WDR5 domain (Fig. 2D). These interactions are stabilized by several hydrogen bonds that KANSL2 forms with Asn225, Lys250, and Gnl289 (Fig. 2E). Similar to RbBP5, the central conserved Asp412 stabilizes the conformation of the peptide by multiple hydrogen bonds with +2 and +3 residues (Val414 and Gly415). To confirm the importance of this motif for the interaction between KANSL2 and WDR5, we mutated Val413 within KANSL2381–492 to aspartate and showed that the corresponding protein no longer coeluted with WDR5 on the S200 gel filtration column (Supplemental Fig. S5A). Using ITC, we showed that WDR5 binds the KANSL2 peptide (406–417) with a Kd of 8.6 μM (Fig. 2F), which is comparable with the WDR5–RbBP5 interaction (Odho et al. 2010; Avdic et al. 2011). No binding was observed for an L411E, V413D KANSL2 mutant even at four times higher ligand concentration (Fig. 2G). While this motif is very well conserved in vertebrate KANSL2 proteins (Fig. 2C), it is absent in the C terminus of Drosophila or Anopheles. Surprisingly, in the Drosophila NSL2 sequence, a similar motif is located between the two sets of the Zn-coordinating motifs (Fig. 2A,C). The corresponding peptide (residues 155–166) interacted with WDS with a Kd of 7.3 μM, while no binding was detectable for an I160E, V162D NSL2 double mutant (Fig. 2H,I). We also determined the crystal structure of the Drosophila NSL1/WDS/NSL2 minimal complex, which showed the WDS/NSL2 interaction to be virtually identical to that in the human complex (Supplemental Fig. S5B,C).
Figure 2.
Structural basis for the WDR5–KANSL2 interaction. (A) Schematic representation of the domain structure of human and Drosophila KANSL2/NSL2. KANSL2 contains four putative Zn-coordinating motifs and an additional WDR5-binding motif (shown in blue) identified in this study. (B) Ribbon diagram of the minimal human KANSL1–WDR5–KANSL2 complex structure. WDR523–334 is shown in yellow, KANSL1 is in red, and KANSL2 is in blue. The key interacting residues are shown as sticks. (C) Sequence alignment of NSL2 proteins. Only the sequence WDR5-binding fragment is shown. The corresponding sequence of the RbBP5- and WDR5-binding motif is aligned with KANSL2. (D) Details of the WDR5–KANSL2 interaction interface. KANSL2 Leu411, Val413, and Val414 insert into a wide hydrophobic pocket on WDR5. (E) Schematic representation of the interactions between KANSL2 and WDR5. Hydrogen bonds are denoted with dotted lines. Hydrogens are not shown for clarity. (F,G) ITC measurement of the interaction between WDR523–334 and KANSL2 WDR5-binding peptide (Y-406-EFSDDLDVVGDG-417) (F) and its mutated version (Y-406-EFSDDEDDVGDG-417) (G). (H,I) ITC measurement of the interaction between WDS50–361 and the NSL2 WDS-binding peptide (Y-155-RDDDEIDVVSPH-166) (H) and its I160E/V162D mutant (Y-155-RDDDEEDDVSPH-166) (I).
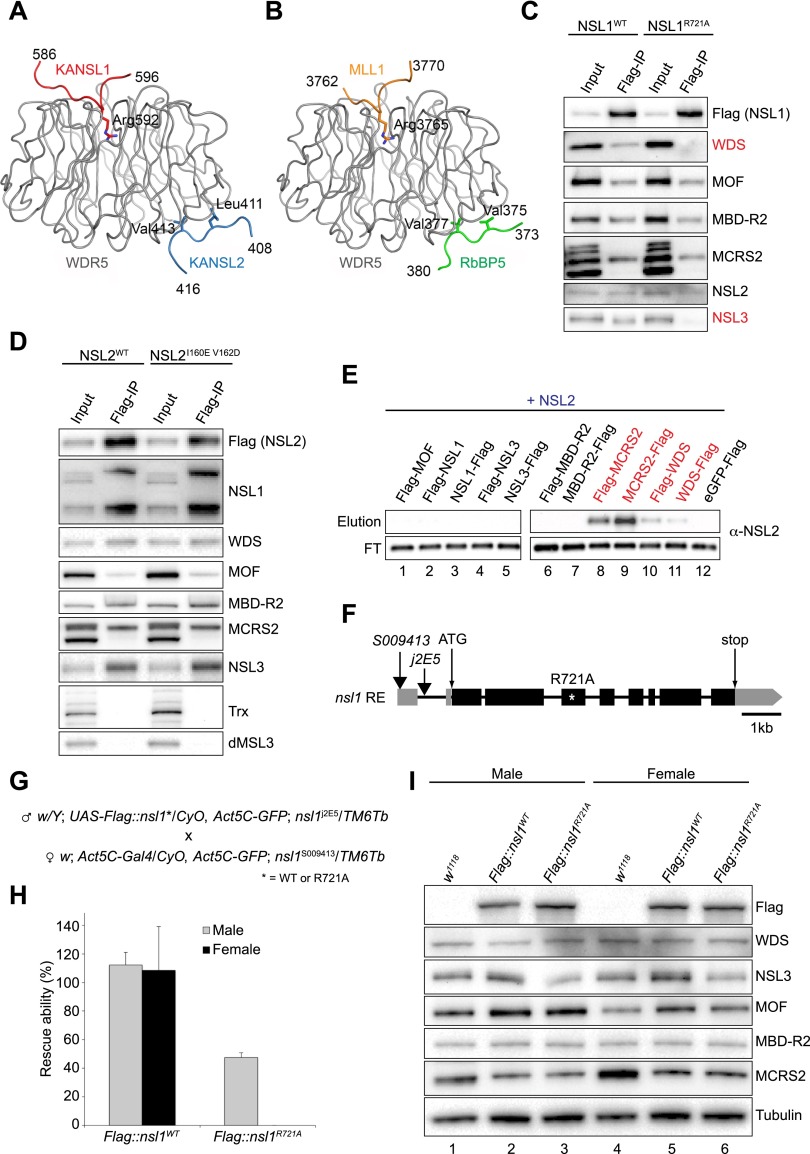
Overall, these biochemical and structural analyses revealed that KANSL1 and KANSL2 subunits contain WDR5-interacting motifs that are similar to the motifs found in the MLL proteins and RbBP5. These interactions are mediated by the same WDR5 residues, indicating that the KANSL1 and MLL as well as KANSL2 and RbBP5 interactions with WDR5 are mutually exclusive (Fig. 3A,B). To further confirm the mutually exclusive nature of the NSL1/MLL binding to WDR5, we performed ITC measurements of the interaction of WDR5 with the MLL4 WIN motif in the absence or presence of saturating amounts of KANSL1. While MLL4 normally interacts with WDR5 with a Kd of 32 nM, no additional binding was observed when WDR5 was saturated with KANSL1 (Supplemental Fig. S6A,B). WDR5 also uses the same surface to interact with the histone H3 N terminus (H3R2). While the H3R2 peptide interacts with WDR5 with a Kd of 46 μM (comparable with the reported value) (Schuetz et al. 2006), no additional binding could be detected in the presence of the KANSL1 WIN motif (Supplemental Fig. S6C,D). These data clearly show that WDR5 cannot interact with KANSL1, MLL, or H3 at the same time.
