tRNAs are encoded by RNA polymerase III-transcribed genes that reside at seemingly random intervals along budding yeast chromosomes. Here, Chen and Gartenberg examined the spatial and temporal aspects of tRNA gene expression. Unexpectedly, they found that tRNA genes are transcribed in a periodic manner during cell cycle progression. Moreover, tRNA genes migrate to nuclear pore complexes when transcription peaks in M phase. This study demonstrates how RNA polymerase III-transcribed genes are gated to nuclear pore complexes in yeast.
Keywords: tRNA gene, RNA polymerase III, nuclear pore complex, cohesin, cell cycle, Los1 exportin
Abstract
tRNAs are encoded by RNA polymerase III-transcribed genes that reside at seemingly random intervals along the chromosomes of budding yeast. Existing evidence suggests that the genes congregate together at the nucleolus and/or centromeres. In this study, we re-examined spatial and temporal aspects of tRNA gene (tDNA) expression. We show that tDNA transcription fluctuates during cell cycle progression. In M phase, when tRNA synthesis peaks, tDNAs localize at nuclear pore complexes (NPCs). Docking of a tDNA requires the DNA sequence of the contacted gene, nucleoporins Nup60 and Nup2, and cohesin. Characterization of mutants that block NPC localization revealed that docking is a consequence of elevated tDNA transcription. NPC–tDNA contact falters in the absence of the principal exportin of nascent tRNA, Los1, and genetic assays indicate that gating of tDNAs at NPCs favors cytoplasmic accumulation of functional tRNA. Collectively, the data suggest that tDNAs associate with NPCs to coordinate RNA polymerase III transcription with the nuclear export of pre-tRNA. The M-phase specificity of NPC contact reflects a regulatory mechanism that may have evolved, in part, to avoid collisions between DNA replication forks and transcribing RNA polymerase III machinery at NPCs.
Nuclear pore complexes (NPCs) form large aqueous channels through the nuclear envelope that permit transfer of materials between the nucleus and cytoplasm. In 1985, Blobel (1985) proposed the gene-gating hypothesis, which stipulated that transcriptionally poised genes associate with NPCs. In this way, tethered genes would be situated to export transcripts directly to the cytoplasm. The original gene-gating scheme also envisioned that genome packaging would influence the distribution of NPCs on the nuclear envelope. A contemporary spin on this idea posits that anchorage of genes to NPCs and other structures at the edge of the nucleus orchestrates the three-dimensional structure of the genome (Taddei and Gasser 2012).
Substantial evidence for gating of RNA polymerase II genes has been obtained primarily from studies of the budding yeast Saccharomyces cerevisiae. Genome-wide approaches revealed that components of the NPC, known as nucleoporins or Nups, associate with many active genes (Casolari et al. 2004; Schmid et al. 2006). Focused analyses of representative examples showed that the genes moved from the nucleoplasm to the nuclear periphery upon induction in a Nup-dependent manner. That transcription and export might be coupled events emerged from studies showing that mRNA export machinery associated with transcribed genes and that the machinery was required for positioning the genes at NPCs (for review, see Dieppois and Stutz 2010). Whether gating of genes at NPCs affects their expression has been less certain. For example, one study showed that artificially targeting a reporter gene to NPCs increased transcriptional output (Menon et al. 2005). A different study, however, found that the level of expression of a prototypical inducible gene was unaffected by mutations that displaced the gene from NPCs (Cabal et al. 2006). Experiments like these and others made it clear that gating of RNA polymerase II genes is not essential for their expression. Instead, recent work suggests that the process fine-tunes expression kinetics by altering the rates of initial transcriptional induction as well as transcriptional reactivation (Hampsey et al. 2011; Texari et al. 2013).
Gating of active genes at NPCs is not strictly conserved. In mice and humans, for example, a number of developmentally regulated genes move away from the nuclear periphery upon induction, producing mRNA protein complexes that travel through the nucleoplasm to NPCs before export (see references in Egecioglu and Brickner 2011; Oeffinger and Zenklusen 2012). Nevertheless, components of the nuclear pore may still play significant roles in transcription. Recent work in Drosophila showed that several Nups migrate from NPCs to the nuclear interior, where they bind to and promote expression of some RNA polymerase II genes and repress the expression of others (Capelson et al. 2010; Kalverda et al. 2010).
RNA polymerase III transcribes small conserved noncoding RNAs, including 274 tRNA genes (termed tDNAs) that are scattered among the chromosomes of yeast. Despite the broad genomic distribution, tDNAs often congregate at or near the nucleolus, where ribosomal RNA is synthesized and assembled into ribosomes (Thompson et al. 2003). Cocompartmentalization, it was argued, might favor coordinated control of both tRNA and ribosome biogenesis (Haeusler and Engelke 2006). Indeed, tRNA synthesis and early processing are thought to occur at the nucleolus (Bertrand et al. 1998). Mutations that attenuate expression of mature ribosomal RNA disperse tDNAs from that location (Thompson et al. 2003).
Cohesin, the protein complex that holds sister chromatids together, was recently shown to also participate in nucleolar events. In mutants that disrupt cohesin activation and utilization, ribosomal RNA production decreases, and tDNA clustering is lost (Gard et al. 2009; Bose et al. 2012). Cohesin is intimately associated with the genes involved. Specifically, the complex binds the array of ribosomal RNA genes (the rDNA), where it suppresses unequal sister chromatid exchange (Kobayashi et al. 2004). The complex also loads onto chromosomal arms at tDNAs (D’Ambrosio et al. 2008).
The yeast tDNA named tT(AGU)C has received substantial attention because of its immediate proximity to HMR, a paradigmatic heterochromatic locus. The gene creates a barrier to propagation of heterochromatin from HMR into adjacent euchromatic domains by a mechanism involving chromatin modification (Kirkland et al. 2013). Barrier function also requires cohesin, which loads onto chromatin at tT(AGU)C to create heterochromatin-dependent cohesion at HMR (Donze et al. 1999; Dubey and Gartenberg 2007). Paradoxically, fluorescence microscopy studies showed that tT(AGU)C localized to the nucleolus infrequently even though the gene is transcribed (Donze and Kamakaka 2001; Valenzuela et al. 2008). Evidence that tT(AGU)C instead associates with NPCs emerged with the discovery that Nups bind the tDNA and direct the gene to the nuclear periphery (Ruben et al. 2011). In this study, we show that NPC tethering is a general feature of yeast tDNAs that occurs in M phase when transcription of the genes elevates. Gating of the genes at NPCs requires the dedicated tRNA exportin Los1, suggesting that tDNA transcription is coordinated with nuclear export of nascent tRNA.
