Abstract
Background
The purpose of this study was to identify the presence of rickettsia and hantavirus in wild rodents and arthropods in response to an outbreak of acute unidentified febrile illness among Indians in the Halataikwa Indian Reserve, northwest of the Mato Grosso state, in the Brazilian Amazon. Where previously surveillance data showed serologic evidence of rickettsia and hantavirus human infection.
Methods
The arthropods were collected from the healthy Indian population and by flagging vegetation in grassland or woodland along the peridomestic environment of the Indian reserve. Wild rodents were live-trapped in an area bordering the reserve limits, due the impossibility of capturing wild animals in the Indian reserve. The wild rodents were identified based on external and cranial morphology and karyotype. DNA was extracted from spleen or liver samples of rodents and from invertebrate (tick and louse) pools, and the molecular characterization of the rickettsia was through PCR and DNA sequencing of fragments of two rickettsial genes (gltA and ompA). In relation to hantavirus, rodent serum samples were serologically screened by IgG ELISA using the Araraquara-N antigen and total RNA was extracted from lung samples of IgG-positive rodents. The amplification of the complete S segment was performed.
Results
A total of 153 wild rodents, 121 louse, and 36 tick specimens were collected in 2010. Laguna Negra hantavirus was identified in Calomys callidus rodents and Rickettsia bellii, Rickettsia amblyommii were identified in Amblyomma cajennense ticks.
Conclusions
Zoonotic diseases such as HCPS and spotted fever rickettsiosis are a public health threat and should be considered in outbreaks and acute febrile illnesses among Indian populations. The presence of the genome of rickettsias and hantavirus in animals in this Indian reserve reinforces the need to include these infectious agents in outbreak investigations of febrile cases in Indian populations.
Keywords: Indian population, Rickettsia bellii, Rickettsia amblyommii, Laguna negra virus
Background
More than 800,000 Indians from 220 ethnic groups speaking 180 languages live in Brazil. Evaluation of indigenous health is very complex, due to demographic and epidemiological differences, especially with some groups that still live in relative isolation without interaction with the prevalent national society [1]. Social changes and economic and environmental factors continue to affect the profile of the Indians’ health, contributing to a higher occurrence of infectious diseases than in other ethnic groups [2,3].
In 2009, the Bureau of Health of the State of Mato Grosso (BHSMT) investigated an outbreak of an unknown acute febrile illness among Indians in the reserve located in the Parecis region, where over 172 cases of hantavirus cardiopulmonary syndrome (HCPS) have been identified since 1999. Blood samples from 59 of 530 apparently healthy Indians were collected; five (8%) were positive for anti-hantavirus IgG antibodies. In addition, as ticks and tick bites were reportedly prevalent in this Indian reserve, serological analysis was performed; IgG antibodies anti-spotted fever group rickettsia (SFGR), Bartonella spp., and Ehrlichia spp. were detected in sera from 12 (8%), five (8%), and four (6.8%) Indians, respectively. Based on these findings, the objective of this study was to collaborate with the health surveillance system and identify SFGR and hantavirus circulating in arthropods and wild rodents collected in this Indian area.
Methods
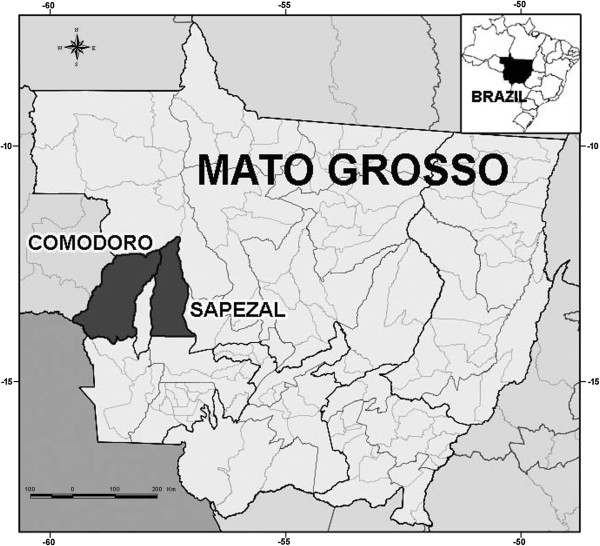
In 2010, in response to an outbreak of acute unidentified febrile illness among Indians in the Halataikwa Indian Reserve, northwest of the Mato Grosso state, a study was carried out in two areas of the Parecis micro-region of the Brazilian Amazon in Mato Grosso - municipalities of Comodoro (13° 39’ 46” S 59° 47’ 09” O) and Sapezal (13° 32’ 33” S 58° 48’ 51” O) (Figure 1).
