Abstract
We previously developed a network phenotyping strategy (NPS), a graph theory-based transformation of clinical practice data, for recognition of two primary subgroups of hepatocellular cancer (HCC), called S and L, which differed significantly in their tumor masses. In the current study, we have independently validated this result on 641 HCC patients from another continent. We identified the same HCC subgroups with mean tumor masses 9 cm×n (S) and 22 cm×n (L), p<10−14. The means of survival distribution (not available previously) for this new cohort were also significantly different (S was 12 months, L was 7 months, p<10−5). We characterized nine unique reference patterns of interactions between tumor and clinical environment factors, identifying four subtypes for S- and five subtypes for L-phenotypes, respectively. In L phenotype, all reference patterns were PVT (portal vein thrombosis) positive, all platelet/AFP levels were high, and all were chronic alcohol consumers. L had phenotype landmarks with worst survival. S phenotype interaction patterns were PVT negative, with low platelet/AFP levels. We demonstrated that tumor-clinical environment interaction patterns explained how a given parameter level can have a different significance within a different overall context. Thus, baseline bilirubin is low in S1 and S4, but high in S2 and S3, yet all are S subtype patterns, with better prognosis than in L. Gender and age, representing macro-environmental factors, and bilirubin, INR and AST levels representing micro-environmental factors, had a major impact on subtype characterization. Clinically important HCC phenotypes are therefore represented by complete parameter relationship patterns and cannot be replaced by individual parameter levels.
Keywords: HCC, NPS, Parameter patterns
Introduction
The idea that tumors grow in part due to the influence of their environment is not new1. We understand tumor clinical environment to be any aspect of the milieu in which a tumor arises, that can potentially influence its behavior. Thus, age2,3 and gender4 can influence the hormonal milieu of the liver. We regard such clinical factors as macro-environmental. The altered liver function that is part of the changed cytokine and inflammatory marker cascade resulting from alcoholism or hepatitis and that is reflected in blood bilirubin, albumin, INR or ASL/SGOT levels, we consider to be clinically micro-environmental5,6. Both the processes of hepatocarcinogenesis and growth of hepatocellular carcinoma (HCC) involve a 2-way influence of the effects of hepatitis viruses, alcohol or carcinogenic mycotoxins on the liver, as well as the reaction of liver components to these chronic and damaging agents. At the level of tissue organization, there are changes in extra cellular matrix components, as well as angiogenesis and chronic inflammation, that are both consequent on the damage and then become necessary components of the developing tumor environment. Some of the biochemical processes have been identified to include oxidative stress, apoptosis, autophagy and the immune system (reviews: refs.7-9). The tumor stroma and micro-environment has been shown to have both characteristic and prognostic molecular signatures10-15, but its components are also seen to be an attractive target for the new molecularly targeted therapies8,9,16. Some of the cell types that are involved, and their products, are now becoming identified16-19. The effects of clinical environment (macro and micro) on tumor biology are not simple, nor are the studies and questions and tools for finding answers. At the same time, novel experimental methods bringing detailed insights about the micro-environmental contributions to disease mechanisms are increasingly powerful. A methodology is needed for finding the optimal intersection of the clinical and molecular directions in tumor-environment research and its clinical interpretation and application. Ideally, tumor-environment models and their diagnostic and prognostic results should use these two information resources simultaneously, in the full mutual context. In this article, we open a clinical direction towards this unification with an approach prepared for incorporating both avenues. Our motivation is that if the standard clinical characterization of the patient in terms of our understanding of micro- and macro environmental clinical factors and the disease status can bring new insights, that would allow direct integration of the results of novel experimental and molecular biology studies with clinical practice data and thus improve the “bedside translation”. We suggest and demonstrate here, that with better characterization of the clinical disease heterogeneity, it is more likely that relevant hypotheses can be formulated and tested through complex studies with better design and patient status identification.
In this paper, we present several novel results. First, we validated the Network Phenotyping Strategy (NPS)-based classification model20, developed by us previously for recognition of HCC subtypes using the extensive screening data on 4139 subjects21. We applied this model without change, to independently collected data from another continent, and confirmed that the same HCC subtypes and the characteristic patterns of relationships were also identified. Since we had survival data for this new data set (which was not available in the previous study), we show next, that the identified HCC subtypes have significantly different survival and thus prognosis. With this additional validation, we then analyzed the clinical and relationship pattern-based characteristics of the identified HCC subtypes and provided their interpretation in terms of the tumor-clinical environmental interactions.
Methods
We approached the extraction of novel information from a standard set of baseline clinical parameter data at diagnosis, used in standard clinical practice clinic of hepatocellular carcinoma (HCC) evaluation, in a way that allows us to better characterize HCC clinical heterogeneity. We have previously demonstrated that this can be done by application of graph theory tools.
Mathematical graphs, when properly selected, can capture what at first sight are complicated relationship patterns, in an elegant and most importantly, in a manageable and clinically understandable way. We call this new graph-based approach the Network Phenotyping Strategy (NPS)20,21. NPS transformation of clinical practice data enabled us to adopt a new paradigm in which we examined the levels of individual typical parameters used in standard baseline evaluation and clinical categorization of HCC within the context of all the other identified clinical parameters.
In the concrete application of this general approach to problems of HCC, the changed paradigm allowed us to use common clinical blood test parameters together with demographic descriptors, and gain novel information from analyzing the relationship patterns by considering their values and levels simultaneously. This novel paradigm is a mathematical incarnation of the common clinical question of the following type: a single 8cm HCC mass in an apparently normal liver carries a quite different prognosis and treatment approach that a similar 8cm mass in the presence of multiple cirrhotic nodules, elevated bilirubin and/or ascites. We considered how to take all these important inter-relationships simultaneously into account and understand their impact on the prognosis or treatment of a concrete patient. We demonstrate here that NPS transformation of the data enables not only high level analysis of information in the relationship patterns between all used clinical variables, but it also provides results in a form that is directly and simply interpretable in clinical terms.
