Abstract
Primary cilia are displayed during the G0/G1 phase of many cell types. Cilia are reabsorbed as cells prepare to re-enter the cell cycle, but the causal and molecular link between these two cellular events remains unclear. We show that phospho(T94)Tctex-1 is recruited to ciliary transition zones prior to S-phase entry and plays a pivotal role in both ciliary disassembly and cell cycle progression. Tctex-1’s role in S-phase entry, however, is dispensable in non-ciliated cells. Exogenously added phosphomimic Tctex-1 T94E accelerates cilium disassembly and S-phase entry. These results support a model in which the cilia act as a brake to prevent cell cycle progression. Mechanistic studies show the involvement of actin dynamics in Tctex-1 regulated cilium resorption. Phospho(T94)Tctex-1 is also selectively enriched at the ciliary transition zones of cortical neural progenitors, and plays a key role in controlling G1 length, cell cycle entry, and fate determination of these cells during corticogenesis.
INTRODUCTION
Primary cilia are microtubule-based, hair-like organelles that extend from the plasma membrane to sense and transduce extracellular signals. Primary cilia are displayed on G0/G1 cells, and are resorbed as the cells re-enter the cell cycle 1. The biological significance of these temporally coupled cellular events and the molecular mechanisms underlying the transition between these processes are poorly understood.
Radial glials (RG) are ciliated neural progenitors in the developing neocortex 2. RG preferentially undergo proliferative division to expand the progenitor population during early corticogenesis, whereas later progenitors preferentially undergo neurogenic division (differentiation). Proliferation and differentiation are modes of division that are characterized by short and long G1 durations, respectively 3. Shortening G1 accelerates cell cycle entry and expands the progenitor population 4,5, while lengthening G1 drives cell cycle exit and differentiation into neurons 6. The mechanisms that drive the molecular switch between self-renewal and neuronal differentiation in neural progenitors of the developing neocortex remain unclear.
Tctex-1 (or DYNLT) is originally described as a light chain subunit of cytoplasmic dynein 7,8. However, Tctex-1 can be uncoupled from the dynein complex to perform dynein-independent functions 9. Tctex-1 is selectively enriched in proliferating neural progenitors of both embryonic (this paper) and adult brains 10. However, its function in these cells is unknown. In this report, we show that Tctex-1, when phosphorylated at Thr94 regulates ciliary resorption and S-phase entry. In the developing neocortex, phospho(T94)Tctex-1 plays an important role in the cell cycle regulation of RG and maintenance of the proliferating progenitor population.
RESULTS
Tctex-1 plays a key role in cilia-dependent S-phase entry
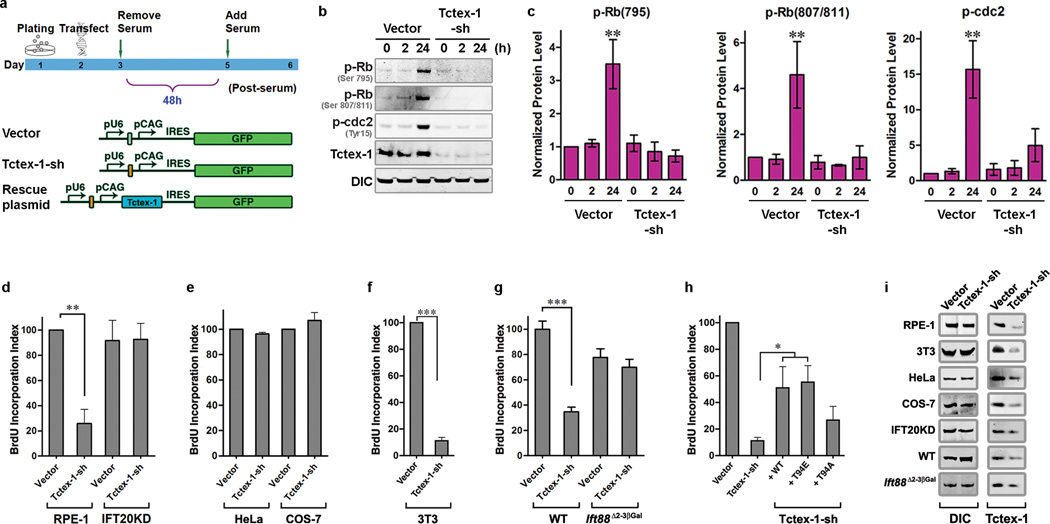
We tested Tctex-1’s role in cell cycle control by performing loss-of-function analysis in diploid human hTERT-RPE-1 (herein RPE-1) cells 11. Shortly prior to serum starvation, each cell was transfected with either a plasmid encoding both Tctex-1-short hairpin (sh) RNA and GFP (Tctex-1-sh), or a plasmid encoding GFP alone (vector) (Fig. 1a). Following 48 hr serum starvation, cell cycle re-entry was induced by serum addition. As predicted, a significant increase in the levels of phospho-(Ser795)Rb, phospho(Ser807/811)Rb and phospho-(Tyr15)cdc2 were observed in the control cells 24 hr after serum treatment (Fig. 1b, c). Rb phosphorylation is required for G0/G1-S transition 12, whereas phospho-(Tyr15)cdc2 is a G2 marker. Serum treatment failed to activate both phospho-Rb and phospho-cdc2 in cells with Tctex-1 suppressed (Fig. 1b, c), indicating that Tctex-1 is required for cell cycle re-entry.