Results
tDNAs associate with NPCs in M phase
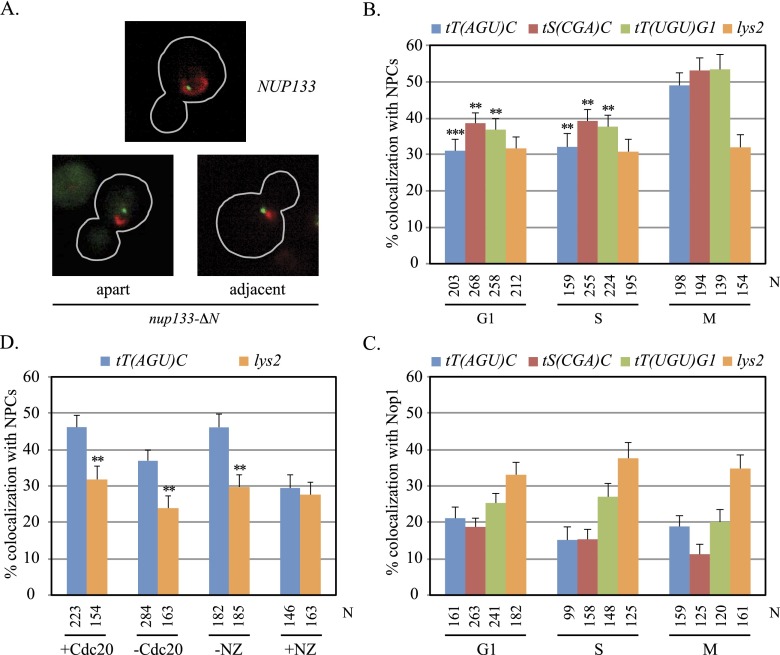
Based on preliminary studies of tT(AGU)C localization, we speculated that the gene and other tDNAs associate with NPCs at the nuclear envelope. This idea was tested by tagging chromosomal domains bearing tDNAs with lac operator arrays (lacop) in cells expressing GFP-tagged lac repressor and Nup Nic96 tagged with mRFP. tDNAs tT(AGU)C and tS(CGA)C on chromosome III were evaluated as well as tT(UGU)G1 on chromosome VII. The strains also contained the nup133-ΔN mutation, which causes NPCs to aggregate on the nuclear membrane without disrupting protein import or polyA+ RNA export significantly (Doye et al. 1994). In nup133-ΔN cells, Nic96-mRFP coalesced into one to three red foci instead of forming a ring of red fluorescence at the nuclear rim (Fig. 1A). With these tools, association of specific chromosomal domains with NPCs could be distinguished from other forms of perinuclear enrichment (Taddei and Gasser 2012).
Figure 1.
Localization of tDNAs at NPCs. (A) Representative images of cells from asynchronously grown cultures. GFP-lac repressor (green) and Nic96-mRFP (red) are shown in nup133-ΔN and NUP133 cells, outlined in gray. (B) tDNA localization at NPCs. Asynchronously grown cultures of strains MC78 [tT(AGU)C∷256lacop], MC197 [tS(CGA)C∷256lacop], MC198 [tT(UGU)G1∷256lacop], and MC180 (lys2∷256lacop) were used. N equals the number of cells examined. Localization of each tDNA at NPCs was significantly higher in M phase than in G1 or S. Pairwise χ2-tests, (***) <5 × 10−4; (**) <5 × 10−3. (C) tDNAs colocalized with the nucleolus rarely. Strains MC250 [tT(AGU)C∷256lacop], MC247 [tS(CGA)C∷256lacop], MC248 [tT(UGU)G1∷256lacop], and MC249 (lys2∷256lacop) bearing a Nop1-CFP expression plasmid (pJW1327) were examined in asynchronous cultures. The specific tDNAs examined here differ from those described in Thompson et al. (2003). (D) NPC-tT(AGU)C colocalization in M phase. Strains MC78 [tT(AGU)C∷256lacop] and MC180 (lys2∷256lacop) bearing MET3p-CDC20 were examined before and after depletion of Cdc20. Strains MC64 [tT(AGU)C∷256lacop] and MC226 (lys2∷256lacop) were examined before and after addition of nocodazole (NZ). In the absence of either arrest, only M-phase cells were scored. NPC localization was significantly higher for tT(AGU)C than lys2 where noted.
Colocalization, defined as adjacent or overlapping tDNA and NPC foci, was scored in asynchronously grown cultures, with each cell categorized morphologically as being in the G1, S, or M phases of the cell cycle (see the Materials and Methods for detailed criteria). Cells with segregating chromosomes in anaphase were excluded from the analysis. Strikingly, all three tDNAs localized at NPCs more frequently in M phase than in either G1 or S phase (Fig. 1B). In the case of tT(AGU)C, colocalization occurred in 50% of M-phase cells yet in only 30% of cells in either G1 or S. As a control, the nuclear position of a chromosomal domain that lacks tDNAs was scored by the same criteria. We used the highly mobile chromosomal domain containing LYS2, referred to here as the lys2 domain (Gartenberg et al. 2004). This tDNA-free region localized at NPCs in only 30% of cells irrespective of whether they were in G1, S, or M phase. Similarly, the tDNAs and lys2 colocalized with a nucleolar marker (Nop1) infrequently and in a cell cycle-independent manner (Fig. 1C). In fact, tDNA–Nop1 colocalization occurred less frequently than lys2–Nop1 colocalization. These results indicate that chromosomal domains bearing tDNAs associate with NPCs more frequently in or near M phase than during other phases of the cell cycle.
Localization studies were repeated for tT(AGU)C in cells arrested at anaphase onset by depletion of Cdc20, a component of the anaphase-promoting complex (see the Materials and Methods). Here, too, positioning of the domain at NPCs was significantly greater than the lys2 control, although the absolute levels of colocalization of both loci were dampened (Fig. 1D). In contrast, M-phase arrest by exposure to the microtubule inhibitor nocodazole abolished the localization of the tDNA at NPCs. Previously, colocalization of tDNA clusters with the nucleolus was blocked by nocodazole (Haeusler et al. 2008). Collectively, these findings implicate microtubules and/or the spindle checkpoint pathway in controlling tDNA positioning. Based on these results, Cdc20 depletion was used to obtain M-phase-arrested cells for most subsequent experiments.
Nups tether tDNAs to NPCs in M phase
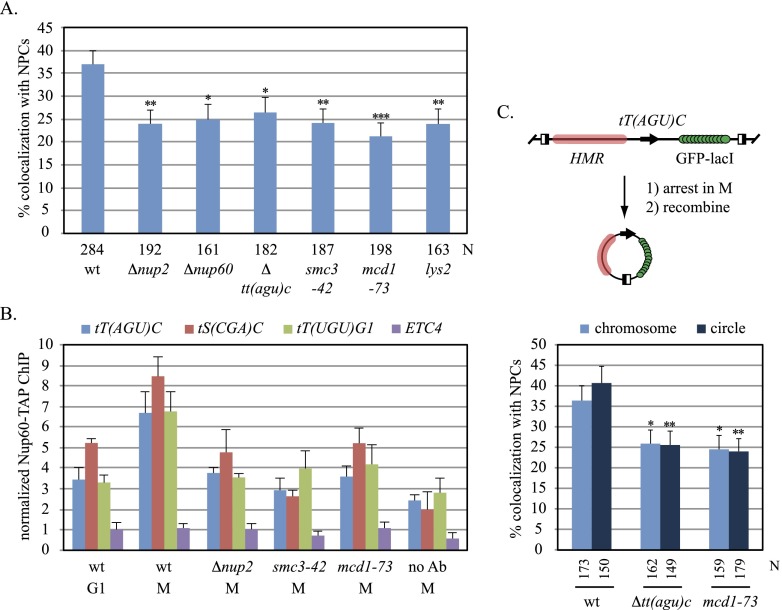
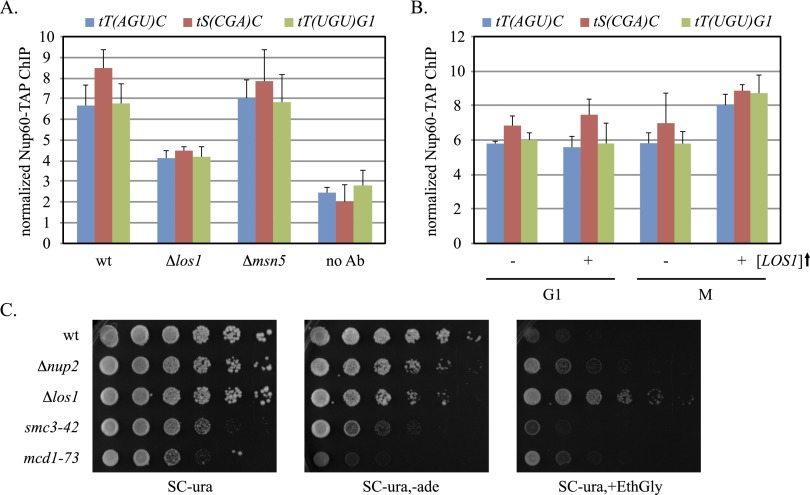
Nup60 is a fixed component of the NPC, binding stably to the nuclear basket that projects into the nucleoplasm. Nup2, in contrast, is a mobile factor that associates with the nuclear basket transiently by binding Nup60 (Denning et al. 2001; Dilworth et al. 2005). Both proteins cross-link efficiently to tT(AGU)C but not to other landmarks of the tT(AGU)C chromosomal domain, like HMR (Ruben et al. 2011). Both proteins also associate with RNA polymerase II genes and have been implicated in repositioning the genes at NPCs upon transcriptional induction (Casolari et al. 2004; Schmid et al. 2006). Figure 2A shows that in M phase-arrested cells, deletion of NUP2 or NUP60 reduced localization of tT(AGU)C at NPCs to the level of the tDNA-free lys2 control. In other phases of the cell cycle, NPC localization was unaffected by the mutations (Supplemental Fig. S1; data not shown). Deletion of tT(AGU)C reduced NPC localization of the GFP-tagged chromosomal domain to a level comparable with those seen in the Nup mutants. Collectively, the results indicate that NUP2 and NUP60 are required for docking tT(AGU)C at NPCs.