Our NPS approach uses the clinical study and/or clinical practice data and with the consideration of all available clinical information that is relevant for the disease. This pre-processing of the data allows us to encode the standard clinical information in a consistent manner for very diverse data types as the partitions and vertices of a network graph, that in turn, represent complete relationship patterns between the clinical data levels and types. Once this is done, it is trivial to represent a personal relationship pattern for every patient, since we generate a k-partite graph in which the actual clinical variable levels, found through the baseline diagnostic tests and data collection for an individual patient, are represented by separate vertices in the respective partitions. These actual levels are then connected by edges (lines), representing all co-occurrences of these levels in the concrete personal clinical profile.
The advantage of this approach is the simplicity of the next step, in which we capture the relationship pattern information from an entire cohort into a “study graph”. The study graph is simply a union (generated by addition of all personal clinical profile relationship networks) of all personal k-partite graphs. In the study graph, the numbers of co-occurrence edges between respective levels of clinical variables carry information about the frequencies of the relationship. In the next step, with the use of graph theory mathematics, we then found the complete decomposition of the study graph into the linear combination of reference relationships patterns (RRP). These RRPs represent unique clinical profiles, which characterize the typical collective relationships between all considered variables, occurring frequently and with clinical significance in the original data. The RRP’s were then used as “landmarks” in the disease clinical profile landscape, relative to which we measured an individual patient’s clinical profiles.
To that effect, in the last NPS step, we extracted the personalized information for characterization of an individual patient’s relationship pattern using the “closeness” between an individual’s clinical pattern and all those RRPs, characterizing the HCC type heterogeneity. This closeness was computed as a vector of graph-graph distances between a personal relationship profile of an individual patient and all respective RRPs. With this definition, the graph distance has simple and clear clinical interpretation, namely, it is the total number of mismatches between the individual patient and the reference relationship profiles. The number of mismatches from one RRP defines one element of the personal distance vector. We found in the previous study that 9 RRPs are necessary for characterization of HCC tumor phenotypes, which we validated here using a different dataset.
The NPS transformation of the HCC patient baseline variables thus constitutes a 9-element vector with element one indicating how a patient actual relationship profile is different from RRP1 and element two indicating how a patient actual relationship profile is different from RRP2 etc. In this way, through the NPS transformation, the original “raw” clinical data are transformed into simple numerical form that unifies encoding of variable levels with the actual pattern of the variable level relationships in every individual patient’s clinical profile. Figure 1 shows an example of how parameter change impacts the relationship pattern. The lines (graph edges) are relationships found in one of the HCC-specific RRPs. While this RRP considers platelets with levels higher than 195×109/L, a concrete patient had platelets level lower than this threshold with all remaining variable levels identical to the RRP. This single parameter level difference between individual and reference clinical profiles (7%) caused a change in 9 out of 45 (20%) relationships captured by NPS transformation of the HCC screening data (see Fig.1).
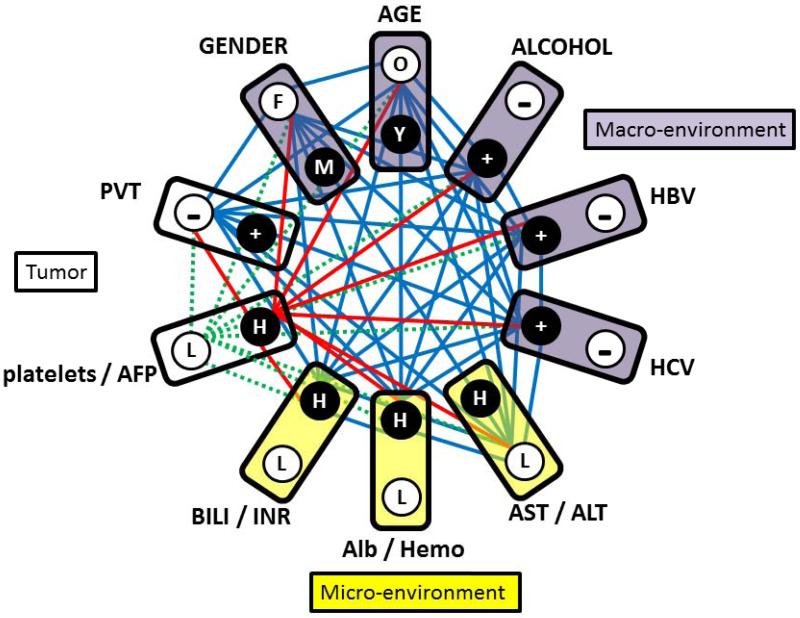
Figure 1.
10-partite graph used in NPS transformation of HCC screening data, which captures patterns of complete relationships between tumor and environmental parameters. Each box represents one partition, which in turn corresponds to one clinical variable. Note that eight blood parameters are represented in 4 pairs, each comprised of two significantly correlated clinical variables. Circles (graph vertices) represent the statuses of categorical variables (+=present,− =absent, F=female, M=male, O=older than 55 years, Y=younger than 55 years) or levels of blood parameter pairs components (L=both parameter levels are from the two low, H=one of both levels in the pair are from the upper terciles in the original study21). Solid lines (graph edges) show the unique information processed by NPS: they represent the complex pattern of relationships between all variable levels for a patient. This relationship pattern represents female, older than 55 years, with self-reported alcoholism, HepB and HepC positive, AST < 4 IU/L & ALT < 3.23 IU/L, albumin > 4.0 g/dL &/OR hemoglobin >14.9 g/dL, bilirubin > 1.5 mg/dL &/OR INR>77, platelets <195 × 103/dL & AFP < 29 000 ng/dL, PVT negative. Red lines show by example that any single parameter (here platelet/AFP levels) contributes to 9 relationships in the complete pattern. By the dotted lines (green) we demonstrate how single parameter change (platelet or AFP are high) results in the more informative change of the 9 components of the original relationship pattern. Because the results of the NPS data transformation are independent of the variable ordering in the 10-partite graph, we grouped the macro-, micro-environmental and tumor factors into adjacent sections as shown.
Clinical data collection
The baseline clinical presentation data of 641 US patients presenting for treatment of biopsy-proven unresectable HCC in an unscreened population was examined. On initial clinical evaluation, all patients had: baseline complete blood count, blood liver function tests, blood alpha fetoprotein (AFP) levels and hepatitis serology, as well as physical examination, liver and tumor biopsy, and a triphasic helical computed axial tomography (CAT scan) scan of the chest, abdomen and pelvis. The data and CT descriptors were prospectively recorded and entered into an HCC database intended for follow-up and analysis. This analysis was done under a university IRB-approved protocol for the retrospective analysis of de-identified HCC patient records.