Figure 1.
Map of the Mato Grosso state, central-western region of Brazil, showing the municipalities of Comodoro and Sapezal.
Arthropod samples
The arthropods were collected from the healthy Indian population and by flagging vegetation in grassland or woodland along the peridomestic environment of the Indian reserve, municipality of Comodoro, by the Indians’ own health service with the collaboration of BHSMT. The identification procedure was done on the basis of morphologic features using standard taxonomic keys [4] and the arthropods were subsequently shipped on dry ice to the Laboratory of Hantaviruses and Rickettsioses, Oswaldo Cruz Institute, FIOCRUZ.
Small-mammals samples
Eco-epidemiological study was conducted in an area bordering the reserve limits in the municipality of Sapezal, due the impossibility of capturing wild animals in the Indian reserve. The wild rodents were identified based on external and cranial morphology and karyotype [5-7]. Voucher specimens were deposited in the collection of the National Museum, Federal Universidad of Rio de Janeiro, RJ, Brazil. Blood and tissue samples from the wild rodents were obtained in accordance with recommended safety procedures [8], after authorization by the Brazilian Institute for the Environment and Renewable Natural Resources (IBAMA) under license number 60 8054/2008.
Rickettsia detection
DNA was extracted from spleen or liver samples of rodents and from invertebrate (tick and louse) pools, ranging from one (engorged adult) to 44 (larvae) specimens, by using a QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) following the instructions of the manufacturer. The polymerase chain reaction (PCR) was performed using oligonucleotide primers to amplify the partial citrate synthase gene (gltA) of genus Rickettsia (RpCS877 and RpCS1258) and outer membrane protein gene (ompA) fragments of spotted fever group Rickettsia—SFGR (Rr190-70 and Rr190-602), published previously [9,10].
Hantavirus detection
In relation to hantavirus, rodent serum samples were serologically screened by IgG ELISA using the Araraquara-N antigen [11]. Total RNA was extracted from lung samples of IgG-positive rodents using Trizol with the Purelink Micro-to-Midi Total RNA Purification System (Invitrogen®, San Diego, CA, USA). In addition, viral RNA was extracted from lung samples of rodents for which there were no blood samples. The amplification of the complete S segment was performed according to Guterres and collaborators [12].
DNA sequencing and phylogenetic analyses
For DNA purification, the Wizard®SV Gel and PCR Clean-Up System kit (Promega, Corp., Madison, WI, USA) was used according to the manufacturer’s recommendations, and strands were directly sequenced. In the sequencing reaction, the BigDye Terminator™ version 3.1 Cycle Sequencing® Kit (Applied Biosystems) was used according to the manufacturer’s recommendations in an automatic sequencer (Applied Biosystems, ABI PRISM 3130X model, Foster City, CA, USA). Nucleotide sequences were analyzed using MEGA5 software [13], and a consensus sequence was derived from contiguous sequences assembled with the same software.
Multiple sequence alignments were done with sequences obtained from this study and sequences from GenBank using the MUSCLE, in the SeaView version 4 software [14]. The rickettsia phylogenetic tree was constructed using the maximum likelihood (ML) implemented in MEGA5 software. The support for the tree nodes was calculated with 1,000 bootstrap replicates.
Phylogenetic relationships among the hantaviruses were estimated by the Bayesian Markov Chain Monte Carlo (MCMC) method implemented in MrBayes version 3.1.2 [15], using the GTR + G model of sequence evolution, as determined by the jModelTest version 2 [16]. The Bayesian analysis consisted of two simultaneous independent runs of 3 million MCMC generations (burn-in of 25%).
Results
Two ticks and 121 louse specimens, removed from the asymptomatic Indian population, were taxonomically classified as Amblyomma nymphs and Pediculus humanus (81 adults and 40 nymphs), respectively. Additionally, 34 ticks were collected by flagging vegetation in grassland or woodland along the peridomestic environment of the Indian reserve—10 Amblyomma cajennense (adults), 12 Amblyomma larvae, and 12 Amblyomma nymphs.