Figure 1.
Tctex-1 is involved in cilium-dependent cell-cycle re-entry. (a) Diagrams depicting the timeline of transfection experiments and the plasmids used for transfection. (b) Representative immunoblots containing lysates of control or Tctex-1-sh transfected RPE-1 cells were probed with indicated Abs. The time points at which cells were harvested after serum re-addition are shown in hrs. (c) Quantification results of (b); the signal levels were normalized with the dynein intermediate chain (DIC) signal (mean ± s.e.m.; n=3 experiments, ** p< 0.01, t-test). (d–h) BrdU incorporation indices of synchronized RPE-1 or IFT20KD-RPE-1 cells (d), HeLa or COS-7 (e), 3T3 cells (f, h), and WT or Ift88 mutant MEF cells (g). BrdU incorporation of the control cells was taken to be 100%. All data shown are mean ± s.e.m.; n= 5, 4, 7, 3, and 7 experiments for figures (d), (e), (f), (g), and (h), respectively; an average of 400 cells were counted in each experiment; ** p< 0.01, *** p< 0.001; t-test for (d)–(g); *p<0.05, one-way ANOVA test for (h). (i) Immunoblotting assays demonstrated the reduction of endogenous Tctex-1 levels in various cell types transfected with Tctex-1-sh plasmid.
To examine S-phase entry, cells were pulse-labeled with BrdU after serum release, and the fraction of BrdU-labeled GFP+-transfected cells (BrdU incorporation index) was scored (Supplementary Information, Fig. S1a). The BrdU incorporation index of Tctex-1-suppressed cells was significantly lower than that of control cells, indicating that reduced Tctex-1 inhibited S-phase entry (Fig. 1d). S-phase entry was also blocked in unsynchronized cells transfected with either Tctex-1-sh or Tctex-1 siRNA (data not shown). In addition, most control cells were Ki67 labeled, whereas only ~30% of Tctex-1-sh transfected cells were Ki67 labeled (Supplementary Information, Fig. S1b). These results imply that Tctex-1 suppression induces cell cycle exit (i.e., G0 arrest).
While a decrease in BrdU incorporation was also seen in Tctex-1-sh transfected 3T3 cells (Fig. 1f), it was not seen in Tctex-1-sh transfected HeLa and COS-7 cells (Fig. 1e). Since both RPE-1 and 3T3, but neither HeLa nor COS-7 cells are ciliated, we postulated that Tctex-1’s role in cell cycle progression is cilia-dependent. Two additional “cilia-free” cell models were employed: (1) a stable RPE-1 cell line in which IFT20 is silenced (i.e., IFT20KD-RPE-1 13), and (2) Ift88 mutant mouse embryonic fibroblasts (MEF) derived from Ift88Δ2–3βGal mice 14,15 (Supplementary Information, Fig. S1c–e). While Tctex-1 knockdown reduced BrdU incorporation in control RPE cells (Fig. 1d) and WT MEF cells (Fig. 1g), it did not affect BrdU incorporation in IFT20KD-RPE-1 (Fig. 1d) and Ift88 mutant (Fig. 1g) cells. Similarly, a significantly smaller fraction of Ki67-labeled cells was found in WT, but not Ift88 mutant MEF cells in which Tctex-1 was suppressed (Supplementary Information, Fig. S1f). Quantification of Tctex-1 level with immunoblots (Fig. 1i) and cell-based immunofluorescence (Supplementary Information, Fig. S1g, h) showed that the degree of Tctex-1-sh-mediated knockdown was comparable among all cell types tested, confirming that differences were not due to differential gene silencing efficacy. Taken together, these results demonstrate that Tctex-1 plays an important role in cilia-dependent S-phase entry.
Tctex-1-sh transfection had no effect on the expression levels of several components of the dynein complex, such as dynein intermediate chain (Fig. 1i) and dynein heavy chain 9. To further test whether the impaired cell cycling mediated by Tctex-1 knockdown was due to its dynein-independent role, we examined whether or not the phosphomimic mutant T94E could rescue the phenotype. Previous studies have shown that T94E fails to be incorporated into the dynein complex, representing the dynein-free pool of Tctex-1 9. The counterpart construct encodes the non-phosphorylable mutant T94A, which binds dynein 9 and is used here as a control. Thr94 of Tctex-1 is a conserved residue, and resides within a consensus motif for protein kinase C and integrin-linked kinase (http://www.cbs.dtu.dk/services/NetPhosK/; http://gps.biocuckoo.org/). T94E and T94A variants were generated in bovine Tctex-1, which is insensitive to shRNA against mouse Tctex-1, allowing us carry out the rescue experiments in mouse 3T3 cells. Impaired S-phase entry caused by Tctex-1 knockdown can be rescued by the re-introduction of WT Tctex-1 and T94E mutant, but to a much lesser extent by the T94A mutant (Fig. 1h). These results not only suggest that the phenotypes caused by Tctex-1-sh were not an “off-target” effect, but also that T94E mimicked the functionally active form of Tctex-1.