RESULTS
HCC heterogeneity: identification of 2 general HCC phenotypes
In this paper we present the first independent validation of the previously published results obtained by NPS analysis of HCC screening data from another HCC cohort, that was not part of a screening program. With these independent clinical data, we therefore followed without any modification all the previously used steps in their preparation for NPS transformation and analysis. The first step was the definition of the partitions in the k-partite graph. That included data-driven approach that simplified the relationship patterns to be analyzed21. For this purpose, we previously used a special algorithm of graph theory, which found the maximal cut sub-graph in the complete weighted graph, representing all possible statistically significant (p<0.01) correlations between eight blood test parameters. This algorithm is mathematically proven22 to find a single unique set of clinical variables that are all statistically significantly correlated and the sum of their correlation coefficients is maximal of all other possible combinations. We used this step to optimally represent by a single partition the two variables that carry equivalent information. These uniquely correlated pairs were: AST/ALT, AFP/platelets, albumin/Hb, and bilirubin/INR.
By repeating the identical procedure with new data, we have shown that this unique, informationally optimal, pairing of clinical variables was found without change and with preserved statistical significance of correlation coefficients also in the cohort studied in this paper. This validated the first design feature of our NPS graph – that it was and is the 10-partite graph (Fig.1), in which each partition represented one clinical component of the analyzed information. In addition to the 4 blood test pairs identified above, the remaining 6 graph partitions represented age, gender, alcoholism, hepatitis and portal vein thrombosis (PVT) statuses.
The next step of the clinical definition of the 10-partite graph needed for the NPS transformation of HCC screening data was defining the thresholds, allowing us to discretize the ranges of real valued clinical variables into low and high categories. In the previous publications, we used a tercile approach to find these thresholds (ref.21 for details). Low category represented two terciles of original cohort patients, all having both levels lower than given threshold, high category represented the upper tercile of patients with at least one variable level above the threshold. With the new data we found that all thresholds from our previous study also sub-divided the US cohort into the terciles. This was evidence that the distributions of the collected clinical variables were equivalent in the two data sets and consequently that parameters we selected for discretization of the clinical information in our NPS analysis are justified and independently valid in both cohorts. With validated thresholds, the high and low levels of clinical variables were then represented by the vertices in the respective partitions (Fig.1). We then used the actual data for every individual from the validation cohort and constructed personal 10-partite graphs, representing the complete patterns of relationships between patient’s clinical profile levels.
With the current validation set, with substantially less patients compared to training set, we implemented the stringent validation approach, confirming the reference relationship patterns (RRPs) which we found by a special graph theory algorithm, which decomposed the training study graph, representing union of 4139 individual relationship patterns into a linear combination of RRP’s are valid landmarks in the HCC clinical landscape of validation cohort. The similarity and dis-similarity of a patient’s individual clinical profile was defined by the number of personal screening data relationships identical to or different from the respective reference relationship patterns. The total number of these differences for each of nine RRPs, defined (through the 9-variable logistic regression model) the odds that a given individual patient’s clinical screening profile is representing an S-tumor or L-tumor HCC subtype21.
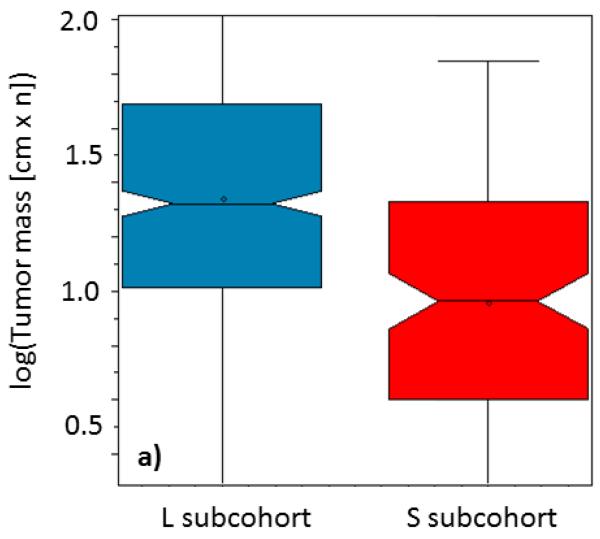
We therefore directly used the RRPs from the training set, generated the input of individual graph-RRP distance vectors with nine components for patients from the validation set. These were used as input into the S- and L-classification logistic regression equation, optimized in the training set. We used the computed odds to recognize two subgroups of the patients with predicted small (S) and large (L) tumor masses. With this strategy, the patients from the validation set were classified into HCC subtypes S and L directly by their relationship patterns derived from personal screening data. This was done independently of the information about the actual tumor masses. We found 80.6% patients in L and 19.4% patients in S subgroup. We then finalized the validation by comparing the distributions of the actual tumor masses in the S and L identified patient sub-group (see. Fig. 2a). The two means of tumor mass distributions were 22 [cm.n] for L and 9 [cm.n] for S patients. These means of tumor mass distributions in the two categories were significantly statistically different (p<10−14).
Figure 2.
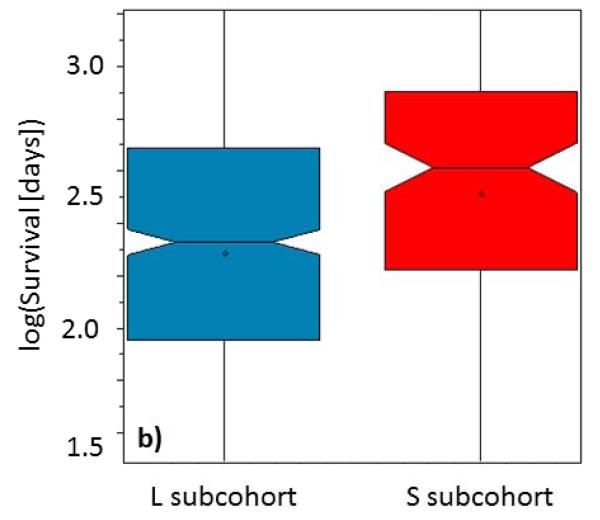
Boxplots representing distributions of the a) tumor masses (logarithm of maximal tumor size in cm × number of nodules) and b) overall survivals (logarithm of survival in days) for the patients, classified by the previously developed 9-variable logistic regression model, computing odds for S (red) and L (blue) from the distances of patients personal relationship profile graphs from graphs of reference relationship patterns. Box notches indicate distribution means.