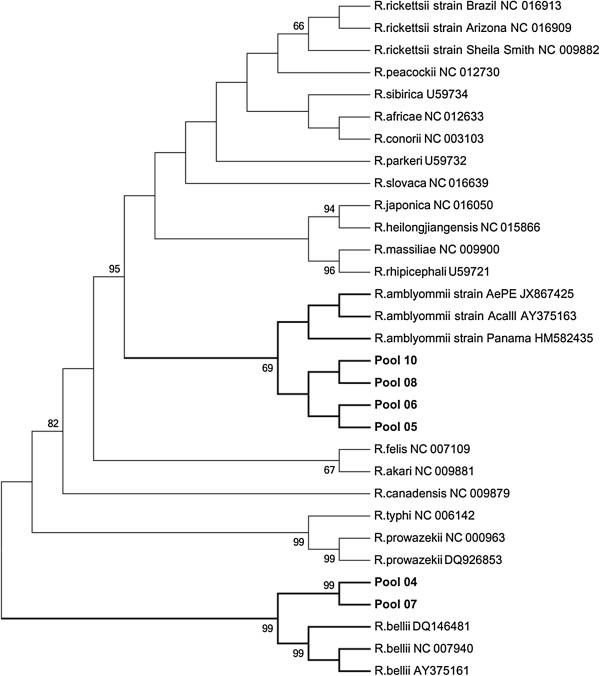
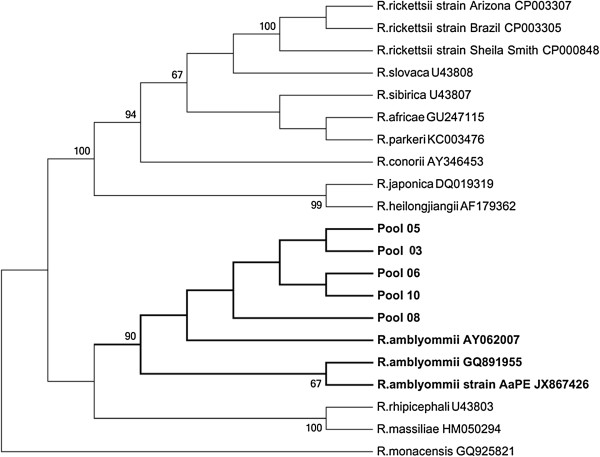
Seven of 16 (44%) tick DNA pools were found to be infected with Rickettsia; one was composed of Amblyomma larvae, two of Amblyomma nymphs, and four of A. cajennense (adults), all were collected on the ground along the peridomestic environment of the Indian reserve (Table 1). The sequences generated for gltA (381bp) and ompA genes (510bp) were analyzed using BLASTn searches of GenBank sequences. The DNA sequence analysis of five tick pools had 99% (ompA) and 99% (gltA) similarity to gene sequences of Rickettsia amblyommii [GenBank: GQ891955 and AY375163, respectively] and DNA of two tick pools had 99% (gltA) similarity to gene sequences of Rickettsia bellii [GenBank: DQ146481]. In the rickettsia phylogenetic tree of the gltA gene, the sequences obtained from pools 05, 06, 08, and 10 formed a monophyletic clade with sequences of R. amblyommii, while the sequences obtained from pools 04 and 07 formed a monophyletic clade with sequences of R. bellii available in the GenBank (Figure 2). In the phylogenetic tree of the ompA gene, five tick DNA pools (05, 06, 08, 10, and 03) formed a monophyletic clade with sequences of R. amblyommii (Figure 3). All 10 louse DNA pools were PCR negative.
Table 1.