Figure 3.
The in vivo function of the NSL1–WDS–NSL2 interactions. (A,B) Comparison of the crystal structures of human ternary KANSL1–WDR5–KANSL2 (A) and MLL1–WDR5–RbBP5 (Protein Data Bank [PDB] code 3P4F) (B) complexes. The two complexes were superimposed using WDR5. (C) Flag immunoprecipitation of wild-type (WT) NSL1 and NSL1R721A in SL-2 cells. Both wild-type nsl1 and nsl1R721A were transiently transfected into SL-2 cells, and immunoprecipitation was performed using Flag-M2 resin. Antibodies used for Western blot analysis are indicated. (D) Flag immunoprecipitation of wild-type (WT) NSL2 and NSL2I160E–V162D in stably expressing SL-2 cells. Both wild-type nsl2 and nsl2I160E–V162D were transfected into SL-2 cells, and stably expressing cells were selected. Immunoprecipitation was performed using Flag-M2 resin. Antibodies used for Western blot analysis are indicated. (E) Western blot analysis of Flag pull-down assays using untagged NSL2 as the prey and either N-terminal or C-terminal 3xFlag-tagged NSLs as bait. (FT) Flowthrough sample. Interacting proteins are highlighted in red. The eluted prey proteins are shown in Supplemental Figure S1D. (F) A schematic representation of the structure of the RE transcript, encoded by nsl1/CG4699, used in this study. Exons (black bars), introns (black lines), and untranslated regions (UTRs) (gray bars) are shown. The ATG start and the STOP codon are indicated as well as the positions of the transposon insertions (black arrows). The site of the introduced mutation disrupting the NSL1 binding to WDS is marked by a white asterisk. The protein alteration is given at the top. (G) Details of the crosses used in the rescue experiments. (H) Relative percentage of adult male (gray bars) and adult female (black bars) viability upon ectopic expression of wild-type nsl1 and mutant nsl1R721A in the absence of endogenous NSL1 at 25°C. The non-CyO; Tb siblings in which endogenous nsl1 is expressed were used as internal controls and scored as 100% viable. The detail of the fly crosses is given in G. The error bars represent standard deviations of three independent crosses. (I) Western blot analysis of proteins extracted from third instar male and female larvae expressing either wild-type or mutant (R721A) NSL1 in the absence of endogenous NSL1. w1118 larvae were used as a wild-type control. Antibodies used for the Western blot analysis are indicated. Tubulin was used as a loading control.
The NSL complex assembly in vivo
To study the impact of NSL1 mutations on the incorporation of WDS into the NSL complex in vivo, we first transiently expressed 3xFlag-tagged wild-type or mutated (R721A) NSL1 proteins in Drosophila SL-2 cells and immunoprecipitated the corresponding NSL complexes using anti-Flag resin (Fig. 3C). While the wild-type full-length NSL1 protein could copurify all NSL subunits, the R721A mutant failed to interact with WDS, whereas the interaction of MOF, MBD-R2, MCRS2, and NSL2 remained unaffected in vivo. Interestingly, R721A also displayed reduced interaction with NSL3, indicating that the NSL1–WDS interaction plays an important role in the integration of this protein into the NSL complex.
To better understand the role of the NSL2–WDS interaction within the NSL complex, we prepared SL-2 cell lines stably expressing 3xFlag-tagged wild-type or mutated (I160E, V162D) NSL2 (Fig. 3D). In contrast to the NSL1 mutant, both wild-type and mutated NSL2 efficiently copurified all NSL subunits but not MSL3 or Trx/MLL, indicating that the NSL2–WDS interaction is not absolutely required for WDS or NSL2 to remain in the complex. The fact that Trx/MLL does not copurify with NSL2 also provides additional evidence for the separate roles of WDS/WDR5 within the NSL and MLL complexes. To test whether another subunit is involved in the recruitment of NSL2, we performed pull-down assays of untagged NSL2 with other subunits fused with a 3xFlag produced in insect cells and showed that in addition to WDS, NSL2 can also directly interact with MCRS2 (Fig. 3E, lanes 8,9; Supplemental Fig. S1D). Since MCRS2 also interacts with NSL1 (Fig. 1A), it is possible that NSL2 is tethered to NSL1 via MCRS2. Thus, the NSL2–WDS interaction seems to have rather a stabilizing role within the complex, being in agreement with its Kd of 8.6 μM, which is relatively weak compared with the 180 nM Kd for the KANSL1–WDR5 interaction.
