Abstract
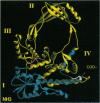
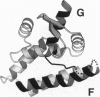
A recently described reverse gyrase from the hyperthermophilic methanogen Methanopyrus kandleri is the only known example of a heterodimeric type I topoisomerase. The enzyme is made up of a 42-kDa subunit which covalently interacts with DNA (RgyA) and a 138-kDa subunit which binds ATP (RgyB). We have now cloned and sequenced the genes for both subunits of this enzyme. Surprisingly, the universally conserved type I topoisomerase domain [Lima, C. D., Wang, J. C. & Mondragon, A. (1994) Nature (London) 367, 138-146] which has been found as a contiguous polypeptide in the prokaryotes and eukaryotes is shared between the protomers. The subdomain with the active-site tyrosine is entirely within RgyA, whereas the subdomain implicated in noncovalent binding of the cleaved DNA strand is contained entirely in RgyB. The appearance of this unique structure in a highly conserved enzyme family supports the hypothesis that the methanogens branched from other prokaryotes and eukaryotes very early in evolution.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrick D., Villanueba K., Childs J., Kalil R., Schneider T. D., Lawrence C. E., Gold L., Stormo G. D. Quantitative analysis of ribosome binding sites in E.coli. Nucleic Acids Res. 1994 Apr 11;22(7):1287–1295. doi: 10.1093/nar/22.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de la Tour C., Portemer C., Huber R., Forterre P., Duguet M. Reverse gyrase in thermophilic eubacteria. J Bacteriol. 1991 Jun;173(12):3921–3923. doi: 10.1128/jb.173.12.3921-3923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de la Tour C., Portemer C., Nadal M., Stetter K. O., Forterre P., Duguet M. Reverse gyrase, a hallmark of the hyperthermophilic archaebacteria. J Bacteriol. 1990 Dec;172(12):6803–6808. doi: 10.1128/jb.172.12.6803-6808.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri F., Elie C., Nadal M., de La Tour C., Forterre P., Duguet M. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGate R. J., Marians K. J. Identification of a potent decatenating enzyme from Escherichia coli. J Biol Chem. 1988 Sep 15;263(26):13366–13373. [PubMed] [Google Scholar]
- DiGate R. J., Marians K. J. Molecular cloning and DNA sequence analysis of Escherichia coli topB, the gene encoding topoisomerase III. J Biol Chem. 1989 Oct 25;264(30):17924–17930. [PubMed] [Google Scholar]
- Forterre P., Mirambeau G., Jaxel C., Nadal M., Duguet M. High positive supercoiling in vitro catalyzed by an ATP and polyethylene glycol-stimulated topoisomerase from Sulfolobus acidocaldarius. EMBO J. 1985 Aug;4(8):2123–2128. doi: 10.1002/j.1460-2075.1985.tb03902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994 Dec;14(12):8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J Mol Biol. 1985 Oct 5;185(3):625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- Kovalsky O. I., Kozyavkin S. A., Slesarev A. I. Archaebacterial reverse gyrase cleavage-site specificity is similar to that of eubacterial DNA topoisomerases I. Nucleic Acids Res. 1990 May 11;18(9):2801–2805. doi: 10.1093/nar/18.9.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima C. D., Wang J. C., Mondragón A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994 Jan 13;367(6459):138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttinger A. The twisted 'life' of DNA in the cell: bacterial topoisomerases. Mol Microbiol. 1995 Feb;15(4):601–606. doi: 10.1111/j.1365-2958.1995.tb02369.x. [DOI] [PubMed] [Google Scholar]
- Marguet E., Forterre P. DNA stability at temperatures typical for hyperthermophiles. Nucleic Acids Res. 1994 May 11;22(9):1681–1686. doi: 10.1093/nar/22.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., O'Dea M. H., Gellert M. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5960–5963. doi: 10.1073/pnas.75.12.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker A. G., Jr, Evans D. R. Isolation of chromosomal DNA from a methanogenic archaebacteria using a French pressure cell press. Biotechniques. 1991 Nov;11(5):612–613. [PubMed] [Google Scholar]
- Rivera M. C., Lake J. A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science. 1992 Jul 3;257(5066):74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- Shibata T., Nakasu S., Yasui K., Kikuchi A. Intrinsic DNA-dependent ATPase activity of reverse gyrase. J Biol Chem. 1987 Aug 5;262(22):10419–10421. [PubMed] [Google Scholar]
- Slesarev A. I., Zaitzev D. A., Kopylov V. M., Stetter K. O., Kozyavkin S. A. DNA topoisomerase III from extremely thermophilic archaebacteria. ATP-independent type I topoisomerase from Desulfurococcus amylolyticus drives extensive unwinding of closed circular DNA at high temperature. J Biol Chem. 1991 Jul 5;266(19):12321–12328. [PubMed] [Google Scholar]
- Tse-Dinh Y. C., Wang J. C. Complete nucleotide sequence of the topA gene encoding Escherichia coli DNA topoisomerase I. J Mol Biol. 1986 Oct 5;191(3):321–331. doi: 10.1016/0022-2836(86)90129-4. [DOI] [PubMed] [Google Scholar]
- Umezu K., Nakayama K., Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989 Jul 28;58(2):409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Watt P. M., Louis E. J., Borts R. H., Hickson I. D. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995 Apr 21;81(2):253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]