Molecular analysis of ectoparasites removed from the asymptomatic Indian population and of the environment
| Pools | Species (amount) | Collected | gltA | OmpA |
|---|---|---|---|---|
| Pool 1 |
A. cajennense (01) |
Environment |
- |
- |
| Pool 2 |
A. cajennense (01) |
Environment |
- |
- |
| Pool 3 |
A. cajennense (01) |
Environment |
- |
Positive |
| Pool 4 |
Amblyomma larvae (12) |
Environment |
Positive |
- |
| Pool 5 |
Amblyomma nymphs (06) |
Environment |
Positive |
Positive |
| Pool 6 |
Amblyomma nymphs (05) |
Environment |
Positive |
Positive |
| Pool 7 |
A. cajennense (01) |
Environment |
Positive |
- |
| Pool 8 |
A. cajennense (01) |
Environment |
Positive |
Positive |
| Pool 9 |
A. cajennense (01) |
Environment |
- |
- |
| Pool 10 |
A. cajennense (01) |
Environment |
Positive |
Positive |
| Pool 11 |
A. cajennense (01) |
Environment |
- |
- |
| Pool 12 |
P. humanus nymphs (08) |
Human (Head) |
- |
- |
| Pool 13 |
P. humanus nymphs (08) |
Human (Head) |
- |
- |
| Pool 14 |
P. humanus nymphs (08) |
Human (Head) |
- |
- |
| Pool 15 |
P. humanus nymphs (08) |
Human (Head) |
- |
- |
| Pool 16 |
P. humanus nymphs (08) |
Human (Head) |
- |
- |
| Pool 17 |
P. humanus (04) |
Human (Head) |
- |
- |
| Pool 18 |
P. humanus (12) |
Human (Head) |
- |
- |
| Pool 19 |
Amblyomma nymphs (01) |
Human (body) |
- |
- |
| Pool 20 |
Amblyomma nymphs (01) |
Human (body) |
- |
- |
| Pool 21 |
Pediculus humanus (10) |
Human (Head) |
|
|
| Pool 22 |
P. humanus (11) |
Human (Head) |
- |
- |
| Pool 23 |
P. humanus (44) |
Human (Head) |
- |
- |
| Pool 24 |
Amblyomma nymphs (01) |
Environment |
- |
- |
| Pool 25 |
A. cajennense (01) |
Environment |
- |
- |
| Pool 26 | A. cajennense (01) | Environment | - | - |
Figure 2.
Maximum-likelihood phylogenetic tree of rickettsia based of the gltA gene partial sequence (381nt) and showing the phylogenetic placement of the novel sequences compared with rickettsia associated sequences. The tree was computed by using MEGA5 (http://www.megasoftware.net). The Tamura 3-parameter model with gamma- distributed rate heterogeneity and a proportion of invariant sites (T92 + G + I) was selected as the best fit evolutionary model according to the Bayesian information criterion calculated with MEGA5. The branch labels include GenBank accession number and species or strain.
Figure 3.
Maximum-likelihood phylogenetic tree of rickettsia based of the ompA gene partial sequence (510nt) and showing the phylogenetic placement of the novel sequences compared with rickettsia associated. The tree was computed by using MEGA5 (http://www.megasoftware.net). The Tamura 3-parameter model with gamma- distributed rate heterogeneity and a proportion of invariant sites (T92 + G + I) was selected as the best fit evolutionary model according to the Bayesian information criterion calculated with MEGA5. The branch labels include GenBank accession number and species or strain.
A total of 153 wild rodents were captured: Calomys callidus (65); Necromys lasiurus (60); Oligoryzomys utiaritensis (9); Rattus rattus (8); Hylaeamys megacephalus (4), Oligoryzomys mattogrossae (1), Oecomys sp. (3), and Calomys sp. (3). In order to investigate the presence of rickettsial infections in these small mammals, DNA was extracted from spleen or liver samples of 136 rodents for which there were tissue samples. Wild rodents were submitted to molecular analysis for genus Rickettsia and were PCR negative to gltA and ompA genes.
One hundred rodents were submitted to serology for anti-hantavirus antibodies, and one O. utiaritensis was reactive against the Araraquara-N antigen. The lung tissue sample of this rodent was submitted to molecular detection (RT-PCR), but it was not possible to recover the viral RNA. In addition, of 53 rodents for which there were no blood samples, one male specimen of the species C. callidus was RT-PCR positive. The comparison of the complete S segment with other known hantaviruses showed the highest degree of similarity, at 99% and 98% [GenBank: JX443686 and FJ816031, respectively], with Laguna Negra virus (LANV) from a human case of HCPS from Mato Grosso [17,18]. The sequence obtained was also similar (86%) to LANV [Genbank: AF005727] identified in Calomys laucha from Paraguay [19].
According to phylogenetic inferences, it was possible to check the formation of a well-supported monophyletic clade of our sequence against sequences of LANV available in GenBank. The sequences of LANV from Brazil are closely related to them (Figure 4).