The NSL1–WDS interaction is important for viability in flies
In order to further assess the functional relevance of the NSL1–WDS interaction in Drosophila in vivo, we generated transgenic flies expressing either the wild-type nsl1 or a mutant nsl1 variant carrying a single point mutation (R721A) (Fig. 3F) that strongly reduces the interaction of NSL1 with WDS (Figs. 1I–L, 3C). The two transgenes were inserted in the same genomic location (VK33 attP-docking site at the polytene location 65B2 on the left arm of chromosome 3) by ϕC31 integrase-mediated transformation to minimize positional effects upon phenotypic comparison (Groth et al. 2004; Venken et al. 2006). Both transgenes carried the Gal4 recognition site UAS upstream of the nsl1 ORF to allow ectopic expression in a spatiotemporally regulated manner (Brand and Perrimon 1993). In addition, a 3xFlag tag sequence was fused to the N terminus to allow easy detection and purification. Strong and ubiquitous ectopic expression of the two transgenes in a wild-type background using the αTub84-Gal4 driver (Lee and Luo 1999) did not affect the viability of the flies (data not shown). To assess the functionality of the ectopically expressed proteins, we used another strong and ubiquitous driver, Act5C-Gal4, to induce expression in the absence of endogenous NSL1 (nsl1S009413/nsl1j2E5-null mutant background) (Fig. 3G). Lack of NSL1 causes 100% lethality in both males and females (Mendjan et al. 2006; Yu et al. 2010). As expected, the ectopically expressed wild-type NSL1 rescued the lethality of the loss-of-function mutants in both males and females (Fig. 3H). Interestingly, ectopic expression of nsl1R721A in the null mutant background could only partially rescue the lethality in males (∼40% survival). However, nsl1R721A caused complete lethality in females (0% of the female animals reached the adult stage) (Fig. 3H). These results clearly showed that the Arg721-mediated interaction between NSL1 and WDS identified in our structural analysis is important for fly viability. Partial rescue in males suggests an attractive possibility that there may be a compensatory function provided by the MOF-containing MSL complex in males, which would be a very interesting avenue to explore in the future.
We next studied the mechanism underlying the strong phenotype observed in the nsl1R721A mutants. We first examined the total protein levels of NSLs in rescued larvae from the crosses in Figure 3G. These larvae all had a nsl1-null background (−/−) and carried either Act5C-Gal4-driven wild-type Flag∷nsl1 (Act5C>Flag∷nsl1WT) or Flag∷nsl1R721A (Act5C>Flag∷nsl1R721A). The Western blot results showed that the expression levels of Flag∷nsl1 transgenes were relatively equal between nsl1WT (Act5C>Flag∷nsl1WT;−/−) and nslR721A (Act5C>Flag∷nsl1R721A;−/−) animals as well as between males and females (Fig. 3I, anti-Flag row), suggesting that both transgenes were well expressed, and the lethal phenotype observed in the nsl1R721A was not caused by changes in the NSL1 levels. Interestingly, while total protein levels of MOF, MBD-R2, and MCRS2 were also very similar between male and female rescued larvae, the NSL3 protein level appeared partially reduced in the nsl1R721A larvae compared with the nsl1WT larvae (Fig. 3I, lanes 3,6). This is consistent with the result of the Flag immunoprecipitation experiments in SL-2 cells when NSL1R721A showed reduced coimmunoprecipitation of NSL3 (Fig. 3C). As a consequence, NSL3 might not be fully integrated into the complex and might therefore be degraded.
The NSL1–WDS interaction is required for the proper targeting of NSL1 to target promoters
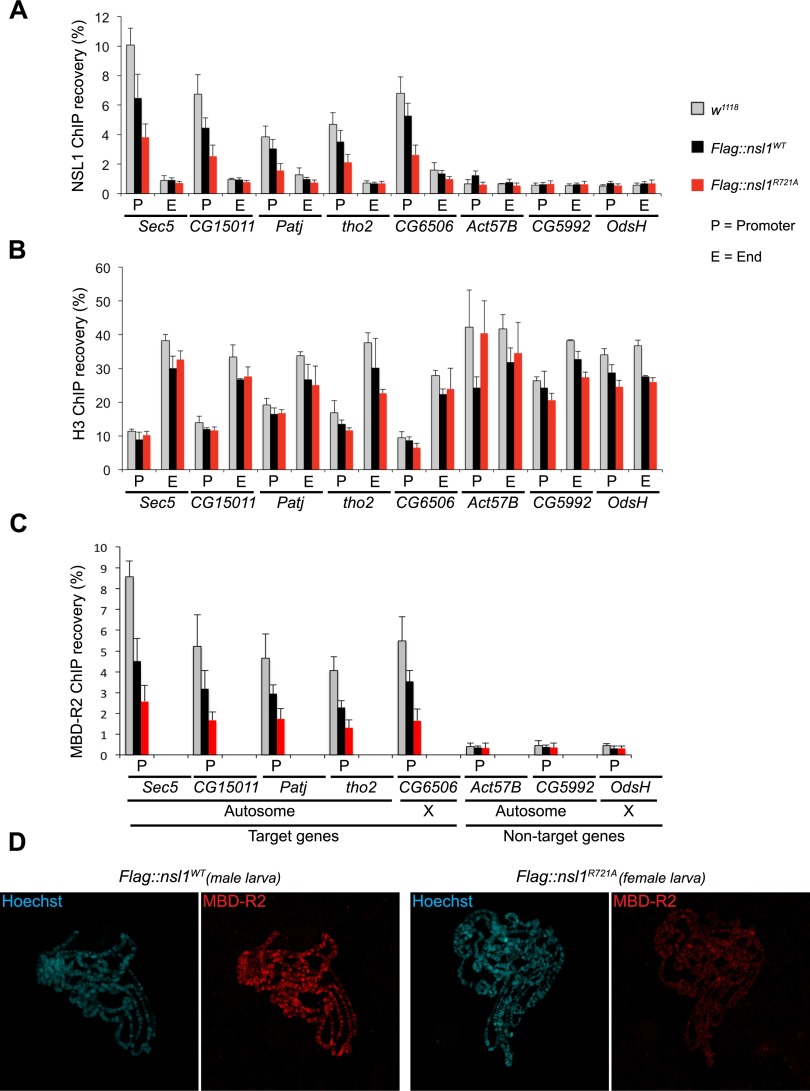
To address the role of the NSL1–WDS interaction in the targeting of NSL1, we performed chromatin immunoprecipitation (ChIP) experiments on chromatins prepared from whole rescued male larvae using both anti-Flag antibody (Supplemental Fig. S7A) and endogenous anti-NSL1 antibody (Fig. 4A). Histone H3 ChIP served as a control for the quality of the chromatin preparation (Fig. 4B). It is important to note that despite overexpression of Flag∷nsl1 (nsl1WT and nsl1R721A) (Supplemental Fig. S7B), NSL1 ChIP signal on promoters of target genes in the nsl1WT larvae was partially reduced compared with the real wild-type (w1118) larvae (Fig. 4A). However, this reduction has no consequence on fly viability, as nsl1WT animals are fully functional and viable (Fig. 3H). In contrast to nsl1WT, binding of NSL1 to the promoters of target genes was further reduced in the mutant nsl1R721A larvae, suggesting that the NSL1–WDS is important for the targeting of NSL1 (Fig. 4A; Supplemental Fig. S7A).
