Abstract
During postnatal development the heart undergoes a rapid and dramatic transition to adult function through transcriptional and post-transcriptional mechanisms, including alternative splicing (AS). Here we perform deep RNA-sequencing on RNA from cardiomyocytes and cardiac fibroblasts to conduct a high-resolution analysis of transcriptome changes during postnatal mouse heart development. We reveal extensive changes in gene expression and AS that occur primarily between postnatal days 1 and 28. Cardiomyocytes and cardiac fibroblasts show reciprocal regulation of gene expression reflecting differences in proliferative capacity, cell adhesion functions, and mitochondrial metabolism. We further demonstrate that AS plays a role in vesicular trafficking and membrane organization, These AS transitions are enriched among targets of two RNA-binding proteins, Celf1 and Mbnl1, which undergo developmentally regulated changes in expression. Vesicular trafficking genes affected by AS during normal development (when Celf1 is down-regulated) show a reversion to neonatal splicing patterns after Celf1 re-expression in adults. Short-term Celf1 induction in adult animals results in disrupted transverse tubule organization and calcium handling. These results identify potential roles for AS in multiple aspects of postnatal heart maturation, including vesicular trafficking and intracellular membrane dynamics.
The heart is the first organ to form and function during vertebrate embryogenesis1. The first four postnatal weeks involve a period of extensive physiological remodeling with dynamic changes as the fetal heart adapts to birth and converts to adult function. This transition occurs through transcriptional and post-transcriptional mechanisms, including coordinated networks of alternative splicing (AS)1–4.
Human and rat hearts are composed of 66% cardiac fibroblasts (CF), 30% cardiomyocytes (CM), and 4% endothelial and vascular smooth muscle cells5–7. Studies differ regarding adult mouse heart composition. While Soonpaa et al. reported that CF account for 86% of cells8, a recent analysis demonstrated a composition of 26% CF, 56% CM, and 18% non-CM and non-CF9. However, CM comprise ~75% of the tissue volume in mammals7. CM generate the contraction force and CF form the mechanical scaffold required for effective pumping10. CM and CF communicate through multiple signaling mechanisms and through extracellular-matrix (ECM)11. Other CF functions include response to cardiac injury12 and electrical isolation of different regions of the cardiac conduction system13. By postnatal day 7 (PN7), CM lose proliferative capacity and heart size increases due to CM hypertrophy14–15. Limited microarray analysis of mRNA expression in freshly isolated CM and CF showed that while certain genes are highly expressed in CM, many growth factors, cytokines, and ECM genes are more highly expressed in CF16. Overall, the published data address a limited number of gene expression changes in CM and CF during development, and notably, do not provide AS information.
High-throughput studies of AS and gene expression regulation have primarily focused on differences between tissues, normal versus pathological conditions, or cultured cells. A small set of reports have addressed AS and gene expression changes during normal physiological transitions17–21. Development provides an outstanding opportunity to identify coordinated AS regulation critical for physiological transitions from embryonic to adult functions. Previously, we showed that genes that undergo AS regulation during heart development produce transitions from embryonic to adult protein isoforms largely without changes in overall transcript levels, presenting a new paradigm for understanding developmentally regulated gene expression in heart3. Nearly half of the AS transitions identified in mouse are conserved during post-hatch chicken heart development, suggesting highly conserved functions for splicing-mediated isoform transitions3.
In the present study, we analyzed AS and gene expression transitions regulated during postnatal mouse heart development using mRNA deep sequencing (RNA-seq)22. To gain insight into the diversity of cell type-specific transitions, we performed RNA-seq using freshly isolated CF and CM from a developmental time course. The results revealed that most gene expression and AS changes occurs within the first four weeks after birth, and that CM and CF exhibit reciprocal transitions in expression of specific functional categories (proliferation, cell adhesion, cytokines-chemotaxis, metabolism, transcription regulation). Interestingly, we found that genes involved in vesicular trafficking and membrane organization are regulated by AS during postnatal CM development. These AS changes are enriched as targets of the CUGBP, ELAV-Like family (Celf) and Muscleblind-like (Mbnl) RNA-binding protein families, both of which are involved in AS and are regulated during postnatal heart development3,23–24. In the heart, vesicular trafficking-related AS transitions likely impact ligand/growth factor uptake, ion channels dynamics, and/or postnatal formation of the sarcoplasmic reticulum (SR) and transverse tubules (T-tubules), crucial processes for excitation-contraction coupling (ECC) that are established by PN3025. We show that re-expression of CELF1 in adults specifically in CM results in altered T-tubule structure and mis-regulated calcium handling consistent with alterations associated with re-expression of fetal splicing patterns.
RESULTS
Extensive transcriptome changes during postnatal development
RNA-seq was performed using RNA from mouse ventricles isolated at five time points: E17, PN1, PN10, PN28, and adult (PN90). cDNA libraries were prepared after ribosomal RNA (rRNA) depletion for 100 bp paired-end reads using the Illumina HiSeq2000. We obtained >150 million read pairs per sample, >80% of which mapped the mouse genome (Supplementary Table 1).
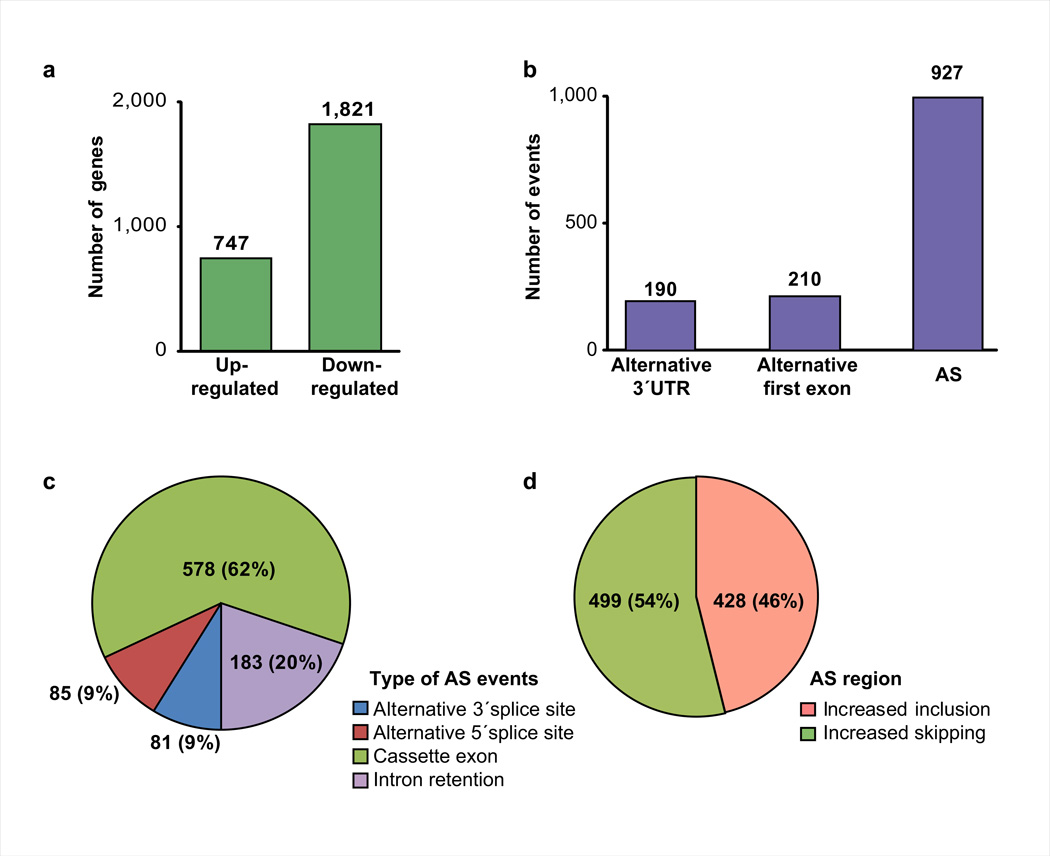
We identified 2,568 differentially expressed genes (≥2.0 fold, FDR 0.01, Supplementary Material) between E17-adult: 747 were up-regulated and 1,821 down-regulated (Fig. 1a). Analysis of mRNA isoforms whose percent spliced in (PSI)26 values changed ≥20% (ΔPSI≥20%) between E17-adult identified 927 AS events, 190 alternative 3´ untranslated regions (UTRs), and 210 alternative first exons (Fig. 1b). Postnatal AS transitions were predominantly cassette exons (62%), while alternative 3´ and 5´ splice sites each represented 9%. We observed a relatively high proportion of transitions (20%) involving intron retention with roughly equal proportions of events exhibiting increased inclusion or exclusion of the variable regions during development (Fig. 1c–d).
Figure 1. Extensive gene expression and RNA processing changes occur during heart development.
a–b. Differentially expressed genes (a) and alternative RNA processing between E17-adult (b). c. AS events categorized by pattern. d. Proportion of skipping/inclusion of the AS regions. AS: alternative splicing.
Extensive remodeling within the first four weeks after birth
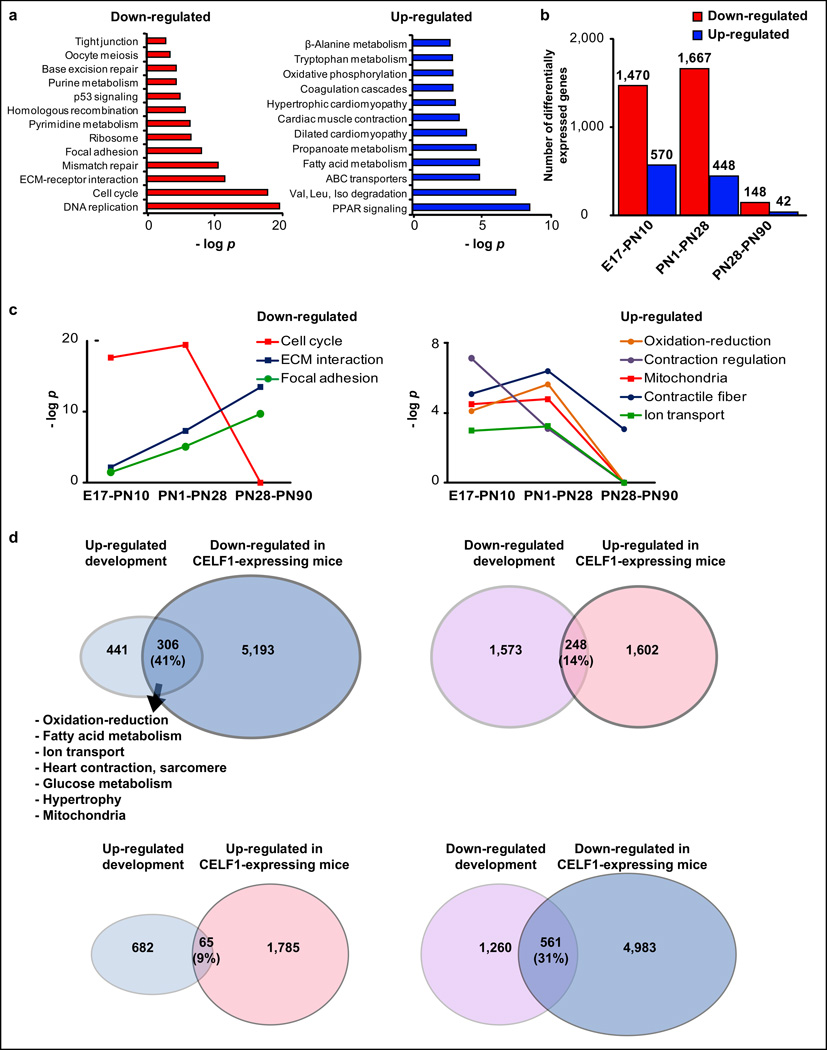
Gene ontology (GO) analysis of down-regulated genes between E17-adult in ventricles showed a clear enrichment (p<1E-10) in categories related with cell cycle and DNA replication (Fig. 2a-left). As ~75% of ventricular RNA originates from CM7, this is consistent with the loss of CM proliferative activity by PN714,15. Up-regulated genes between E17-adult showed enrichment in categories related to mitochondria, fatty acid metabolism, and oxidation-reduction processes (Fig. 2a-right) consistent with increased mitochondrial number and the metabolic switch from carbohydrates to fatty acids, a hallmark of the fetal to adult CM metabolic transition27,28.
