Abstract
Epstein-Barr Virus Nuclear Antigen (EBNA) 2 features an Arg-Gly repeat (RG) domain at amino acid positions 335-360, which is a known target for protein arginine methyltransferaser 5 (PRMT5). In this study, we performed protein affinity pull-down assays to demonstrate that endogenous PRMT5 derived from lymphoblastoid cells specifically associated with the protein bait GST-E2 RG. Transfection of a plasmid expressing PRMT5 induced a 2.5- to 3-fold increase in EBNA2-dependent transcription of both the LMP1 promoter in AKATA cells, which contain the EBV genome endogenously, and a Cp-Luc reporter plasmid in BJAB cells, which are EBV negative. Furthermore, we showed that there was a 2-fold enrichment of EBNA2 occupancy in target promoters in the presence of exogenous PRMT5. Taken together, we show that PRMT5 triggers the symmetric dimethylation of EBNA2 RG domain to coordinate with EBNA2-mediated transcription. This modulation suggests that PRMT5 may play a role in latent EBV infection.
Keywords: EBV, EBNA2-dependent transcription, PRMT5, Ariginine symmetric dimethylation
1. Introduction
Epstein-Barr Virus (EBV) is an oncogenic virus that is associated with a broad spectrum of human malignancies. EBV is capable of converting human B lymphocytes into indefinitely transformed lymphoblastoid cells lines (LCLs) in vitro. Maintenance of EBV-immortalized LCLs requires the expression of a cluster of latency-specific viral genes. EBNA2 is expressed immediately after EBV infection and plays an essential role in EBV-mediated transformation of primary B cells by activating transcription. Transcription of the BamHI C promoter (Cp), the latent membrane proteins LMP1, LMP2A, and LMP2B, and cellular genes including c-myc, CD21, CD23 and c-fgr appears to be partially dependent on EBNA2. Due to the lack of a classical DNA binding domain, EBNA2 is tethered to target promoters through interactions with cellular transcription factors, such as RBP-Jκ, PU.1, AUF1, and CRE [For reviews see [1]].
EBNA2 contains a C-terminal activation domain (AD) that induces the assembly of the basal transcription machinery and an N-terminal activation domain (AD2) that is involved in transcriptional regulation [2,3]. The intermediate conserved WWP motif confers binding to RBP-Jκ. Mutations in these residues lead to the abrogation of EBNA2-dependent transcription and EBV-transforming capacity [4]. EBNA2 contains an arginine-glycine (RG) repeat spanning the amino residues 337 to 352. This RG repeat has been implicated in protein arginine methyltransferase 5 (PRMT5)-mediated symmetrical dimethylation [5]. The symmetrically dimethylated arginine (sDMA) residues have been shown to trigger EBNA2 binding to the Tudor domain of the survival motor neuron protein (SMN). Although deletion of the RG domain does not affect EBNA2-dependent transcription of target promoters in cell-based reporter assays, a recombinant EBV that contains this deletion exhibits substantially reduced transforming activity [6].
PRMTs catalyze the methylation of the terminal nitrogen atoms in guanidium side chains of arginine residues in proteins [7]. Like all type II PRMTs, PRMT5 triggers the symmetric dimethylation of arginine residues that are involved in different cellular processes [8]. Induction of interleukin-2 by PRMT5 was observed in mitogen activated Jurkat T cells and human peripheral blood lymphocytes [9]. In contrast, methylation of H3 at Arg-8 and H4 at Arg-3 by PRMT5 leads to the repression of both ST7 and NM23 [10]. In addition to EBNA2, EBNA1 is also a substrate for PRMT5 [11], which suggests that PRMT5 plays a role in latent EBV infection. An advanced understanding of the effects that PRMT5 has on EBNA1 and EBNA2 function remains to be elucidated.
In this study, we demonstrated that PRMT5 binds to the EBNA2 RG domain and catalyzes the symmetric dimethylation of the arginine residue(s). This biochemical event increases the occupancy of EBNA2 on target promoters and leads to enhanced transcriptional activity. Our data suggest that PRMT5 plays a role in the maintenance of transformation in EBV-infected B cells.