Figure 4.
The NSL1–WDS interaction is required for proper targeting of NSL1 to the promoters of target genes. (A) NSL1 ChIP from whole male larvae. qPCR was performed using primers corresponding to the promoter (P) and end (E) regions of genes. The list of target and nontarget genes was chosen based on the genome-wide data (Raja et al. 2010; Lam et al. 2012). ChIP recovery was calculated over the input DNA (shown as percentage [%]). The end of target genes and both the promoter and the end of nontarget genes were used as negative controls. The error bars represent standard deviations of three biological replicates. (B) As in A, but this time Histone H3 ChIP from whole male larvae. This provides a control for the quality of the chromatin preparations. (C) As in A, but this time MBD-R2 ChIP from whole male larvae. (D) Polytene chromosome immunostaining of Flag∷nsl1WT male and Flag∷nsl1R721A female third instar larvae. Chromosomal squashes were performed on the same slide; shown images of single nuclei were cropped from the same tile scan image. Immunofluorescence staining of MBD-R2 (red) is shown; DNA is counterstained with Hoechst (cyan).
Additionally, we wanted to test whether reduction in NSL1 chromatin targeting also affects chromatin targeting of other complex members in vivo. We therefore performed ChIP using the MBD-R2 antibody since there were no differences in total protein levels of endogenous MBD-R2 in the nsl1WT or nsl1R721A larvae (Fig. 3I). Indeed, consistent with NSL1 reduction (Fig. 4A), we also observed a similar reduction in targeting of MBD-R2 to the promoters of target genes (Fig. 4C). To address whether this targeting defect was also observed globally in the Drosophila genome, we performed polytene chromosomal staining in nsl1WT versus nsl1R721A larvae. To be able to directly compare the staining of nsl1WT (wild-type) and mutant larvae, the chromosomal squashes were prepared on the same slide, and images of wild-type and mutant polytene chromosomes were taken in the same tile scan. The male sample was distinguished from the female sample using MSL1 as an X-chromosomal marker (Supplemental Fig. S7C). Consistent with the reduction of MBD-R2 in ChIP analyses, we observed a striking reduction of MBD-R2 staining on the mutant chromosomes in comparison with wild type in male and female larvae (Fig. 4D; Supplemental Fig. S7C). These data clearly show that the NSL1–WDS interaction is essential for the viability of Drosophila and the proper targeting of the NSL complex to the promoters of target genes in vivo.
Discussion
The subunits of the NSL complex play important roles in various cellular processes, including transcription regulation and stem cell identity maintenance or reprogramming (Raja et al. 2010; Li et al. 2012; Zhao et al. 2013a), and are misregulated in various diseases, including cancer (Fraga et al. 2005; Gupta et al. 2008; Yoshida et al. 2013). The composition of the NSL complex has been analyzed by mass spectrometry (Mendjan et al. 2006; Cai et al. 2010), but essentially nothing is known about its biochemistry, molecular structure, and mode of action. In this study, we report the first detailed biochemical and structural analyses of the NSL complex architecture. We show that NSL1 acts as the scaffold for the complex assembly interacting with MOF, WDR5/WDS, MBD-R2, and MCRS2. The high-resolution crystal structures of the NSL1/WDS/NSL2 complexes revealed that NSL1 interacts via a short linear motif around Arg721 with WDS, which also recognizes another short motif of NSL2. Previously, we showed that another interacting motif in the NSL1 C terminus is involved in the interaction with MOF (Kadlec et al. 2011). The interacting regions for MBD-R2 and MCRS2 remain unknown, but given the high predicted disorder content of NSL1, it is possible that, similar to the MSL1 subunit of the MSL complex (Kadlec et al. 2011; Hallacli et al. 2012), NSL1 uses short conserved motifs separated by long flexible regions to interact with its partners, providing the NSL complex with a certain level of plasticity. Together, our data suggest that NSL1 bridges the MOF acetyltransferase with WDR5/WDS, which in turn brings together the putative Zn finger of NSL1 with four Zn-coordinating motifs of NSL2. We speculate that these putative Zn-binding modules might be involved in the interaction with the target DNA. Additionally, NSL1 helps NSL2 recruitment via the interaction with MCRS2 (Fig. 5).
Figure 5.
A summary model. (A) Schematic model of the NSL complex architecture based on our structural and protein interaction studies. KANSL1 is likely to be a scaffold molecule of the complex, interacting with MOF, WDR5, MCRS1, and PHF20. KANSL1 connects the MOF acetyltransferase (Kadlec et al. 2011) with WDR5 using conserved, short binding motifs. Similarly, WDR5 recognizes short motifs of KANSL1 and KANSL2 bringing in proximity the putative Zn finger of KANSL1 with four Zn-coordinating motifs of KANSL2, possibly involved in the interaction with the target DNA. KANSL2 is incorporated into the complex by additional interaction with MCRS1. (CD) Chromo-barrel domain; (CC) coiled coil; (WIN) WDS-interacting motif; (ZN) Zn finger. The arrows indicate interactions of MCRS1 and PHF20 with KANSL1. (B) Summary of known interactions within the NSL complex in humans and Drosophila. (C) Summary model. WDR5 serves a binding platform mediating mutually exclusive interactions essential for efficient promoter targeting of the NSL HAT complex and stimulating the H3K4 methyltransferase activity of MLL proteins within the MLL/COMPASS complexes (Patel et al. 2008b; Odho et al. 2010; Avdic et al. 2011; Zhang et al. 2012).