Figure 2. Gene expression regulation occurs primarily before PN28.
a. GO analysis (KEGG-pathways) of down- and up-regulated genes between E17-adult (ventricles). b. Differentially expressed genes between E17-PN10, PN1–PN28, and PN28–PN90 (ventricles). c. Significance of KEGG-pathways (−log p) were plotted against time windows for specific categories. d. Venn diagrams with developmentally regulated genes and those regulated after CELF1 re-expression on adult mice (RNA-seq data). GO analysis of the intersected genes (summary). ECM: extracellular-matrix.
Analysis of three time windows (E17-PN10, PN1-PN28, PN28-PN90) revealed that the majority of gene expression transitions occurred before PN28 (Fig. 2b). Enrichment analysis in each period showed that down-regulation of cell cycle related genes occurred before PN28 (−log p dropped off after PN28) (Fig. 2c-left) in agreement with the loss of CM proliferative capacity early after birth. Similarly, up-regulated genes related to mitochondria, fatty acid metabolism, and oxidation-reduction were more enriched before PN28 (Fig. 2c-right). In contrast, categories such as focal adhesion and ECM-receptor interaction were more enriched after PN28 in down-regulated genes (-log p increased after PN28) (Fig. 2c-left). Detailed analysis of individual cell cycle and ECM genes confirmed temporal differences in down-regulation. The majority of ECM related genes maintained high mRNA levels within the first ten days after birth, dropped off at PN28 and decreased further in adults, while 81% of cell cycle genes were strongly down-regulated before PN10 (Supplementary Fig. 1a–b). The 3´ UTRs of down-regulated ECM- and cell cycle-related genes were computationally analyzed for putative microRNA (miRNA) miR-15 and miR-29 seed sequences. While 41% of the down-regulated ECM genes were predicted to be miR-29 targets, 33% of the down-regulated cell cycle genes were predicted to be miR-15 targets. These results suggest that a possible mechanism for the distinct patterns of cell cycle and ECM down-regulation could involve miRNAs since these transcripts are well established targets of developmentally up-regulated miRNAs such as miR-15 and miR-29 families29–32.
Celf1 loss correlates with increased expression of gene sets
To investigate a mechanism of up-regulation during postnatal development, we explored a potential role for Celf1, an RNA-binding protein down-regulated ten-fold during the first two weeks after birth3. Celf1 binds to GU-rich motifs within introns regulating AS3,33 and within 3' UTRs promoting mRNA decay34,35. To identify genes that are responsive to Celf1 expression during postnatal development, we used previously described tetracycline-inducible transgenic mice (TRECUGBP1/MHC) to induce human CELF1 expression in CM33. RNA-seq was performed using ventricles of adult TRECUGBP1/MHC and control (MHC) littermates after 12, 24, 72 hours or 7 days of induction. A high fraction (41%) of the developmentally up-regulated genes was down-regulated within 72 hours of exogenous CELF1 re-expression in adults (Fig. 2d-left). While a large number of genes are down-regulated after CELF1 induction (5,499), the majority (93%) of them shows a relatively small change (≤2.0 fold, after 72 hours). In contrast, a high fraction (78%) of the 306 overlapping genes (up-regulated during development and down-regulated by CELF1 expression) shows a fold change ≥2.5, suggestive of a physiological response. Developmentally up-regulated genes responsive to CELF1 (the 306 genes) were enriched in oxidation-reduction pathways, fatty acid metabolism, mitochondria, and sarcomere related categories. By contrast, only 14% of developmentally down-regulated genes were affected by CELF1 re-expression (Fig. 2d), suggesting that a percentage of the developmentally up-regulated mRNAs are destabilized by CELF1. A high proportion of the developmentally down-regulated genes overlapped with the down-regulated genes after CELF1 induction (Fig. 2d, bottom right). However, CELF1 down-regulated genes show only 31% overlap with developmental down-regulated genes compared to 41% overlap with developmentally up-regulated genes suggesting that Celf1 postnatal down-regulation stabilizes a large subset of mRNAs. It is also likely that there are secondary consequences to CELF1 re-expression (such as effects on transcription factor expression) resulting in both positive and negative effects on mRNA levels. To further investigate the potential CELF1 role in regulating mRNA stability during postnatal development, we analyzed the 3´ UTRs of 58 genes in the oxidation-reduction and mitochondria categories for CELF1-binding motifs (Supplementary Fig. 1c–d). The GU-rich motif shown in Supplementary Fig. 1c was significantly enriched (E=2.5E-05) and present in 24% of the analyzed 3´ UTRs. A similar GU-rich motif was found by CLIPZ with a high level of enrichment (Supplementary Fig. 1d). CLIPZ and MEME analysis also identified enrichment of polyA and GGA motifs (Supplementary Fig. 1d, and data not shown). These results suggest that postnatal Celf1 down-regulation promotes up-regulation of genes within specific functional pathways through mRNA stabilization.
Transcriptome changes in CM and CF during development
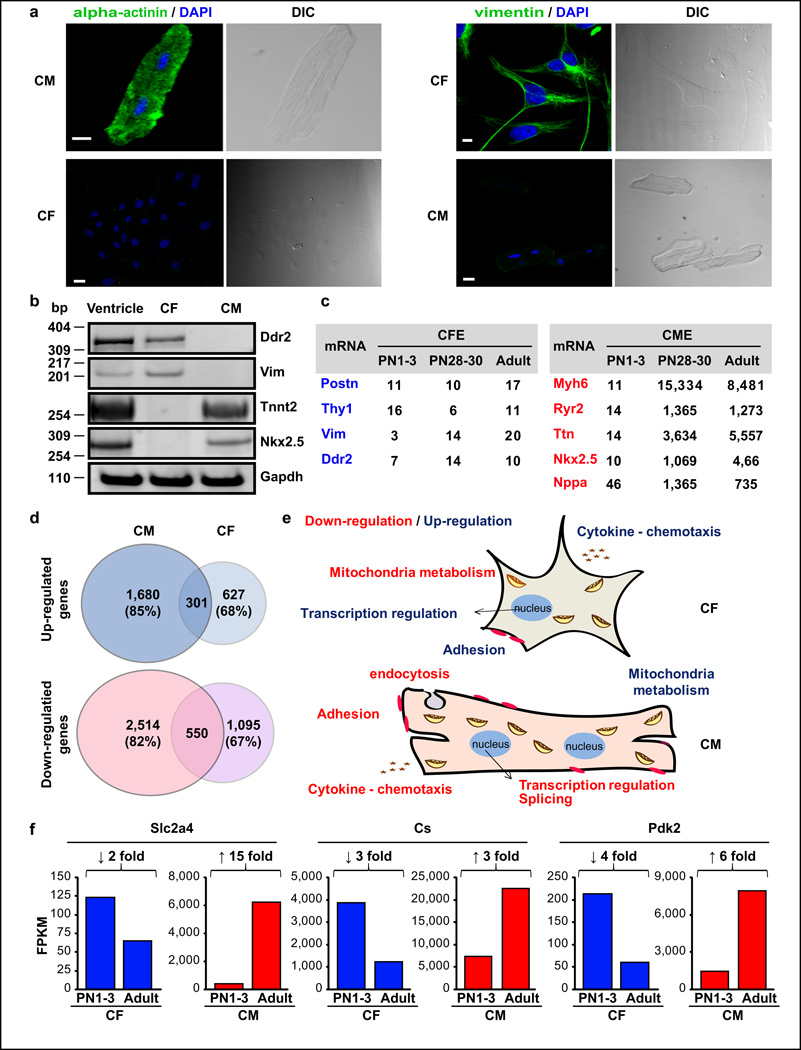
To identify cell-type specific transitions, we isolated CM and CF from ventricles of PN1–3, PN28–30, and adult (PN60–67) animals. RNA was extracted from CM and CF cell populations within three hours of animal sacrifice to reduce post-mortem and cell manipulation effects. CF from all developmental stages and neonatal CM were isolated by an enzymatic digestion/pre-plating method. Adult and PN30 CM were isolated by Langendorff perfusion (see Methods). Morphology, binucleation (CM), and positive immunostaining for alpha-actinin (CM) and vimentin (CF) from adult animals were confirmed by confocal and differential interference contrast microscopy (DIC) (Fig. 3a). Reverse transcription PCR (RT-PCR) analysis of cell-specific markers in adult CM and CF demonstrated a high degree of purity (Fig. 3b). Analysis of cell-specific markers in RNA-seq data confirmed these results demonstrating similar purity levels for early and late postnatal stages (see below).
Figure 3. Reciprocal gene expression transitions between CM and CF.
a. Isolated adult ventricular CM and CF stained with anti-vimentin or anti-alpha-actinin. Scale bars: 10 µm. b. RT-PCR assays of CF (Vim, Ddr2) and CM (Tnnt2, Nkx2.5) markers on RNA from adult ventricles, CM and CF. Experiments were repeated with at least 3 independent samples. c. CF enrichment (CFE) for CF-enriched transcripts (blue) and CM enrichment (CME) for CM-enriched transcripts (red) calculated from RNA-seq data. d. Postnatal gene expression transitions in CM and CF (PN1–3 versus adult) by RNA-seq. e. GO analysis of down- and up-regulated genes in CM and CF (Supplementary Table 2). f. Postnatal expression of three metabolic genes (RNA-seq data) showing reciprocal CM-CF regulation. bp: base pairs. FPKM: fragments per kilobase per million mapped.