2. Materials and methods
2.1 Plasmids and DNA recombination
The expression vectors for the GST-EBNA2 (GST-E2) acidic domain (AD) and GST-E2 1-103 were described previously [12]. The remaining GST-E2 expression plasmids were generated by amplifying the sequences flanking the EBNA2 gene by polymerase chain reaction (PCR) using the appropriate pairs of primers and then subcloning the amplicons into the BamHI and EcoRI sites of the plasmid pGEX-2TK (GE Healthcare). The expression vectors for pSG5-EBNA2 (E2) and pSG5-EBNA2 ΔRG have been previously described [2]. The vector containing PRMT5 cDNA was purchased from GeneDiscovery, Taiwan. The PRMT5 flanking sequences were amplified by PCR and then subcloned into the XhoI and BglII sites of the pSG5-flag [13] or pTrcHis vector (Invitrogen). The expression vector for the PRMT5 368R/A mutant was generated by PCR mutagenesis according to the manufacturer's protocol (Stratagene). All of the resulting expression vectors were verified by DNA sequencing (Genomics BioSci & Tech Taiwan).
2.2 Cell culture and cell-based transcription reporter assays
BJAB is an EBV-negative cell line, and AKATA is a type I B-lymphoma cell line that is latently infected with EBV [14,15]. Cells were cultured in RMPI 1640 medium (Gibco BRL) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. BJAB cells (1 × 107) were transfected with 5 μg of the Cp reporter plasmid [13,16], 1 μg of the CMV-β gal control plasmid, 10 μg of either the EBNA2 or ΔRG expression vector and 30 μg of either the FPRMT5 or 368R/A expression plasmid using the BTX820 electroporator. Luciferase and β-galactosidase reporter assays were then performed on the cells 18 hours (hrs) post transfection. A similar experiment was also conducted using AKATA cells without the reporter plasmids. Expression levels of LMP1 from the endogenous EBV genome were measured to look EBNA2-dependent transcription and the co-activating effects of the transfected plasmids.
2.3 Immunoblotting analyses and chromatin immunoprecipitation (ChIP) assays
Cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then immunoblotted using antibodies for EBNA2 (Millipore), PRMT5 (Santa Cruz), or flag-epitope (M2, Sigma). For the in vitro methylation assay, the methylated arginine residues in the GST-E2 RG fusion protein were identified with an anti-symmetric-dimethyl-arginine antibody (SYM10). The proteins of interest were detected and visualized using an ECL detection kit (Millipore).
BJAB cells (1 × 107) co-transfected with the appropriate amounts of the EBNA2, FPRMT5 and LMP1-Luc expression vectors were subjected to an enzymatic shearing protocol (Active motif) followed by a subsequent ChIP analysis using antibodies for EBNA2 (PE2; from Abcam) and PRMT5 (from SantaCruz). The abundance of EBNA2 and PRMT5 on the LMP1 promoter was monitored by quantitative real time PCR (qPCR) using the following primers: 5′- TCCCACAACACTACTCACT-3′ (forward) and 5′- AGCGTCAGAGGAAATGGAAAG-3′ (reverse). The ChIP assay kit for the GAPDH promoter was purchased from Millipore. The protocol for qPCR was described previously [17]. The promoter occupancy of the indicated proteins and an IgG control was normalized to the input DNA, and the resulting promoter abundance for each sample was expressed as the relative intensity compared to the IgG control.
2.4 Immunofluorescence microscopy
Immunofluorescence (IF) analysis was performed according to the immunostaining protocol described previously [18] using specific antibodies for EBNA2 (Millipore) and PRMT5 (Santa Cruz). In co-immunostainings, rhodamin-conjugated goat anti-mouse and FITC-conjugated donkey anti-goat (Kirkegaard & Perry Laboratories, Inc) antibodies were used as fluorochromes. In addition, DNA was counterstained with DRAQ5 (Bio Status). The nuclear co-localization of EBNA2 and PRMT5 was visualized by confocal microscopy (LEICA TCS SP2 AOBS).
2.5 Statistic analysis
The data that were obtained from the cell-based reporter assays and ChIP analyses are represented as the mean ± the standard error of the mean (SEM) from three independent experiments. Whenever necessary, statistical comparisons were performed by a one-way ANOVA. A p-value of less than 0.05 was considered to be statistically significant.