Mutating NSL1 Arg721 is sufficient to eliminate WDS from the complex, indicating that its interaction with NSL1 is absolutely required for its recruitment into the NSL complex. Interestingly, removal of WDS from the complex is accompanied by the loss of NSL3. Additionally, the overall NSL3 protein level was also partially reduced in the nsl1R721A larvae compared with the nsl1WT. As we did not observe a direct interaction between NSL3 and WDS (data not shown), NSL3 might require possible WDS-induced conformational changes within the complex or the presence of a composite binding site, including WDS and other complex subunits. Substitutions in the lower-affinity WDS-binding motif of NSL2 seem to have no effect on WDS levels, consistent with the major role of NSL1 in WDS recruitment.
We show that the NSL1–WDS interface mutagenesis triggered a decrease of NSL1 binding to promoters of the NSL target genes, resulting in fly lethality. In addition, ChIP and polytene chromosome immunostaining revealed a similar reduction of the MBD-R2 subunit on mutant chromosomes compared with wild type. Interestingly, while the binding of the Flag∷nsl1WT transgene was also partially reduced, it could still efficiently rescue nsl1-deficient flies. The observed phenotype thus seems critically dependent on the amount of the chromatin-bound NSL complex. This observation is supported by the fact the KANSL1 haploinsufficiency results in the 17q21.31 microdeletion multisystem syndrome (Koolen et al. 2012; Zollino et al. 2012).
The reduced NSL complex binding to promoters might be a result of an altered NSL2 configuration within the complex (in the wild type, it is connected to NSL1 via WDS), since NSL2 possesses four Zn-coordinating motifs possibly involved in the recognition of target DNA sequences. The strong lethal phenotype could also (at least partially) originate from the lack of NSL3 within the mutated complex, induced by the absence of WDS. NSL3 possesses a putative enzymatic domain of the α/β hydrolase family, but its function remains unknown. Nevertheless, a severe reduction in transcript levels of most NSL target genes was observed upon depletion of NSL3 (Raja et al. 2010). In addition NSL3 is able to activate transcription in a luciferase reporter-tethering assay (Raja et al. 2010). It is thus possible that the lack of NSL3 within the complex directly affects the established role of the NSL complex in transcription regulation.
WDR5 was originally shown to bind H3R2 peptides and proposed to present H3K4 for methylation by MLL proteins (Couture et al. 2006; Han et al. 2006; Ruthenburg et al. 2006; Schuetz et al. 2006). Later, the crystal structure of the MLL1-SET domain in complex with the H3K4 peptide revealed that H3 is unlikely to be bound by WDR5 (H3R2) and MLL1 (H3K4) at the same time (Southall et al. 2009; Trievel and Shilatifard 2009). In accord with this structure, WDR5 was shown to recognize a similar sequence (the WIN motif) of the MLL proteins themselves as well as RbBP5 (Patel et al. 2008a; Song and Kingston 2008; Odho et al. 2010; Avdic et al. 2011). However, while the structure of the WDR5 β-propeller domain and its ability to bind H3, the WIN motif, and RbBP5 within MLL complexes are well documented, it remains unclear how WDR5 presents itself within other complexes, such as NSL or ATAC. WDR5 was proposed to be a bridging or cross-shared subunit between the NSL and MLL/COMPASS complexes, forming complexes with both histone methyltransferase and acetyltransferase activities (Dou et al. 2005; Li et al. 2009; Zhao et al. 2013b). However, we and others could not confirm the existence of such a complex (Cai et al. 2010; Raja et al. 2010). Another possibility is that WDR5 targets its complexes to histone H3 (via H3R2 binding). It could also interact within these complexes in a manner similar to or different from that of the MLL/COMPASS complexes. Since other WDR5-binding partners as well as the molecular details of their interactions were not known, the role of WDR5 within could not be investigated. We now, for the first time, identified proteins directly interacting with WDR5 within the NSL complex and mapped the interacting regions. Our crystal structures clearly show that WDR5 regions interacting with KANSL1 and KANSL2 are the same as within the MLL/COMPASS complexes and that these interactions are mutually exclusive. This study thus provides compelling evidence that WDR5 cannot be a cross-shared subunit between MLL and NSL complexes and explains how WDR5 can serve as a binding platform in distinct complexes such as NSL, MLL/COMPASS, and possibly ATAC. Analogously, the NSL complex cannot use WDR5 to recognize H3R2, since its binding site is occupied by KANSL1. The interaction between WDR5 and the MLL WIN motif has recently been proposed to be a promising new cancer therapeutic target (Karatas et al. 2013; Senisterra et al. 2013; Cao et al. 2014). While the developed small molecule inhibitors interfere efficiently with MLL binding to WDR5, possible side effects on the essential WDR5 interactions within other complexes, such as NSL, should be considered.
Interestingly, WDR5 has also very recently been shown to interact with hundreds of long noncoding RNAs (lncRNAs), including HOTTIP and NeST (Wang et al. 2011; Gomez et al. 2013; Yang et al. 2014), that presumably target the MLL complex to its target genes in embryonic stem cells. The RNA recognition site on WDR5 was mapped by random mutagenesis to a site overlapping with that responsible for the interaction with RbBP5 and NSL2 (Yang et al. 2014). Indeed, the presence of RbBP5 prevented HOTTIP lncRNA binding to WDR5 (Yang et al. 2014). While the role of lncRNA in the targeting of the NSL has not yet been studied, it should be noted that the way WDR5 is recruited into the NSL complex is compatible with the RNA binding as reported for the MLL complex. It is tempting to speculate that the recruitment of the NSL and MLL complexes to their target genes via WDR5-bound lncRNAs could possibly explain observed functional cross-talk between them that was originally assigned to WDR5 itself.
