Abstract
Previously, we showed that ADAM10 is necessary for HIV-1 replication in primary human macrophages and immortalized cell lines. Silencing ADAM10 expression interrupted the HIV-1 life cycle prior to nuclear translocation of viral cDNA. Furthermore, our data indicated that HIV-1 replication depends on the expression of ADAM15 and γ-secretase, which proteolytically processes ADAM10. Silencing ADAM15 or γ-secretase expression inhibits HIV-1 replication between reverse transcription and nuclear entry. Here, we show that ADAM10 expression also supports replication in CD4+ T lymphocytes. The intracellular domain (ICD) of ADAM10 associates with the HIV-1 pre-integration complex (PIC) in the cytoplasm and immunoprecipitates and co-localizes with HIV-1 integrase, a key component of PIC. Taken together, our data support a model whereby ADAM15/γ-secretase processing of ADAM10 releases the ICD, which then incorporates into HIV-1 PIC to facilitate nuclear trafficking. Thus, these studies suggest ADAM10 as a novel therapeutic target for inhibiting HIV-1 prior to nuclear entry.
Keywords: ADAM10, ADAM15, γ-Secretase, HIV-1 replication, Macrophages, CD4+ T lymphocytes, HIV-1 pre-integration complex (PIC), HIV-1 integrase
Introduction
While numerous host factors involved in HIV-1 replication have been identified and their roles cataloged, our understanding of how the virus appropriates cellular machinery is still incomplete. Although the role of HIV-1 enzymes have been well characterized, processes and associated cellular factors adjacent to viral life cycle steps such as integration may reveal potential sites to target new therapy. One such example is the delineation of the path for the lentivirus pre-integration complex (PIC) to reach the nucleus. After entry, the HIV-1 reverse transcriptase converts viral RNA into cDNA in the reverse transcription complex (RTC), a nucleoprotein complex derived from the core of the infecting virion (Suzuki and Craigie, 2007). Thereafter, the newly synthesized viral cDNA is enzymatically processed by integrase (IN) and transitions from the RTC to PIC, a large nucleoprotein complex containing both viral and cellular proteins (Suzuki and Craigie, 2007). The viral IN protein is a key component of the PIC that mediates proviral integration into the host genome, and is also implicated in facilitating nuclear PIC import through its karyophilic properties (Depienne et al., 2001; Gallay et al., 1997; Suzuki and Craigie, 2007).
A widely accepted view is that the PIC harbors determinants that promote active nuclear import because it is larger (~28 nm) than the passive diffusion limit for nuclear pores (~9 nm) (Mattaj and Englmeier, 1998; Miller et al., 1997; Woodward et al., 2009). Numerous studies have documented the importance of HIV-1 capsid (CA), importins, and nuclear transport proteins, including NUP153, NUP358, CPSF6, and/or TNPO3 in PIC nuclear import (Ao et al., 2010; Hearps and Jans, 2006; Lee et al., 2010; Matreyek and Engelman, 2011; Schaller et al., 2011). The CA protein is essential for HIV-1 infection of nondividing cells, suggesting that viral core uncoating or CA interactions with nuclear pore proteins regulate lentiviral PIC nuclear import (Ambrose et al., 2012; Dismuke and Aiken, 2006; Lee et al., 2010; Yamashita and Emerman, 2006; Yamashita et al., 2007). Indeed, a single mutation in the CA protein (N74D) can alter HIV-1 dependence on NUP153, NUP358 and CPSF6 (Lee et al., 2010; Matreyek and Engelman, 2011; Schaller et al., 2011). In a recent review (Matreyek and Engelman, 2013) the authors proposed a model which elegantly accounts for the multiple CA dependencies during nuclear import. Briefly, initial CA binding to NUP358 docks the PIC to the cytoplasmic surface of the nuclear pore complex (NPC). At the NPC, CA interactions with CPSF6 facilitate transport through the NPC and subsequent CA-dependent transfer to NUP153 located on the nuclear surface of the NPC. This model is further supported by recent studies that identified the MX2 protein as a potent HIV-1 restriction factor that blocks nuclear import in a CA dependent manner. This interaction was disrupted by the N74D mutation (Goujon et al., 2013; Kane et al., 2013), similar to interactions with other nucleoproteins.
In previous work, our lab identified A Disintegrin and Metallo-protease (ADAM)10 as a vital host factor involved in HIV-1 replication in primary human macrophages, at the level of nuclear trafficking (Friedrich et al., 2011). In support, other groups have also implicated ADAM10 as important for HIV-1 replication (Brass et al., 2008; Swaminathan et al., 2012). ADAM10 is a cellular metalloprotease that activates numerous and diverse cellular proteins via proteolytic cleavage. ADAM10 itself is also proteolytically cleaved by regulated intramembrane processing, which occurs through the sequential actions of ADAM15 (which cleaves the extracellular domain of ADAM10) and by γ-secretase which releases the 6 kDa intracellular domain (ICD) (Tousseyn et al., 2009). Following cleavage, the ICD migrates to the nucleus, where it is thought to induce gene expression (Arima et al., 2007). We showed that siRNAs targeting ADAM15 and three subunits of γ-secretase (presenilin-2, presenilin enhancer protein 2, and nicastrin) inhibited HIV-1 replication similarly to ADAM10 siRNA (Friedrich et al., 2011). We also demonstrated that two inhibitors of γ-secretase (DAPT and L-685,458) inhibited HIV-1 replication, whereas inhibitors of the ADAM10 extracellular domain did not (Friedrich et al., 2011). Collectively, these data raised the possibility that the ADAM10 ICD, once released, may recruit the HIV-1 PIC during normal transit to the nucleus to promote PIC docking to the NPC or translocation into the nucleoplasm.