2.6 In vitro methylation assay
The protocol for the arginine methylation assay has been described previously [19]. Briefly, 293T cells (2 × 106) that were transfected with 10 μg of either the FPRMT5 or 368R/A expression vector were harvested 36 hrs post transfection. Anti-flag antibody-conjugated agarose beads were used to purify either PRMT5 or the catalytic mutant (368R/A). Five micrograms of either GST or GST-E2 335-360 and the appropriate amounts of purified PRMT5 or 368R/A were incubated in a 40-μl reaction buffer (25 mM Tris-HCl (pH:7.5), 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, and 45 mM AdoMet) at 30°C for 2 hrs. Methylation reactions were stopped by the addition of 2× SDS sample buffer (50 mM Tris-HCl (pH: 6.8), 100 mM DTT, 2% SDS, 10% glycerol, and 0.1% bromophenol blue).
3. Results
3.1 PRMT5 specifically binds to the EBNA2 RG motif
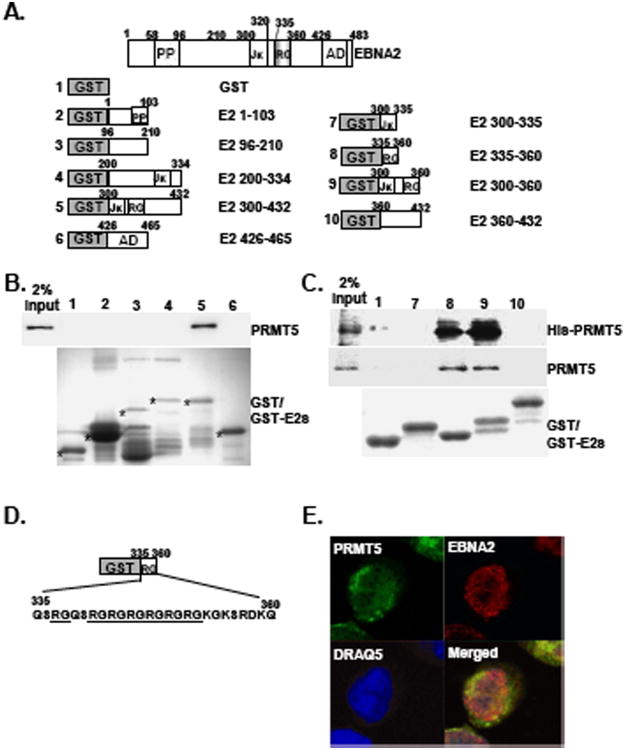
PRMT5 has been implicated in the methylation of arginines in the EBNA2 RG repeat domain [5], suggesting that there is likely a direct or indirect physical interaction between EBNA2 and PRMT5. To explore the mechanism of PRMT5-mediated methylation of the EBNA2-RG domain, we first characterized the potential protein-protein interactions between EBNA2 and PRMT5. GST and GST-fusion proteins with the overlapping EBNA2 open reading frames (ORFs) (GST-E2s), which included the amino acids (aa) 1-103, 96-210, 200-334, 300-432, and 426-465 (AD) and covered almost the entire EBNA2 protein, were purified from E. coli and used as the affinity matrices to pull down cellular proteins from the IB4 cell lysates (Fig. 1 A). Endogenous PRMT5 was pulled down by the protein bait GST-E2 300-432 but not the GST control or other GST-E2 fusion proteins (Fig. 1B). These results suggest that the amino acid region between residues 300 and 432 of EBNA2 contains an intact PRMT5 binding domain.
Fig. 1.
The EBNA2 RG repeat domain is the binding site for PRMT5 Schematic depiction of the GST-EBNA2 fusion proteins (GST-E2s). The amounts of the GST or GST-E2 fusion proteins were quantitated by coomassie blue staining following SDS-PAGE analysis. Each protein is marked with an individual asterisk (A). GST or GST-E2 fusion proteins were used as protein baits to pull down cell lysates from IB4 LCL (B) or the his-PRMT5 recombinant protein (C). The amount of endogenous PRMT5 or his-PRMT5 that was pulled down was determined by immunoblot analysis. Two percent input of PRMT5 or his-PRMT5 is shown. The coding sequences of the EBNA2 RG domain are shown (D). The immunofluorescence (IF) analysis was carried out using antibodies for EBNA2 (PE2) and PRMT5 (C-20) followed by counterstaining with a goat anti-mouse antibody conjugated to Rodamin (Red) or a donkey anti-goat antibody conjugated to FITC (Green). Confocal images for EBNA2 (Red) and PRMT5 (Green) from IB4 cells are shown. Nuclei were counterstained with DRAQ5 (Blue)(E).