In addition, since we had survival data for this set that was not available for the training dataset, we did additional independent validation of clinical relevance of the NPS-recognized S and L subgroups of HCC patients. We found (Fig. 2b) that there was significantly different survival between the 2 groups, S and L (mean survival in L subgroup was 7 months, mean survival in the S subgroup was 12 months. This 1.7 fold difference in means of the survival distributions was again strongly statistically significant, p < 10−5).
Influence of single clinical environmental parameter change on phenotypes
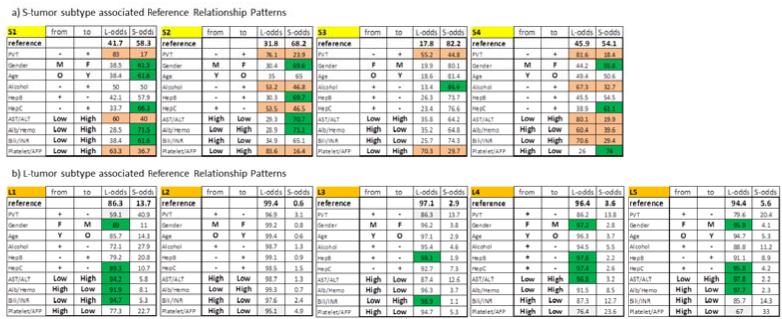
With the above results validating the NPS results and identifying HCC subtypes in terms of the clinical tumor biology and disease outcome characteristics (prognosis), we gained more detailed insight into the HCC heterogeneity, its factors and the role of the clinical environment interaction with the tumor by probing the clinically relevant details of the NPS classification model. The actual patient allocation into S or L HCC subtypes and the survival prognosis were derived by the 9-variable logistic regression model, which processed differences between the patients’ relationship profiles and respective RRPs. Our previous NPS analysis of the tumor and clinical environment interaction had shown that the HCC clinical landscape is separated into two main regions. One was (S), with smaller tumors having more variable relationship patterns of clinical parameters, and the other was (L), with more stringent clinical parameter relationship characteristics. In addition to this basic differentiation, our analysis provides further differentiation of these two subcategories, which is defined by the clinical landmark statuses of two classes of the nine RRPs. There are four RRP’s, which are located in the S-tumor region (Table 1a) and five RRPs (Table 1b) associated with L-tumors. Here “location” or “association” of RRP with a particular clinical state of RRPs is understood as the data-driven feature of the patient’s clinical data, revealed by the pattern-based information processing we introduced through NPS. The actual clinical profiles of HCC patients are unevenly distributed for subjects with typically S- or L-data patterns. In addition to the primary S/L division, there are additional heterogeneities in patient clinical data relationship patterns within these two main sub-categories and RRP’s represent the “focal points” of them. This role of RRPs, as validated here to be well-defined and characteristic descriptors of the tumor and clinical environment, allows us to obtain further insight by detailed analysis of the response to single parameter changes, into the NPS-derived subtype assignment of ‘S’ or ‘L’.
Table 1.
Results of systematic analysis of single-parameter change in a) S-tumor and b) L-tumor associated reference relationship patterns on the odds used to diagnose S or L HCC phenotype. In all panels, orange highlights the forbidden parameter changes, green highlights the changes that improve and white reduces, but not changing the odds for respective phenotypes after the parameter change (top row), all relatively to reference relationship patterns (top row). See text for details.
|
The advantage (see Fig.1) of this novel pattern-based NPS analysis paradigm is that single parameter changes induce extensive variation of the original relationship pattern, which brings a new level of clinical information and can be traced to the functional aspects of the environment-tumor interactions. Full results of this analysis are summarized in Table 1. We next give 2 representative examples demonstrating how we arrived at the main result of this analysis, namely, that there are three basic types of clinical consequences of these variations in patient tumor phenotype subgroup and hence in prognosis.
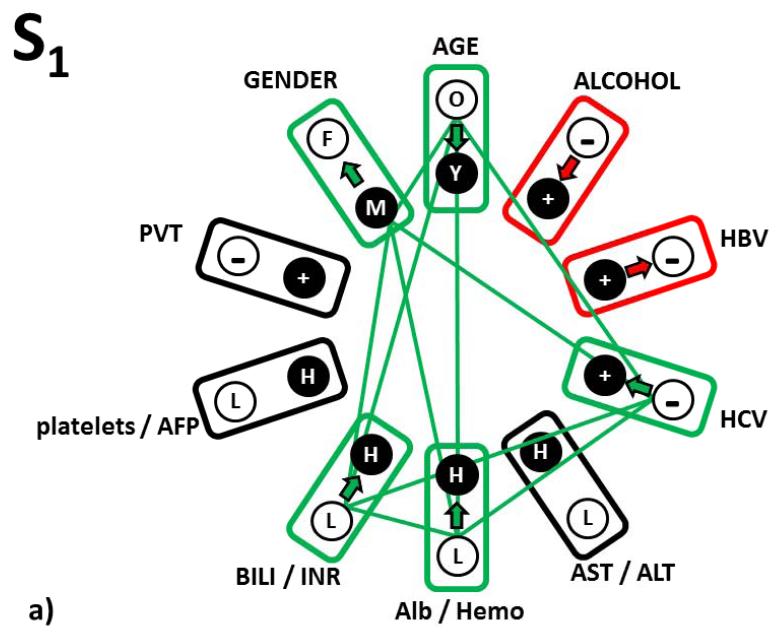
In a first example, the first panel of Table 1a defines the S1-subtype of the S-tumor phenotype. The clinical meaning of the RRP, that is the clinical pattern serving as a focus for the cluster of patients with this HCC subtype is found in the first (“from”) column of the S1 sub-table. This is a male patient, older than 55 years, with no self-reported alcoholism, Hep B antigen positive, Hep C antigen negative, AST < 4 IU/L and ALT < 3.23 IU/L, albumin < 4.0 g/dL, hemoglobin <14.9 g/dL, bilirubin < 1.5 mg/dL, INR<77 , platelets <195 × 103/dL and AFP < 29 000 ng/dL, PVT negative. The “reference” column of the panel indicates that for a patient whose actual clinical profile would be exactly identical with this S1 RRP, the odds for diagnosis of the S- and L- HCC tumor types are 58.3% for S and 41.7% for L, respectively (top, Reference row). This parameter pattern defines the association of S1 with S-tumor phenotype.