Given the mutually exclusive binding of WDR5 to KANSL1/MLL and KANSL2/RbBP5, it will be important to understand how the proper pairing of the interactions in distinct complexes is achieved. For example, we could show that NSL2 pulls down the entire NSL complex but not Trx, indicating that such mixed complexes (e.g., Trx/WDS/NSL2), which might be inactive or have aberrant activity, do not exist in vivo. However, the reported minimal H3/WDR5/RbBP5 structure indicates that such complexes can indeed be formed in vitro (Odho et al. 2010). We show that both MLLs and KANSL1 as well as KANSL2 and RbBP5 interact with WDR5 with similar affinities. Interestingly, in both the NSL and MLL complexes, the two WDR5-binding proteins are linked by other interactions/proteins. While RbBP5 was proposed to directly interact with MLL1 (Odho et al. 2010), NSL2 is linked to NSL1 via MCRS2. It is thus likely that the proper combinations of WDR5 binders connected together by additional interactions will have a higher affinity for WDR5 than the incorrect combination of WDR5-binding proteins that cannot be physically linked. Future structural studies will be very important in unraveling how other components of the NSL complex shape its molecular interaction network and cross-talk with other chromatin-modifying complexes.
Materials and methods
Flag pull-down assays
All protein extracts used for binding assays were produced by baculoviruses in the Sf21 cells (see the Supplemental Material for details of generation of baculoviruses and protein extraction). For each binding assay, 1.5 mL of the bait protein extract was mixed with 5 mL of the prey protein extract. The mixture was incubated for 2 h at 4°C while rotating. About 50 µL of equilibrated Flag resin (Sigma) was added to each mixture and then incubated for 3 h at 4°C while rotating. After five washes with HEMGT250 buffer (25 mM HEPES at pH 7.6, 12.5 mM MgCl2, 10% glycerol, 0.2% Tween 20, 250 mM KCl), the beads were either boiled in 125 µL of 2× Roti-Load buffer (Carl-Roth) or eluted overnight in 500 µL of HMGT250 buffer containing 250 µg/mL 3xFlag peptide (Sigma).
Protein expression and purification for structural studies
6xHis-WDR523–334 was produced by expression in Escherichia coli BL21Star (DE3, Invitrogen) from the pProEXHTb (Invitrogen) expression vector and affinity-purified on Ni2+ chelating Sepharose (GE Healthcare). After His tag cleavage with TEV protease, the protein was further purified by a second Ni2+ column and subsequent size exclusion chromatography on a Superdex 75 (GE Healthcare) gel filtration column pre-equilibrated with 20 mM Tris (pH 7.0), 200 mM NaCl, and 5 mM β-mercaptoethanol. 6xHis-WDS 50-361 was produced by expression in Hi5 insect cells from the pFastBacHTb (Invitrogen) vector and purified as for WDR5.
KANSL1–WDR5 and KANSL2–WDR5 interaction analysis
Four constructs of untagged KANSL1 (residues 1–233, 262–537, 537–773, and 777–1105) were each coexpressed with 6xHis-WDR5 (residues 23–334) in E. coli BL21Star (DE3, Invitrogen) from pRSFDuet-1 (Novagen) and pProEXHTb (Invitrogen) expression vectors, respectively. 6xHis-WDR5 was affinity-purified on Ni2+ chelating Sepharose (GE Healthcare) in 20 mM Tris (pH 7.0), 200 mM NaCl, and 5 mM β-mercaptoethanol. Proteins were eluted with an increasing concentration of imidazole. Samples were analyzed on SDS-PAGE. A His-tagged minimal interacting region of KANSL1 (residues 584–690; wild type or R592A mutant) was coexpressed with untagged WDR5 (residues 23–334) in E. coli BL21Star (DE3, Invitrogen) from pProEXHTb (Invitrogen) and pRSFDuet-1 (Novagen) expression vectors, respectively. Pull-down experiments were performed as above.
Constructs of KANSL2 were produced as 6xHis-GST fusion proteins in pETM30 expression vector. KANSL2381–492 and mutated KANSL2381–492 L411E,V413D were expressed in E. coli BL21Star (DE3, Invitrogen) and purified as for WDR5. Pure wild-type KANSL2381–492 or its V413D mutant were mixed with pure WDR523–334 and loaded onto a Superdex 200 gel filtration column. Fractions containing protein were analyzed on SDS-PAGE.
Crystallization
All of the crystals of the WDR5/WDS-containing complexes were grown using pure WDR5 or WDS concentrated to 12–16 mg/mL supplemented with a threefold molar excess of peptide of individual peptides. The best diffracting crystals of the binary complex of WDR523–334 with KANSL1 peptide (585-DGTCVAARTRPVLS-598-Y; Y was added to facilitate quantification) were obtained in a condition containing 26% (w/v) PEG 3350, 0.2 M ammonium sulphate, and 0.1 M Bis-Tris (pH 6.5). Crystals of the binary complex of WDS50–361 with NSL1 peptide (714-GSDYLCSRARPLVLSE-729) grew in 20% (w/v) PEG 3350 and 0.2 M potassium sodium tartrate (pH 7.2). Crystals of the ternary complex of WDR523–334 with KANSL1 peptide (585-DGTCVAARTRPVLSY-598) and KANSL2 peptide (Y-406-EFSDDLDVVGDG-417) grew in 20% (w/v) PEG 3350 and 0.2 M potassium nitrate (pH 6.9). The best crystals of the ternary complex of WDS50–361 with NSL1 peptide (714-GSDYLCSRARPLVLSE-729) and NSL2 peptide (Y-155-RDDDEIDVVSPH-166) were obtained in 20% (w/v) PEG 3350 and 0.2 M di-ammonium tartrate (pH 6.6). For data collection at 100 K, crystals were snap-frozen in liquid nitrogen with a solution containing mother liquor and 30% (v/v) glycerol.