To identify the specific PRMT5-binding domain located within the region spanning residues 300 to 432, protein affinity pull-down assays were performed using similar amounts of GST and GST-E2 fusions that contained smaller portions of the EBNA2 ORF, including aa 300-335, 335-360, 300-360, and 360-432. Approximately 5% of endogenous PRMT5 was pulled down by GST-E2 335-360 and GST-E2 300-360 (Fig. 1C). In contrast, neither GST nor any of the remaining GST-E2s were able to bind to PRMT5. Similarly, the in vitro protein affinity binding assay further indicated that recombinant his-tagged PRMT5 specifically bound to GST-E2 335-360, which contains an intact RG tail (Fig. 1D). This result revealed that the EBNA2 RG domain is critical for PRMT5 binding, although it is possible that the region spanning aa 353-360 may also play a role. To explore the in vivo interactions between EBNA2 and PRMT5 in lymphoblastoid cells, we used confocal immunofluorescence microscopy to visualize this biochemical event. Similar to previous studies [20,21], we showed that the localization of PRMT5 is dispersed throughout the whole cell, while EBNA2 is localized to the nucleus (Fig 1E). Our results further indicate that nuclear co-localization of EBNA2 and PRMT5 takes place in IB4 LCL, suggesting that protein-protein interactions took place in vivo.
3.2 PRMT5 potentiates EBNA2-dependent transcription
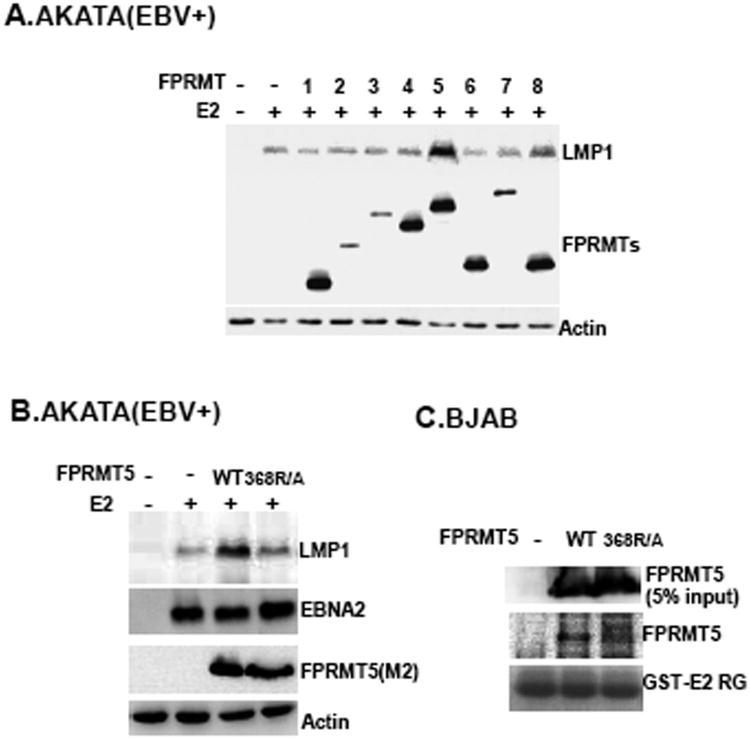
To assess whether the biochemical interactions between PRMT5 and EBNA2 are correlated with EBNA2-mediated transcription, PRMTs, including 1-8, were ectopically expressed in type I AKATA BL cells, which are latently infected with EBV, and EBNA2-induced LMP1 expression from the endogenous EBV genome was assayed. Each PRMT was expressed as a flag-tagged protein that could be recognized by a flag-epitope specific antibody. Among those ectopically expressed PRMTs, only PRMT5 induced about a 3.5-fold increase in LMP1 expression in the presence of EBNA2 compared to LMP1 expression induced by EBNA2 alone (Fig. 2A). The expression levels of plasmid-expressed PRMT- 1, 4, 5, 6, and 8 were similar but were relatively higher than the levels of PRMT- 2, 3, and 7. This result suggested that PRMT5 could trigger the upregulation of EBNA2-mediated transcription. The expression vector for the PRMT5 catalytic mutant (368R/A) was used next to determine if the enzymatic activity of PRMT5 is required for co-stimulation with EBNA2. We found that although ectopically expressed 368R/A was similar to PRMT5, transcriptional upregulation was not observed when EBNA2 and 368R/A were co-expressed (Fig. 2B). This result suggests that PRMT5 induces methylation to modulate EBNA2-dependent transcription. A protein pull-down assay further verified that GST-E2 335-360 associates with ectopically expressed PRMT5 rather than the 368R/A mutant (Fig. 2C). Our data indicate that the physical interaction between PRMT5 and EBNA2 is essential for PRMT5 co-stimulation with EBNA2.