Here, we demonstrate that ADAM10 also supports HIV-1 replication in primary human CD4+ T lymphocytes. ADAM15 and γ-secretase were found to act at the same stage of the HIV-1 life cycle as ADAM10, suggesting that the proteolytic modification of ADAM10 by ADAM15 and γ-secretase is required for HIV-1 nuclear trafficking. In support of this model, we show that the ADAM10 ICD associates with the HIV-1 PIC during active infection and that the ICD immunoprecipitates with HIV-1 IN.
Results
ADAM10 is required for HIV-1 replication in Human CD4+ T lymphocytes
The requirement of ADAM10 expression during HIV-1 infection was further studied in CD4+ T lymphocytes, the primary circulating target cell in vivo. Transfecting CD4+ T lymphocytes with ADAM10 reduced ADAM10 expression by ~70% at the mRNA (Fig. 1A) and protein levels (Fig. 1B).
Fig. 1.
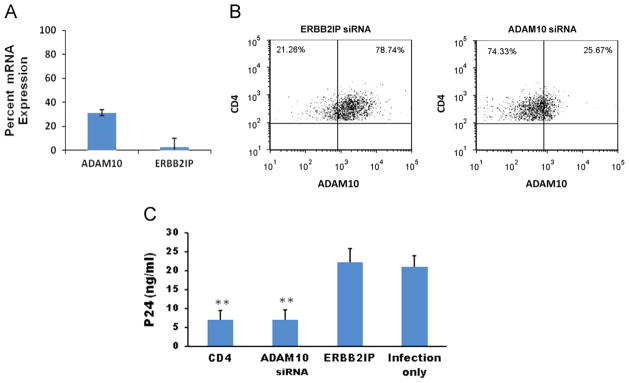
Inhibition of ADAM10 expression and HIV-1 production in CD4+ T lymphocytes by RNA interference (RNAi). (A) Non-adherent CD4+ T lymphocytes were transfected with ADAM10 or control ERBB2IP siRNAs using an AMAXA nucleofector system. Forty-eight hours after transfection, cells were harvested and total mRNA was isolated for cDNA synthesis. Target gene mRNA expression levels were quantitated by real time RT-PCR. Results are presented as percent expression compared to non-transfected cells. Experiments were performed in triplicate, and error bars represent standard deviations. (B) Flow cytometry data showing expression of ADAM10 in CD4+ T lymphocytes at 48 h post-transfection with control ERBB2IP or ADAM10 siRNAs. (C) Primary CD4+ lymphocytes (5 × 105 cells/24 well) were transfected via an AMAXA nucleofector system with 50 nM siRNAs (CD4, ADAM10, or ERBB2IP) 48 h prior to infection with dual-tropic HIV-189.6 (m.o.i. =0.02). At 7 days post-infection, Gag p24 was detected in culture supernatants by ELISA assays. The infection only control reflects HIV-1 production in untransfected cells that were infected in parallel. Data presented represent averages from four independent experiments. Error bars represent standard error. **p<0.01, compared with ERBB2IP control group.
CD4+ T lymphocytes were infected with a dual-tropic strain of HIV-1 (89.6) at 48 h post-transfection. At 7 days post-infection, supernatants were assayed for HIV-1 p24 production. Fig. 1C shows that silencing ADAM10 expression inhibited HIV-1 replication equally well as silencing CD4, the primary cellular receptor for HIV-1 (Dalgleish et al., 1984). ERBB2IP was used as a cellular target negative control siRNA in virus production assays because it exhibits no effect on HIV-1 replication (Murray et al., 2005). Reducing ADAM10 expression in CD4+ T lymphocytes did not affect viability or function (data not shown).