Figure 4.
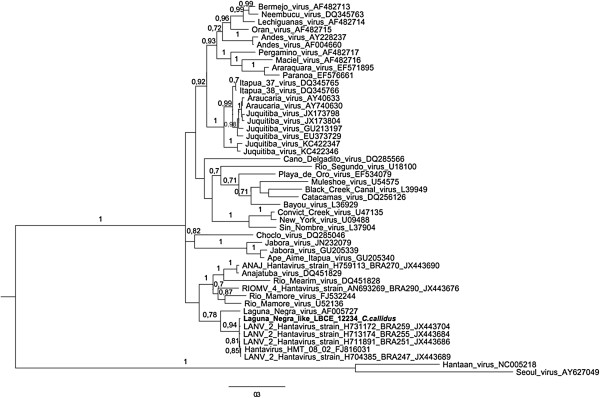
Phylogenetic relationships among hantaviruses based on Bayesian analysis of genetic distances generated from comparisons of a 950nt fragment of the nucleocapsid gene sequences. The scale bars indicate an evolutionary distance of 0.3 substitutions per position in the sequence. The numerical value ≥ 0.7 at the node indicates the posterior probability (pp) replicates that supported by the interior branch. The branch labels include GenBank accession number and viral species or strain.
Discussion
The role of ticks in the transmission of Rocky Mountain spotted fever (RMSF) was first documented by King and also by Ricketts in 1906, and since then several species of ticks have been identified as reservoirs of rickettsiae that are pathogenic and non-pathogenic to humans. In this study, R. bellii and R. amblyommii genomes were detected in A. cajennense ticks collected from the Indian environment. These species of the Rickettsia were previously identified in several species of Amblyomma sp. ticks in northern Brazil, Peru, and Argentina’s Chaco province [20-23]. Although R. amblyommii is considered a non-pathogenic rickettsia, a recent study suggested that this species might be responsible for several cases of RMSF-like disease [24]. As unknown acute febrile illness has been continually reported among Indians in the Brazilian northern region, and as serological tests may have detected cross-reactive antibodies to several SFGR, the possibility of occurrence of human spotted fever in this Indian area should be considered.
Rickettsia belli, a member of the ancestral rickettsiae group with unrecognized pathogenicity for humans, have been reported in larvae of Amblyomma varium found in infested Amazonian birds caught in Peru [25] and in a host-seeking male of A. tigrinum in Argentina, where ixodid ticks collected from vegetation and from humans and wild and domestic mammals in a rural area in the semi-arid Chaco province were found infected with an SFGR of unknown pathogenicity [22]. In this scenario, even if the pathogenicity of R. bellii and R. amblyommii has not been demonstrated or is unknown, the presence of ectoparasites infected with two species of rickettsiae in the Indian reserve may be a threat.
In relation to HCPS, since its first description in Navajo Indians in the United States, several studies have demonstrated the high incidence of hantavirus infection in the Indian populations throughout the American continent [26]. In 1998, a study conducted in Indian communities in northern Argentina and western Paraguay showed seroreactivity to hantavirus antibodies of 17.1% and 40.4%, respectively [27]. Later, two additional studies on hantavirus infection in the Indian population, also conducted in Paraguay, one in the western region and another in the eastern region, showed prevalence rates of 45.2% and 17.8%, respectively, higher rates than are usually observed in the general population (3.5%, 4.7%). In this sense, their habits, the forests or wild environments in which they dwell, their precarious housing with food storage in places that promote contact with animals and their excreta, among other factors, subject Indians to a greater exposure to hantavirus [28-31].
Our study also revealed the presence of LANV in a C. callidus rodent captured in an area bordering the reserve limits in the Sapezal municipality. Genetic analysis revealed the highest degree of similarity, at 99% and 98% with LANV (GenBank: JX443686 and FJ816031, respectively), with human cases of HCPS from Mato Grosso [17,18,32]. The LANV had been first described as a cause of HCPS in the Chaco region, Paraguay, in 1997 in a study that also identified the Calomys laucha rodent as the primary reservoir of this virus in Paraguay [19]. In 2004, LANV was identified for the first time in Argentina, recovered from human cases and from C. callosus samples. The high sequence identity between human and rodent samples implicated C. callosus as the primary rodent reservoir for LANV in Argentina [33]. Subsequently, in 2005, in the city of Concepción, Bolivia, where a non-fatal case of HCPS was reported, LANV was identified in C. callosus rodents. The sequencing reaction of the amplified segment showed an 87-88% similarity with LANV and a 99% match with the viral sequences obtained from the patient with HCPS in that region [18]. A recently published study associated LANV with HCPS in Mato Grosso state, Brazil, and cited a previously unidentified potential host, the C. callidus rodent corroborating our findings.