Temporo-spatial activation of phospho(T94)Tctex-1 at transition zone is required for ciliary resorption
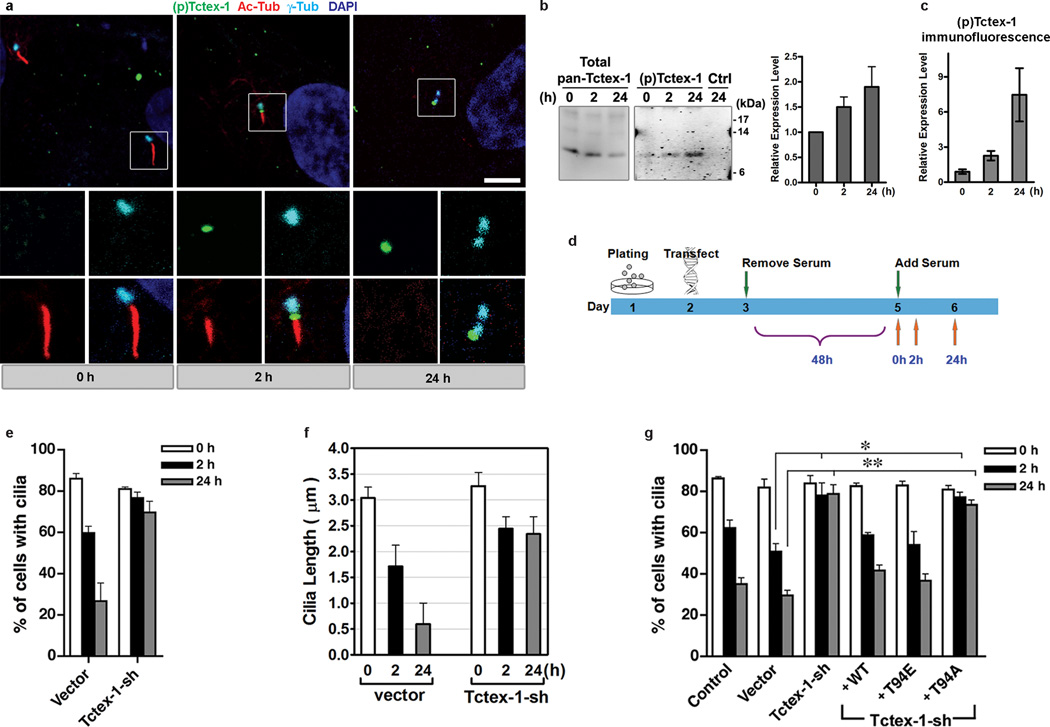
The kinetics of cilium assembly/disassembly, as well as the temporal relationship between cilium dynamics and cell cycle progression, have been characterized in RPE-1 and 3T3 cells 11,16,17. In both cell types, serum starvation induces quiescence and cilium formation. The return of serum triggers biphasic ciliary resorption, which peaks at 2hr and 24 hr post-serum treatment. The first wave of cilium shortening occurs at mid/late G1-phase preceding S phase entry, while the second wave occurs as cells are preparing to enter the G2/M phase 16. Immunolableing using an antibody (Ab) specifically recognizing phospho(T94)Tctex-1 show it was rarely seen in quiescent cells (0 hr), whereas prominent phospho(T94)Tctex-1 signals were consistently detected at the ciliary base immediately adjacent to the shortened cilia in 2-hr serum treated cells (Fig. 2a).
Figure 2.
Temporal activation of phospho(T94)Tctex-1 at the transition zone and its function in ciliary disassembly. (a) Immunofluorescence of phospho(T94)Tctex-1 (green), acetylated α-tubulin (red), and γ-tubulin (cyan) in quiescent RPE-1 cells or cells treated with serum for 2 hr or 24 hr. DAPI: nuclei (blue). Bar= 5 µm. (b) Representative immunoblots show the levels of (pan)Tctex-1 and phospho-Tctex-1 in cells harvested after serum addition. To improve the detection of phospho-Tctex-1, immunoprecipitation was first carried out using a saturated amount of anti-(pan)Tctex-1 Ab, and the immunoprecipitates were electrophoresed and immunoblotted with phospho(T94)Tctex-1 Ab. Anti-HA Ab was used as an immunoprecipitation Ab control (Ctrl). The relative expression level of phospho(T94)Tctex-1 level was normalized with the total amount of immunoprecipitated Tctex-1. (c) Relative phospho(T94)Tctex-1 immunofluorescence intensity in RPE-1 cells post serum re-addition. The ratios of intensity of phospho(T94)Tctex-1:γ-tubulin were quantified by MetaMorph software (means±s.e.m.; n=3 experiments). (d) Diagram depicting the timeline of cilium disassembly experiments. (e) The fractions of transfected RPE-1 cells containing a cilium harvested at various time points after serum addition are shown. An average of 500 cells were counted for each experiment (means ± s.e.m.; n=3 experiments). (f) Quantification of the lengths of cilia in transfected 3T3 cells harvested after serum addition (means ± s.e.m.; n=3 experiments). (g) The fractions of transfected cells containing a cilium harvested at various time points after serum addition are scored and statically analyzed as described above (mean ± s.e.m.; n=5 experiments; *p<0.05, p**<0.01; one-way ANOVA).