The second column (“to”) identifies a single clinical parameter state change from the original S1 level to the level indicated in this column, which we tested. Note that all remaining nine parameters were kept at their original levels in the S1 RRP. In the last two columns we reported the odds for L and S tumor subtypes, as they are computed after the tested clinical variable level change. This process was repeated systematically and independently for all ten single clinical variable level changes (these results are shown in the variable-labeled rows, below the Reference row of Table 1). Three types of response of the HCC classification to any single parameter change were found. We explain these three categories below and show them in graphical representation in Fig. 3
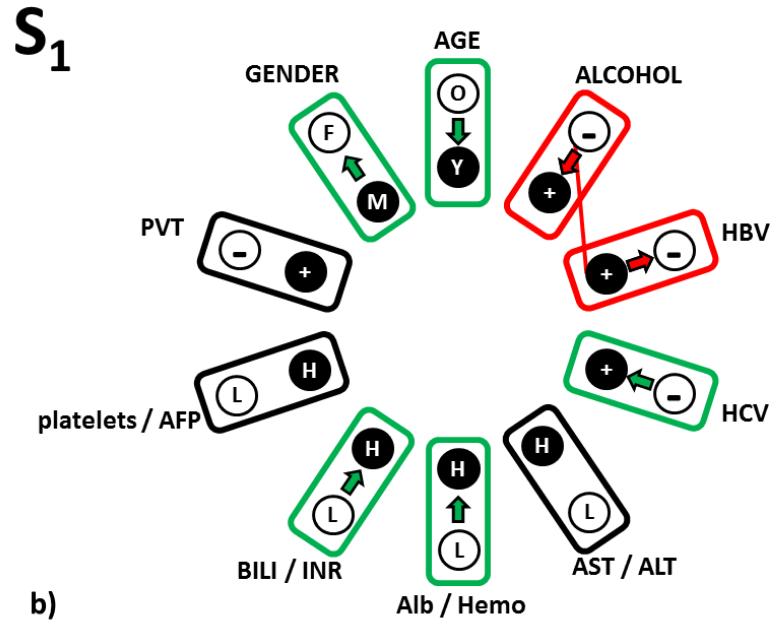
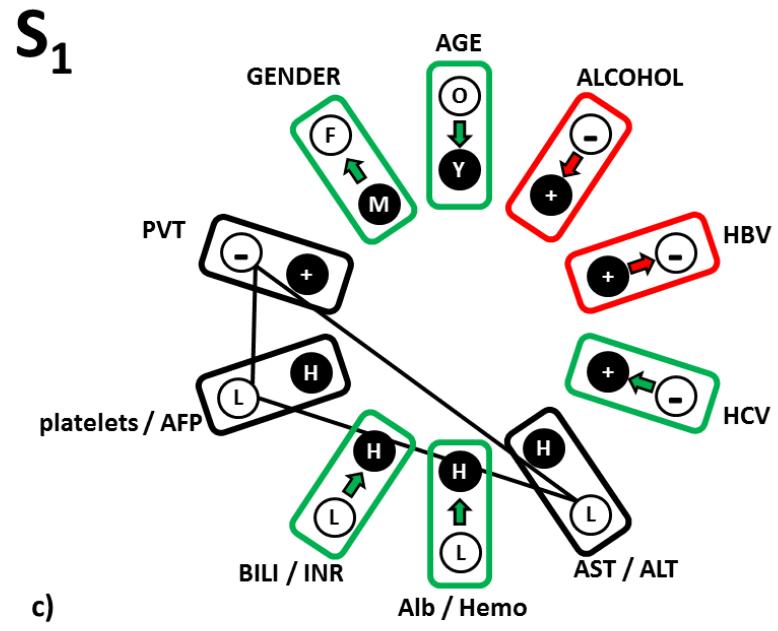
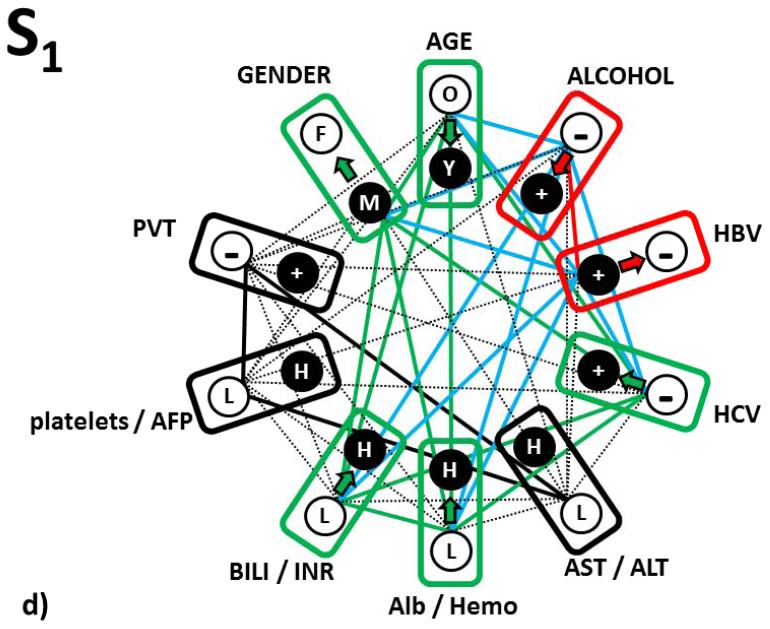
Figure 3.