Fig. 2.
PRMT5 is able to augment EBNA2-dependent transcription. AKATA cells (1 × 107) co-transfected with 10 μg of the EBNA2 expression vector (E2) and 30 μg of the indicated FPRMT expression plasmids (1 to 8) were subjected to immunoblot analysis using an LMP1 specific antibody (S12) 48 hours (hrs) post transfection. Actin was used a loading control. The ectopically expressed FPRMT plasmids contained an N-terminal flag-tagged epitope that was identified by an anti-flag M2 antibody. An actin specific antibody was used to visualize actin in each sample (A). The immunoblots for LMP1, EBNA2, FPRMT5, and actin are shown (B). Protein affinity pull-down assays using GST E2 RG as bait were performed on cell lysates from BJAB cells that were transfected with either PRMT5 or the 368R/A expression vector. The amount of ectopically expressed FPRMT5 or 368R/A that was pulled down by the protein bait was determined by immunoblot analysis. The input of transfected FPRMT5 or 368R/A is shown (C).
3.3 PRMT5 and EBNA2 co-stimulation occurs through the RG repeat domain
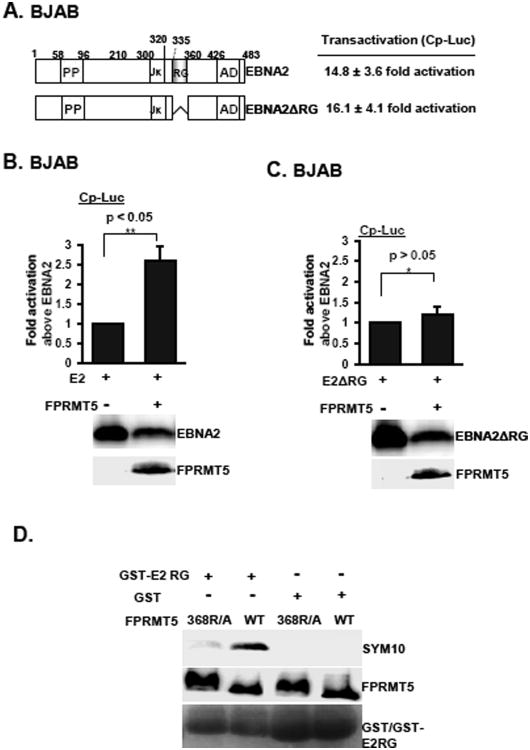
Because it is known that the EBNA2 RG-repeat domain is a unique PRMT5 binding site, we determined if PRMT5 co-stimulation with EBNA2 is an RG-dependent process. The co-stimulating effects that were produced by PRMT5 and either EBNA2 or EBNA2 ΔRG were compared using the data from the cell-based reporter assays. In multiple experiments, EBNA2 ΔRG displayed wild-type activity on the Cp reporter plasmid (Cp-Luc) (Fig. 3A). PRMT5 consistently induced a 2.5-fold increase in EBNA2-induced Cp-Luc reporter activity (Fig. 3B). In contrast, plasmids expressing PRMT5 were unable to co-stimulate with the EBNA2 ΔRG-induced reporter activity (Fig. 3C). To demonstrate that the EBNA2 RG tail serves as a substrate for PRMT5, an in vitro methylation assay was performed to compare the enzymatic activity of PRMT5 and 368R/A in triggering the symmetric dimethylation of GST-E2 RG (Fig. 3D). As expected, the GST protein alone did not serve as a substrate for PRMT5. Although similar amounts of the GST-E2 RG substrates were used in each assay, we found that only wild-type PRMT5 could actively mediate the symmetric dimethylation of the target protein; the catalytic activity was almost completely abrogated in the PRMT5 368R/A mutant. These data suggest that PRMT5-dependent symmetric dimethylation of the RG repeat domain is responsible for EBNA2-mediated transcriptional upregulation.
Fig. 3.