Phospho(T94)Tctex-1 was specifically distributed to the transition zone between the γ-tubulin-labeled basal bodies and acetylated α-tubulin-labeled cilia. Consistent with the cilia loss and cell cycle re-entry in cells after 24 hr serum treatment, phospho(T94)Tctex-1 signals were found on centrosomes in interphase cells (Fig. 2a) and mitotic poles in dividing cells (Supplementary Information Fig. S2a). Quantitative immunoblotting and immunofluorescence assays showed that the level of phospho(T94)Tctex-1 gradually increases with serum re-addition (Fig. 2b, c).
Preabsorption with the antigen peptide, but not the control peptide (corresponding to the antigen except lacking the phospho(T94) modification), effectively removed the phospho(T94)Tctex-1 signals (Supplementary Information, Fig. S2b). Phospho(T94)Tctex-1 immunofluorescence was greatly reduced in the RPE-1 cells transfected with Tctex-1-sh (Supplementary information, Fig. S2c). Furthermore, phospho-Tctex-1 signal on immunoblots was sensitive to the treatment of alkaline phosphates (Supplementary information, Fig. S2d). These results confirm the specificity of phospho(T94)Tctex-1 immunolabeling.
The appearance of phospho(T94)Tctex-1 at the ciliary base shortly preceding the disassembly of cilia indicates its functional involvement in this cellular event. To directly test Tctex-1’s role in ciliary disassembly, serum-starved Tctex-1-sh transfected cells were harvested at 0 hr, 2 hr, and 24 hr after serum return (Fig. 2d). The fraction of GFP+ cells displaying a cilium and the length of the cilia in transfected cells were scored. In RPE-1 (Fig. 2e), 3T3 (Fig. 2f, g), and MEF (Supplementary Information, Fig. S2e) cells, controls and Tctex-1-suppressed cells had similar abilities to form cilia after serum starvation. The lengths of their cilia were also similar (Fig. 2f). However, serum-induced cilium disassembly was significantly blocked by Tctex-1 suppression, suggesting that Tctex-1 is not required for ciliogenesis, but is critical for cilium disassembly. Furthermore, co-transfection of FLAG-tagged WT Tctex-1, or T94E, but not T94A, with Tctex-1-sh effectively reversed the inhibition of cilium disassembly caused by Tctex-1 silencing (Fig. 2g).
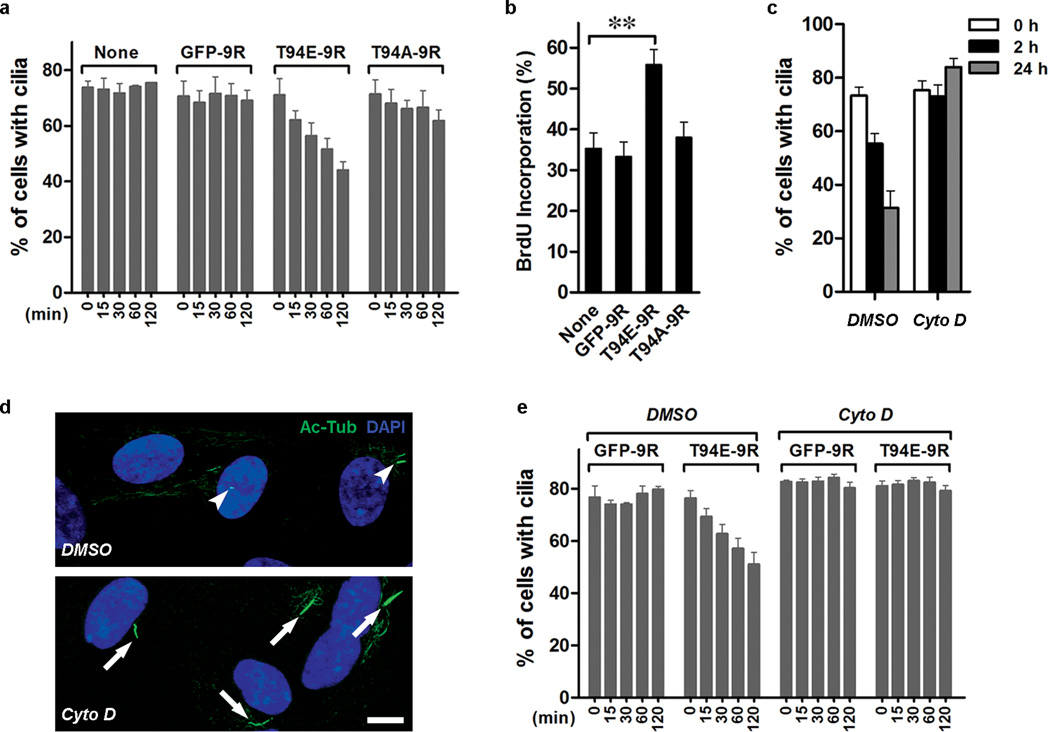
To further investigate the causal relationship between ciliary disassembly and S-phase entry, we investigated whether or not exogenously added T94E can trigger rapid cilium disassembly of RPE-1 cells, and thus accelerate their S-phase entry. We employed a high-efficiency protein transduction system 18 to deliver Tctex-1 peptides into RPE-1 cells with pre-formed cilia. Purified recombinant Tctex-1 (or GFP) containing a 9-arginine (9R) entered cells effectively, as confirmed by both immunoblotting and immunostaining assays (Supplementary Information, Fig. S2f, g). The addition of control peptides GFP-9R and T94A-9R had no effect on cilia disassembly (Fig. 3a). By contrast, cells treated with T94E-9R underwent significantly faster cilium resorption even when cultured in the absence of serum (Fig. 3a). Cells treated with T94E-9R, but not control peptides, also exhibited significantly higher BrdU incorporation (Fig. 3b).