ADAM15 and γ-secretase act concordantly with ADAM10
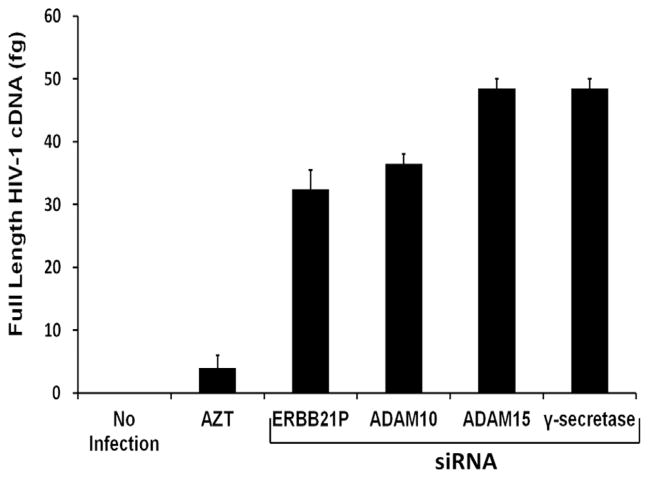
Previously, we demonstrated that knockdown of either ADAM15 or γ-secretase subunits significantly inhibited HIV-1 replication (Friedrich et al., 2011). These two proteases cooperate to sequentially cleave ADAM10 from the cell membrane (Tousseyn et al., 2009). Therefore, we hypothesized that ADAM15 and γ-secretase processing of ADAM10 may support HIV-1 replication at the same step as ADAM10. To test this hypothesis, we first determined whether ADAM15 and γ-secretase are required for completing HIV-1 reverse transcription. We transfected U373-MAGI-CCR5 cells with siRNAs specific for ADAM10, ADAM15, γ-secretase or ERBB2IP control and then infected with the R5-tropic HIV-1 strain SF162. Cells were harvested at 24 h and small, non-genomic DNA was isolated for quantification by real-time PCR. The PCR assay for full-length viral cDNA (Zack et al., 1990) showed that knockdown of ADAM15 or γ-secretase did not interfere with reverse transcription (Fig. 2), similar to results obtained by knocking down ADAM10 (Friedrich et al., 2011). As expected, viral cDNA formation was markedly inhibited by the reverse transcriptase inhibitor AZT. These data indicate that ADAM15 or γ-secretase function in HIV-1 replication following the completion of reverse transcription.
Fig. 2.
HIV-1 reverse transcription is unaffected by ADAM10, ADAM15 or γ-secretase downregulation. The U373-MAGI-CCR5 cell line was transfected with ADAM10, ADAM15, γ-secretase subunit presenilin enhancer protein 2, or ERBB21P siRNAs prior to infection with HIV-1 SF162. DNA was isolated 24 h post-infection and the formation of full length HIV-1 cDNA in the indicated transfectants was measured by quantitative real time PCR. The reverse transcription inhibitor AZT was used as a control.
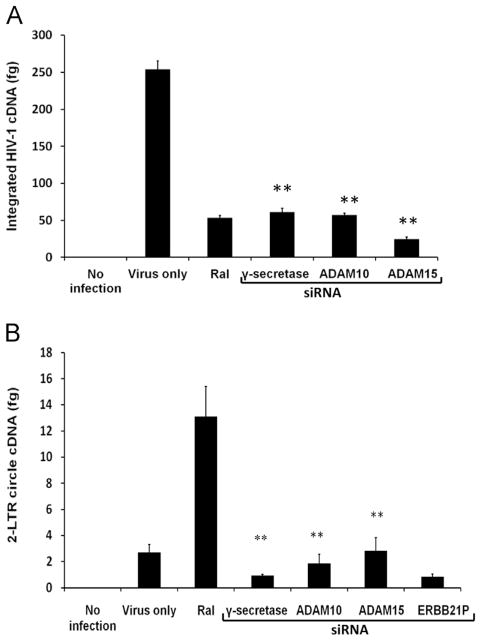
Experiments were performed to determine if ADAM15 or γ-secretase are required for proviral DNA integration or 2-LTR circle formation. Once viral cDNA enters the nucleus, it has two alternative fates: integration or 2-LTR circle formation (Bukrinsky et al., 1993; Farnet and Haseltine, 1991). U373-MAGI-CCR5 cells were transfected with siRNAs targeting ADAM10, ADAM15, γ-secretase or ERBB2IP, and infected after 48 h with the R5-tropic stain HIV-1 SF162. At 48 h post-infection, cells were harvested and genomic or small non-genomic cDNA were isolated to study the effects of gene silencing on integration or 2-LTR circle formation, respectively. Real-time PCR was used to quantify differences in HIV-1 integration or 2-LTR circle formation between control cells and ADAM15- or γ-secretase siRNA transfectants. Silencing ADAM15 or γ-secretase expression significantly inhibited viral integration with no corresponding increase in 2-LTR circle formation (Fig. 3). In contrast, the integrase inhibitor raltegravir blocked integration and caused a ~4-fold increase in 2-LTR circle formation, relative to untreated infected cells. The observation that both HIV-1 integration and 2-LTR circle formation are inhibited by ADAM15 or γ-secretase silencing suggests both enzymes participate in the nuclear translocation of viral cDNA. These findings are consistent with ADAM15 and γ-secretase acting concordantly with ADAM10 in HIV-1 replication, and lend support to the model previously proposed (Friedrich et al., 2011).
Fig. 3.
HIV-1 nuclear entry is inhibited by ADAM10, ADAM15 and γ-secretase downregulation. U373-MAGI-CCR5 cells were transfected with siRNAs specific for ADAM10, ADAM15, and γ-secretase, or ERBB2IP control and then infected after 48 h with R5-tropic HIV-1 strain SF162. At 48 h post-infection, cells were harvested and genomic and non-genomic DNA was isolated for quantification by real-time PCR. (A) Genomic DNA and real-time PCR primers recognizing integrated HIV-1 DNA were used to quantitate integrated proviral genomes. Raltegravir (Ral), an IN inhibitor was used as a control. (B) Formation of 2-LTR circles was quantitated by real-time PCR. All experiments were performed in triplicate. **p<0.01, compared with virus only (untransfected) control cells in panel A, or raltegravir-treated in panel B.