RNA-seq (paired-end 100 nt reads) was performed on CM and CF polyA-selected RNA producing >160 million read pairs per sample with 85–90% mapping to the mouse genome (Supplementary Table 1). Biological replicates generated highly reproducible results in neonatal CF and CM populations with high Pearson coefficients (R) for both gene expression (RCM=0.98, RCF=0.99) and AS (RCM=0.96, RCF=0.94). In addition, gene expression from CM and ventricle RNA-seq data highly correlated (R=0.81) while CF-ventricle correlation was low (R=0.33) (Supplementary Fig. 2a–c). These results are consistent with the fact that ~75% of the ventricular volume is formed by CM7.
To evaluate CM and CF separation at all developmental stages, we used the fragments per kilobase per million mapped (FPKM) to estimate “CM-enrichment” (CME) and “CF-enrichment” (CFE) values defined as the ratios CMexpression/CFexpression and CFexpression/CMexpression respectively16,36 (Fig. 3c). This analysis provided several conclusions. First, while several CF-enriched transcripts showed constant ratios during development, CM-enriched transcripts showed a dramatic increase in CME values from PN1–3 to PN28. This reflects an active burst of CM maturation after birth compared to a less robust change during CF maturation (see below). Second, PN1–3 CME values were ≥10 for CM-enriched transcripts in PN1–3 and much higher for PN30 and adult indicating highly pure CM populations at all developmental stages. Third, the lower CFE values for some CF-enriched mRNAs at PN1–3 compared to PN30 and adult could reflect a low level of contamination as well as incomplete cell maturation. To evaluate possible contamination of the CF population with CM in PN1–3, we examined Nppa which is highly expressed in neonatal CM28. We found Nppa to be highly enriched in CM compared to CF populations and estimate contamination of CF preparations (PN1–3) with CM to be <2%.
Reciprocal gene expression transitions between CM and CF
RNA-seq revealed 3,064 down-regulated genes in CM and 1,645 in CF from PN1–3 to adult. Up-regulated genes numbered 1,981 in CM and 928 in CF. Interestingly, >82% of the differentially expressed genes in CM were not differentially expressed in CF (Fig. 3d). Postnatal gene expression transitions in CM and CF showed a reciprocal regulation of enriched functional categories such as mitochondrial metabolism, chemotaxis, cell adhesion, and proliferation, among others (Fig. 3e, Supplementary Table 2). To validate reciprocal expression at the level of individual genes, we analyzed expression of three previously published CM-specific genes involved in mitochondrial metabolism. RNA-seq data from CM and CF for Slc2a4 (solute carrier family 2, facilitated glucose), Cs (citrate synthase), and Pdk2 (pyruvate dehydrogenase kinase 2) transcripts showed postnatal up-regulation for CM and down-regulation for CF (Fig. 3f). The fold induction observed by RNA-seq in CM for these metabolic transcripts was comparable to quantitative PCR data previously reported in total mouse heart28.
AS transitions in ventricles occur mainly before PN28
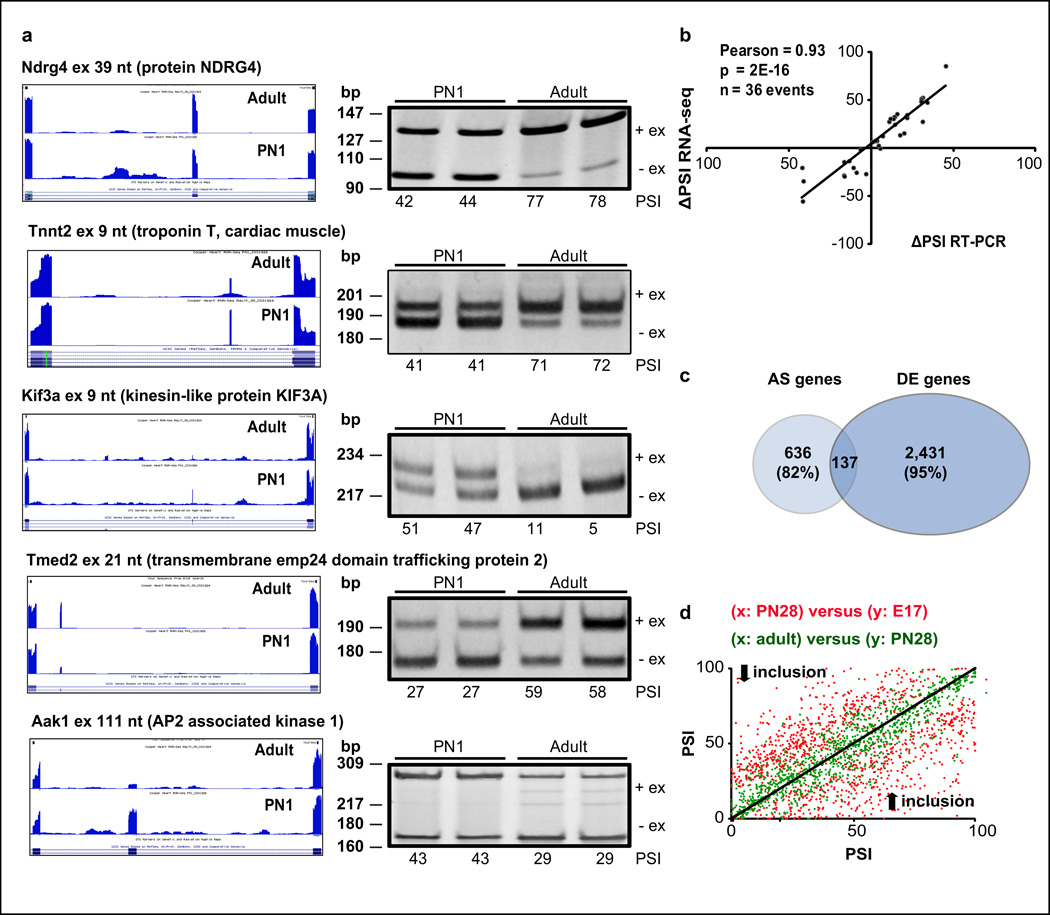
We next focused on AS transitions during postnatal heart development. AS assayed by either RNA-seq or RT-PCR was quantified as the PSI of the variable region and the change in splicing is ∆PSI26. To validate postnatal transitions detected by RNA-seq in ventricular RNA, we performed RT-PCR analysis of 36 events, 17 with ΔPSI≥20% predicted from RNA-seq, 8 with ΔPSI≤-20%, and 11 with intermediate changes. A high Pearson correlation (R=0.93) between ΔPSI values from RT-PCR and RNA-seq experiments demonstrated the accuracy of our RNA-seq data for both strongly and weakly regulated transitions (Fig. 4a–b, Supplementary Fig. 3, and Supplementary Table 3).
Figure 4. Extensive postnatal AS transitions occur primarily without changes in gene expression.
a. RNA-seq data (UCSC browser) and RT-PCR assays (n=2 biological replicates) for AS events developmentally regulated (+ex: exon included; −ex: exon skipped). See Supplementary Fig. 3, and Supplementary Table 3. b. Correlation between RNA-seq and RT-PCR ΔPSI values. c. Genes undergoing postnatal AS transitions (|ΔPSI|≥20%) were intersected with differentially expressed (DE) genes in ventricles (E17-adult). d. AS transitions (|ΔPSI|≥20%; E17-adult) were plotted comparing their change between E17-PN28 (red) and PN28-adult (green). Diagonal line: no difference between time points. Dots above/below the diagonal: increased/decreased inclusion. AS: alternative splicing. bp: base pairs. PSI: percent spliced in.
We identified 927 AS transitions during development (|ΔPSI|≥20%) in 773 genes, 82% occurring in genes for which transcript levels changed less than 2-fold (Fig. 4c). These results are consistent with previous evidence from a smaller set of AS transitions3. Therefore, a substantial impact of AS during development occurs through changes in protein isoform levels rather than total gene expression variations. As observed for gene expression transitions, the majority of the developmental AS changes occurred before PN28 (Fig. 4d).
Temporal dynamics of AS transitions in CM and CF
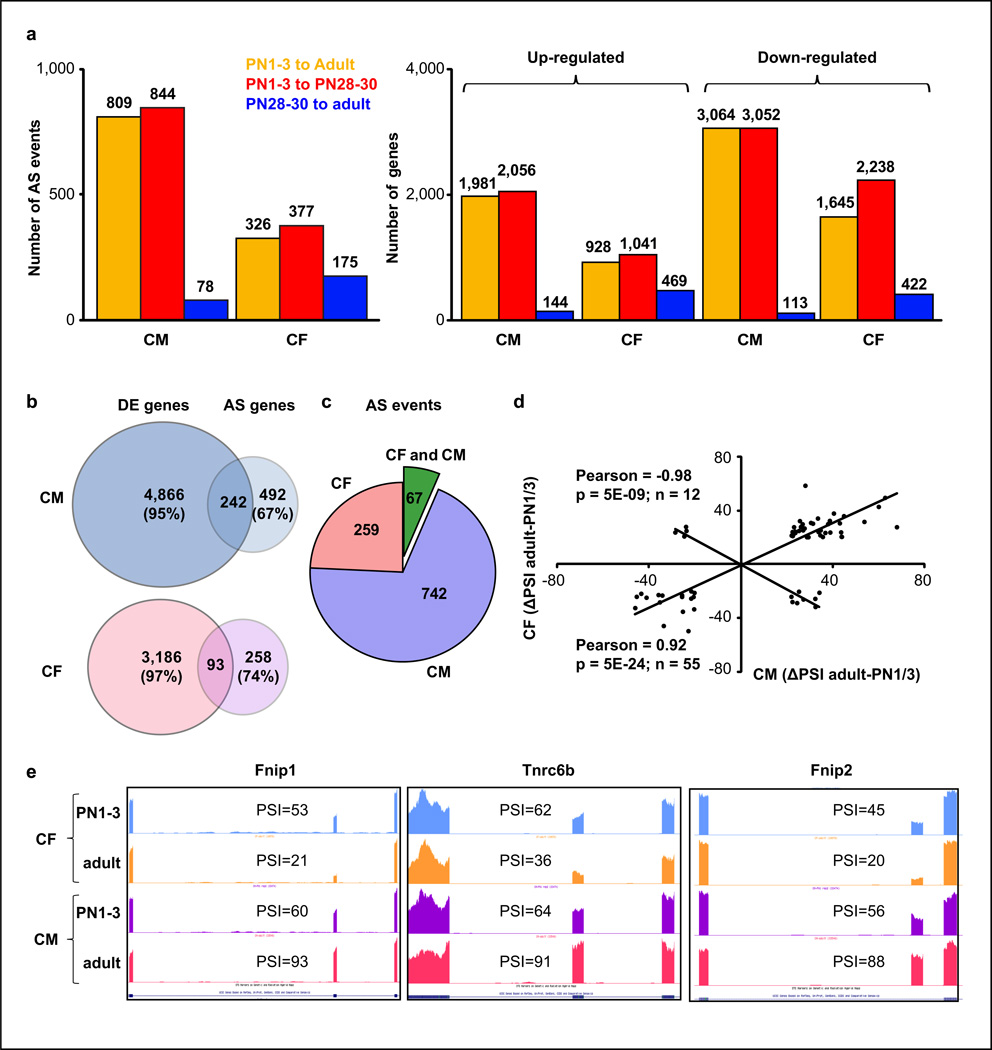
RNA-seq revealed that during postnatal development more AS events changed in CM (809) than in CF (326) between neonatal and adult stages (Fig. 5a). Six transitions were validated in neonatal and adult CM and CF populations, showing high correlation (R=0.84) between RT-PCR and RNA-seq ΔPSI values (Supplementary Fig. 2d–e). In both cell types the majority were cassette exons (82–83%) and the rest were alternative 3´ splice sites (6%), 5´ splice sites (7–9%) and intron retention (3–4%). There was a slight preference for inclusion of the variable region in both CM (65%) and CF (59%) during development (Supplementary Fig. 2f–g).