Figure 3.
Phospho(T94)Tctex-1 and actin dynamics participate in ciliary resorption. (a) Fractions of cells displaying cilia in serum-starved RPE-1 cells, either untreated (none) or treated with GFP-9R, T94E-9R, or T94A-9R, harvested at indicated time points after the treatments. All data shown are mean ± s.e.m.; n=4 experiments, except untreated control, where n=3 experiments. (b) Cells in serum-free medium were treated with various peptides followed by the incubation with BrdU for additional 16 hr. Fractions of BrdU-labeled cells are shown (mean ± s.e.m.; n=7 experiments; **p< 0.01; one-way ANOVA). (c) Fraction of ciliated cells in control (DMSO) and CytoD (0.5 µM) treated cells post serum re-addition (mean ± s.e.m.; n=3 experiments). (d) Representative image of cilia displayed in control cells vs. CytoD treated cells 24-hr after serum re-addition. (e) Fractions of cells that had cilia after the addition of GFP-9R or T94E-9R for indicated time in the presence of DMSO (n=3 experiments) or CytoD (n= 5 experiments). All data shown are mean ± s.e.m.
Phospho(T94)Tctex-1 has a reported ability to modulate actin dynamics 9. To investigate the role of actin in cilium resorption, we treated quiescent RPE-1 cells with an actin polymerization inhibitor, cytochalasin D (CytoD), 2 hr prior to serum addition. CytoD effectively blocked serum-mediated cilium disassembly (Fig. 3c, d). Furthermore, CytoD inhibited T94E-peptide mediated acceleration of ciliary resorption (Fig. 3e). These results imply that phospho(T94)Tctex-1 modulates cilium disassembly through a process involving actin polymerization.
Previous studies have shown that blocking Aurora A kinase (AurA) phosphorylation-mediated histone/tubulin deacetylase-6 (HDAC6) activation suppresses cilium resorption in RPE-1 cells 11. We sought out to test whether AurA- or HDAC6-depleted cells also exhibit impaired S-phase entry. Our results showed that suppression of both AurA and HDAC6 via sh- or siRNA transfection were able to inhibit both ciliary resorption and BrdU incorporation (Supplementary information, Fig. S3a–c).