Figure 2.
Requirements for NPC–tT(AGU)C contact. (A) Localization of tT(AGU)C in mutants. Strains MC78 (wt), MC210 (Δnup2), MC131 (Δnup60), MC105 [Δtt(agu)c], MC83 (smc3-42), MC82 (mcd1-73), and MC180 (lys2∷256lacop) were evaluated after Cdc20 depletion. NPC colocalization was significantly lower in mutants. Pairwise χ2-tests, (***) <5 × 10−4; (**) <5 × 10−3; (*) < 5 × 10−2. (B) Nup60 occupancy at tDNAs. ChIP-qPCR of Nup60-TAP was performed with strains MC177 (wt), MC207 (Δnup2), MC178 (smc3-42), and MC179 (mcd1-73) after arrest in G1 or M phase. (No Ab) No TAP antibody. Nup60 was enriched at tDNAs significantly during M-phase arrest in the wild-type strain but not in mutants (P-values in Supplemental Table S4). (C) Localization of extrachromosomal tT(AGU)C. The half-filled boxes of the diagram represent recombinase target sites that were used to form DNA circles inducibly after arrest in M phase (Dubey and Gartenberg 2007).
If tDNA association with NPCs oscillates during the cell cycle, then binding of Nups to the genes should also vary with cell cycle phase. Chromatin immunoprecipitation (ChIP) followed by quantitative PCR (ChIP-qPCR) was used to evaluate binding of TAP-tagged Nup60 at the three tDNAs described in Figure 1. Experiments were performed in cells arrested in M phase by Cdc20 depletion or in G1 by exposure to the α-factor mating pheromone. Values were normalized to the GIT1 promoter, where Nup60 binds negligibly (Ruben et al. 2011). Figure 2B shows that all three tDNAs cross-linked efficiently to Nup60 in M-phase-arrested cells but not in G1-arrested cells. Nup2 was required for the enhanced binding of Nup60 in M phase, suggesting that the mobile Nup mediates contact between tDNAs and NPCs. In this set of experiments, Nup60 binding was also measured at ETC4. ETC sites bind transcription factor TFIIIC but not the rest of the RNA polymerase III transcriptional machinery (for review, see Kirkland et al. 2013). Notably, ETC sites contribute to genome organization in both S. cerevisiae and Schizosaccharomyces pombe by tethering chromosomal domains at the nuclear periphery (Noma et al. 2006; Hiraga et al. 2012). Figure 2B shows that Nup60 did not associate with ETC4 in either M or G1. Taken together, these experiments show that tDNAs but not ETC sites associate with an integral feature of NPCs in a Nup2-dependent manner during M phase. The data support and extend the microscopy results in Figure 1 with an independent set of strains that lack artificially clustered NPCs.
To determine whether other sequences on chromosome III contribute to tT(AGU)C positioning, the chromosomal domain bearing the tDNA spanning ∼7 kb from HMR to GIT1 was excised from the chromosome and ligated into a circle by inducible site-specific recombination (Fig. 2C; Dubey and Gartenberg 2007). The excision product was enriched at NPCs in M-phase-arrested cells but not when tT(AGU)C was deleted. These data indicate that positioning of tT(AGU)C at NPCs does not require cis-linkage to neighboring domains, like telomeres, which anchor chromosomes to the nuclear membrane (Taddei and Gasser 2012).
The ability of yeast heterochromatin to sequester repressed domains at the nuclear periphery prompted us to evaluate NPC–tDNA contact in mutants that block heterochromatin assembly (Taddei and Gasser 2012). Localization of tT(AGU)C at NPCs was reduced somewhat but not by a measure that was statistically significant (Supplemental Fig. S2). In agreement, binding of Nup60 at tT(AGU)C was not found to be heterochromatin-dependent (Ruben et al. 2011). We conclude that the tDNA is the dominant factor that positions the tT(AGU)C chromosomal domain at NPCs in M phase.
NPC–tDNA contact requires cohesin
The numerous links between tDNAs and cohesin led us to test whether the complex plays a role in NPC–tDNA contact. To this end, the microscopy and ChIP-qPCR assays described above were repeated in conditional mutants of cohesin subunits Smc3 and Mcd1. The strains were cultured at 30°C, an intermediate temperature that leads to nonlethal defects in sister chromatid cohesion during mitotic growth (data not shown). Strikingly, localization of tT(AGU)C at clustered NPCs dropped below 25% in both M-phase-arrested mutants (Fig. 2A). Similar results were obtained when the tT(AGU)C domain was uncoupled from the chromosome by recombination (Fig. 2C). Correspondingly, ChIP-qPCR experiments with TAP-tagged Nup60 showed that binding of the Nup to representative tDNAs required cohesin (Fig. 2B). Together, these findings define an additional role for cohesin in the behavior of tDNAs in yeast. With regard to NPC tethering, we do not know whether cohesin acts directly at tDNAs or the complex influences tDNAs indirectly through actions within the nucleolus or elsewhere (Gard et al. 2009; Bose et al. 2012). Either way, the ability to disrupt tDNA positioning with these mutants provides a tool to probe the causes and consequences of NPC–tDNA contact.