Figure 5. Postnatal gene expression and AS transitions in CM and CF.
a. AS transitions (|ΔPSI|≥20%) and gene expression changes in CM and CF. b. Genes developmentally regulated by splicing (|ΔPSI|≥20%) in CM and CF were intersected with differentially expressed (DE) genes (PN1–3 versus PN28–30). c. Postnatal AS transitions (between PN1–3 and adult) specifically in CM, CF or both. d. Analysis of the 67 events regulated in CF and CM. e. Three postnatal AS transitions occurring in opposite directions in CM and CF. AS: alternative splicing. PSI: percent spliced in.
Consistent with the AS analysis from ventricular RNA, >90% of postnatal transitions in CM occurred by PN30 (844 versus 78 after PN30). In CF, less than half of the postnatal transitions occurred before PN28 (377 versus 175 after PN28) (Fig. 5a-left). These results suggest that AS primarily impacts CM maturation during the first four weeks after birth while in CF splicing changes are distributed throughout postnatal life. Gene expression patterns showed a similar burst of expression before PN28–30 that was more pronounced in CM than in CF (Fig. 5a-right).
More than half of the genes undergoing postnatal AS transitions in either CM (67%) or CF (74%) did not change expression ≥2.0 fold and the majority of postnatal transitions were specific to either CM or CF populations (Fig. 5b–c). Only 67 events were developmentally regulated in both cell types, 82% of them (55 events) occurred in the same direction, and 12 events showed opposite transitions in CF and CM with a strong negative correlation (R=−0.98) (Fig. 5d–e).
Taken together these data confirmed that, as in ventricles, AS transitions occur primarily by PN28 in CM and are mainly cell-type specific.
Postnatal regulation of vesicular trafficking genes by AS
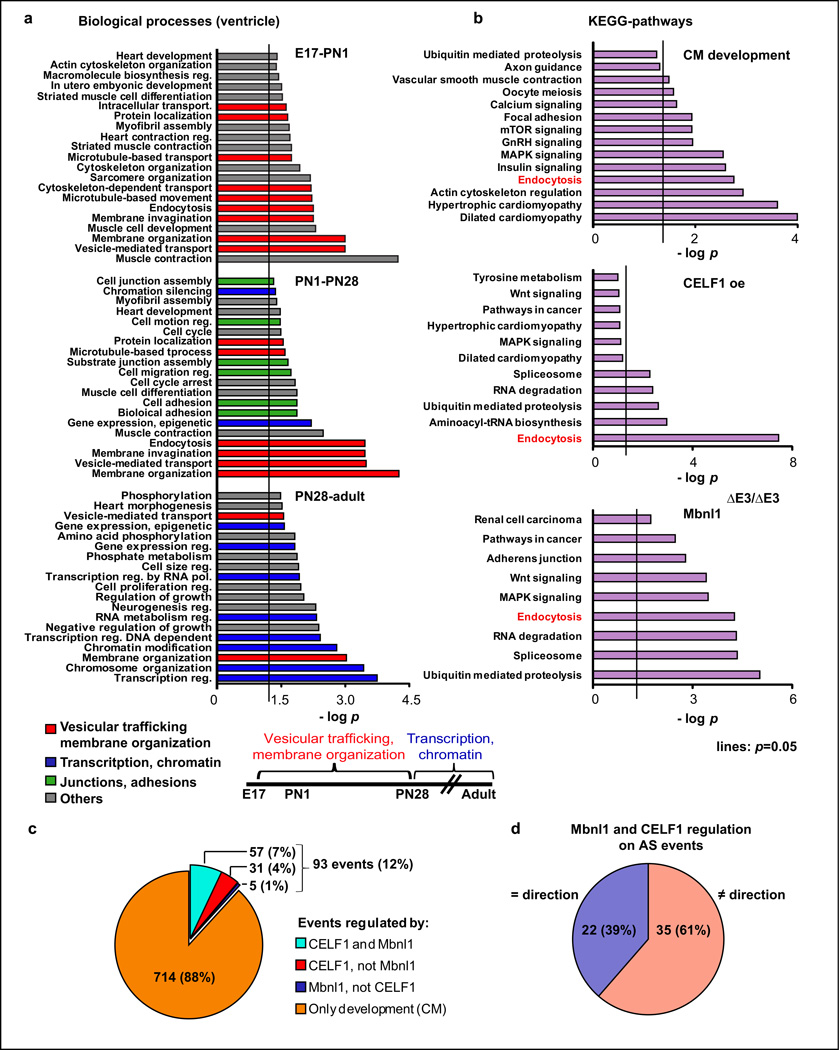
The AS transitions in ventricles (E17-adult) showed that vesicular trafficking, protein localization-transport, endocytosis, membrane organization-invagination were the most represented categories (p<5E-03) including the largest numbers of genes (10 to 27). Analysis of three time windows (E17-PN1, PN1–PN28, and PN28-adult) revealed that categories related to vesicular trafficking and membrane organization were enriched and highly significant before PN28. In contrast, categories related to transcription regulation and chromatin organization were predominant after PN28 (Fig. 6a). In general, GO analysis of AS and differentially expressed genes showed little overlap, further suggesting independence of AS and gene expression regulation (Fig. 2a, 2c, 3e, 6a–b).
Figure 6. Vesicular trafficking genes are regulated by AS during development.
a. GO analysis on AS genes (|ΔPSI|≥20%) in three time windows: E17-PN1, PN1–PN28, PN28-adult (ventricles). b. KEGG-pathway analysis on AS genes detected by RNA-seq during wild type CM development (|ΔPSI|≥20%), in adult CELF1-expressing hearts (CELF1 oe) and adult Mbnl1∆E3/∆E3 hearts (|ΔPSI|≥10%). c. AS transitions (|ΔPSI|≥20%) during CM development (between PN1–3 and adult): regulation by Mbnl1 and/or CELF1. d. The 57 events regulated by Mbnl1 and CELF1 were analyzed in terms of antagonistic/not-antagonistic effects (Supplementary Table 4). AS: alternative splicing.
Vesicular trafficking categories were also highly predominant during CM development (Fig. 6b) while transitions during CF development were enriched for other KEGG-pathways (adherens junction, p=1E-02; actin-cytoskeleton regulation, p=5E-02; ECM-receptor interaction, p=8E-02). Therefore, AS regulates different processes in each cell type.
Vesicular trafficking genes AS regulation by Celf1 and Mbnl1
Celf1 and Mbnl1 have been shown to regulate AS during mouse heart development3,37–38. We used the RNA-seq data obtained from adult TRECUGBP1/MHC animals after 12, 24, 72 hours or 7 days of doxycycline induction and MHC littermates (72 hours on doxycycline) to identify CELF1-responsive AS transitions. Published RNA-seq data from hearts of Mbnl1 knock-out mice (Mbnl1∆E3/∆E3)39 allowed the identification of Mbnl1-sensitive events among postnatal AS transitions. From the 88 developmental transitions responsive to CELF1 re-expression in adults, 71 (81%) reverted to neonatal patterns. Of the 62 Mbnl1-sensitive transitions, 38 (61%) reverted to neonatal patterns in adult Mbnl1∆E3/∆E3 animals. Overall, 93 postnatal AS transitions (12%) were regulated by CELF1 and/or Mbnl1 and more than half of them (57 events) were regulated by both (35 events antagonistically, 22 events in the same direction) (Fig. 6c–d, Supplementary Table 4).
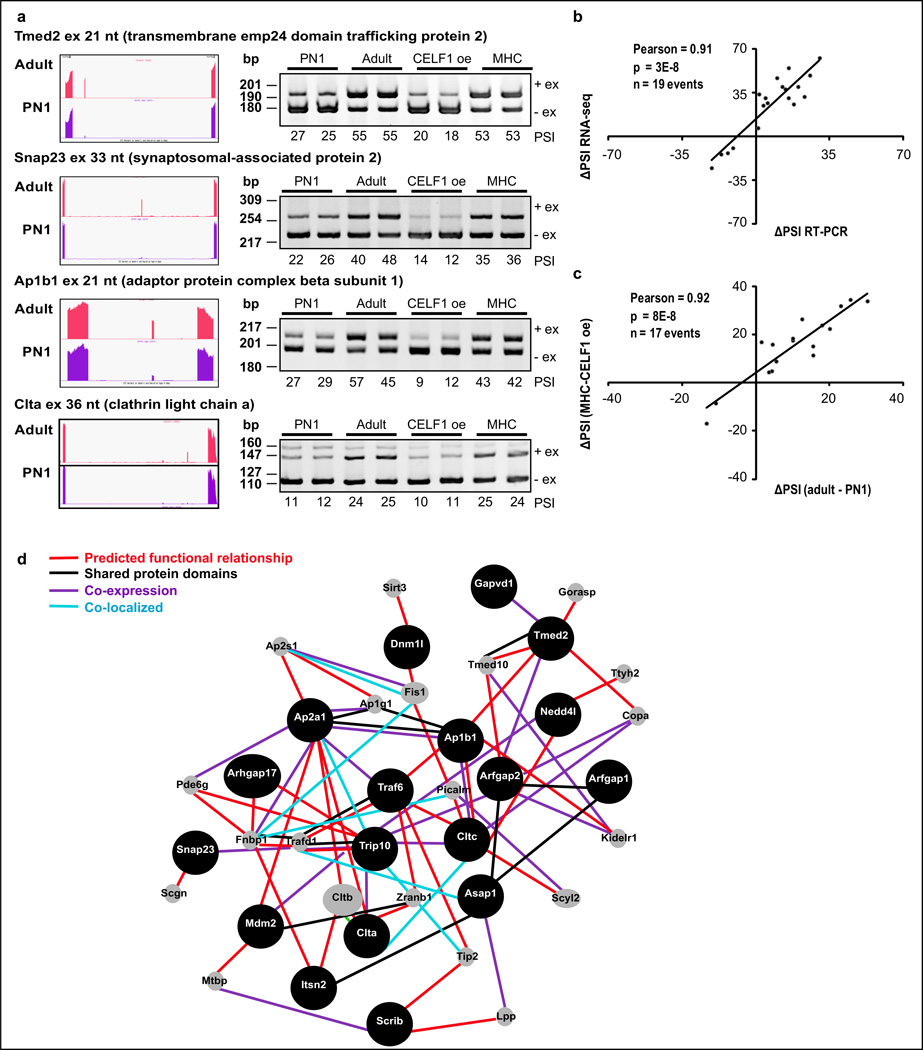
Genes with postnatal AS transitions affected by CELF1 and/or Mbnl1 were enriched for vesicular trafficking, endocytosis, and membrane organization-invagination processes (Fig. 6b). Reversion to neonatal splicing pattern in trafficking genes by CELF1 expression was confirmed by RT-PCR analysis of 17 events (Fig. 7a–c, Supplementary Fig. 4, data not shown). A high correlation was observed between RNA-seq and RT-PCR ΔPSI values and between developmental change (ΔPSIadult-PN1) and reversion after CELF1 expression (ΔPSIMHC-CELF1oe) (Fig. 7b–c). The vesicular trafficking genes regulated by AS during development and responsive to CELF1 re-expression in adults are linked with each other after testing networks connections using GeneMANIA software (Fig. 7d).