Data collection and structure determination
Crystals of the binary complex WDR523–334 with KANSL1 peptide (585-DGTCVAARTRPVLSY-598) belong to the space group P21, with unit cell dimensions of a = 39.8 Å, b = 92.5 Å, c = 81.2 Å, and β = 90.2°. The asymmetric unit contained two complexes and had a solvent content of 45%. A complete data set was collected to a resolution of 1.5 Å on beamline ID14-EH4 at the European Synchrotron Radiation Facility (ESRF). Crystals of the binary complex WDS50–361 with NSL1 peptide (714-GSDYLCSRARPLVLSE-729) belong to the space group C2221, with unit cell dimensions a = 78.7 Å, b = 98.6 Å, and c = 80 Å. A complete data set was collected to a resolution of 1.4 Å on the ESRF beamline ID23-EH1. Crystals of the ternary complex WDR523–334 with KANSL1 peptide (585-DGTCVAARTRPVLSY-598) and KANSL2 peptide (406-YEFSDDLDVVGDG-417) belong to the space group P21212, with unit cell dimensions a = 81.6 Å, b = 86.6 Å, and c = 45.1 Å. A complete data set was collected to a resolution of 2 Å on the ESRF beamline ID23.2. Crystals of the ternary complex WDS50–361 with NSL1 peptide (714-GSDYLCSRARPLVLSE-729) and NSL2 peptide (155-YRDDDEIDVVSPH-166) belong to the space group P21212, with unit cell dimensions a = 81.5 Å, b = 86.2 Å, and c = 47.6 Å. A complete data set was collected to a resolution of 2.3 Å on beamline BM14 at the ESRF. The data were processed using XDS (Kabsch 2010). The structures were solved by molecular replacement with Phaser (McCoy et al. 2005) using the structure of WDR5 (Protein Data Bank [PDB] code 2G99) as a search model. The structures were manually rebuilt in Coot (Emsley and Cowtan 2004) and refined in REFMAC5 (Murshudov et al. 1997) to the final R factors indicated in Table 1 with all residues in allowed (94%–96% in favored) regions of the Ramachandran plot, as analyzed by MolProbity (Davis et al. 2004). Representative parts of the Fo–Fc electron density maps calculated using the refined models are shown in Supplemental Figure S8.
Table 1.
Data collection and refinement statistics
ITC
ITC experiments were performed at 25°C using an ITC200 microcalorimeter (MicroCal). Experiments included 26 injections of 1.5 μL of 0.5 mM wild-type or 2 mM mutant peptide solution into the sample cell containing 50 μM WDR523–334 or WDS50–361 in 20 mM Tris (pH 7.0) and 200 mM NaCl. Peptides used were KANSL1 wild type (585-DGTCVAARTRPVLS-598-Y), KANSL1 R592A (585-DGTCVAAATRPVLSY-598), KANSL2 wild type (406-YEFSDDLDVVGDG-417), KANSL2 L411E,V413D (406-YEFSDDEDDVGDG-417), NSL2 wild type (155-YRDDDEIDVVSPH-166), and NSL2 I160E V162D (155-YRDDDEEDDVSPH-166). The initial data point was deleted from the data sets. Binding isotherms were fitted with a one-site binding model by nonlinear regression using Origin software version 7.0 (MicroCal).
To confirm that KANSL1 and MLL4 bind to the same binding site on WDR5, 0.56 and 0.35 mM MLL4 peptide (2504-LNPHGAARAEVYLRK-2518) was injected into the sample cell containing 50–60 μM WDR523–334 with or without 200 μM KANSL1 peptide, respectively. To test the mutually exclusive binding of KANSL1 and histone H3, 1.9 and 1.8 mM H3 peptide (1-ARTKQTARK-9-Y) was injected into the sample cell containing 120 μM WDR523–334 with or without 400 μM KANSL1 peptide, respectively.
Flag immunoprecipitation from SL-2 cells
Flag immunoprecipitation from transiently or stably expressing SL-2 cells were performed as previously described (Hallacli et al. 2012).
Analysis of lethality rescue and rate of adult eclosion
To determine the relative viability upon ectopic expression of Flag∷nsl1 (wild type or R721A), w; Act5C-Gal4/CyO, Act5C-GFP; nsl1S009413/TM6BTb virgin females were crossed to either w/Y; UAS-Flag∷nsl1WT; nsl1j2E5/TM6BTb or w/Y; UAS-Flag∷nsl1R721A; NLS1j2E5/TM6BTb males. Male and female adult flies from at least three independent crosses were counted every other day for a period of 10 d from the start of eclosion. The total number of non-CyO, non-Tb males and females was divided by the total number of non-CyO, Tb males and females, respectively, which were used as an internal control with 100% viability. Details of fly genetics are available in the Supplemental Material.
ChIP
ChIP from male whole larvae was carried out as previously described (Raja et al. 2010) using primers listed in Supplemental Table S1.
Coordinates
The atomic coordinates and structure factors of the human KANSL1/WDR5 and KANSL1/WDR5/KANSL2 complexes and Drosophila NSL1/WDS and NSL1/WDS/NSL2 complexes have been deposited under the PDB accession codes 4CY1, 4CY2, 4CY3, and 4CY5, respectively.
Acknowledgments
We thank the Grenoble European Molecular Biology Laboratory (EMBL)–European Synchrotron Radiation Facility (ESRF)–Institut Laue-Langevin (ILL)–Institut de Biologie Structurale (IBS) Partnership for Structural Biology (PSB) for access to structural biology instrumentation—notably L. Signor of the IBS for his help with mass spectrometry, and the High-Throughput Crystallisation Laboratory (HTX) group of EMBL Grenoble for performing initial screening crystallization trials. We thank A. Palencia for help with the ITC, and K. Dzeyk of EMBL Heidelberg for help with mass spectrometry. We thank M. Shvedunova and H. Holz for the initial help with baculoviruses. We thank S. Chlamydas and T. Lingg for technical support, and other members of both laboratories and Ramesh Pillai for useful advice and fruitful discussions. We thank R. Sawarkar and R. Paro for providing the Trx antibody. We thank the ESRF–EMBL Joint Structural Biology Group for access to and assistance on the ESRF synchrotron beamlines. This work was supported by the EU FP7-funded Network of Excellence EpiGeneSys awarded to S.C. and A.A., and SFB746 and SFB992 awarded to A.A. A.A. is also part of the BIOSS II excellence initiative.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.240200.114.