Phospho(T94)Tctex-1 is expressed in the ciliary transition zone of RG in developing neocortex
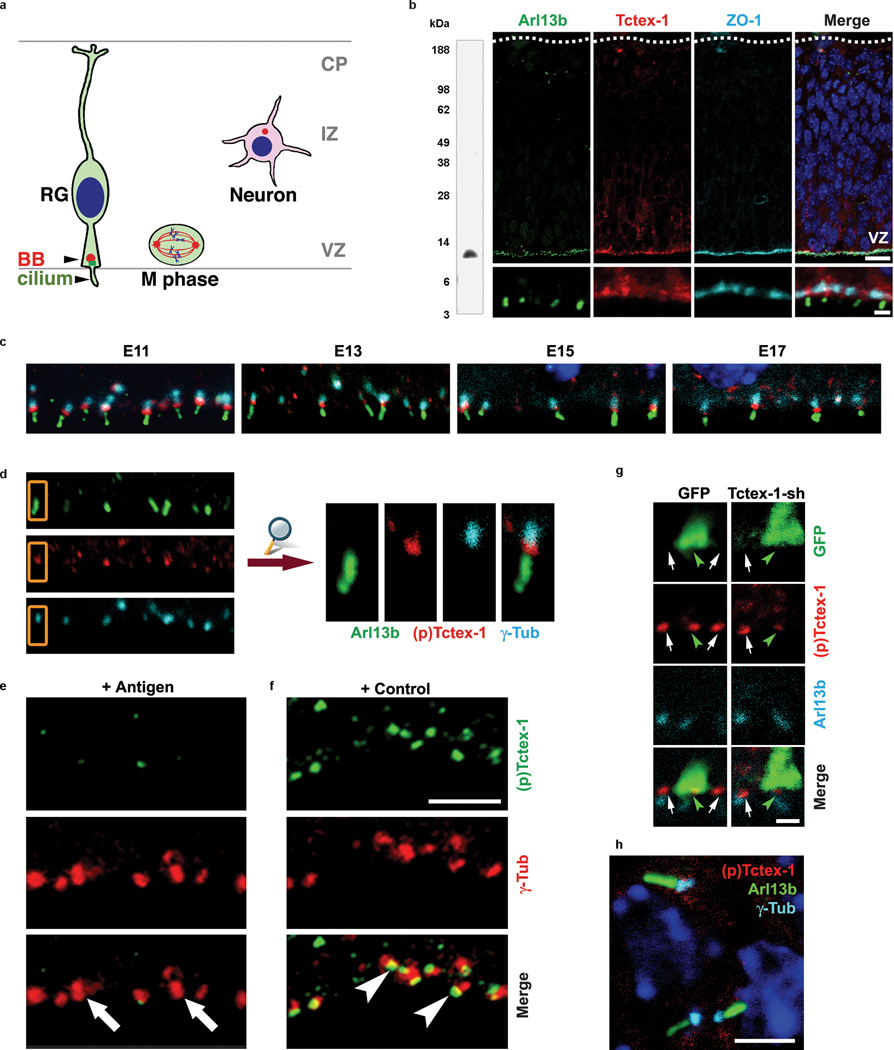
The developing neocortex is an ideal model system for studying the physiological relevance of phospho(T94)Tctex-1 in vivo because the cortical progenitor RG also display primary cilia (Fig. 4a), and the relationship between the cell cycle regulation and the fate control of RG has already been established 19,20.
Figure 4.
Phospho(T94)Tctex-1 is specifically expressed at the transition zone of RG in developing neocortex. (a) Drawing depicting the radially elongated RG with their endfeet contacting both the ventricular and pial surfaces. The primary cilium, anchored on a basal body (BB), extends into ventricular spaces. The large majority of mitosis (M phase) occurs at the ventricular surfaces. Cells leave the cell cycle, migrate away from the ventricular zone (VZ), form multipolar post-mitotic neurons, and pause their migration in the intermediate zone (IZ) prior to reaching their cortical location. CP: cortical plate. (b) Anti-Tctex-1 mouse Ab recognized a single band of ~12-kDa on an immunoblot containing embryonic mouse brain lysates. Confocal images of E13 mouse cortical slice co-labeled for primary cilium marker Arl13b 42 (green), Tctex-1 (red), and cell-cell junction marker ZO-1 (cy5). Bars= 20 µm (top panel); 2 µm (bottom panel). (c) Confocal images of the ventricle surfaces of E11-E17 neocortical slices that were triple-labeled for Arl13b (green), phospho(T94)Tctex-1 (red), and γ-tubulin (cyan). In these experiments, cortical slices were first incubated with phospho(T94)Tctex-1 Ab, followed by excess biotinylated goat-anti-rabbit Ab. The sections were then PFA fixed, incubated with anti-Arl13b and γ-tubulin Abs, followed by detection by using Alexa488-conjugated anti-rabbit Ab, Cy5-conjugated anti-mouse Ab, and Alexa568-conjugated streptavidin. (d) Representative enlarged view of (c) demonstrating that phospho(T94)Tctex-1 (red) was specifically located in the transition zone between Arl13b-labeled primary cilia (green) and γ-tubulin-labeld basal body (cyan) of the RG. (e, f) Confocal images of the ventricular zone double labeled with γ-tubulin Ab and phospho(T94)Tctex-1 Ab preabsorbed with the phosphopeptides corresponding to the antigen (e) or control peptides (f). Arrows in (e) point to the γ-tubulin labeled basal bodies and centrosomes, which lacked phospho(T94)Tctex-1 signal. Bar=5 µm. (g) Co-labeling of phospho(T94)Tctex-1 and Arl13b in 24-hr transfected cortical slices. Note that cells transfected with GFP control plasmid (left panel, green arrowhead) and neighboring non-transfected cells (arrows) displayed similar levels of phospho(T94)Tctex-1 at the endfeet. However, cells transfected with Tctex-1-sh had reduced phopsho(T94)Tctex-1 immunolabeling. Bar= 2 µm. (h) Confocal image of the post-mitotic neurons located in the intermediate zone region displayed no detectable phospho(T94)Tctex-1 (red) between the Arl13-labeled primary cilia (green) and γ-tubulin-labeled basal bodies (cyan). Bar= 5 µm.