Figure 7. Vesicular trafficking genes revert to neonatal splicing patterns after CELF1 re-expression in adult hearts.
a. RT-PCR (ventricles) of vesicular trafficking AS events during development and in CELF1-expressing hearts. Bar graphs: mean±s.e.m (n=2 biological replicates). b. Correlation between RNA-seq and RT-PCR data in vesicular trafficking genes (PN1-adult). c. Correlation between developmental transitions in vesicular trafficking genes and reversion after CELF1 re-expression in adults (RT-PCR). d. Network of vesicular trafficking genes developmentally regulated by splicing and responsive to CELF1 (black circles). Grey circles: linking-genes. bp: base pairs. CELF1 oe: TRECUGBP1/MHC animals. MHC: control animals. PSI: percent spliced in.
We analyzed the direct involvement of CELF1 in regulating AS during CM development by performing a motif analysis within the up- and downstream flanking introns (300 bp) of the exons regulated during development that responded to CELF1 re-expression in adults (88 events). GU rich motifs were enriched (p≤1E-08) in downstream regions of AS exons (Supplementary Fig. 5a). Upstream flanking regions of AS exons were enriched in U or C rich motifs (p=1E-14) when the variable regions were more skipped after CELF1 induction and GU rich sequences (p≤1E-09) when the exons were more included in response to CELF1 (Supplementary Fig. 5b). iCLIP-seq data are available for Celf1 in C2C12 muscle cell differentiation40 and were used for evidence of direct binding of Celf1 to the 71 transcripts regulated by AS during heart development that show a reversion to the neonatal pattern after CELF1 re-expression in adults. We evaluated the presence of iCLIP-tags within the up- and/or downstream 500 bp of the intron sequences flanking the AS exons and observed that 54% of them (38 out of 71) contain iCLIP-tags within these regions (Supplementary Table 5) (Fisher test: 1.7E-11 compared with whole gene exon iCLIP binding). When iCLIP-tags were evaluated for AS exons of vesicular trafficking genes we found that 50% of them contain iCLIP-tags within 500 bp of their flanking regions, two examples are shown in Supplementary Fig. 5c.
Taken together these results support the hypothesis that vesicular trafficking is regulated at the splicing level during postnatal CM development and that CELF1 directly regulates a substantial fraction of these transitions.
T-tubule and calcium release disruption by CELF1 induction
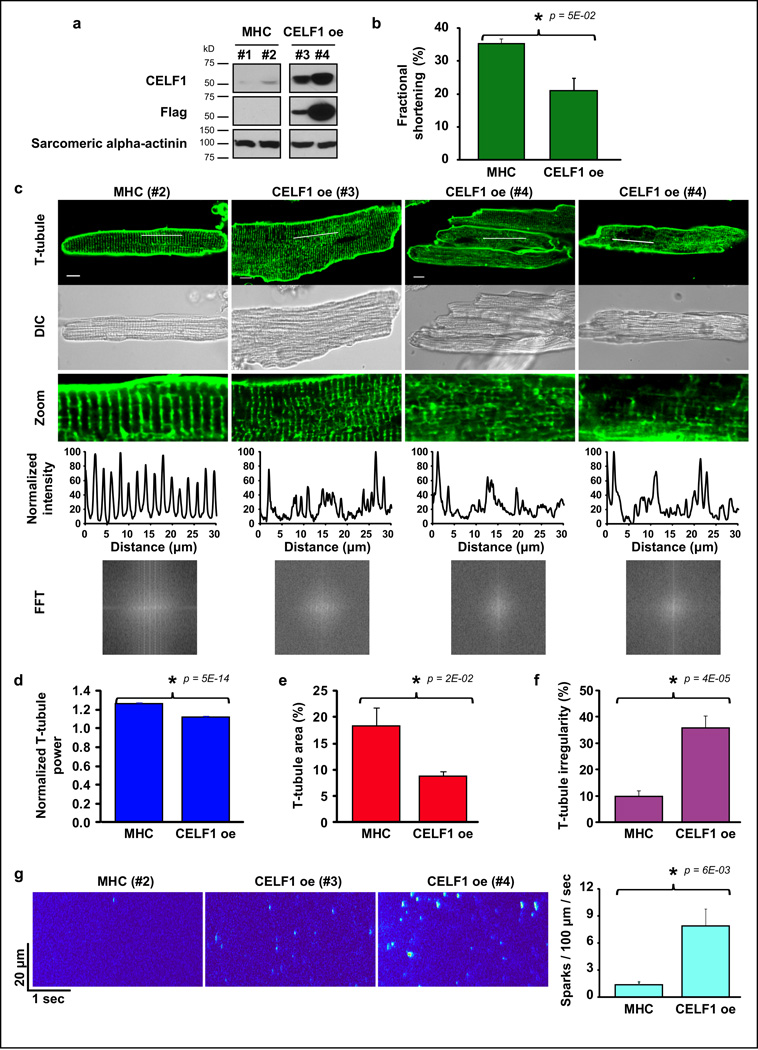
Membrane organization and vesicular trafficking are particularly dynamic during postnatal CM development. The assembly of the excitation-contraction apparatus starts at birth and ends within four weeks involving T-tubules invagination from the sarcolemma, SR formation and vesicular trafficking of ion channels and adaptor proteins to specialized membrane regions41,42. We hypothesized that the splicing transitions regulated by Celf1 play a role in assembly and maintenance of these structures. To test this hypothesis, we determined the effect of CELF1 induction on adult T-tubule structure and function. CELF1 was re-expressed in adult mice for four days specifically in CM (Fig. 8a). CELF1-expressing animals showed altered electro- and echocardiograms exhibiting lower fractional shortening, ejection fraction, and heart rate compared with MHC controls (Fig. 8b, Supplementary Tables 6 and 7). Animals with higher exogenous CELF1 expression (#4) showed more affected cardiac parameters than animals with lower exogenous CELF1 expression (#3). T-tubules were stained and imaged in living CM (Fig. 8c) and T-tubule structure was studied by three approaches. First, we delimited a rectangular region within the cell excluding nuclei and we performed the Fast Fourier Transform (FFT), an algorithm to convert a function of time/space into a frequency function and vice versa. With the FFT we obtained the normalized T-tubule power by measuring the ratio between the pick power (first pick of the FFT) and the baseline power. Normalized T-tubule power after FFT measures the global regularity of the T-tubule network, and densities of transverse and longitudinal elements of T-tubules41,43 (see Methods). CELF1-expressing CM showed lower T-tubule power than MHC controls (Fig. 8d). Second, we quantified the average T-tubule area within CM28. T-tubule area is defined as the percentage of the area containing T-tubule staining28 (see Methods). While MHC mice showed 18±3% (n=12 cells) of CM area as T-tubules, CELF1-expressing CM showed 9±1% (n=16 cells) (p=0.02) (Fig. 8e). When analyzed separately, CM from mice expressing higher CELF1 levels showed less T-tubule area compared to mice expressing lower CELF1 levels (mouse #3: 12±1%, mouse #4: 7±1%). Third, we devised an approach to estimate the proportion of individual cells with disorganized T-tubule structure (T-tubule irregularity) (see Methods). Control CM showed lower T-tubule irregularity (10±2%; n=13 cells) than CELF1-expressing CM (36±5%; n=16 cells) (p=4E-05) (Fig. 8f).
Figure 8. T-tubule disorganization after CELF1 re-expression in adults.
a. Western blot analysis from adult hearts after CELF1 induction. b. Fractional shortening was measured in MHC (n=6 animals) and TRECUGBP1/MHC mice (n=3 animals) given doxycycline (four days). Results are expressed as the mean±s.e.m. c. Confocal imaging of T-tubules on living CM from MHC (#1–2; n= 2 animals) and CELF1-expressing (#3–4, n= 2 animals) mice. Scale bars: 10 µm. Third row: fluorescence plot over the line (first row). d–g. T-tubule and calcium spark analysis: normalized T-tubule power (d) (n=13 cells for MHC animals, n=16 cells for TRCUGBP1/MHC animals), T-tubule area (e) (n=12 cells for MHC animals, n=16 cells for TRCUGBP1/MHC animals), T-tubule irregularity (f) (n=13 cells for MHC animals, n=16 cells for TRCUGBP1/MHC animals), calcium spark frequency (g) (n=11 cells for each genotype). Asterisks show statistical differences (p≤0.05). Results are expressed as the mean±s.e.m. CELF1 oe: TRECUGBP1/MHC animals. MHC: controls. FFT: Fast Fourier Transform T-tubules: transverse tubules.
Next, we addressed whether T-tubule disruption correlated with defective calcium release. Calcium spark frequency was higher in CELF1-expressing CM than in controls (7.9±1.9 versus 1.4±0.4 sparks (100 µm)−1 sec−1; n=11 cells each genotype; p=6E-03) (Fig. 8g). Similarly to T-tubule disruption, CELF1-expressing mouse #4 (sickest) presented more frequent calcium sparks than CM from mouse #3 (healthiest) (10±3 versus 6±2 sparks (100 µm) −1 sec−1). Spontaneous calcium sparks are due to the opening rate of ryanodine receptors and frequency changes reflect alterations in ryanodine receptor itself, its association with other proteins, receptor cluster spatial organization, inter-cluster distance, and/or calcium content in the SR44. Overall, T-tubule organization observed in CELF1-expressing CM resembles PN10–15 structure41, suggesting reversion to earlier developmental stages.