Three types of response of the S1 reference relationship pattern to the single parameter change. a) Pattern relationships that responded to change by improving S-type diagnosis (green relationship edges); b) Pattern relationship that responded to change by moderately reducing the odds for S-type diagnosis (red relationship edge); c) Pattern relationships that are forbidden (see text for explanation – black relationship edges); d) Complete picture of all tree relationship types in the complete relationship pattern S1.
The first category is highlighted in green (Fig. 3a). These are changes that strengthen the identification of the S-tumor relative to the clinical pattern status identical to the RRP S1. For patients similar to S1 subtype of S-tumor and in the order of improving the odds of an Sdiagnosis, these changes are a) either increase hemoglobin level above 14.9 g/dL, or increase of albumin level above 4.0 g/dL or having both these levels high; b) hepatitis C antigen changing from negative to positive; c) individual changes from male to female, from patient older than 55 years to younger and levels of bilirubin and INR from levels lower than the above presented thresholds to levels higher in either of them or both, also increase the S-diagnosis odds in comparable manner. Thus, both macro- and micro-environmental changes in a single parameter, change the odds of such a patient being in the S or L phenotype.
The second category of odds responses to a single parameter change were left white in Table 1a (Fig 3b). If, in contrast to the original relationship pattern in S1, patient reports alcoholism or hepatitis B is not diagnosed, that decreases the odds but still identifies a patient as having S-tumor subtype.
The third category was highlighted in orange in Table 1a (Fig 3c). These are changes, which we interpret as forbidden, because they change the reference relationship pattern S1 in the HCC clinical landscape populated by actual personal clinical relationship patterns characteristic to S-tumor in such a way that the distances from this altered RRP are suddenly wrongly described by higher odds for the L-tumor subtype. These three “forbidden” single parameter changes (from absence to presence of PVT, the AST/ALT inflammation markers and platelet/AFP markers increasing from low to high levels) thus have to stay in the original S1-RRP defined levels. This category of single parameter changes therefore represents a relationship sub-pattern, which is the most characteristic for the S1 subtype of S-tumor.
In Fig. 3d we integrated these responses of the S1-S-tumor subtype prognosis changes into a complete scheme and also added relationships that go beyond the single-parameter ones to pairwise and higher order ones (this is possible because of additive terms in logistic regression equation computing the odds).
A second example of the influence of a single parameter change is from the third panel in Table 1a, showing properties of the S3 subtype of S-tumors. For this S-tumor subtype, there are only two characteristic parameters which are forbidden (from absence to presence of PVT and platelet/AFP markers changing from low to high, shown in red). Only a change of reported alcoholism to no alcoholism increases the S-tumor odds relatively to original S3-pattern result. All remaining allowed changes decreased the S-tumor odds, with micro-environmental inflammatory marker AST/ALT change from low to high and albumin/hemoglobin change from high to low, having the largest impact. Note that absence of PVT is a common non-variable and therefore the most characteristic feature of S-tumors (this emerged directly from the data relationship analysis). The low platelet/AFP levels are required for three out of four S-tumor subtypes.
Results of the systematic single parameter level variations for the five L-tumor subtypes (Table 1b) can be summarized as follows: the 10-variable relationship patterns associate with these more aggressive and worst survival prognosis tumors are very characteristic, resulting in a very clear diagnosis (all reference odds are close to 90 % and higher). With such very structured relationship patterns (out of many and many theoretically possible) that we identified in our NPS analysis, single parameter changes do not induce significant changes in the L-tumor characterization. Thus, in contrast to S-tumors, L-tumors are not amenable to subtype change as a result of single parameter changes.
DISCUSSION
It has been long-recognized in HCC studies that unlike many other tumors, prognosis depends upon both tumor and micro-environment factors (liver inflammation), as well as macro-environmental factors such as age and gender. In order to discuss these combinations of various factors, a variety of approaches have been taken23, such as multivariable regression, principal component analysis24-28 or neural networks27,29-32. Regression methods become too complicated for considering complete tumor-environment interactions; and the principal component and neural network analyses provide statistically significant associations for diagnosis and prognosis, which are difficult to interpret in simple clinical terms.
The motivation of our approach was to concentrate on tumor-environment interactions by a) designing an NPS to characterize these interactions using the correct number of RRP’s and b) extracting the diagnostic and prognostic information individually and quantitatively by comparing personal patterns of these interactions for individual patients to reference relationship patterns that have clear clinical interpretation. This approach was developed on a previous cohort showing that the HCC patients in that cohort could be described within 2 broad phenotypes, S and L, which differed significantly with respect to tumor mass. The significance of these two HCC phenotypes was validated here in two ways. First, without any change of the NPS model, the means of tumor mass distributions in this cohort were different with significance p<10−14. Second, the clinical importance of this is that overall survival was also significantly different between the 2 groups p<10−5. This significance of the two phenotypes for disease outcome allowed us to begin to interpret the results in more details.
Within these 2 phenotypes, we recognized 4 patterns within S and 5 patterns within L phenotype. Each phenotypic pattern comprised unique combination of relationship between the levels of 10 clinical parameters, which in turn represent different interactions between the tumor and environment factors. The NPS results shown that patient relationship patterns are unevenly distributed in the tumor-environment landscape with RRPs’ landmarks, such that there are patient that are by the nature of the complete relationship between their tumor and clinical environment interactions are related to one, but distant to most or all other reference relationship patterns. It is this heterogeneity that underpins the functionality of our approach and allows for even more detailed testing of the clinical significance of this result. We systematically changed individual single parameters in each of the 9 subgroups and then we examined the clinical consequences in terms of the resultant phenotypic assignation that resulted from the complex change in pattern of relationships between all the tumor and clinical environment parameters, consequent on that minimal level change in any one of them. This computational exercise provided interesting insight into the different nature of the two tumor phenotypes.
For each of the S phenotype patterns we found clinical parameters that cannot be changed from the original, reference levels defined in RRPs, together with clinical parameters whose leves can be varied to alternative levels. For example, presence of PVT+ was invariable or inadmissible in all four S subtypes, since its presence resulted in a relationship between the tumor and environmental parameters that were not observed within the training nor validation HCC patterns. By contrast, change in micro environmental parameters such as bilirubin (S1, S2 and S3) or AST (S2, S3), resulted in a change in the odds ratio within the S phenotype in some of the 4 S phenotypes (Table 1a).