ADAM10 ICD associates with HIV-1 IN
To determine which component(s) of the PIC that the ADAM10 ICD may associate with during HIV-1 infection, we examined the reported properties of known PIC proteins. HIV-1 IN contains an SH3-like fold in its C-terminal end (Eijkelenboom et al., 1995). Interestingly, the ADAM10 ICD contains two proline-rich putative SH3-binding regions (Epis et al., 2010), leading us to hypothesize that ADAM10 ICD may interact with the PIC through IN. To test this hypothesis, we infected U373-MAGI-CCR5 cells by co-culture with HIV-1 producing U1 cells, and performed proximity ligation assays (PLA) using primary antibodies to IN and ADAM10 ICD. The PLA is an immunofluorescence assay based on the de novo synthesis of a fluorescent protein that occurs when two proteins colocalize. Cells are stained with primary antibodies specific to the two proteins of interest (ADAM10 ICD and IN), one raised in mice and one in rabbits. Primary antibodies are then labeled with secondary anti-rabbit and anti-mouse antibodies, each of which are conjugated to a DNA oligo comprising opposite strands of the coding sequence for red fluorescent protein (RFP). When the two secondary antibodies are within 40 nm of each other (reflecting juxtaposition of the proteins of interest), the oligos hybridize to form a double-stranded RFP expression cassette. The cells are then washed in a buffer containing RNA polymerase II, nucleotides, ribosomes and amino acids that drives rolling-circle amplification and the production of RFP, resulting in punctate patterns of red fluorescence. Results from the PLA assay showed that the ADAM10 ICD colocalizes with the viral IN (Fig. 4B).
Fig. 4.
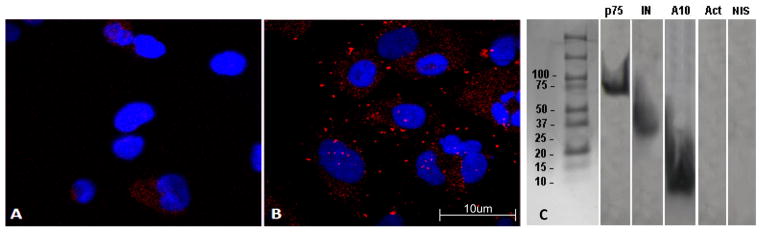
Colocalization and coimmunoprecipitation of the ADAM10 ICD and HIV-1 IN. U373 cells were co-cultured with the HIV-producing cell line, U1/HIV-1. Cells were stained with anti-IN and anti-ADAM10 ICD primary antibodies, followed by PLA secondary antibodies, and then fixed and imaged for immunofluorescence. Representative experiments are shown. (A) PLA results using an isotype-matched negative control anti-biotin antibody as the primary antibody. (B) Colocalization between the ADAM10 ICD and HIV-1 IN as revealed by PLA with punctate patterns of red fluorescence. Cell nuclei are stained with DAPI for contrast. (C) U373 cells were co-cultured for 6 h with an HIV-1 producing cell line, U1/HIV-1. Subsequently, cells were harvested and cytoplasmic fractions were collected. Co-immunoprecipitation was performed using an anti-IN mAb. Precipitates were separated by SDS-PAGE, transferred to a poly-vinyl membrane, and analyzed in Western blots. The membranes were cut into strips and probed with the indicated monoclonal antibodies. p75/LEDGF and HIV-1 IN were used as positive controls to confirm that whole PICs were precipitated. Actin and non-immune serum (NIS) were used as negative controls for non-specific precipitation of cellular proteins.
Coimmunoprecipitation studies were performed to confirm that the ADAM10 ICD associates with IN and/or the PIC during infection. U373 cells were co-cultured with HIV-producing U1 cells, and the cytoplasmic fraction was immunoprecipitated with anti-IN antibody and then studied by Western blot analysis. The blot was probed for p75/LEDGF as a positive control for PIC immunoprecipitation, HIV-1 IN, ADAM10 ICD, actin or pre-immune serum as negative controls for non-specific cellular proteins. ADAM10 ICD was found to coimmunoprecipitate with HIV-1 IN during active infection (Fig. 4C), confirming the results of the PLA.
Discussion
Previously, we described ADAM10 as a critical host factor involved in HIV-1 replication in immortalized cell lines and primary human macrophages (Friedrich et al., 2011). Here, we show that ADAM10 is also important for HIV-1 replication in CD4+ T lymphocytes, indicating that ADAM10 is broadly utilized in primary cell types of clinical relevance. These results were anticipated, given that ADAM10 functions in HIV-1 replication at a step downstream of viral entry.