PRMT5 triggers the symmetric dimethylation of arginine residues to stimulate EBNA2-dependent transcription. The schematic diagram of EBNA2 and EBNA2 ΔRG is shown (A left panel). BJAB cells that were co-transfected with either EBNA2 or EBNA2 ΔRG, the Cp-Luc reporter plasmid, and the β-Gal internal control were collected for luciferase and β-Gal activity assays 18 hrs post transfection. The luciferase activity produced by each transfectant was normalized to β-Gal activity (A right panel). The same experiment as described in A was performed with the addition of the expression vector of FPRMT5. The fold induction of activity in cells co-transfected with EBNA2 and FPRMT5 (B) or EBNA2-ΔRG and 368R/A (C) was expressed relative to the basal activity induced by EBNA2 or EBNA2-ΔRG. Data are represented as the mean ± the standard deviation from three independent experiments. P* value>0.05. P** value<0.05. The expression levels of EBNA2, EBNA2-ΔRG, and FPRMT5 are shown. The FPRMT5 or 368R/A plasmids were purified from 293T cells. The GST or GST-E2 RG recombinant proteins were used for the in vitro methylation assay. The symmetric dimethylated arginines were visualized by immunoblotting with the SYM10 antibody (D).
3.4 PRMT5 enhances EBNA2-dependent transcription by increasing the promoter occupancy of EBNA2 on target promoters
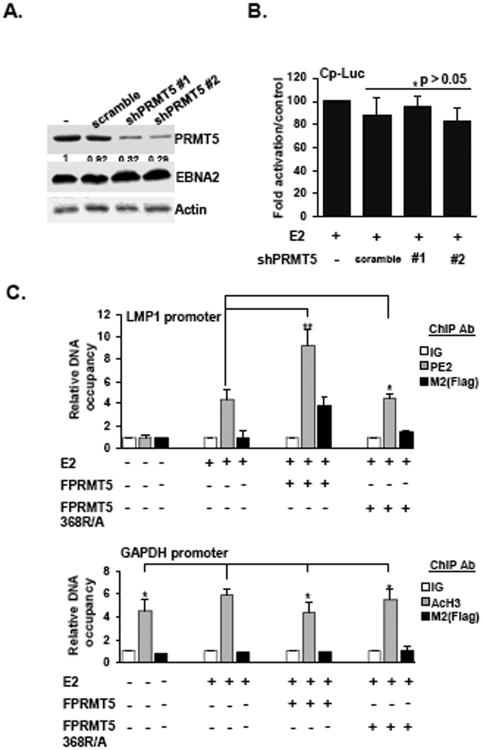
To test whether PRMT5 is essential for basal levels of EBNA2-dependent transcription, EBNA2-induced Cp-Luc activity was monitored in BJAB cells where PRMT5 was knocked down by an shRNA as well as the scrambled shRNA. The expression levels of PRMT5 in shPRMT5 #1 and shPRMT5 #2 expressing cells were reduced to 32% and 29%, respectively, while the scrambled shRNA did not alter expression (Fig. 4A). Our results indicate that EBNA2-induced Cp-Luc activity was similar in each shRNA-expressing cell line and that this activity was indistinguishable from Cp-Luc activity in the presence of EBNA2 alone (Fig. 4B). Similar to the results obtained in the EBNA2 ΔRG cell-based reporter assays, our data indicate that PRMT5 is not required for basal levels of EBNA2-mediated transcription.
Fig. 4.
Overexpression of PRMT5 causes enrichment of EBNA2 occupancy on target promoters. The amount of PRMT5 expressed in each shRNA knockdown cell line was determined by immunoblot analysis. The expression level of PRMT5 in each shRNA expressing cell line is expressed relative to the control group (A). Actin was used as the loading control. Transfection-mediated reporter assays were performed on BJAB cells and the shRNA-expressing cell lines as described in Fig. 3A. The EBNA2-induced Cp-Luc reporter activity in each cell line was expressed relative to the Cp-Luc activity with EBNA2 alone that was observed in BJAB cells (B). ChIP assays were performed on BJAB cells (1 × 107) that were co-transfected with the EBNA2 expression vector, LMP1-Luc reporter plasmid, and either the FPRMT5 or 368R/A expression vector using M2 or EBNA2 antibodies. The abundance of each assayed protein or IgG control on the LMP1 promoter (C upper panel) or GAPDH promoter (C lower panel) is shown. P* value >0.05. P** value<0.05.