In our study, it was possible to perform the sequencing of a complete S segment of LANV in Brazil. The identification of LANV associated with another rodent species of the genus Calomys reinforces the idea that many hantaviruses, including LANV, cannot be strictly associated with only one species of rodent reservoir. The capacity of C. callidus to harbor LANV and its role as a reservoir are still unclear and new studies need to be conducted in order to better understand the relationship dynamics involving C. callidus and LANV.
Conclusions
Zoonotic diseases such as HCPS and spotted fever rickettsiosis are a public health threat and should be considered in outbreaks and acute febrile illnesses among Indian populations. The interrelationship of the Indian population with rodents and arthropods in forests or wild environments and their precarious housing, among other factors, increases the risk of occurrence of this zoonosis with high mortality rates. Therefore, our study reinforces the importance of the knowledge concerning the geographical distribution and prevalence of zoonotic diseases in Indian reserve areas.
Competing interests
No competing financial interests exist. The authors have no conflicts of interest or disclosures to make concerning this work.
Authors’ contributions
JFF, IA, SJ, ABMVA, VCS, AVGMV performed tick and rodents field study and revised the manuscript. LBL and AG lab experiments, processed the data, and drafted the manuscript. TR, RCO, MAMG and JF conducted lab experiments, processed the data, and revised the manuscript. CRB and PSD performed identification of the rodents and revised the manuscript. JDB performed identifications of the arthropods and revised the manuscript. ERSL contributed to study design, field study, data analysis and interpretation, and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Lívia de Barros Lopes, Email: liviabl@ig.com.br.
Alexandro Guterres, Email: guterres@ioc.fiocruz.br.
Tatiana Rozental, Email: rozental@ioc.fiocruz.br.
Renata Carvalho de Oliveira, Email: reoliveira@ioc.fiocruz.br.
Maria Angélica Mares-Guia, Email: amguia@ioc.fiocruz.br.
Jorlan Fernandes, Email: jorlan@ioc.fiocruz.br.
José Ferreira Figueredo, Email: figueredocase@gmail.com.
Inês Anschau, Email: ines_sanschau@yahoo.com.br.
Sebastião de Jesus, Email: lnccbio@hotmail.com.
Ana Beatriz M V Almeida, Email: anabiavasconcelos@gmail.com.
Valéria Cristina da Silva, Email: guterres_rj@yahoo.com.br.
Alba Valéria Gomes de Melo Via, Email: albagomes@gmail.com.
Cibele Rodrigues Bonvicino, Email: cibelerb@inca.fiocruz.br.
Paulo Sérgio D’Andrea, Email: dandrea@ioc.fiocruz.br.
Jairo Dias Barreira, Email: jairobarreira@bol.com.br.
Elba Regina Sampaio de Lemos, Email: elemos@ioc.fiocruz.br.
Acknowledgements
The authors thank the Public Health Secretary of the state o Mato Grosso for their assistance in the rodent capture and Indian blood collection. Financial support: Fiocruz and CAPES.
References
- National Foundation of Indian. [ http://www.funai.gov.br/index.php/indios-no-brasil/quem-sao]
- Coimbra JCEA, Santos RV. In: Lost Parad Ethics Res Publ. Salzano FM, Hurtado AM, editor. New York, USA: Oxford University Press; 2004. Emerging health needs and epidemiological research in indigenous peoples in Brazil; pp. 89–109. [Google Scholar]
- Santos RV, Coimbra CEA. , JrAs causas sociais das iniquidades em saúde no Brasil. Relatório final da Comissão Nacional sobre Determinantes Sociais da Saúde. 2013. pp. p. 121–p. 124. Available from: http://www.cndss.fiocruz.br/pdf/home/relatorio.pdf.