By contrast, the characterization of L tumor phenotype did not require any invariable levels of any clinical parameter. It is solely the unique relationship patterns between majority of the clinical parameters, captured by the unique patterns L1-L5, that characterize the L-phenotype. This importance of tumor-clinical environment relationship pattern is such that change in an individual parameter had no significant effect on the L phenotype recognition.
A main result of our approach, by examining micro- and macro-environmental interactions using not arbitrary but the data- and disease defined RRPs, allowed us to through clinically exactly defined impact on the large number of tumor-clinical environment interactions just in the limited number of patterns. Most importantly, we demonstrated that tumor-clinical environment interaction patters explained how the same level of an individual parameter can have a different diagnostic and/or prognostic meaning within a different overall context. As an example, the baseline bilirubin level is low in S1 and S4, but high in S2 and S3, yet all of them are S subtype patterns. By careful analysis of these different relationship contexts, it is obvious that there is no simple (binary) clinical association to other parameters that would link the both high and low bilirubin level to the same tumor phenotype. We therefore believe that minimal meaningful clinical information for recognizing HCC phenotypes needs to use the complete parameter patterns and not individual parameter levels their simple combinations.
Returning back to the actual reference patterns, they also comprise previously known facts. For example, in the L phenotype, all reference patterns L1-L5 are PVT+, all platelet/AFP levels are high, and are all alcohol self-reporting, so that the tumor factors contributing to these phenotype landmarks with the worst survival prognosis are the most aggressive in the conventional clinical sense. Another example uses the fact that it has previously been shown the female gender macro-environmental influence is associated with a less aggressive HCC phenotype. This is fully compatible with our pattern based result for S phenotype RRPs single-parameter changes. In the S phenotype, only S3 incorporated female gender and the reference odds for S phenotype were the highest among all four S sub-phenotypes. In the remaining S characteristic patterns S1, S2 and S4, the impact of change in gender from male to female on the complete pattern of the tumor – clinical environment relationship pattern resulted in increasing S odds. Thus, PVT, platelets and female gender seem to have an overwhelming influence on phenotype.
If the biology underlying the levels of the assayed screening panel of clinical parameters includes processes that are clinically relevant for the status of the tumor microenvironment, then the change of paradigm in how the “conventional” information is processed will change how these apparently “simple” but extensively used information resources can start contributing to a “higher level” tumor microenvironment understanding. Using clinical profile patterns, removing the obscurity from them, having a non-statistical tool for how to take the standard screening data and convert them (without losing the study statistical power) into a new form of information, where the internal tumor growth factors are directly considered in the deterministic number of clinically well-characterized microenvironment context, provides just that necessary clinical landscape, from which more detailed and complex tumor microenvironment research (and its translation to bedside) can benefit most. After validating these insights in terms of the patient’s clinical relationship profile distances from the RRP’s landmarks, more detailed analysis of just that limited, but optimal number of landmark clinical statuses, around which patients with a given HCC subtypes are clustered, is possible. We can then, instead of reporting on the significance of age or gender or alcoholism, investigate the different prognostic and clinical impact on the patterns of all of these 3 parameters together as contributors to tumor biology. We have done this systematically, which has led to the identification of specific collections of clinical states and relationship sub-patterns that characterize individual sub-states of HCC and the characteristics of associated clinical environments.
There were clinical differences between the training set and the current US set. Most of patients from the validation study cohort were not diagnosed through screening, so that they tended to have more advanced disease than the training set patients. This clinical difference between the training and validation cohorts is seen in our results. Besides other evidence, we can see it primarily in the numbers of the recognized the S and L tumor subtypes. In the training set, we found nearly balanced fractions of patients in the L and S sub-cohorts (50.8% in L and 49.1 in S). In the current US patient validation cohort, we have identified 80.6% patients with L and 19.4% with S-tumor subtypes.
At the same time, in current validation US patients, we used the same thresholds for dichotomization of blood parameter pairs into low and high levels. In the training study, the definition of these parameters was strictly by tumor size terciles. After looking at the distributions of the patients with the high and low levels of all blood test pairs, we found that with this clinically more progressed disease cohort, the distribution of these low and high levels of the blood parameters are very close to tercile – related proportions of 2/3 patients with low parameter levels and 1/3 of patients with high clinical variable levels for all 4 blood test parameter pairs.
In one clinical micro/macro environment, a single parameter level can have one diagnostic meaning, while in another clinical environment the meaning of the same parameter level is completely different. Only by analyses such as this, which brings the blood test levels into the proper context of the data-captured clinical environment interactions, we can properly interpret all these levels, This in turn is one of the main reasons behind the ability of this approach in identifying S and L HCC tumor subtypes.
Acknowledgments
Grant support: ERZ-CZ LL1201 (CORES) to PP and NIH grant CA 82723 to BIC.