To explore the mechanism by which PRMT5 enhances EBNA2-depedent transcription, we determined if overexpression of PRMT5 affected the accumulation of EBNA2 on the target promoters. When EBNA2 alone was expressed, the promoter occupancy of this protein was 4.5-fold higher than the IgG control in a ChIP assay (Fig. 4C upper panel). In multiple experiments, we found that ectopically expressed FPRMT5 could also accumulate on the LMP1 promoter, but this accumulation was not observed in the presence of 368R/A. Strikingly, FPRMT5 substantially increased promoter occupancy of EBNA2 by approximately 2-fold, whereas the 368R/A mutant did not alter occupancy. In addition, a parallel ChIP assay was performed to monitor the enrichment of acetylated H3 (AcH3) on the GAPDH promoter using the same cell lysates. We found that the promoter occupancy of AcH3 was 4 to 6 -fold higher than that of the IgG control (Fig. 4C lower panel). Both FPRMT5 and 368R/A failed to bind to the GAPDH promoter, and neither affected the promoter occupancy of AcH3. Our results demonstrate that PRMT5 is involved in EBNA2-mediated transcription by modulating the promoter occupancy of EBNA2.
4. Discussion
Although the EBNA2 RG repeat domain has been implicated in PRTM5-medated methylation, it is unknown if PRMT5 is involved in EBNA2-dependent transcription. The involvement of PRMTs in transcriptional regulation has been extensively studied during the past decade. PRMT5-dependent post-translational modifications include the methylation of H3 at Arg 8 and H4 at Arg 3, which leads to the down-regulation of the ST7 and NM23 tumor suppressor genes [10]. In contrast, it has been shown that PRMT5 is capable of forming a repressive complex with E2F on the cyclin E1 promoter [22]. Moreover, it has been shown that PRMT5-mediated methylation of FCP1, a phosphatase that targets the carboxy terminus of the large subunit of RNA polymerase II, promotes transcriptional elongation [23]. In this study, we highlighted a positive role played by PRMT5 in EBNA2-mediated transcription of EBV latency specific promoters, including the LMP1 and C promoters. Plasmids expressing PRMT5 induced a 2.5- to 3-fold increase of LMP1 expression or the Cp-Luc reporter activity compared to the basal activity of EBNA2 alone in the transfection-mediated reporter assays. It should be noted that the enhancement of EBNA2-dependent transcription of LMP1 by PRMT5 in AKATA cells directly relies on the measurement of LMP1 expression from the endogenous EBV genome, which could largely reduce the artifacts resulting from transfection-mediated reporter assays.
In this study, we performed protein affinity pull-down assays and confocal immunofluorescence microscopy to validate the substantial protein-protein interactions between EBNA2 and PRMT5 both in vivo and in vitro. In addition, we showed that the EBNA2 RG domain was sufficient for PRMT5 binding, and recombinant PRMT5 possessed catalytic activity to mediate the symmetric dimethylation of the EBNA2 RG tail. In contrast, the catalytic mutant 368R/A failed to bind to the EBNA2 RG tail and did not trigger the methylation process. Most importantly, we were able to demonstrate that the biochemical interaction between PRMT5 and EBNA2 is the major factor affecting PRMT5-mediated transcriptional upregulation. This result was supported by the finding that EBNA2ΔRG-induced transcription is no longer co-stimulated by ectopically expressed PRMT5. Furthermore, the catalytic mutant 368R/A was neither able to upregulate EBNA2-induced LMP1 expression nor co-stimulate Cp-Luc reporter activity in the presence of EBNA2.
Although the RG repeat domain of EBNA2 is dispensable for the basal level of EBNA-mediated transcription, recombinant EBV lacking this specific motif had a substantially reduced ability to transform B cells [6]. Our data demonstrate that PRMT5 plays a crucial role in the enhancement of EBNA2-mediated transcription. Interestingly, an increase of EBNA2 occupancy in the target promoters was correlated with ectopic expression of PRMT5. Our data suggest that PRMT5 binds to the EBNA2 RG domain and triggers symmetric dimethylation to increase the abundance of EBNA2 on promoters. We speculate that the biochemical interaction between PRMT5 and EBNA2 could additionally affect other signaling pathways involved in EBV-mediated transformation of B cells.
Highlights.