Nascent tRNA increases when tDNAs associate with NPCs
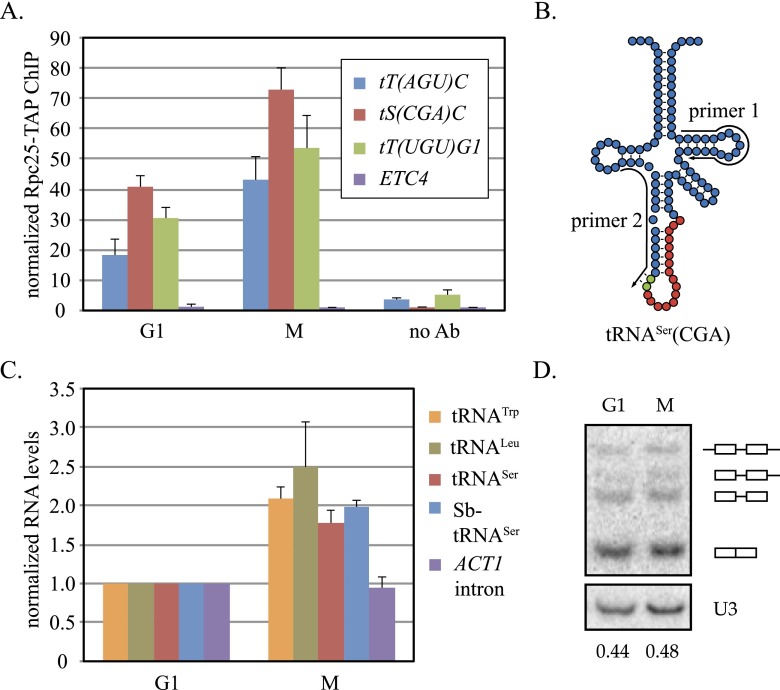
Based on the precedent of activated RNA polymerase II genes associating with NPCs, we hypothesized that localization of tDNAs at NPCs was related to their transcription. To examine this possibility, ChIP-qPCR was used to measure the binding of RNA polymerase III to representative tDNAs. A TAP tag was appended to Rpc25, an RNA polymerase III subunit not found in either RNA polymerase I or II, and ChIP-qPCR was performed as described above. Figure 3A shows that binding of Rpc25 to all three tDNAs was greater during M-phase arrest than during G1 arrest. Nonetheless, binding in G1 cells was higher than the no antibody control, suggesting that some transcription persists when tDNAs are not at NPCs. No binding was recorded at ETC4. We conclude that RNA polymerase III occupancy on tDNAs increases when the genes associate with NPCs in M phase.
Figure 3.
tRNA synthesis during the cell cycle. (A) Rpc25 occupancy at tDNAs. ChIP-qPCR of Rpc25-TAP was performed with strain MC195 arrested in M phase or G1. Rpc25 binding at tDNAs was enriched significantly during M-phase arrest (P-values are in Supplemental Table S4). (B) Pre-tRNASer(CGA) of S. cerevisiae and S. bayanus. Red circles represent the conserved intronic nucleotides, and green circles represent intronic nucleotide polymorphisms. Primers for RT (primer 1) and qPCR (primers 1 and 2) are shown. (C) Nascent tRNA levels. RT-qPCR was performed with strain MC204 [Δtt(agu)c∷Sb-tS(CGA)C]. tRNA levels were significantly higher in M phase (P-values are in Supplemental Table S4). (D) Northern blot analysis of tRNA precursors and products. Radiolabeled oligonucleotide probes were hybridized to unspliced and spliced tRNASer(CGA), albeit with different affinities, in extracts of strain MC172. Unprocessed, 5′ processed, end processed, and mature species are labeled graphically. The ratio of end processed/spliced species is indicated below each lane.
To determine whether the levels of nascent tRNA transcripts fluctuate during the cell cycle, we measured the appearance of select short-lived, intron-bearing pre-tRNAs. Our analysis focused on (1) tRNATrp(CCA) and tRNALeu(CAA), which are encoded by families of six and 10 yeast tDNAs, respectively, and (2) tRNASer(CGA) encoded by either the single yeast gene tS(CGA)C described above or an orthologous gene from Saccharomyces bayanus that we integrated in place of the intron-free tT(AGU)C. In preliminary studies, we verified that the ectopic S. bayanus gene [referred to here as Sb-tS(CGA)C] complimented deletion of the essential tS(CGA)C gene, demonstrating that it produced functional tRNASer(CGA) in S. cerevisiae (Supplemental Fig. S3). Unspliced transcripts from the two genes can be distinguished from one another by a dinucleotide polymorphism within their introns (Fig. 3B; Supplemental Fig. S3). Pre-tRNA levels were measured by reverse transcription (RT) followed by qPCR (RT-qPCR) and normalized to mature ACT1 mRNA. We found that the levels of the four representative pre-tRNAs were roughly twofold higher in cells arrested in M phase than in cells arrested in G1 (Fig. 3C). In contrast, the level of nascent ACT1 mRNA, which also bears an intron, did not vary between the two cell cycle arrests. These findings indicate that the levels of unspliced pre-tRNAs rise in M phase when tDNAs associate more frequently with RNA polymerase III and NPCs.
If tDNA transcription during M phase exceeds the capacity of some subsequent step in tRNA biogenesis, such as splicing, then the levels of pre-tRNA measured in Figure 3C might reflect diminished maturation instead of increased production. In yeast, pre-tRNA splicing occurs on the cytoplasmic surface of the mitochondria (Yoshihisa et al. 2003). Failure to export nascent tRNA during M-phase arrest could result in an abnormal accumulation of unspliced intermediates. To determine whether pre-tRNA levels rise disproportionately relative to the mature tRNA pool, we measured the distribution of processing intermediates of the endogenous tRNASer(CGA) by Northern blot analysis. The U3 snoRNA served as an RNA polymerase II-generated loading control. Figure 3D shows that the ratio of precursor to mature tRNASer(CGA) is equivalent in both M- and G1-arrested cultures. We conclude that the increase in pre-tRNA level in M phase when tDNAs associate with NPCs is the consequence of increased transcription, not decreased maturation. Curiously, the cell cycle oscillations of tRNA synthesis in budding yeast contrasts those in fission yeast and mammals, where RNA polymerase III transcription drops during mitosis (Gottesfeld and Forbes 1997; Iwasaki et al. 2010).
NPC contact is not required for tDNA transcription
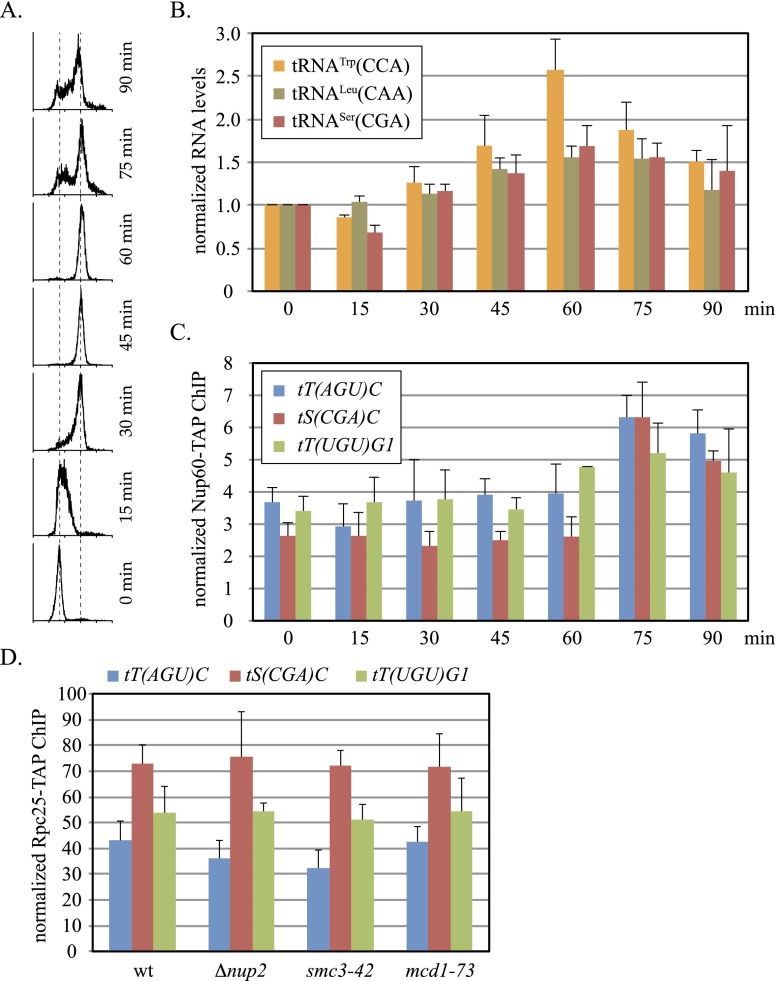
To determine whether association of tDNAs with NPCs is the cause or consequence of transcription, we measured more precisely when pre-tRNA levels and Nup60 binding elevate during the cell cycle. To this end, cells were released from α-factor arrest in G1, and aliquots of culture were harvested at timed intervals following the resumption of synchronous growth (Fig. 4A). RT-qPCR of RNA extracted from the cell pellets showed that levels of three different unspliced pre-tRNAs increased incrementally after release and peaked at 60 min when the cultures consisted entirely of cells with 2N DNA content (Fig. 4B). Pre-tRNATrp(CCA) provides the most dramatic example, with a 2.5-fold rise from trough to peak. Using the same cell pellets, Nup60-TAP binding was evaluated by ChIP-qPCR. Binding remained low and fairly constant at three select tDNAs for 60 min after release (Fig. 4C). Strikingly, Nup60 binding rose sharply at 75 min, the first time point measured following the peak in pre-tRNA levels. Based on the delay between pre-tRNA production and NPC binding, we infer that RNA polymerase III transcription increases first, and then tDNAs migrate to NPCs.