Staining with anti-(pan)Tctex-1 Ab in E11-E17 embryonic cortices showed that Tctex-1 was specifically concentrated at the apical endfeet of RG, from which Arl13b-labeled cilia project into the ventricles (Fig. 4b; Supplementary Information, Fig. S4a). However, phospho(T94)Tctex-1 appeared as prominent punctas lining the ventricular surfaces. Co-localization studies confirmed that phospho(T94)Tctex-1 specifically resides in the transition zone between the γ-tubulin-labeled basal bodies and the primary cilia (Fig. 4c, d). The transition zone-labeling of phospho(T94)Tctex-1 was universal in all RG regardless of developmental stage, and despite the fact that there were fewer RG in later stages of neurogenesis (i.e., E17) (Fig. 4c). Preabsorption with the antigen, but not the control, peptide effectively removed the phospho(T94)Tctex-1 signals (Fig. 4e, f). Phospho(T94)Tctex-1 labeling was also greatly reduced in RG transfected with Tctex-1-sh (Fig. 4g). In stark contrast, no phospho(T94)Tctex-1 signal was detected at the bases of primary cilia in the post-mitotic neurons located at the intermediate zone or cortical plate (Fig. 4h), indicating that phospho(T94)Tctex-1 is present in G1, but not G0 cells.
Tctex-1 knockdown triggers premature neuronal differentiation at the expense of the neural progenitor pool
The similar transition zone location of phopsho(T94)Tctex-1 in RG and cultured ciliated cells prompted us to hypothesize that this molecule may also regulate the cell cycling of RG during corticogenesis. If this were the case, the depletion of Tctex-1 would drive RG to exit the cell cycle and differentiate into neurons. The post-mitotic neurons typically migrate away from the ventricular zone where the cell division takes place, halt temporarily at the intermediate zone (Fig. 4a), and are eventually incorporated into laminated cortical plates according to their birthdates 21. Conversely, we predicted that excess phospho(T94)Tctex-1 would shorten the G1 phase, accelerate S-phase entry, and expand the progenitor population.
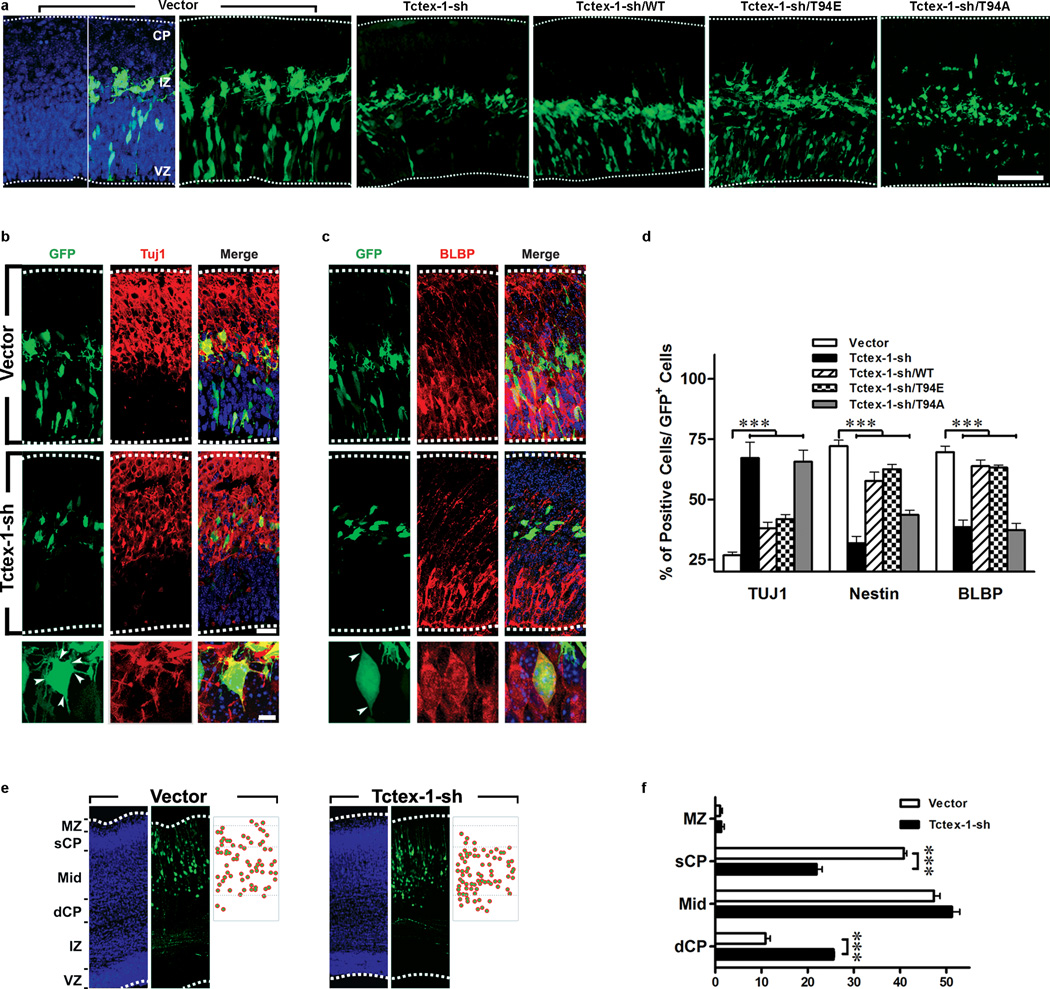
To test our model, we employed in utero electroporation (IUE), an efficient gene delivery method for RG to perform loss-of-function and gain-of-function analyses. To test the first part of our hypothesis, Tctex-1-sh plasmid was transfected into an E13.5 neocortex. Immunolabeling of cells dissociated from brains harvested as early as 24 hr and as late as 5 days after IUE verified the specific knockdown of endogenous Tctex-1 in Tctex-1-sh, but not control, plasmid transfected cells (Supplementary Information, Fig. S4b). We subsequently conducted phenotypic characterization of Tctex-1-sh transfected cells in situ. There were several lines of evidence suggesting that Tctex-1 silencing in RG caused their premature neuronal differentiation. First, in cortical slices harvested 40 hr post electroporation, cells transfected with control and Tctex-1-sh plasmids displayed strikingly different distribution patterns. Cells transfected with either vector alone (Fig. 5a) or control sh-plasmid (Supplementary Information, Fig. S4c) were distributed throughout the area between the ventricular zone, subventricular zone, and intermediate zone. The large majority of these cells had bipolar shapes and expressed RG markers Nestin and brain lipid binding protein (Fig. 5c, d), and a much smaller fraction of them displayed post-mitotic neuronal marker Tuj1 (Fig. 5b, d). Subventricular zone-localized GFP+ control cells also displayed intermediate progenitor marker-Tbr2 (Supplementary Information, Fig. S4d). In contrast, Tctex-1-silenced GFP+ cells were predominantly concentrated in the intermediate zone (Fig. 5a); these cells bore tangentially radiating multi-polar processes and expressed neuronal marker Tuj1 (Fig. 5b,d). Significantly fewer Tctex-1-sh targeted cells displayed RG markers compared to the controls (Fig. 5c, d), and almost none of them displayed Tbr2 (Supplementary Information, Fig. S4d).