► Catalytic active PRMT5 substantially binds to the EBNA2 RG domain
► PRMT5 augments the EBNA2-dependent transcription
► PRMT5 triggers the symmetric dimethylation of the EBNA2 RG domain
► PRMT5 enhances the promoter occupancy of EBNA2 on its target promoters
Acknowledgments
This work was supported by grants NSC 101-2320-B-320-005-MY3 and NHRI-EX-101-9910BC from the NSC and the NHRI to C.-W. P. and National Institutes of Health Grants R01CA047006, R01CA131354, and R01CA085180 to E.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ea R, Kieff A. Epstein-Barr Virus and Its Replication. In: Knipe DaHP., editor. Fields Virology. Fifth. Lippincott, Williams, and WIlkins; Philadelphia: 2007. pp. 2603–2700. [Google Scholar]
- 2.Peng CW, Zhao B, Kieff E. Four EBNA2 domains are important for EBNALP coactivation. J Virol. 2004;78:11439–11442. doi: 10.1128/JVI.78.20.11439-11442.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen JI, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65:2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling PD, Ryon JJ, Hayward SD. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barth S, Liss M, Voss MD, et al. Epstein-Barr virus nuclear antigen 2 binds via its methylated arginine-glycine repeat to the survival motor neuron protein. J Virol. 2003;77:5008–5013. doi: 10.1128/JVI.77.8.5008-5013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong X, Yalamanchili R, Harada S, et al. The EBNA-2 arginine-glycine domain is critical but not essential for B- lymphocyte growth transformation; the rest of region 3 lacks essential interactive domains. J Virol. 1994;68:6188–6197. doi: 10.1128/jvi.68.10.6188-6197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedford JL, Childs PJ, Hansen VN, et al. Treatment of extensive scalp lesions with segmental intensity-modulated photon therapy. Int J Radiat Oncol Biol Phys. 2005;62:1549–1558. doi: 10.1016/j.ijrobp.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Pahlich S, Zakaryan RP, Gehring H, et al. Protein arginine methylation: Cellular functions and methods of analysis. Biochim Biophys Acta. 2006;1764:1890–1903. doi: 10.1016/j.bbapap.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Richard S, Morel M, Cleroux P, et al. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5) Biochem J. 2005;388:379–386. doi: 10.1042/BJ20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal S, Vishwanath SN, Erdjument-Bromage H, et al. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shire K, Kapoor P, Jiang K, et al. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J Virol. 2006;80:5261–5272. doi: 10.1128/JVI.02682-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong X, Wang F, Thut CJ, et al. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng CW, Zhao B, Chen HC, et al. Hsp72 up-regulates Epstein-Barr virus EBNALP coactivation with EBNA2. Blood. 2007;109:5447–5454. doi: 10.1182/blood-2006-08-040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menezes J, Leibold W, Klein G, et al. Establishment and characterization of an Epstein-Barr virus (EBC)- negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV- genome-negative African Burkitt's lymphoma. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- 15.Takada K, Horinouchi K, Ono Y, et al. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes. 1991;5:147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Johannsen E, Robertson E, et al. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J Virol. 2002;76:232–242. doi: 10.1128/JVI.76.1.232-242.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YL, Tsai HL, Peng CW. EGCG debilitates the persistence of EBV latency by reducing the DNA binding potency of nuclear antigen 1. Biochem Biophys Res Commun. 2012;417:1093–1099. doi: 10.1016/j.bbrc.2011.12.104. [DOI] [PubMed] [Google Scholar]
- 18.Cooper A, Johannsen E, Maruo S, et al. EBNA3A association with RBP-Jkappa down-regulates c-myc and Epstein-Barr virus-transformed lymphoblast growth. J Virol. 2003;77:999–1010. doi: 10.1128/JVI.77.2.999-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rho J, Choi S, Seong YR, et al. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem. 2001;276:11393–11401. doi: 10.1074/jbc.M008660200. [DOI] [PubMed] [Google Scholar]
- 20.Pal S, Baiocchi RA, Byrd JC, et al. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Maruo S, Cooper A, et al. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proc Natl Acad Sci U S A. 2006;103:1900–1905. doi: 10.1073/pnas.0510612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabbrizio E, El Messaoudi S, Polanowska J, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amente S, Napolitano G, Licciardo P, et al. Identification of proteins interacting with the RNAPII FCP1 phosphatase: FCP1 forms a complex with arginine methyltransferase PRMT5 and it is a substrate for PRMT5-mediated methylation. FEBS Lett. 2005;579:683–689. doi: 10.1016/j.febslet.2004.12.045. [DOI] [PubMed] [Google Scholar]