Figure 4.
tDNA transcription without NPC contact. (A) Flow cytometry of synchronized cell culture. Strain MC230 was released from α-factor arrest at time 0. (B) Unspliced tRNA levels at 15-min intervals after release. Values at 60 min were significantly higher than those at time 0 (P-values are in Supplemental Table S4). (C) Levels of Nup60-TAP binding at representative tDNAs at intervals after release. (D) RNA polymerase III binding in mutants. ChIP-qPCR of Rpc25-TAP at the tDNAs was performed in Cdc20-depleted strains MC195 (wt), MC211 (Δnup2), MC214 (smc3-42), and MC212 (mcd1-73). Rpc25 occupancy was not altered in these mutants.
If tDNA transcription precedes NPC–tDNA contact, then transcription of tDNAs should persist in mutants that abolish contact. To test this notion, binding of Rpc25-TAP was measured under conditions that block association of tDNAs with Nup60 and NPCs. The ChIP-qPCR experiments in Figure 4D show that binding of the RNA polymerase III subunit to the tDNAs in M-phase-arrested cells persisted in strains either lacking Nup2 or bearing cohesin mutation smc3-42 or mcd1-73. These results indicate that NPC–tDNA contact is not required for binding of RNA polymerase III to tDNAs in M phase.
The use of synchronously growing cultures in the experiments of Figure 4, A–C, allays potential concerns about measuring nascent tRNA levels during cell cycle arrests. For example, Figure 4B showed that tRNA precursors accumulated as cells approached M phase even when M-phase arrest by Cdc20 depletion was omitted from the protocol. Similarly, tRNA production dropped between successive M phases in cells synchronized by transient Cdc20 depletion (Supplemental Fig. S4). This second result showed that nascent tRNA levels oscillated during cell cycle progression even in the absence of α-factor arrest. The pheromone is a convenient tool but is known to trigger a host of physiological responses in yeast.
Unrestricted RNA polymerase III transcription yields NPC–tDNA contact in G1 and S
If transcription is a prerequisite for association of tDNAs with NPCs, then conditions that elevate tDNA transcription can be expected to increase NPC–tDNA contact. This hypothesis was tested by deleting MAF1, the central negative regulator of RNA polymerase III transcription (Boguta 2013). Upon exposure to stressors, such as nutrient deprivation or DNA damage, the protein is dephosphorylated, whereupon it interferes with binding of RNA polymerase III at tDNAs. Even under relatively robust growth conditions, Maf1 exerts a measurable level of transcriptional inhibition (Roberts et al. 2006; Karkusiewicz et al. 2011).
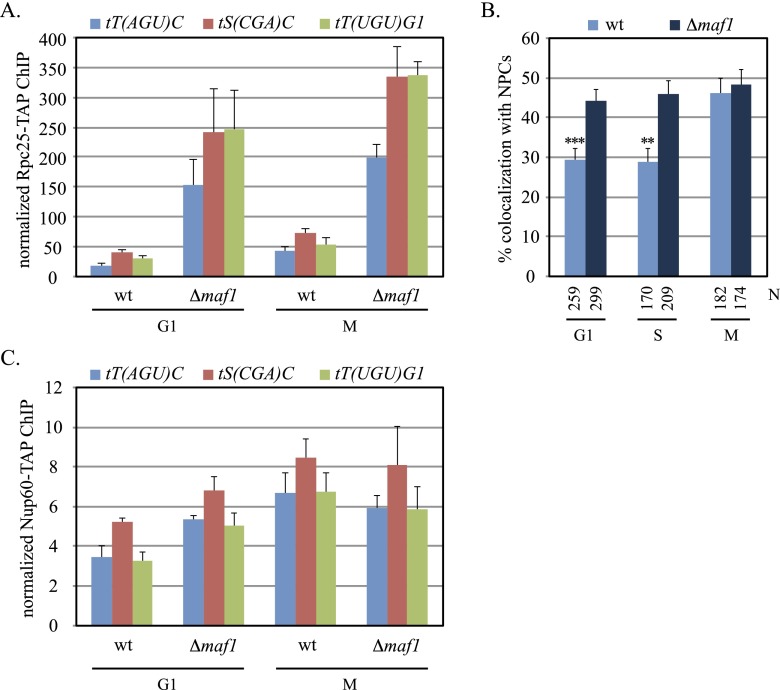
ChIP-qPCR of the Rpc25 subunit was used to evaluate the influence of Maf1 on RNA polymerase III binding to representative tDNAs. In Figure 5A, elimination of the regulator elevated levels of Rpc25 at all three genes. Notably, binding increased in both M- and G1-phase-arrested cells. The results indicate that MAF1 attenuates tDNA transcription even during the cell cycle arrests used in this study. Importantly, changes in RNA polymerase III activity were often accompanied by changes in NPC–tDNA contact. Figure 5B shows that localization of tT(AGU)C at clustered NPCs increased in G1 and S phases to the level only seen in M phase of wild-type cells. However, deletion of MAF1 did not enhance localization of tDNAs with NPCs in M phase. In a complimentary approach, ChIP-qPCR was used to measure the influence of MAF1 on the association of Nup60-TAP with tDNAs. In agreement, deletion of the regulator yielded an increase in Nup60 binding only in G1 cells (Fig. 5C). During M-phase arrest, the level of bound Nup60 remained constant despite the increase in Rpc25 occupancy. The data are revealing in two ways. Foremost, they indicate that transcription by RNA polymerase III is limiting for NPC–tDNA contact during G1 and S phase. At these stages, artificial stimulation of transcription increases tethering of tDNAs at NPCs. Conversely, they indicate that NPC docking is maximal in M phase under the conditions tested. The data are consistent with NPC–tDNA contact being limited by a factor other than transcription at this stage of the cell cycle.
Figure 5.
tDNA transcription and NPC contact in the absence of MAF1. (A) Rpc25 occupancy. ChIP-qPCR of Rpc25-TAP at representative tDNAs was evaluated during M and G1 arrest in strains MC195 (wt) and MC217 (Δmaf1). Rpc25 binding was enhanced significantly in the maf1-null (P-values are in Supplemental Table S4). (B) Localization of tT(AGU)C at NPCs. Asynchronous cultures of MC78 (wt) and MC208 (Δmaf1) were used, as in Figure 1, A and B. Colocalization was significantly higher in the maf1 strain in G1 and S phase. Pairwise χ2-tests, (***) <5 × 10−4; (**) <1 × 10−3. (C) Nup60 binding at tDNAs. ChIP-qPCR of Nup60-TAP was performed in strains MC177 (wt) and MC213 (Δmaf1) during arrest in M or G1 phase. Nup60 binding was enriched significantly in the maf1 mutant in G1.