ADAM15 and γ-secretase support HIV-1 replication following the completion of reverse transcription and before nuclear entry, as observed with ADAM10 (Friedrich et al., 2011). Thus, ADAM10, ADAM15 and γ-secretase potentially serve supportive roles in cytoplasmic trafficking, nuclear docking, or nuclear translocation. Given that ADAM15 and γ-secretase regulate the proteolytic release of the ADAM10 ICD, we considered the possibility that the ICD binds HIV-1 PICs during its normal trafficking to the nucleus (Arima et al., 2007). Proximity ligation assays and coimmunoprecipitation experiments were performed to explore potential interactions between the ADAM10 ICD and the viral IN or PIC. These studies demonstrated that the ADAM10 ICD does stably associate with HIV-1 IN in the PIC during infection, consistent with the possibility that the ICD tethers the PIC complex en route from the cytoplasm to the nuclear membrane.
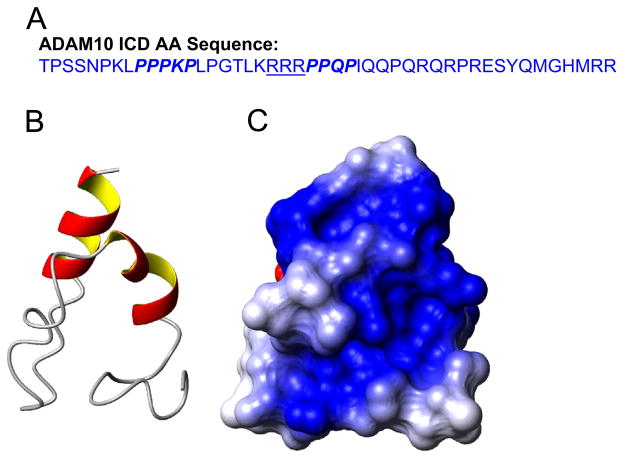
The ADAM10 ICD has two proline-rich SH3-binding motifs (Fig. 5A) that bind directly to SH3 domains of other signaling proteins (Epis et al., 2010). The C-terminal DNA-binding domain of HIV-1 IN consists of two domains that are similar to SH3-binding domains (Eijkelenboom et al., 1995). From our data, we predict that ADAM10 ICD and HIV-1 IN are interacting through these proline-rich sequences. Protein structure homology modeling predicts that the ADAM10 ICD contains a highly positively charged groove (Fig. 5B–C), reminiscent of the DNA binding domain of HIV-1 IN that associates with negatively charged HIV-1 cDNA (Eijkelenboom et al., 1995). This raises the possibility that ADAM10 ICD binds to the PIC through interaction with the HIV-1 DNA, as well as with integrase. The ADAM10 ICD possesses a classical KRRR nuclear localization signal (NLS) located between the two proline-rich regions (Fig. 5A), that we suggest is involved in transporting HIV-1 PIC into the nucleus. Although further studies are needed to verify how ADAM10, ADAM15 and γ-secretase mediate nuclear trafficking, this study represents some of the first evidence for a potential mechanism by which the HIV-1 PIC traverses the cytoplasm to reach the nuclear membrane.
Fig. 5.
Predicted tertiary structure of the ADAM10 ICD. (A) Amino acid sequence of the C-terminal 46 amino acids of the ADAM10 ICD. Putative SH-3 binding domains are shown in italics and the putative nuclear localization signal is underlined. Panels (B) and (C) show the ribbon diagram and surface electrostatics, respectively, of the ITASSER model of the ADAM10 ICD, in the same orientation. Blue shading indicates positively charged residues and red shading indicates negatively charged residues.
Their key roles in the early life cycle of HIV-1 make ADAM10 and ADAM15 very attractive targets for anti-retroviral therapy. The recently published finding that microRNA-155 targets ADAM10, TNPO3 and other cellular proteins necessary for HIV-1 replication implicates ADAM10 down regulation as part of an established antiviral response (Swaminathan et al., 2012). Given the evolutionarily recent introduction of HIV-1 into the human population, this is likely not an HIV-specific response. This suggests that ADAM10 may be involved in the infection and replication of other retroviruses, and that an antiviral drug therapy targeting ADAM10 may be broadly active.
Materials and methods
Cells, viruses and reagents
The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: U373-MAGI-CCR5 contributed by Drs. Michael Emerman and Adam Geballe (Harrington and Geballe, 1993). This is a glioblastoma cell line stably expressing human CD4, CCR5 and a HIV-1 Tat-responsive LTR-β-galactosidase, allowing for measurement of infection by β-galastosidase assays (McDonald et al., 2002). The U1/HIV-1 cell line (also obtained from the NIH AIDS Research and Reference Reagent Program) is a latently-infected promonocytic cell line that contains two integrated copies of the HIV-1 proviral genome, and which produces HIV-1 upon PMA stimulation (Folks et al., 1988, 1986). Primary human leukocytes were obtained from buffy coats from normal, healthy donors from the UTMB Blood Center. HIV-1 stocks of SF162 (4.2 × 105 TCID50/ml) and 89.6 (1 × 105 TCID50/ml) were purchased from the Virology Core Facility, Center for AIDS Research at Baylor College of Medicine, Houston, TX, and propagated and titered in human PBMCs.
siRNA transfections
siRNA SMARTpools targeting ADAM10 (M-004503-02), ERB-B2IP (M-031861-01), ADAM15 (M-004505-01), CD4 (M-005234-02), and γ-secretase component presenilin enhancer protein 2 (M-008057-02) were purchased from Dharmacon (Lafayette, CO). Cell lines were transfected with 50 nM siRNA using Oligofectamine (Invitrogen) 48 h prior to infection, according to manufacturer’s instructions. Primary CD4+ T lymphocytes were transfected with 50 nM siRNA using an Amaxa Nucleofector (Lonza, Walkersville, MD) 48 h prior to infection, following the manufacturer’s protocol.