References
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdic V, Zhang P, Lanouette S, Groulx A, Tremblay V, Brunzelle J, Couture JF 2011. Structural and biochemical insights into MLL1 core complex assembly. Structure 19: 101–108 [DOI] [PubMed] [Google Scholar]
- Badeaux AI, Yang Y, Cardenas K, Vemulapalli V, Chen K, Kusewitt D, Richie E, Li W, Bedford MT 2012. Loss of the methyl lysine effector protein PHF20 impacts the expression of genes regulated by the lysine acetyltransferase MOF. J Biol Chem 287: 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC 2010. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem 285: 4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Townsend EC, Karatas H, Xu J, Li L, Lee S, Liu L, Chen Y, Ouillette P, Zhu J, et al. 2014. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell 53: 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A 2011. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet 13: 123–134 [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Trievel RC 2006. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol 13: 698–703 [DOI] [PubMed] [Google Scholar]
- Cui G, Park S, Badeaux AI, Kim D, Lee J, Thompson JR, Yan F, Kaneko S, Yuan Z, Botuyan MV, et al. 2012. PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53. Nat Struct Mol Biol 19: 916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32: W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmarajan V, Lee JH, Patel A, Skalnik DG, Cosgrove MS 2012. Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J Biol Chem 287: 27275–27289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Feller C, Prestel M, Hartmann H, Straub T, Soding J, Becker PB 2012. The MOF-containing NSL complex associates globally with housekeeping genes, but activates only a defined subset. Nucleic Acids Res 40: 1509–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400 [DOI] [PubMed] [Google Scholar]
- Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B 2013. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500: 468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K 2013. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 152: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage ϕC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, et al. 2008. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol 28: 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallacli E, Lipp M, Georgiev P, Spielman C, Cusack S, Akhtar A, Kadlec J 2012. Msl1-mediated dimerization of the dosage compensation complex is essential for male X-chromosome regulation in Drosophila. Mol Cell 48: 587–600 [DOI] [PubMed] [Google Scholar]
- Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J 2006. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell 22: 137–144 [DOI] [PubMed] [Google Scholar]
- Kabsch W 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Hallacli E, Lipp M, Holz H, Sanchez-Weatherby J, Cusack S, Akhtar A 2011. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol 18: 142–149 [DOI] [PubMed] [Google Scholar]
- Karatas H, Townsend EC, Cao F, Chen Y, Bernard D, Liu L, Lei M, Dou Y, Wang S 2013. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J Am Chem Soc 135: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Kramer JM, Neveling K, Nillesen WM, Moore-Barton HL, Elmslie FV, Toutain A, Amiel J, Malan V, Tsai AC, et al. 2012. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat Genet 44: 639–641 [DOI] [PubMed] [Google Scholar]
- Lam KC, Muhlpfordt F, Vaquerizas JM, Raja SJ, Holz H, Luscombe NM, Manke T, Akhtar A 2012. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet 8: e1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461 [DOI] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CA, Kunkel S, Dou Y 2009. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell 36: 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y 2012. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell 11: 163–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21: 811–823 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Odho Z, Southall SM, Wilson JR 2010. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J Biol Chem 285: 32967–32976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, Cosgrove MS 2008a. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem 283: 32158–32161 [DOI] [PubMed] [Google Scholar]
- Patel A, Vought VE, Dharmarajan V, Cosgrove MS 2008b. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem 283: 32162–32175 [DOI] [PubMed] [Google Scholar]
- Raja SJ, Charapitsa I, Conrad T, Vaquerizas JM, Gebhardt P, Holz H, Kadlec J, Fraterman S, Luscombe NM, Akhtar A 2010. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell 38: 827–841 [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL 2006. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol 13: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J 2006. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J 25: 4245–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senisterra G, Wu H, Allali-Hassani A, Wasney GA, Barsyte-Lovejoy D, Dombrovski L, Dong A, Nguyen KT, Smil D, Bolshan Y, et al. 2013. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J 449: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Kingston RE 2008. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem 283: 35258–35264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR 2009. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell 33: 181–191 [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A 2005. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 25: 6798–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G, Eskeland R, Hekimoglu-Balkan B, Pradeepa M, Bickmore WA 2013. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res 23: 2053–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Dixon MP, Kueh AJ, Voss AK 2008. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol 28: 5093–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A 2009. WDR5, a complexed protein. Nat Struct Mol Biol 16: 678–680 [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY 2014. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife 3: e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, Sanada M, Park MJ, Terui K, Suzuki H, et al. 2013. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet 44: 1293–1299 [DOI] [PubMed] [Google Scholar]
- Yu L, Song Y, Wharton RP 2010. E(nos)/CG4699 required for nanos function in the female germ line of Drosophila. Genesis 48: 161–170 [DOI] [PubMed] [Google Scholar]
- Zhang P, Lee H, Brunzelle JS, Couture JF 2012. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res 40: 4237–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q, Chen Y, Wang HY, Wang RF 2013a. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell 152: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Su J, Wang F, Liu D, Ding J, Yang Y, Conaway JW, Conaway RC, Cao L, Wu D, et al. 2013b. Crosstalk between NSL histone acetyltransferase and MLL/SET complexes: NSL complex functions in promoting histone H3K4 di-methylation activity by MLL/SET complexes. PLoS Genet 9: e1003940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollino M, Orteschi D, Murdolo M, Lattante S, Battaglia D, Stefanini C, Mercuri E, Chiurazzi P, Neri G, Marangi G 2012. Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype. Nat Genet 44: 636–638 [DOI] [PubMed] [Google Scholar]