DISCUSSION
We used RNA-seq to identify transcriptome dynamics from late embryonic to adult mouse heart development. While multiple studies have focused on specific genes or gene sets during heart development, this study presents a global analysis of postnatal gene expression and AS transitions in ventricles and freshly isolated CM and CF. The strong correlations of global postnatal changes between CM and ventricles flash-frozen immediately after sacrifice supports our contention that transcriptome transitions identified in isolated CM and CF are largely representative of those occurring in tissue. The similarity between ventricle and CM RNA-seq data is particularly striking given that CM RNA was polyA selected and ventricular RNA was prepared by rRNA depletion. Overall, we revealed novel insight into strikingly different postnatal gene expression and AS transitions that occur in distinct but highly interacting cell populations within ventricles.
The largest transitions between E17-adult occur before PN28, a period in which extensive physiological changes take place as the fetal heart adapts to birth and converts to adult function1,2. In CM, 90% of the gene expression and AS transitions occur before PN28 while in CF nearly 50% occur later. Gene expression and AS follow the same patterns in each cell type suggesting a general tendency for CM to set an adult pattern of gene output within the first four weeks after birth while CF undergo a more gradual transition. We hypothesize that this reflects the maturation required in CM in response to cell proliferation loss, cellular hypertrophy, and rapidly increasing workload during the first four weeks after birth. By contrast, CF would not require as dramatic physiological maturation.
Our data reveal a high level of specificity in CM and CF gene expression and a reciprocal expression changes in the two cell types. While genes involved in critical CM functions such as mitochondrial metabolism are developmentally up-regulated, they are down-regulated in CF. Similarly, focal adhesions and chemotaxis genes are up-regulated during CF development but down-regulated in CM.
We examined potential mechanisms for post-transcriptional regulation of gene expression during development and identified a likely role for the 10-fold down-regulation of Celf1 in the developmentally up-regulation of a subset of genes. While our previous analysis focused on the Celf1 role in a set of splicing transitions3, Celf1 has also been shown to destabilize mRNAs by binding to GU-rich motifs within 3´ UTRs34,35. We showed that 41% of the genes normally up-regulated during development were down-regulated upon CELF1 induction and from those GU-rich motifs are enriched in 3´ UTRs of mitochondrial metabolism genes. Therefore, our results implicate Celf1 in coordinated up-regulation of genes involved in the postnatal metabolic transition of CM.
Consistent with our previous smaller scale study3, the majority of AS transitions detected during postnatal heart development occurs without changes in total gene output, indicating that protein isoform switches are important regulatory components during postnatal development. Categories highly regulated by AS in heart, specifically in CM, are vesicular trafficking, endocytosis, membrane organization-invagination. A link between trafficking and AS is novel although several individual AS events in trafficking genes (e.g., Snap25, Dab2, Mdm2) have been reported45,46. Additionally, categories relevant to trafficking were enriched among genes modulated by AS by CELF1 and Mbnl1 suggesting that these developmentally regulated RNA-binding proteins play a role in vesicle mediated transport and membrane remodeling in CM.
Membrane remodeling and endocytosis are crucial in eukaryotic cells for a variety of functions, some of which are cell type specific47. Recently, it was reported that AS impacts endocytosis during brain development, suggesting that vesicular trafficking functions are coordinated by AS21. Our results are similar in heart and the impact of vesicular trafficking on cardiac functions is not known. Although many differences exist between heart and brain, parallels between neuroscience and cardiac fields are growing, particularly with regard to ion channels and vesicular trafficking processes48.
We hypothesize several non-exclusive scenarios for vesicular trafficking roles during postnatal heart development. First, AS regulation of vesicle-mediated transport could reflect changes in ligand/growth factor uptake during development. Second, after PN7, CM stop dividing and undergo cellular hypertrophy14,15. The most rapid increase in CM volume occurs between PN4-PN20, after which CM volume increases only slightly49. Therefore, cells must maintain the appropriate internalization-recycling balance while membrane exposure to the extracellular environment is rapidly increasing. Third, vesicular trafficking regulation by AS could control the dynamics of ion channel production and localization. Ion channels are continuously formed, trafficked within vesicles for insertion and anchoring to specific sub-regions of the plasma membrane, and removed for degradation or recycling. Ion channel function is, therefore, dependent in part on surface density which is tightly regulated by vesicular trafficking. The last scenario is supported by our analysis of T-tubule organization and function after CELF1 re-expression in adults. Maturation and assembly of SR and T-tubules in CM occur within the three-four weeks after birth41 producing the machinery for ECC. These maturation processes involve extensive cell architecture transitions, including membrane re-organization and vesicular-mediated protein transport. Membrane invagination and vesicular trafficking is regulated by AS during postnatal development and this may impact CM functions such as ECC and uptake, distribution, and recycling of key molecules (ion channels, growth factors, receptors). Importantly, both up regulation of CELF1 and loss of MBNL1 function are involved in the pathogenic mechanisms of myotonic dystrophy type 1 (DM1) in which the best characterized molecular feature is expression of fetal AS splicing patterns50. The alterations in T-tubule structure and function we demonstrated strongly correlate with the finding that eighty percent of individuals with DM1 exhibit cardiac arrhythmias51.
METHODS
Materials
Cell culture reagents were obtained from GIBCO, Life Technologies. Liberase TH Research Grade and Collagenase/Dispase enzymes were from Roche Applied Science.
Animals
Ventricles, CM and CF were isolated from FvB wild type mice. TRECUGBP1/Myh6-rtTA (TRECUGBP1/MHC) mice were used for human CELF1 over-expression in CM (tetracycline-inducible) as described3,33. Bitransgenic and Myh6-rtTA (MHC) control adult animals were fed 2 g/Kg doxycycline (BioServ) for 12, 24, 72 hours and 7 days (RNA-seq), 8 days (RT-PCR), or 4 days (T-tubule/calcium experiments). We followed NIH Guidelines for use and care of laboratory animals approved by BCM Institutional Animal Care and Use Committee.
Ventricle heart isolation
Animals were anesthetized and after cervical dislocation (older than PN10) or decapitation (neonatal) hearts were removed. Blood and atria were removed; ventricles were frozen in liquid nitrogen or freshly used for CM and CF isolation.
CF and CM isolation
PN28 and adult CF isolation by pre-plating method. Minced ventricles were digested in Dulbecco’s modified Eagle’s medium (DMEM) containing 0.1 U/ml Collagenase, 0.8 U/ml Dispase, and 0.1% EDTA-free Tripsin for 10 min at 37°C (with stirring). After sedimentation the remaining tissue was similarly digested for 10 min. The suspension was centrifuged for 5 min at 3,000 × g, cells were resuspended in Minimum Essential Medium α (MEM-α) supplemented with 10% fetal bovine serum (FBS) and kept on ice. This procedure was repeated three to four times, cells were centrifuged for 5 min at 3,000 × g, the pellet was resuspended in MEM-α/20% FBS and cells were seeded on cell culture dishes. Cells were plated for 2 hours at 37°C under 5% CO2. Adherent cells were washed ten times with PBS and RNA was immediately extracted. Neonatal CF and CM isolation. The neonatal CM isolation kit (Cellutron Life Technology) was used with 20–35 ventricles accordingly to manufacturer´s protocols. PN30 and adult CM isolation by Langendorff perfusion. The heart was removed with the lungs without touching the heart, rinsed in Ca2+ free Tyrode´s solution (140 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 10 mM Glucose, 5 mM HEPES pH 7.4) and cleaned from fat tissue. The heart was cannulated through the aorta and perfused on a Langendorff apparatus with Ca2+ free Tyrode´s solution for 3–5 min, then with Ca2+ free Tyrode´s solution containing 20 µg/ml (0.104 Wünsch units/ml) Liberase for ~15–25 min at 37°C and then with KB solution (90 mM KCl, 30 mM K2HPO4, 5 mM MgSO4, 5 mM pyruvic acid, 5 mM β-hydroxybutyric acid, 5 mM creatinine, 20 mM taurine, 10 mM glucose, 0.5 mM EGTA, 5 mM HEPES pH 7.2) to wash out the enzyme. Atria were removed, the heart was minced in KB solution, cells were gently pipeted up and down (on ice) and filtered through Nylon mesh 210 µm open 155 µm thread, 24"×12" mash (Smallparts inc). The sample was centrifuged for 5 min at 3,000 × g and RNA was immediately extracted, or fresh CM were used for T-tubule experiments or immunofluorescences, or were frozen in liquid nitrogen for Western blot assays.
RNA extraction
RNeasy fibrous tissue mini-kit (ventricles and CM) and RNeasy micro-kit (CF) were used (QIAGEN).
RNA-seq analysis of CM and CF during development
RNA-seq samples showed the following parameters: A260nm/A280nm≥1.8, A260nm/A230nm≥1.4, r28S:16S≥1.5, and RNA integrated number (RIN)≥8.6. Illumina TruSeq RNA sample preparation protocols were used for CM (PN1–2, PN1, PN30, and PN67) and CF (PN1–3, PN1–2, PN28, and PN60). Briefly, a double-stranded DNA library was created using 1 µg total RNA. First, cDNA was created using the fragmented 3’ poly(A) selected portion of total RNA and random primers. Libraries were generated from the cDNA by blunt ending the fragments, attaching an adenosine to the 3’ end and finally ligating unique adapters to the ends. The ligated products were amplified by PCR (15 cycles). Libraries were quantified using the NanoDrop spectrophotometer and fragment size assessed with the Agilent Bioanalyzer. A qPCR was performed on the libraries to determine the concentration of adapter ligated fragments using a Bio-Rad iCycler iQ Real-Time PCR Detection System and a KAPA Library Quant Kit. Using the concentration from the Bio-Rad qPCR, 11 pM library was loaded onto a flow-cell and amplified by bridge amplification using the Illumina cBot equipment. A paired-end 100 cycle run was used to sequence the flow-cell on a HiSeq Sequencing System.