Real time quantitative PCR
Real time PCR primers and probes were custom ordered from Sigma-Aldrich and Applied Biosystems. Genomic DNA was isolated using the Qiagen Blood and Tissue Kit (Qiagen, Valencia, CA) for detection of integrated viral genomes. Non-genomic small DNA was isolated using a Qiagen Miniprep kit for detection of viral cDNA and 2-LTR circles. Full length HIV-1 cDNA was assayed for with primers M667 (5′-GGC TAA CTA GGG AAC CCA CTG-3′) and M661 (5′-CCT GCG TCG AGA GAG CTC CTC TGG-3′), in conjunction with probe MH603 (5′-(FAM)-ACA CTA CTT GAA GCA CTC AAG GCA AGC TTT-(TAMRA)-3′) (Zack et al., 1990; Butler et al., 2001). We assayed for the presence of 2-LTR circles with primers MH535 (5′-AAC TAG GGA ACC CAC TGC TTA AG-3′) and MH536 (5′-TCC ACA GAT CAA GGA TAT CTT GTC-3′) in conjunction with the MH603 probe. To detect the presence of integrated viral genomes, we used a nested PCR strategy. The initial PCR amplification used Alu forward, (5′-GCC TCC CAA AGT GCT GGG ATT ACA G-3′) and HIV-1 gag reverse (5′-GCT CTC GCA CCC ATC TCT CTC C-3′) primers. The second step real-time PCR used LTR forward, (5′-GCC TCA ATA AAG CTT GCC TTG A-3′) and LTR reverse (5′-TCC ACA CTG ACT AAA AGG GTC TGA-3′) primers with an LTR probe (5′-FAM-GCG AGT GCC CGT CTG TTG TGT GAC TCT GGT AAC TAG CTC GC-DBH1-3′). A standard curve was established using serial dilutions of HIV-SX plasmid. Copy numbers of HIV-1 cDNA in each sample were calculated based on the resulting curve, and are expressed in femtograms (fg) of cDNA. Total mRNA was isolated using Qiagen RNeasy Mini Kits. All reactions were performed using Applied Biosystems TaqMan Universal Master Mix and run on an Applied Biosystems 7500 Fast Real Time PCR system and 7500 Fast System Software. Expression of all genes was normalized to GAPDH expression, and all reactions were performed in triplicate.
HIV-1 p24 assays
To determine the effect of silencing cellular genes on HIV-1 replication, non-adherent primary human PBMCs were transfected with siRNA SMARTpools for 48 h prior to infection. PBMCs were then infected in 24-well plates with HIV-89.6 (m.o.i=0.2), and p24 production was assayed from culture supernatants at 7 days post-infection using the p24 Capture ELISA kit (ImmunoDiagnostics, Woburn, MA).
Flow cytometry
Cells were harvested 48 h post-transfection and washed twice in FACS buffer (1% BSA, 0.5 mM EDTA, 0.1% Sodium Azide). Cells were then surface stained with mouse anti-human ADAM10-APC (R&D Systems) and mouse anti-human CD4-PE/Cy7 (R&D) for 1 h. After staining, cells were washed twice in FACS buffer and fixed in 2% paraformaldehyde from a 16% stock (Electron Microscopy Sciences, Hatfield, PA). Cells were visualized on a BD FacsCanto (UTMB Flow Cytometry and Cell Sorting Core Facility), and analysis was performed with FCS Express (DeNovo Software, Los Angeles, CA).
Co-immunoprecipitation assay
U373-MAGI-CCR5 cells were grown on cover slips and infected by co-culture for 6 h with activated U1/HIV-1 cells (PMA 1 μg/ml, 48 h). Subsequently, U1/HIV-1 cells were washed off, and cytoplasmic fractions of U373 cells were collected using a Nuclear Extraction Kit (Millipore, Temecula, CA), according to the manufacturer’s instructions. Proteins were labeled with the following biotinylated antibodies: p75/LEDGF was labeled with rabbit anti-LEDGF (Bethyl Laboratories, Montgomery, TX); IN was labeled with anti-IN IgG (NIH AIDS Research and Reference Reagent Program); ADAM10 ICD was labeled with goat anti-ADAM10 (T-17, Santa Cruz Biotech); actin was labeled with mouse anti-actin IgG (R&D Systems, Minneapolis, MN). Non-immune serum control was labeled with Human AB type serum (Lonza). Labeled proteins were then precipitated using Immunopure immobilized avidin (Pierce, Rockford, IL) by centrifugation at 1000g for 20 min at 4 °C. Proteins were dissociated from antibody bead complexes by boiling in SDS buffer for 5 min and centrifugation at 10,000g for 20 min, resolved by acrylamide gel electrophoresis, and transferred to PVDF membranes for Western blot analysis.