- Aragão H Fonseca F Notas de Ixodologia. VIII. Lista e chave para os representantes da fauna ixodológica brasileira Mem Inst Oswaldo Cruz 196159115–130.13861962 [Google Scholar]
- Bonvicino CR, D’Andrea PS, Cerqueira R, Seuánez HN. The chromosomes of Nectomys (Rodentia, Cricetidae) with 2n = 52, 2n = 56, and interspecific hybrids (2n = 54) Cytogenet Cell Genet. 1996;73:190–193. doi: 10.1159/000134337. [DOI] [PubMed] [Google Scholar]
- Bonvicino CR, Otazu IB, D’Andrea PS. Karyologic evidence of diversification of the genus Thrichomys (Rodentia, Echimyidae) Cytogenet Genome Res. 2002;97:200–204. doi: 10.1159/000066613. [DOI] [PubMed] [Google Scholar]
- Bonvicino CR, Moreira MA. Molecular phylogeny of the genus Oryzomys (Rodentia: Sigmodontinae) based on cytochrome b DNA sequences. Mol Phylogenet Evol. 2001;18:282–292. doi: 10.1006/mpev.2000.0878. [DOI] [PubMed] [Google Scholar]
- Mills JN, Childs JE, Ksiazek TG, Peters CJ. Methods for Trapping and Sampling Small Mammals for Virologic Testing. Atlanta, GA: Departament of Health and Human Services; 1995. p. 71. [Google Scholar]
- Wood DO, Williamson LR, Winkler HH, Krause DC. Nucleotide sequence of the Rickettsia prowazekii citrate synthase gene. J Bacteriol. 1987;169:3564–3572. doi: 10.1128/jb.169.8.3564-3572.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnery RL, Spruill CL, Plikaytis BD. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol. 1991;173:1576–1589. doi: 10.1128/jb.173.5.1576-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo LTM, Moreli ML, Borges AA, de Figueiredo GG, Badra SJ, Bisordi I, Suzuki A, Capria S, Padula P. Evaluation of an enzyme-linked immunosorbent assay based on Araraquara virus recombinant nucleocapsid protein. Am J Trop Med Hyg. 2009;81:273–276. [PubMed] [Google Scholar]
- Guterres A, de Oliveira RC, Fernandes J, D’Andrea PS, Bonvicino CR, Bragagnolo C, Guimarães GD, Almada GL, Machado RR, Lavocat M, Elkhoury MDR, Schrago CG, de Lemos ERS. Phylogenetic analysis of the S segment from Juquitiba hantavirus: Identification of two distinct lineages in Oligoryzomys nigripes. Infect Genet Evol. 2013;18:262–268. doi: 10.1016/j.meegid.2013.05.027. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Raboni SM, De Borba L, Hoffmann FG, De Noronha L, Azevedo, Marin Luize Viola Carstensen S, Mazzarotto GACA, Bordignon J, Nunes C. Evidence of circulation of Laguna Negra-like hantavirus in the Central West of Brazil: Case report. J Clin Virol. 2009;45:153–156. doi: 10.1016/j.jcv.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Firth C, Tokarz R, Simith DB, Nunes MRT, Bhat M, Rosa EST, Medeiros DBA, Palacios G, Vasconcelos PFC, Lipkin WI. Diversity and distribution of hantaviruses in South America. J Virol. 2012;86:13756–13766. doi: 10.1128/JVI.02341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Peters CJ, Nichol ST. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238:115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- Labruna MB, Whitworth T, Bouyer DH, McBride J, Camargo LMA, Camargo EP, Popov V, Walker DH. Rickettsia bellii and Rickettsia amblyommii in Amblyomma ticks from the State of Rondônia, Western Amazon, Brazil. J Med Entomol. 2004;41:1073–1081. doi: 10.1603/0022-2585-41.6.1073. [DOI] [PubMed] [Google Scholar]
- Ogrzewalska M, Uezu A, Labruna MB. Ticks (Acari: Ixodidae) infesting wild birds in the Atlantic Forest in northeastern Brazil, with notes on rickettsial infection in ticks. Parasitol Res. 2011;108:665–670. doi: 10.1007/s00436-010-2111-8. [DOI] [PubMed] [Google Scholar]