NPC–tDNA contact and the nuclear export of nascent tRNA
We reasoned that active tDNA genes associate with NPCs to facilitate export of pre-tRNA from the nucleus. It is well established that tRNAs transit from the nucleus to the cytoplasm via a Ran-GTPase-mediated pathway (Hopper 2013). Only recently has it become clear that tRNAs travel in both directions. Cytoplasmic tRNAs return to the nucleus by a constitutive process only to be re-exported (Shaheen and Hopper 2005; Takano et al. 2005). In yeast, Los1 is the principal exportin for intron-containing pre-tRNA (Hopper et al. 1980; Hellmuth et al. 1998; Sarkar and Hopper 1998; Murthi et al. 2010). Los1 is joined by a second exportin, Msn5, for the re-export of tRNAs (Shaheen and Hopper 2005; Takano et al. 2005). Msn5 contributes little to the export of intron-containing tRNA (Murthi et al. 2010). It is not certain whether the protein contributes to the initial export of intron-free tRNAs. Curiously, neither exportin is essential, and the double mutant is viable. These observations indicate that at least one additional route for tRNA export from the nucleus must exist.
To investigate whether tRNA export influences an upstream event like NPC–tDNA contact, we measured Nup60-TAP binding to tDNAs in strains that lack either LOS1 or MSN5. Figure 6A shows that deletion of LOS1 hindered association of Nup60 with three representative tDNAs, including one with an intron [tS(CGA)C] and two without introns [tT(AGU)C and tT(UGU)G1]. In contrast, deletion of MSN5 did not alter binding of Nup60 to any of the tDNAs evaluated in this study (Fig. 6A). We surmise that Los1 alone couples transcribing tDNAs to NPCs irrespective of whether the genes contain or lack introns.
Figure 6.
A role for LOS1 in NPC–tDNA contact. (A) Nup60 occupancy at tDNAs. ChIP-qPCR of Nup60-TAP was performed with strains MC177 (wt), MC237 (Δlos1), and MC253 (Δmsn5) during M-phase arrest. Nup60-TAP binding was significantly diminished in the los1 mutant (P-values are in Supplemental Table S4). (B) NPC–tDNA contact with increased LOS1 dosage. ChIP-qPCR of Nup60-TAP was performed with strain MC213 (Δmaf1) transformed with either YEpFAT4-LOS1 or YEpFAT4. Strains were grown in SC-ura,met prior to arrest in M or G1. (C) Diminished nonsense suppression in the absence of NPC–tDNA contact. Fivefold serial dilutions of strains W303-1A (wt), MC206 (Δnup2), MC232 (Δlos1), K5824 (smc3-42), and K5832 (mcd1-73) bearing plasmid pUN60 were spotted on selective plates to measure activity of the plasmid-borne nonsense suppressor SUP11°. (EthGly) 1.5 M EthGly. To compensate for slower growth of strains K5824 and K5832 at 30°C, the cultures were concentrated fivefold and 25-fold, respectively, before diluting serially. Changes in growth of strains K5824 and K5832 in medium containing EthGly is not due to osmotic remediation of the cohesin mutations (Supplemental Fig. S5).
If Los1 is required for NPC–tDNA contact and contact is maximal in M phase (Fig. 5B,C), then native levels of the protein might be a limiting factor in positioning transcribed tDNAs at NPCs. To test whether NPC–tDNA contact is limited by Los1, ChIP-qPCR was used to measure binding of Nup60 at tDNAs in a strain bearing additional copies of a plasmid-borne LOS1 gene. To sensitize the assay, a maf1 mutant was used because elevated production of pre-tRNAs in this genetic background is known to exceed the capacity for their export (Karkusiewicz et al. 2011). During M-phase arrest, binding of Nup60-TAP to two representative tDNAs was significantly higher in cells with increased LOS1 gene dosage (Fig. 6B). Binding of Nup60-TAP to a third tDNA, tS(CGA)C, was also increased, but the significance was obscured by variability in the empty vector samples. Extra LOS1 did not increase binding in G1-arrested cells, consistent with the observation in Figure 5 that NPC contact is limited at this stage of the cell cycle by an upstream event; specifically, transcription by RNA polymerase III. Taken together, these experiments show not only that Los1 promotes NPC–tDNA contact but that maximal contact in M phase can be limited by the available pool of the exportin.
The LOS1 gene was originally cloned by virtue of its role in tRNA-mediated nonsense suppression: los1 mutants blocked suppression by hindering nuclear export of tRNA suppressors (Hopper et al. 1980; Hurt et al. 1987). We employed a pair of nonsense suppression assays to determine whether NPC–tDNA contact was required for efficient pre-tRNA export. The first is based on an early observation that nonsense tRNA suppressors such as the SUP11° ochre allele of the tY(GUA)F1 tDNA impart sensitivity to high osmolarity (Singh 1977). In the second assay, SUP11° suppresses the premature stop codon in ade2-1, thereby restoring adenine biosynthesis. In the presence of plasmid-borne SUP11°, a LOS1 ade2-1 strain grew well without exogenous adenine but poorly in the presence of a concentrated osmolyte, 1.5 M ethylene glycol (EthGly) (Fig. 6C). Absence of LOS1, in contrast, hindered growth when adenine was omitted and improved growth in the presence of EthGly. Importantly, strains lacking NUP2 or bearing the cohesin mutations displayed phenotypes similar to the los1 strain on the tester plates, albeit to different degrees. The combination of both positive and negative selections ensured that true suppression was not confused with the intrinsic growth behavior of each particular mutant. The simplest explanation is that NPC–tDNA contact facilitates nonsense suppression, presumably by the export of the SUP11° gene product.
Discussion
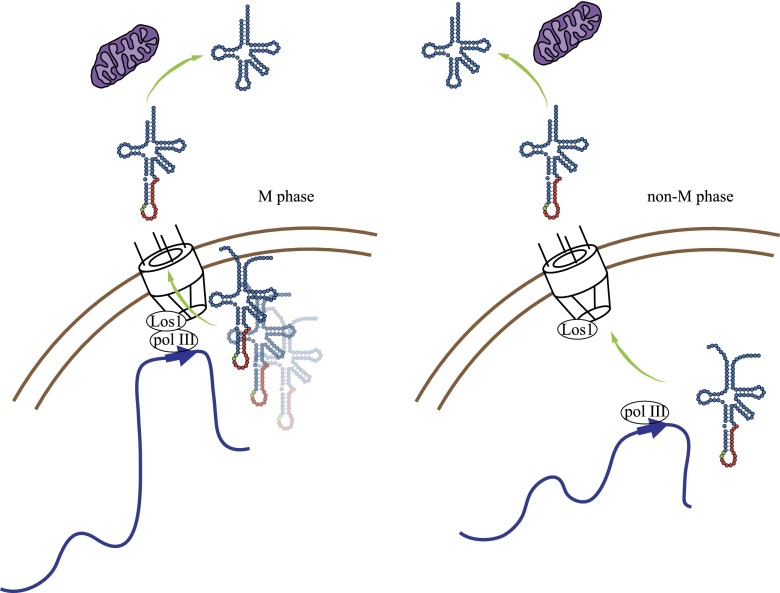
The central findings of this study are that (1) tDNAs migrate to NPCs as a consequence of increased transcription in M phase; (2) association of tDNAs with NPCs requires Los1, the pre-tRNA exportin; and (3) NPC–tDNA contact contributes to nonsense suppression by a mutant tRNA. Taken together, these findings suggest that tRNA synthesis and nuclear export are coordinated events when tDNAs localize at NPCs in yeast (Fig. 7).