Proximity ligation assay
U373-MAGI-CCR5 cells were grown on cover slips and infected by 6 h co-culture with activated U1/HIV-1 cells (PMA 1 μg/ml, 48 h). Cells were then fixed in 4% formaldehyde, permeablized with methanol and washed three times. Cells were incubated with primary antibodies to ADAM10 (R&D Systems), IN (AIDS Reagent Program) or biotin (rabbit anti-biotin, Bethyl Laboratories) and washed three times in PBS. Cells were then stained with PLA probes (mouse- or rabbit-specific secondary antibodies labeled with DNA tails), provided as part of the PLA kit (Olink Bioscience, Uppsala, Sweden). When these DNA tails of the secondary antibodies are in close proximity (<40 nm), they hybridize, indicating the primary antibody targets are in close proximity. Cells are then washed in a buffer containing an RNA polymerase, ribosomes, nucleotides and amino acids, and the hybridized probes undergo rolling-circle amplification and express red-fluorescent protein.
PLA imaging was performed by the UTMB Cellular Microscopy Core. Images were collected using a Zeiss LSM-510 META confocal microscope with a 100 × 1.40 numerical aperture oil immersion objective (Optical Microscopy Core at UTMB) at a zoom of 0.7. The images were obtained using two different lines of excitation (364, 543 nm) and two different channels of emission with sequential acquisition. Emission was measured with a 435–485 nm filter following excitation at 364 nm. Emission was measured with a 560–615 nm filter following excitation at 543 nm. All images were collected using 8-frame-Kallman-averaging with a pixel time of 1.03 μs, a pixel size of 110 nm and frame size of 512 × 512 pixels and an optical slice of 0.8 μm. Z-stack acquisition was done at z-steps of 0.6 μm.
Homology modeling of ADAM10 ICD
Widely used depositories of homology models such as MOD-BASE (Pieper et al., 2011) and SWISSMODEL (Kiefer et al., 2009) did not generate models with a high degree of confidence. However, the homology modeling suite ITASSER (Wu et al., 2007) did generate a molecular model of the ADAM10 ICD, using all 46 amino acids of the ADAM10 C-terminus. The model C-score was −2.08 on a scale of −5 to 2 (2 being perfect model). It had two small helices of 8 amino acids. We used this top model to show the topology (ribbon plot) and global charge distribution on the surface. All model visualizations were performed with the program Molmol (Koradi et al., 1996).
Acknowledgments
This work was partly supported by the Public Health Service Grant HL088999 from the National Heart, Lung, and Blood Institute. MAE was also supported by the Public Health Service-NIAID-NIH T32 Training Grant AI007536. We would like to thank Merck & Co., Inc. for generously providing raltegravir, the NIH AIDS Research and Reference Reagent Program for providing U1/HIV-1 and U373-MAGI-CCR5 cell lines, Adriana Paulucci-Holthauzen, Ph. D. (Manager of the Optical Microscopy Core at the University of Texas Medical Branch, Galveston, TX) for assistance with confocal microscopy imaging, and Edward Siwak, Ph.D. (Associate Director of Virology Core Facility, Center for AIDS Research at Baylor College of Medicine, Houston, TX) for providing HIV-1SF162 and HIV-189.6. This project was also partially supported by the Division of Infectious Diseases, Department of Internal Medicine, University of Texas Medical Branch, Galveston, TX.
Contributor Information
Mark A. Endsley, Email: maendsle@utmb.edu.
Anoma D. Somasunderam, Email: asomasun@utmb.edu.
Guangyu Li, Email: LIG001@mail.etsu.edu.
Numan Oezguen, Email: numan.oezguen@bcm.edu.
Varatharasa Thiviyanathan, Email: Varatharasa.Thiviyanathan@uth.tmc.edu.
James L. Murray, Email: jmurray100@yahoo.com.
Donald H. Rubin, Email: don.h.rubin@vanderbilt.edu.
Thomas W. Hodge, Email: twhodge3@gmail.com.
William A. O’Brien, Email: william.obrien@boehringer-ingelheim.com.
Briana Lewis, Email: br3lewis@utmb.edu.