RNA-seq
Illumina TruSeq protocols were used for wild type CM (PN1–2, PN1, PN30, and PN67) and CF (PN1–3, PN1–2, PN28, and PN60), and ventricles from TRECUGBP1/MHC or MHC animals (n=3 animals each time point). For developmental ventricle analysis, RNA was rRNA depleted and libraries were prepared by Ribo-Zero Magnetic Gold and ScriptSeq-v2 RNA-seq library preparation kits (Epicentre Biotechnologies). Briefly total RNA (2 µg) from ventricles of E17, PN1, PN10, PN28, and PN90 (adult) animals was rRNA depleted using the Ribo-Zero Magnetic Gold Kit following manufacturer protocols. The rRNA-reduced samples (~30 ng) were fragmented and primed with random hexamers containing a 5’-tagging sequence for RT priming. The 5’-tagged first strand cDNA was then generated. During the second strand cDNA generation the RNA templates were removed leaving the 5’-tagged strand. Terminal-tagging oligos (TTO) were annealed to the 5’-tagged strand and terminal-tagging oligos were blocked at the 3’ end preventing the synthesis of a secondary strand. Resulting cDNA strand was single-stranded and tagged at both ends. The di-tagged cDNA was purified using Agencourt AMPure XP system (Beckman Coulter). Single-stranded cDNA generated for each sample was used for library construction. By PCR amplification (10 cycles), the second cDNA strand was generated and the Illumina adapter sequences were added simultaneously to the single-stranded di-tagged cDNA fragments. Illumina sequencing libraries were purified using Agencourt AMPure XP system. Libraries were validated on an Agilent Bio-analyzer High Sensitivity DNA chip checking the purity of the sample and the size of the insert. Libraries were within 150–2000 bp on the electropherogram. A second round of purification using Agencourt AMPure XP system was required for removal of primer-dimer and adapter-dimer contamination. Libraries were quantified using KAPA SYBR Fast qPCR kit from KAPA Biosystem. Libraries were diluted to 2 nM (based on cT values), alkali denatured for 5 min, diluted to a final concentration of 11 pM, and dispensed into thin-walled PCR tubes to be loaded into the Illumina cBot. Clusters were generated on the flow-cell by PCR with the Illumina cluster generation kit. The flow-cell containing clustered libraries were loaded on the Illumina HiSeq instrument along with the kitted sequencing reagents for a PE 100 bp run. During each sequencing cycle, a dye-terminated nucleotide was incorporated into the single-stranded DNA strand and fluorescence was monitored. Fluorescent dye was cleaved to allow next nucleotide incorporation. CASAVA software converted the fluorescence measurements into sequence files.
Computational processing of RNA-seq data from wild type CM, CF, and ventricles
RNA-seq Alignment. Paired-end RNA-seq reads were aligned to the mouse genome (mm9) using TopHat 2.0.552. Differentially expressed gene analysis and FPKM. RSEM53 was used to count the number of fragments mapped into Ensembl gene models, followed by edgeR54 to call differentially expressed genes with false discovery rate less than 0.01. The gene expression was quantified by FPKM55. Differential AS events. Based on Ensembl 65 gene model, SpliceTrap56 was employed to identify differential exon skipping, intron retention events, alternative 5’ or 3´splice site. AS was quantified by the percentage of mRNAs that contain an alternative region as PSI value. The events with PSI changes between two conditions |ΔPSI|≥20% were called differential splicing events.
Computational processing of RNA-seq data from CELF1 over- expressing and control animals
Estimation of gene and isoform expression levels was performed similarly to previously described39. Briefly, reads were mapped to the mouse genome (mm9) and a database of splice junctions using the Bowtie tool57. Gene expression. Gene expression was measured as following: i) the number of reads mapping to constitutive exons for each gene was measured, ii) that measurement was divided by the number of kilobases of constitutive exon model per million uniquely mapped reads. AS events. Isoform levels and Bayes factors were measured by MISO58 (single end mode).
Bioinformatic analysis of 3´ UTRs
The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.759–60 was used for GO analysis considering p≤0.05 significant. miRNA prediction analysis was performed using TargetScan (http://www.targetscan.org/) and motif analysis with CLIPZ (SIB) and MEME version 4.9.061 (motif>6–18 bp) softwares. The E-value (MEME) is the enrichment of a motif based on its log likelihood ratio, width, sites, back-ground frequencies, and the training set size. The p-value of a site is computed from the the match score of the site with the position specific scoring matrix for the motif. The p-value gives the probability of a random string (generated from the background letter frequencies) having the same match score or higher. Gene network of AS genes involved in vesicular traffic, developmentally regulated, and sensitive to CELF1 induction was performed using GeneMANIA62.
Bioinformatic analysis of AS events
The analysis involved the 88 developmental transitions responsive to CELF1 re-expression in adults. The 300 nt upstream and downstream of the alternative spliced exons were used for de novo motif analysis, excluding the last 30 nt of the upstream intron and first 9 nt of the downstream intron of the alternative exons which contain the conserved 5’ and 3’ splice sites, respectively. Then motif analysis tool HOMER63 (v4.3) was employed for RNA motif analysis with the background sequences generated by first-order Markov model whose parameters were estimated based on hg19.
Celf1 HITS-Clip data analysis
C2C12 Celf1 HITS-CLIP data was downloaded from European Nucleotide Archive (ENA; accession code ERP000789)64. As described by authors64, the 4-bp tags were trimmed and sequences composed primarily of Illumina adapter were removed. The pre-processed reads were mapped to the mouse genome (mm9) using the alignment tool Bowtie57 with allowed 2-mismatch. Finally, reads with identical 5’ starts were further collapsed into a single read to avoid potential PCR duplicates effects and only uniquely mapped reads were kept as final Celf1 binding tags.
AS validations by RT-PCR
RT-PCR were performed (High Capacity cDNA RT Kit, Applied Biosystems; and GoTaq DNA Polymerase, Promega) in biological duplicates and products were analyzed by 6% PAGE. PCR reactions involved the following steps: i) 95°C for 3 min, ii) 20–27 cycles of 95°C for 30 sec, 58°C for 30 sec, and 72°C for 30 sec, iii) 72°C for 7 min, iv) 25°C for 5 min. RNA-seq data were used to design primers annealing in the constitutive flanking exons of the AS region. Sequences of the primers (SIGMA) used for validation in ventricles are shown in Supplementary Table 8.Sequences of the primers (SIGMA) used for validation of AS events on vesicular trafficking related genes are shown in Supplementary Table 9. Densitometry measurements were performed using Kodak Gel logic 2200 and Molecular Imaging Software. Percent spliced in (PSI)26 was calculated by densitometry following equation (1).
| equation (1) |
Analysis of CM and CF markers by RT-PCR
RT-PCR assays were performed using the following primers (SIGMA): Vimentin-F (5´-tgaaggaagagatggctcgt-3´), Vimentin-R (5´-ttgagtgggtgtcaaccaga-3´), Ddr2-F (5´-caagatcatgtctcggctca-3´), Ddr2-R (5´-gccctggatccggtagtaat-3´), Nkx2.5-F (5´-aagcaacagcggtacctgtc-3´), Nkx2.5-R (5´-gggtaggcgttgtagccata-3´), Tnnt2-F (5´-cggaagagtgggaagagaca-3´), Tnnt2-R (5´-ttcccacgagttttggagac-3´), mGadph-F (5´-cgtcccgtagacaaaatggt-3´) and mGadph-R (5´-ttgatggcaacaatctccac-3´). PCR reactions involved the following steps: i) 95°C for 75 sec, ii) 20–27 cycles of 95°C for 45 sec, 57°C for 45 sec, and 72°C for 1 min, iii) 72°C for 10 min, iv) 25°C for 5 min. PCR products were analyzed by 6% PAGE.
Immunofluorescence
After adult CM and CF isolation, cells were seeded onto coverslips overnight (CM) or for 70 hours (CF). Coverslips were pre-coated with 20 µg/ml laminin for 1 hour at 37°C for CM. Cells were fixed in 3.7% paraformaldehyde on ice for 20 min, blocked with PBS/0.3% TritonX-100/1% bovine serum albumin (BSA) for 1 hour at 37°C, and incubated overnight at 4°C with primary antibodies (rabbit monoclonal anti-vimentin (D21H3) (Cell Signaling, #5741) (1:100) or mouse monoclonal (clone BM-75.2) anti-alpha-actinin (SIGMA, #A5044) (1:50)). After washes samples were incubated with secondary antibodies conjugated with Alexa-fluor-488 (Invitrogen) (1:500) for 1 hour at 37°C, washed, stained with 2 µM DAPI for 5 min, and mounted (Slow Fade Gold Antifade Reagent, Invitrogen). Confocal microscopy was performed with a Nikon A1-Rs inverted laser scanning microscope with a 40× Plan-Fluor/1.3 NA oil-immersion objective. Excitation: argon multi-line laser at 488 nm (Alexa-fluor-488) or a 404 nm diode at 38 mW (DAPI). Emission filters: band-pass 525/50 nm (Alexa-fluor-488), band-pass 450/50 nm (DAPI). Confocal images were processed with ImageJ. Background was subtracted and a median filter was applied (radius=1 pixel) for presentation.
T-tubules analysis
Isolated CM (adult TRECUGBP1/MHC or MHC doxycycline fed, n= 2 animals each) were stained with 10 µM Di-8-ANEPPS (Invitrogen, #D3167) in KB-solution for 15 min, washed and imaged by confocal microscopy (Zeiss LSM-510 microscope, 40× oil-immersion objective). Excitation: argon multi-line laser, 488 nm. Emission filter: long-pass 505 nm. Quantifications were performed using ImageJ: 1) a region of interested was selected covering longitudinally the CM avoiding the nuclei, Fast Fourier Transform (FFT) was obtained, power spectrum was computed and normalized (normalized T-tubule power)41; 2) a threshold (MaxEntropy) was applied (ImageJ), cell interior was manually delimitated (excluding plasma membrane), areas with green signal were summed within the threshold, and normalized to the total area (T-tubule area); 3) CM from MHC animals showed 4–6 T-tubules (10 µm)−1 (ImageJ), therefore, regions with 4–6 T-tubules (10 µm)−1 were considered “regular-regions” otherwise “irregular-regions”. Three lines were drawn along each CM, plots were obtained, normalized to its maximum, a cut off (15 units above the baseline) was determined and each line was analyzed by the number of picks (T-tubules) above the cut-off (between 0–10, 5–15, 10–20, 15–25 µm, etc) computing the “regular-regions” and the “irregular-regions” (in µm). For each CM, the total length of the three lines and the total “irregular-regions” were summed and T-tubule irregularity was calculated.
Calcium sparks analysis
The same samples were incubated with 3.5 µM Fluo-4-acetoxymethyl ester (Invitrogen, #F14201) in Tyrode´s solution (1.8 mM Ca2+) (1 hour, room temperature), washed (15 min) and transferred to a chamber containing parallel platinum electrodes. Fluorescence images were recorded after pacing (10 V, 0.5 msec) from CM showing clear striation and normal contractility. Sparks were analyzed using SparkMaster plugin (ImageJ).