Abbreviations
- HCC
hepatocellular carcinoma
- AST/ALT
aspartate amino transferase/alanine amino transferase
- AFP
alpha-fetoprotein
- Hb
hemoglobin
- INR
international normalized ratio for prothrombin
- PVT
portal vein thrombosis
- NPS
Network Phenotyping Strategy
- RRP
reference relationship patterns
- Hep B
hepatitis B
- Hep C
hepatitis C
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer metastasis reviews. 1989;8:98–101. [PubMed] [Google Scholar]
- 2.Carr BI, Pancoska P, Branch RA. HCC in young adults. Hepato-gastroenterology. 2010;57:436–40. [PubMed] [Google Scholar]
- 3.Carr BI, Pancoska P, Branch RA. HCC in older patients. Digestive diseases and sciences. 2010;55:3584–90. doi: 10.1007/s10620-010-1177-6. [DOI] [PubMed] [Google Scholar]
- 4.Buch SC, Kondragunta V, Branch RA, Carr BI. Gender-based outcomes differences in unresectable hepatocellular carcinoma. Hepatology international. 2008;2:95–101. doi: 10.1007/s12072-007-9041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr BI, Pancoska P, Branch RA. Tumor and liver determinants of prognosis in unresectable hepatocellular carcinoma: a large case cohort study. Hepatology international. 2010;4:396–405. doi: 10.1007/s12072-009-9157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr BI, Guerra V. HCC and its microenvironment. Hepato-gastroenterology. 2013;60:1433–7. doi: 10.5754/hge121028. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez-Gea V, Toffanin S, Friedman SL, Llovet JM. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512–27. doi: 10.1053/j.gastro.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu SD, Ma YS, Fang Y, Liu LL, Fu D, Shen XZ. Role of the microenvironment in hepatocellular carcinoma development and progression. Cancer treatment reviews. 2012;38:218–25. doi: 10.1016/j.ctrv.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Yang JD, Nakamura I, Roberts LR. The tumor microenvironment in hepatocellular carcinoma: current status and therapeutic targets. Seminars in cancer biology. 2011;21:35–43. doi: 10.1016/j.semcancer.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 11.Chew V, Tow C, Teo M, et al. Inflammatory tumour microenvironment is associated with superior survival in hepatocellular carcinoma patients. Journal of hepatology. 2010;52:370–9. doi: 10.1016/j.jhep.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. The New England journal of medicine. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurokawa Y, Matoba R, Takemasa I, et al. Molecular-based prediction of early recurrence in hepatocellular carcinoma. Journal of hepatology. 2004;41:284–91. doi: 10.1016/j.jhep.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 14.Schrader J, Gordon-Walker TT, Aucott RL, et al. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011;53:1192–205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Utsunomiya T, Shimada M, Imura S, Morine Y, Ikemoto T, Mori M. Molecular signatures of noncancerous liver tissue can predict the risk for late recurrence of hepatocellular carcinoma. Journal of gastroenterology. 2010;45:146–52. doi: 10.1007/s00535-009-0164-1. [DOI] [PubMed] [Google Scholar]
- 16.Kuang DM, Zhao Q, Wu Y, et al. Peritumoral neutrophils link inflammatory response to disease progression by fostering angiogenesis in hepatocellular carcinoma. Journal of hepatology. 2011;54:948–55. doi: 10.1016/j.jhep.2010.08.041. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JP, Yan J, Xu J, et al. Increased intratumoral IL-17-producing cells correlate with poor survival in hepatocellular carcinoma patients. Journal of hepatology. 2009;50:980–9. doi: 10.1016/j.jhep.2008.12.033. [DOI] [PubMed] [Google Scholar]
- 18.Zhu AX, Duda DG, Sahani DV, Jain RK. HCC and angiogenesis: possible targets and future directions. Nature reviews Clinical oncology. 2011;8:292–301. doi: 10.1038/nrclinonc.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capece D, Fischietti M, Verzella D, et al. The inflammatory microenvironment in hepatocellular carcinoma: a pivotal role for tumor-associated macrophages. BioMed research international. 2013;2013:187204. doi: 10.1155/2013/187204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pancoska P, Carr BI, Branch RA. Network-based analysis of survival for unresectable hepatocellular carcinoma. Seminars in oncology. 2010;37:170–81. doi: 10.1053/j.seminoncol.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Pancoska P, Lu SN, Carr BI. Phenotypic Categorization and Profiles of Small and Large Hepatocellular Carcinomas. Journal of gastrointestinal & digestive system. 2013;(Suppl 12) doi: 10.4172/2161-069X.S12-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diestel R. Graph theory. 4th ed. Springer; Heidelberg; New York: 2010. [Google Scholar]
- 23.Dvorchik I, Demetris AJ, Geller DA, et al. Prognostic models in hepatocellular carcinoma (HCC) and statistical methodologies behind them. Current pharmaceutical design. 2007;13:1527–32. doi: 10.2174/138161207780765846. [DOI] [PubMed] [Google Scholar]
- 24.Cao W, Li J, Hu C, et al. Symptom clusters and symptom interference of HCC patients undergoing TACE: a cross-sectional study in China. Supportive care in cancer: official journal of the Multinational Association of Supportive Care in Cancer. 2013;21:475–83. doi: 10.1007/s00520-012-1541-5. [DOI] [PubMed] [Google Scholar]
- 25.Liu SY, Zhang RL, Kang H, Fan ZJ, Du Z. Human liver tissue metabolic profiling research on hepatitis B virus-related hepatocellular carcinoma. World journal of gastroenterology: WJG. 2013;19:3423–32. doi: 10.3748/wjg.v19.i22.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taleb I, Thiefin G, Gobinet C, et al. Diagnosis of hepatocellular carcinoma in cirrhotic patients: a proof-of-concept study using serum micro-Raman spectroscopy. The Analyst. 2013;138:4006–14. doi: 10.1039/c3an00245d. [DOI] [PubMed] [Google Scholar]
- 27.Virmani J, Kumar V, Kalra N, Khandelwal N. A comparative study of computer-aided classification systems for focal hepatic lesions from B-mode ultrasound. Journal of medical engineering & technology. 2013;37:292–306. doi: 10.3109/03091902.2013.794869. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Thiefin G, Gobinet C, et al. Profiling serologic biomarkers in cirrhotic patients via high-throughput Fourier transform infrared spectroscopy: toward a new diagnostic tool of hepatocellular carcinoma. Translational research: the journal of laboratory and clinical medicine. 2013 doi: 10.1016/j.trsl.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Chiu HC, Ho TW, Lee KT, Chen HY, Ho WH. Mortality predicted accuracy for hepatocellular carcinoma patients with hepatic resection using artificial neural network. The Scientific World Journal. 2013;2013:201976. doi: 10.1155/2013/201976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ying X, Han SX, Wang JL, et al. Serum peptidome patterns of hepatocellular carcinoma based on magnetic bead separation and mass spectrometry analysis. Diagnostic pathology. 2013;8:130. doi: 10.1186/1746-1596-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M, Yin F, Chen B, et al. Mortality risk after liver transplantation in hepatocellular carcinoma recipients: a nonlinear predictive model. Surgery. 2012;151:889–97. doi: 10.1016/j.surg.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 32.Shi HY, Lee KT, Lee HH, et al. Comparison of artificial neural network and logistic regression models for predicting in-hospital mortality after primary liver cancer surgery. PloS one. 2012;7:e35781. doi: 10.1371/journal.pone.0035781. [DOI] [PMC free article] [PubMed] [Google Scholar]