References
- Ambrose Z, Lee K, Ndjomou J, Xu H, Oztop I, Matous J, Takemura T, Unutmaz D, Engelman A, Hughes SH, KewalRamani VN. Human immunodeficiency virus type 1 capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection. J Virol. 2012;86 (8):4708–4714. doi: 10.1128/JVI.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Z, Danappa Jayappa K, Wang B, Zheng Y, Kung S, Rassart E, Depping R, Kohler M, Cohen EA, Yao X. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J Virol. 2010;84 (17):8650–8663. doi: 10.1128/JVI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima T, Enokida H, Kubo H, Kagara I, Matsuda R, Toki K, Nishimura H, Chiyomaru T, Tatarano S, Idesako T, Nishiyama K, Nakagawa M. Nuclear translocation of ADAM-10 contributes to the pathogenesis and progression of human prostate cancer. Cancer Sci. 2007;98 (11):1720–1726. doi: 10.1111/j.1349-7006.2007.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319 (5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Bukrinsky M, Sharova N, Stevenson M. Human immunodeficiency virus type 1 2-LTR circles reside in a nucleoprotein complex which is different from the preintegration complex. J Virol. 1993;67 (11):6863–6865. doi: 10.1128/jvi.67.11.6863-6865.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7 (5):631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312 (5996):763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Depienne C, Mousnier A, Leh H, Le Rouzic E, Dormont D, Benichou S, Dargemont C. Characterization of the nuclear import pathway for HIV-1 integrase. J Biol Chem. 2001;276 (21):18102–18107. doi: 10.1074/jbc.M009029200. [DOI] [PubMed] [Google Scholar]
- Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80 (8):3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom AP, Lutzke RA, Boelens R, Plasterk RH, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2 (9):807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- Epis R, Marcello E, Gardoni F, Vastagh C, Malinverno M, Balducci C, Colombo A, Borroni B, Vara H, Dell’Agli M, Cattabeni F, Giustetto M, Borsello T, Forloni G, Padovani A, Di Luca M. Blocking ADAM10 synaptic trafficking generates a model of sporadic Alzheimer’s disease. Brain. 2010;133 (11):3323–3335. doi: 10.1093/brain/awq217. [DOI] [PubMed] [Google Scholar]
- Farnet CM, Haseltine WA. Circularization of human immunodeficiency virus type 1 DNA in vitro. J Virol. 1991;65 (12):6942–6952. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks TM, Kessler SW, Orenstein JM, Justement JS, Jaffe ES, Fauci AS. Infection and replication of HIV-1 in purified progenitor cells of normal human bone marrow. Science. 1988;242 (4880):919–922. doi: 10.1126/science.2460922. [DOI] [PubMed] [Google Scholar]
- Folks TM, Powell D, Lightfoote M, Koenig S, Fauci AS, Benn S, Rabson A, Daugherty D, Gendelman HE, Hoggan MD, et al. Biological and biochemical characterization of a cloned Leu-3-cell surviving infection with the acquired immune deficiency syndrome retrovirus. J Exp Med. 1986;164 (1):280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich BM, Murray JL, Li G, Sheng J, Hodge TW, Rubin DH, O’Brien WA, Ferguson MR. A functional role for ADAM10 in human immunodeficiency virus type-1 replication. Retrovirology. 2011;8:32. doi: 10.1186/1742-4690-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94 (18):9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, Hue S, Barclay WS, Schulz R, Malim MH. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502 (7472):559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington RD, Geballe AP. Cofactor requirement for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67 (10):5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearps AC, Jans DA. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem J. 2006;398 (3):475–484. doi: 10.1042/BJ20060466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502 (7472):563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Gr. 1996;14 (1):51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7 (3):221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Engelman A. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J Virol. 2011;85 (15):7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Engelman A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses. 2013;5 (10):2483–2511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159 (3):441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71 (7):5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JL, Mavrakis M, McDonald NJ, Yilla M, Sheng J, Bellini WJ, Zhao L, Le Doux JM, Shaw MW, Luo CC, Lippincott-Schwartz J, Sanchez A, Rubin DH, Hodge TW. Rab9 GTPase is required for replication of human immunodeficiency virus type 1, filoviruses, and measles virus. J Virol. 2005;79 (18):11742–11751. doi: 10.1128/JVI.79.18.11742-11751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, Datta RS, Sampathkumar P, Madhusudhan MS, Sjolander K, Ferrin TE, Burley SK, Sali A. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–D474. doi: 10.1093/nar/gkq1091. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. HIV-1 capsid–cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7 (12):e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Craigie R. The road to chromatin—nuclear entry of retroviruses. Nat Rev Microbiol. 2007;5 (3):187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- Swaminathan G, Rossi F, Sierra LJ, Gupta A, Navas-Martin S, Martin-Garcia J. A role for microRNA-155 modulation in the anti-HIV-1 effects of Toll-like receptor 3 stimulation in macrophages. PLoS Pathog. 2012;8 (9):e1002937. doi: 10.1371/journal.ppat.1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tousseyn T, Thathiah A, Jorissen E, Raemaekers T, Konietzko U, Reiss K, Maes E, Snellinx A, Serneels L, Nyabi O, Annaert W, Saftig P, Hartmann D, De Strooper B. ADAM10, the rate-limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS-9, ADAMS-15, and the gamma-secretase. J Biol Chem. 2009;284 (17):11738–11747. doi: 10.1074/jbc.M805894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward CL, Prakobwanakit S, Mosessian S, Chow SA. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J Virol. 2009;83 (13):6522–6533. doi: 10.1128/JVI.02061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Retroviral infection of non-dividing cells: old and new perspectives. Virology. 2006;344 (1):88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3 (10):1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack J, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61 (2):213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]