Western blots
After perfusion, a portion of the ventricles from TRECUGBP1/MHC or MHC control animals were lysed in HEPES-sucrose buffer (10 mM HEPES pH 7.4, 0.32 M sucrose, 1 mM EDTA, and proteases inhibitors) using Bullet blender (Next Advance) equipment and SDS was added (final concentration: 1% SDS). Samples were sonicated (3 min at 75 V: 30 sec ON and 30 sec OFF) and centrifuged for 10 min at 14,000 r.p.m at 4°C. Supernatants were transferred to new tubes and protein concentration was estimated with Pierce BCA protein assay kit (Thermo Scientific). Samples were diluted in loading buffer (100 mM Tris-HCl pH 6.8, 4% SDS, 0.2% Bromophenol blue, 20% glycerol, 200 mM β-mercaptoethanol), boiled for 5 min, and total proteins (40 µg) were assayed by 10% SDS-PAGE. After transfer, membranes were blocked in 5% non-fat dried milk/0.1% Tween-TBS buffer (TTBS) for 1 hour, washed and incubated overnight at 4°C with primary antibodies diluted in 5% milk/TTBS: mouse monoclonal anti-CUG-BP1, clone 3B1 (Milipore, #05–621) (1:1,000), rabbit polyclonal anti-sarcomeric alpha actinin (Abcam, #ab72592) (1:2,000). The following day, membranes were incubated with secondary antibodies (1:5,000) for 1 hour at room temperature: peroxidase-conjugated goat anti-mouse IgG light chain specific (Jackson Immunoresearch, #115-035-174), goat anti-rabbit IgG horseradish peroxidase-conjugated (Invitrogen, # 621234). Flag-tag was detected by monoclonal anti-FLAG M2 peroxidase (Sigma, #A8592) (1:1,000; 1 hour at room temperature). Super Signal West Pico Chemiluminiscent Substrate kit (Thermo Scientific) was used for developing.
ECG and echocardiogram recording
Echocardiograms were performed using a Vevo 770 Ultrasound equipment with a 707B probe for the cardiac analysis. The animals were sedated with 2% isoflurane for imaging. The m-mode images were analyzed for data acquisition using the Vevo analysis package. The ECG's were recorded using a Mouse Monitor made by Indus Instruments. Mice were lightly anesthetized using 1.5% isoflurane and the paws were taped to the ECG leads. ECGs were recorded for leads I, II, and III for each animal and analyzed. Both ECGs and ultrasounds were performed in the Mouse Phenotyping Core at BCM.
Statistic
Results were expressed as the mean±s.e.m, p-values were estimated using Student´s T-test (two tails), p≤0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Donnie Bundman (Baylor College of Medicine) for technical assistance, Rebecca L. Thornton (Baylor College of Medicine) for help with RNA-seq details, Corey L. Reynolds and Mouse Phenotyping Core (Baylor College of Medicine) for electro- and echocardiograms, Gloria Vittone-Echeverria, Simona Pedrotti, and Ravi Singh (Baylor College of Medicine) for critical reading of the manuscript. This project was funded by the NIH (R01HL045565, R01AR060733, and R01AR045653) and Muscular Dystrophy Association (MDA 276796) grants to TAC. RNA-seq was performed in the Genomic and RNA Profiling Core (Baylor College of Medicine) with the assistance of the Core Director (L.D. White, PhD). JG is a Pew Latin American Fellow in the Biomedical Sciences supported by The Pew Charitable Trusts (#2933). ETW was funded by a post-doctoral fellowship from Myotonic Dystrophy Foundation. AK is funded by the American Heart Association (scientist development grant-11SDG4980011). AJW was funded by National Institute of Neurological Disorders and Stroke (NINDS-F31NS067740). WL is funded by CPRIT (RP110471), DOD (W81XWH-10-1-0501), and NIH (R01HG007538) grants. XHTW is funded by NIH (HL089598, HL091947, and HL117641), American Heart Association (13EIA14560061), Muscular Dystrophy Association (186530), and Fondation Leducq (08CVD01) grants.
Footnotes
AUTHORS´ CONTRIBUTIONS
JG designed research, performed the experiments, analyzed the data and wrote the manuscript. ZX, WL, performed computational analysis of sequencing reads from RNA-seq data, iCLIP and motif analysis of splicing events, and contributed to the manuscript. ETW, CBB performed computational analysis of sequencing reads from RNA-seq data and contributed to the manuscript. MAS performed RT-PCR validations and contributed with manuscript suggestions. AJW isolated CELF1-expressing hearts and controls for RNA-seq. AK provided suggestions for initial cell isolation set-up and contributed to the manuscript. WL, XHTW collaborated in T-tubule/calcium experiments. TAC supervised and designed research, analyzed the data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
NONE
ACCESSION Codes
RNA-Seq data have been deposited at NCBI Gene Expression Omnibus under accession code GSE49906.
REFERENCES
- 1.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X, et al. ASF/SF2-regulated CaMKII-delta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 3.Kalsotra A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zak R. Development and proliferative capacity of cardiac muscle cells. Circ. Res. 1974;32:17–26. [PubMed] [Google Scholar]
- 6.Nag A. Study of non-muscle cells of the adult mammalian heart: a fine structural analysis and distribution. Cytobios. 1980;28:41–61. [PubMed] [Google Scholar]
- 7.Camelliti P, Borg TK, Kohl P. Structural and functional characterization of cardiac fibroblasts. Cardiovasc. Res. 2005;65:40–51. doi: 10.1016/j.cardiores.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H1883–H1891. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
- 10.Miragoli M, Gaudesius G, Rohr S. Electrotonic modulation of cardiac impulse conduction by myofibroblasts. Circ. Res. 2006;98:801–810. doi: 10.1161/01.RES.0000214537.44195.a3. [DOI] [PubMed] [Google Scholar]
- 11.Tirziu D, Giordano FJ, Simons M. Cell communications in the heart. Circulation. 2010;122:928–937. doi: 10.1161/CIRCULATIONAHA.108.847731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeisberg EM, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 13.Takeda N, Manabe I. Cellular interplay between cardiomyocytes and nonmyocytes in cardiac remodeling. Int. J. Inflam. 2001;2011:535241. doi: 10.4061/2011/535241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahmoud AI, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ieda M, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1-integrin signaling. Dev. Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagnani M, et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ames EG, Lawson MJ, Mackey AJ, Holmes JW. Sequencing of mRNA identifies re-expression of fetal splice variants in cardiac hypertrophy. J. Mol. Cell Cardiol. 2013;62C:99–107. doi: 10.1016/j.yjmcc.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dillman AA, et al. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat. Neurosci. 2013;16:499–506. doi: 10.1038/nn.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Met. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 24.Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 over-expression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziman AP, Gómez-Viquez NL, Bloch RJ, Lederer WJ. Excitation-contraction coupling changes during postnatal cardiac development. J. Mol. Cell Cardiol. 2010;48:379–386. doi: 10.1016/j.yjmcc.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santalucía T, et al. Developmental regulation of GLUT-1 (erythroid/HepG2) and GLUT-4 (muscle/fat) glucose transporter expression in rat heart, skeletal muscle, and brown adipose tissue. Endocrinology. 1992;130:837–846. doi: 10.1210/endo.130.2.1370797. [DOI] [PubMed] [Google Scholar]
- 28.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001;104:2923–2931. doi: 10.1161/hc4901.100526. [DOI] [PubMed] [Google Scholar]
- 29.Van Rooij E, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda S, Pu WT. Expression and function of microRNAs in heart disease. Curr. Drug Targets. 2010;11:913–925. doi: 10.2174/138945010791591304. [DOI] [PubMed] [Google Scholar]
- 31.Porrello ER, et al. miR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao X, et al. microRNA profiling during rat ventricular maturation: a role for miR-29a in regulating cardiomyocyte cell cycle re-entry. FEBS Lett. 2013;587:1548–1555. doi: 10.1016/j.febslet.2013.01.075. [DOI] [PubMed] [Google Scholar]
- 33.Koshelev M, Sarma S, Price RE, Wehrens XH, Cooper TA. Heart-specific over-expression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlasova IA, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rattenbacher B, et al. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol. Cell Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda N, et al. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J. Clin. Invest. 2010;120:254–265. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terenzi F, Brimacombe KR, Penn MS, Ladd AN. CELF-mediated alternative splicing is required for cardiac function during early, but not later, postnatal life. J. Mol. Cell Cardiol. 2009;46:395–404. doi: 10.1016/j.yjmcc.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Dasgupta T, Stillwagon SJ, Ladd AN. Gene expression analyses implicate an alternative splicing program in regulating contractile gene expression and serum response factor activity in mice. PLoS One. 2013;8:e56590. doi: 10.1371/journal.pone.0056590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, Ohno K. CUGBP1 and MBNL1 preferentially bind to 3' UTRs and facilitate mRNA decay. Sci. Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds JO, et al. Junctophilin-2 is necessary for T-tubule maturation during mouse heart development. Cardiovasc. Res. 2013;100:44–53. doi: 10.1093/cvr/cvt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haddock PS, et al. Subcellular [Ca2+]i gradients during excitation-contraction coupling in newborn rabbit ventricular myocytes. Circ. Res. 1999;85:415–427. doi: 10.1161/01.res.85.5.415. [DOI] [PubMed] [Google Scholar]
- 43.Song LS, Sobie EA, McCulle S, Lederer WJ, Balke CW, Cheng H. Orphaned ryanodine receptors in the failing heart. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4305–4310. doi: 10.1073/pnas.0509324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng H, Lederer MR, Lederer WJ, Cannell MB. Calcium sparks and [Ca2+]i waves in cardiac myocytes. Am. J. Physiol. 1996;270:C148–C159. doi: 10.1152/ajpcell.1996.270.1.C148. [DOI] [PubMed] [Google Scholar]
- 45.Bark C, Bellinger FP, Kaushal A, Mathews JR, Partridge LD, Wilson MC. Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission. J. Neurosci. 2004;24:8796–8805. doi: 10.1523/JNEUROSCI.1940-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okoro DR, Rosso M, Bargonetti J. Splicing-up mdm2 for cancer proteome diversity. Genes Cancer. 2012;3:311–319. doi: 10.1177/1947601912455323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 48.Akhavan A. Motorized traffic of a cardiac ion channel: implication of conventional kinesin in transport of Kv1.5 channels to the plasma membrane. J. Physiol. 2010;588:903–904. doi: 10.1113/jphysiol.2009.186692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leu M, Ehler E, Perriard JC. Characterisation of postnatal growth of the murine heart. Anat. Embryol. 2001;204:217–224. doi: 10.1007/s004290100206. [DOI] [PubMed] [Google Scholar]
- 50.Udd B, Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012;11:891–905. doi: 10.1016/S1474-4422(12)70204-1. [DOI] [PubMed] [Google Scholar]
- 51.Groh WJ, et al. Electrocardiographic abnormalities and sudden death in myotonic dystrophy type 1. N. Engl. J. Med. 2008;358:2688–2697. doi: 10.1056/NEJMoa062800. [DOI] [PubMed] [Google Scholar]
- 52.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, Akerman M, Sun S, McCombie WR, Krainer AR, Zhang MQ. SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics. 2011;27:3010–3016. doi: 10.1093/bioinformatics/btr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, Ohno K. CUGBP1 and MBNL1 preferentially bind to 3' UTRs and facilitate mRNA decay. Sci. Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.