Figure 7.
A model for coordinated transcription and export of pre-tRNAs from the nucleus. Increased transcription in M phase by RNA polymerase III is accompanied by repositioning of tDNAs at NPCs in a manner that depends on select Nups and the Los1 exportin. The model demands that nascent tRNAs mature at or near NPCs or that the transcripts synthesized at NPCs return to the nucleoplasm before export. Transcription outside of M phase when tDNAs are not enriched at NPCs requires that pre-tRNA transit through the nucleoplasm before export. Pre-tRNAs that leave the nucleus are spliced on the cytoplasmic surface of mitochondria.
Our results might seem at odds with an earlier report of tDNA clustering at the nucleolus and a more recent report of tDNA clustering with centromeres (Thompson et al. 2003; Duan et al. 2010). A parsimonious explanation would be that tDNAs associate simultaneously with NPCs and either the nucleolus or centromeres at the edge of the nucleus. For the genes tested, however, NPC contact increased during M phase, whereas colocalization with the nucleolus remained low and constant throughout the cell cycle (Fig. 1). Similarly, colocalization of the genes with spindle pole bodies, where centromeres cluster at the nuclear membrane, was also rare (data not shown). If tDNAs migrate to and from NPCs dynamically, transient positioning of individual genes outside the nucleolus or away from centromeres might have been missed with the technologies used earlier to monitor their positions.
Potential advantages of tDNA transcription at NPCs
Transcription of tDNAs at NPCs might have evolved for several reasons. Foremost, NPC localization might expedite export of pre-tRNA, thereby avoiding an otherwise rate-limiting step in tRNA biogenesis. A corresponding advantage of accelerated export would be a decrease in the time that potentially deleterious pre-tRNAs spend in the nucleus. Mutations that increase the nuclear accumulation of mRNA, for example, pose a threat to genome stability by favoring the formation of R loops (Aguilera and Garcia-Muse 2012). According to this view, tRNAs that return to the nucleus must somehow evade R-loop formation.
tDNAs might also associate with NPCs to fine-tune tRNA production by means unrelated to export. For example, a recent study showed that Ulp1, an NPC-associated SUMO protease, accelerates derepression of the prototypical GAL1 gene, which migrates to NPCs during induction (Texari et al. 2013). This scenario and others in which tDNAs associate with NPCs for reasons other than export are distinct because, in these cases, pre-tRNAs generated at the NPC might venture elsewhere in the nucleus before subsequent export.
Los1 and the coordination of tRNA transcription and export
The main argument for coordination between tRNA synthesis and export stems from the unexpected role for a pre-tRNA exportin in an upstream event in tRNA biogenesis; namely, docking of tDNAs at NPCs. In gating of RNA polymerase II genes, the Mex67 export factor binds transcribed loci in an RNA-independent manner and then transfers to the assembling mRNP complex (Dieppois and Stutz 2010). How Los1 mediates NPC–tDNA contact is not yet known. One model holds that the exportin bridges RNA polymerase III to NPCs directly. A less direct model, however, cannot be discounted: In the absence of Los1, accumulation of tRNA in the nucleus could trigger a regulatory response that releases all RNA polymerase III-transcribed genes from NPCs.
Coordinated transcription and nuclear export of pre-tRNA cannot be an essential function of the cell because mutants lacking NPC–tDNA contact are viable (Figs. 2, 6). We surmise that transcription of tDNAs at NPCs facilitates export of pre-tRNAs that would eventually escape the nucleus by a less efficient process. Only when survival requires nonsense suppression can the defect in NPC–tDNA contact be detected (Fig. 6C).
Los1 versus Msn5 in the initial export of pre-tRNA
Los1 was defined as the principal exportin for intron-bearing pre-tRNAs based on assays that monitored the fate of tRNA introns, which in yeast can only be spliced upon arrival in the cytoplasm (Hopper 2013). Due to the challenge of tracking the directional movement of tRNAs without introns, assigning the division of labor between LOS1 and MSN5 during the initial nuclear export phase has not been definitive. Our work now provides an unanticipated criterion for distinction. LOS1 alone mediates NPC–tDNA contact (Fig. 6A,B). That the gene facilitates tethering of tDNAs with introns as well as those without suggests that Los1 acts globally in the export of pre-tRNA. The results are consistent with Msn5 acting only on a subsequent step in tRNA trafficking, such as re-export.
Spatial considerations of nuclear tRNA processing events
Before exiting the nucleus, pre-tRNAs undergo significant processing. Most primary transcripts are trimmed, and numerous bases are modified chemically. Current thinking holds that sequential steps toward maturation occur at different locations in yeast (Hopper 2013). In order to reconcile this expectation with coordinated synthesis and export of tRNA at NPCs, two possibilities can be considered. First, pre-tRNAs synthesized at NPCs might venture back into the nuclear interior and to the sites of modification before subsequent export. Alternatively, tRNA processing events might actually occur at the NPC. A precedent for this scenario recently emerged from studies in Caenorhabditis elegans that showed that integral Nups of the NPC associate with numerous RNA polymerase III-transcribed genes to regulate 3′ end processing of the gene products (Ikegami and Lieb 2013). Based on these observations, the question of where tRNA modifications occur in the yeast nucleus warrants closer inspection.
Cell cycle considerations of tDNA transcription
Mature tRNAs are highly abundant and typically long-lived, yet the gene products are produced periodically during the cell cycle (Figs. 3, 4; Supplemental Fig. S4). The logic behind such regulated synthesis might not be intuitive. Two recent discoveries provide conceptual frameworks for what might be occurring in budding yeast. First, a pair of research teams (Brickner and Brickner 2010; Bermejo et al. 2011) showed that gated RNA polymerase II-transcribed genes transiently release from NPCs during S phase. Bermejo et al. (2011) argued that the process, which is regulated by the replication stress checkpoint pathway, avoids deleterious topological consequences of replicating immobilized DNA. Association of tDNAs with NPCs might create a similar impediment to replication fork progression and thus might also be subject to a similar form of regulation. Second, Nguyen et al. (2010) showed that RNA polymerase III transcription is down-regulated by the replication stress checkpoint during S phase, presumably to avoid the deleterious consequences of blocked replication forks that accumulate at tDNAs (Deshpande and Newlon 1996; Ivessa et al. 2003; Clelland and Schultz 2010; Szilard et al. 2010). Exactly how RNA polymerase III regulation occurs during normal proliferation in budding yeast and why attenuation also occurs in G1 prior to DNA replication will require further study.
Materials and methods
Strains and plasmids
Supplemental Table S1 lists the yeast strains used in this study. Complete ORF deletions and gene fusions were generated by PCR-mediated gene replacement and confirmed by PCR. Additional chromosome modifications were made with the integrating plasmids listed in Supplemental Table S2, as described in the Supplemental Material. nup133 deletions were covered with plasmids bearing either the full-length gene or the nup133-ΔN allele (pNUP133-URA3 or pMC3, respectively).