- Tomassone L, Nuñez P, Ceballos LA, Gürtler RE, Kitron U, Farber M. Detection of “Candidatus Rickettsia sp. strain Argentina” and Rickettsia bellii in Amblyomma ticks (Acari: Ixodidae) from Northern Argentina. Exp Appl Acarol. 2010;52:93–100. doi: 10.1007/s10493-010-9339-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labruna MB, Pacheco RC, Nava S, Brandão PE, Richtzenhain LJ, Guglielmone AA. Infection by Rickettsia bellii and Candidatus “Rickettsia amblyommii” in Amblyomma neumanni ticks from Argentina. Microb Ecol. 2007;54:126–133. doi: 10.1007/s00248-006-9180-3. [DOI] [PubMed] [Google Scholar]
- Apperson CS, Engber B, Nicholson WL, Mead DG, Engel J, Yabsley MJ, Dail K, Johnson J, Watson DW. Tick-borne diseases in North Carolina: is “Rickettsia amblyommii” a possible cause of rickettsiosis reported as Rocky Mountain spotted fever? Vector Borne Zoonotic Dis. 2008;8:597–606. doi: 10.1089/vbz.2007.0271. [DOI] [PubMed] [Google Scholar]
- Ogrzewalska M, Literak I, Cardenas-Callirgos JM, Capek M, Labruna MB. Rickettsia bellii in ticks Amblyomma varium Koch, 1844 from birds in Peru. Ticks Tick Borne Dis. 2012;3:254–256. doi: 10.1016/j.ttbdis.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- Ferrer JF, Jonsson CB, Esteban E, Galligan D, Basombrio MA, Peralta-Ramos M, Bharadwaj M, Torrez-Martinez N, Callahan J, Segovia A, Hjelle B. High prevalence of hantavirus infection in Indian communities of the Paraguayan and Argentinean Gran Chaco. Am J Trop Med Hyg. 1998;59:438–444. doi: 10.4269/ajtmh.1998.59.438. [DOI] [PubMed] [Google Scholar]
- Ferrer JF, Galligan D, Esteban E, Rey V, Murua A, Gutierrez S, Gonzalez L, Thakuri M, Feldman L, Poiesz B, Jonsson C. Hantavirus infection in people inhabiting a highly endemic region of the Gran Chaco territory, Paraguay: association with Trypanosoma cruzi infection, epidemiological features and haematological characteristics. Ann Trop Med Parasitol. 2003;97:269–280. doi: 10.1179/000349803235001903. [DOI] [PubMed] [Google Scholar]
- Chu Y-K, Owen RD, Gonzalez LM, Jonsson CB. The complex ecology of hantavirus in Paraguay. Am J Trop Med Hyg. 2003;69:263–268. [PubMed] [Google Scholar]
- Souza WM, Machado AM, Figueiredo LT, Boff E. Serosurvey of hantavirus infection in humans in the border region between Brazil and Argentina. Rev Soc Bras Med Trop. 2011;44:131–135. doi: 10.1590/S0037-86822011005000013. [DOI] [PubMed] [Google Scholar]
- Mendes WS, da Silva AA, Neiva RF, Costa NM, Assis MS, Vidigal PM, Leite M, Rosa ES, Medeiros DB, Simith D, Vasconcelos PF. Serologic survey of hantavirus infection, Brazilian Amazon. Emerg Infect Dis. 2010;16:889–891. doi: 10.3201/eid1605.090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis S, Garcia J, Pini N, Calderón G, Ramírez J, Bravo D, St Jeor S, Ripoll C, Bego M, Lozano E, Barquez R, Ksiazek TG, Enria D. Hantavirus pulmonary syndrome in northwestern Argentina: circulation of Laguna Negra virus associated with Calomys callosus. Am J Trop Med Hyg. 2004;71:658–663. [PubMed] [Google Scholar]
- Travassos Da Rosa ES, Medeiros DBA, Nunes MRT, Simith DB, Pereira ADS, Elkhoury MR, Santos ED, Lavocat M, Marques AA, Via AVG, Kohl VA, Terças ACP, D’Andrea P, Bonvícino CR, Sampaio De Lemos ER, Vasconcelos PFC. Molecular epidemiology of Laguna Negra virus, Mato Grosso State, Brazil. Emerg Infect Dis. 2012;18:982–985. doi: 10.3201/eid1806.110948. [DOI] [PMC free article] [PubMed] [Google Scholar]