Cell growth
Asynchronous cultures were grown to mid-log in SC medium. For M-phase arrest, Cdc20 was depleted from strains carrying MET3p-CDC20 by adding methionine (Cf = 2 mM) to mid-log cultures pregrown in SC-met. Cells were harvested 2.5 h later after ∼80%–90% cells displayed a dumbbell-shaped morphology. For arrest with nocodazole (Cf = 10 µg/mL), cells were grown in YPDA. Cells were arrested in G1 with α factor (Cf = 1 × 10−5 M). To remove α factor, cells were washed twice and resuspended in medium containing pronase E (Cf = 100 µg/mL). Flow cytometry was performed at the Rutgers Environmental and Occupational Health Sciences Institute (EOHSI) core facility. All experiments were performed at 30°C except the nonsense suppression assays in which strains were pregrown at 25°C in SC-ura before plating on selective medium at 30°C.
Microscopy
Paraformaldehyde fixation, slide preparation, fluorescence microscopy, and error analysis were described previously (Chang et al. 2005). All data sets were based on at least three independent trials for a total of 100–300 cells per condition. Cell cycle stage was defined as follows: G1, no bud; S, small bud; and M, large bud with nucleus at the bud neck. Cells that had begun anaphase with single dots on either side of the bud neck were excluded from the analysis. Cohesin mutants yielded pairs of dots that were scored independently. GFP-tagged loci were defined as colocalized with an mRFP- or CFP-tagged protein if the fluorescent signals were separated by no more than the width of the GFP focus within the same or adjacent image planes.
ChIP
Cross-linking, extract preparation, and sonication were performed as in Ausubel et al. (2010). Subsequent procedures with anti-TAP antibody (Thermo Scientific) and protein A-coated Dynabeads (Life Technologies) were performed as in van Attikum et al. (2004). qPCR was performed with a Rotor-Gene Q (Qiagen) using the primers listed in Supplemental Table S3. Reported values correspond to the signal for each site relative to an internal control (GIT1) divided by the same ratio of sites within input. The mean and standard deviation of three or more biological replicates are presented. Statistical significance was determined by pairwise Student’s t-tests.
RNA analysis
RNA was extracted with hot acidic phenol (Ausubel et al. 2010) and treated with DNase I (Roche). Reverse transcription was performed using SuperScript III First Strand Synthesis Supermix (Life Technologies) followed by qPCR using the primers listed in Supplemental Table S3. Values for each tRNA were normalized to an internal control (ACT1 mRNA) and are reported relative to the ratio in G1. The mean and standard deviation of three or more biological replicates are presented. Statistical significance was determined by pairwise Student’s t-tests. Northern blots were hybridized with the γ-32P-labeled oligonucleotides listed in Supplemental Table S3 according to Karkusiewicz et al. (2011) and were quantified with a Storm 840 PhosphorImager (Molecular Dynamics).
Acknowledgments
We thank Giulia Ruben, Rohinton Kamakaka, and Rudra Dubey for critical materials, discussion, and complementary experiments at the beginning of this study. We thank Eric Phizicky, Ian Willis, Rodney Rothstein, Andrés Aguilera, Chi Kwan Tsang, Nancy Woychik, and Steve Zheng for constructive criticism, technical advice, and materials. We thank Ruiheng Yin for expert experimental assistance, and Mike Hampsey for comments on the manuscript. This work was funded by National Institutes of Health grant R01GM51402.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.236729.113.
References
- Aguilera A, Garcia-Muse T 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, ed. 2010. Current protocols in molecular biology. John Wiley & Sons, New York [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B, et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR 1998. Nucleolar localization of early tRNA processing. Genes Dev 12: 2463–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G 1985. Gene gating: a hypothesis. Proc Natl Acad Sci 82: 8527–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguta M 2013. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta 1829: 376–384 [DOI] [PubMed] [Google Scholar]
- Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, Unruh J, Garrett A, McDowell W, Box A, et al. 2012. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet 8: e1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Brickner JH 2010. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol Biol Cell 21: 3421–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773 [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 [DOI] [PubMed] [Google Scholar]
- Chang CR, Wu CS, Hom Y, Gartenberg MR 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev 19: 3031–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clelland BW, Schultz MC 2010. Genome stability control by checkpoint regulation of tRNA gene transcription. Transcription 1: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22: 2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D, Mykytka B, Allen NP, Huang L, Al B, Rexach M 2001. The nucleoporin Nup60p functions as a Gsp1p–GTP-sensitive tether for Nup2p at the nuclear pore complex. J Cell Biol 154: 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dieppois G, Stutz F 2010. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J Cell Sci 123: 1989–1999 [DOI] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD 2005. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol 171: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J 20: 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev 13: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC 1994. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J 13: 6062–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS 2010. A three-dimensional model of the yeast genome. Nature 465: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey RN, Gartenberg MR 2007. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev 21: 2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D, Brickner JH 2011. Gene positioning and expression. Curr Opin Cell Biol 23: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, Fleharty B, Seidel C, Brickner JH, Gerton JL 2009. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol 187: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR, Neumann FN, Laroche T, Blaszczyk M, Gasser SM 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955–967 [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ 1997. Mitotic repression of the transcriptional machinery. Trends Biochem Sci 22: 197–202 [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Engelke DR 2006. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res 34: 4826–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22: 2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Singh BN, Ansari A, Laine JP, Krishnamurthy S 2011. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv Enzyme Regul 15: 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18: 6374–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Botsios S, Donze D, Donaldson AD 2012. TFIIIC localizes budding yeast ETC sites to the nuclear periphery. Mol Biol Cell 23: 2741–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194: 43–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA 1980. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19: 741–751 [DOI] [PubMed] [Google Scholar]
- Hurt DJ, Wang SS, Lin YH, Hopper AK 1987. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol 7: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Lieb JD 2013. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell 51: 840–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K 2010. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell 21: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371 [DOI] [PubMed] [Google Scholar]
- Karkusiewicz I, Turowski TW, Graczyk D, Towpik J, Dhungel N, Hopper AK, Boguta M 2011. Maf1 protein, repressor of RNA polymerase III, indirectly affects tRNA processing. J Biol Chem 286: 39478–39488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland JG, Raab JR, Kamakaka RT 2013. TFIIIC bound DNA elements in nuclear organization and insulation. Biochim Biophys Acta 1829: 418–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453 [DOI] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci 102: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK 2010. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol Biol Cell 21: 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VC, Clelland BW, Hockman DJ, Kujat-Choy SL, Mewhort HE, Schultz MC 2010. Replication stress checkpoint signaling controls tRNA gene transcription. Nat Struct Mol Biol 17: 976–981 [DOI] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125: 859–872 [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Zenklusen D 2012. To the pore and through the pore: a story of mRNA export kinetics. Biochim Biophys Acta 1819: 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR 2006. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell 22: 633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben GJ, Kirkland JG, Macdonough T, Chen M, Dubey RN, Gartenberg MR, Kamakaka RT 2011. Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS ONE 6: e21923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK 1998. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell 9: 3041–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK 2006. Nup-PI: the nucleopore–promoter interaction of genes in yeast. Mol Cell 21: 379–391 [DOI] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 11290–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A 1977. Nonsense suppressors of yeast cause osmotic-sensitive growth. Proc Natl Acad Sci 74: 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard RK, Jacques PE, Laramee L, Cheng B, Galicia S, Bataille AR, Yeung M, Mendez M, Bergeron M, Robert F, et al. 2010. Systematic identification of fragile sites via genome-wide location analysis of γ-H2AX. Nat Struct Mol Biol 17: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Gasser SM 2012. Structure and function in the budding yeast nucleus. Genetics 192: 107–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T 2005. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142 [DOI] [PubMed] [Google Scholar]
- Texari L, Dieppois G, Vinciguerra P, Contreras MP, Groner A, Letourneau A, Stutz F 2013. The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol Cell 51: 807–818 [DOI] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT 2008. Long-range communication between the silencers of HMR. Mol Cell Biol 28: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788 [DOI] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T 2003. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]