Figure 5.
Suppression of Tctex-1 in RG induced premature neuronal differentiation. (a) Cortical slices harvested 40 hr after electroporation with the indicated plasmids. GFP was detected by direct green immunofluoresence; DAPI: blue. The dashed lines depict the borders of the cortexes. The intermediate zone (IZ) is defined by the presence of tangentially oriented cells. (b, c) Representative images showing the labeling of Tuj1 (b) and brain lipid binding protein (BLBP) (c) of vector- and Tctex-1-sh transfected brain slices. Arrowheads point to the sites where the cell processes project from the cell bodies. (d) Percentage of total transfected GFP+ cells that were Tuj1+, Nestin+, or BLBP+. All data shown are mean ± s.e.m.; n=4 experiments, except BLBP staining, where n=5 experiments; an average of 600 cells were counted in each experiment; ***p<0.001; one-way ANOVA. (e) Representative images depicting the cortical distribution of GFP cells in the E18.5 brains (i.e., 5-days post electroporation). The bins of marginal zone (MZ), superficial (sCP), mid-, and deeper cortical plate (dCP) are defined, as described 43. (f) The numbers of cell bodies located in each bin were scored, and quantification of the cortical location of transfected cells (mean ± s.e.m.; n=3 experiments; ***P<0.001, t-test).
Second, to corroborate the cell-based phenotypic analysis in situ, the fraction of GFP+ cells dissociated from the electroporated brains that also expressed Tuj1 was scored (Supplementary Information, Fig. S4e). These results suggested that a significantly greater number of Tctex-1-sh transfected cells were neurons. Third, quantitative RT-PCR assays using RNAs isolated from the flow cytometer-sorted GFP+ cells showed that a significantly higher fraction of Tctex-1-silenced cells expressed Tuj1+ in comparison with control cells (data not shown).
A neuron’s birthdate is correlated with its laminar fate in the cortical plate as corticogenesis progresses; cells that differentiate at later time points tend to be found in the upper (more superficial) layers of the cortical plate 21. In agreement with the proposed idea that Tctex-1-sh targeted cells differentiate and migrate earlier, these cells were found to be located in a relatively deeper cortical layer compared with the control cells in brains harvested 5 days after IUE (Fig. 5e, f). It was also noted that neurons derived from Tctex-1-sh targeted cells, like the controls, displayed neuronal markers (e.g., HuD and doublecortin) and extended axons (data not shown). These results suggest that Tctex-1 expression is not critical for cell migration. Previous studies have shown that cytoplasmic dynein is important for cortical cell migration 22. The differences in phenotype seen between the Tctex-1-suppressed and cytoplasmic dynein-suppressed cells further support the idea that Tctex-1's role in corticogenesis is independent of its dynein-related activity.
Tctex-1 suppression drives RG exit cell cycle
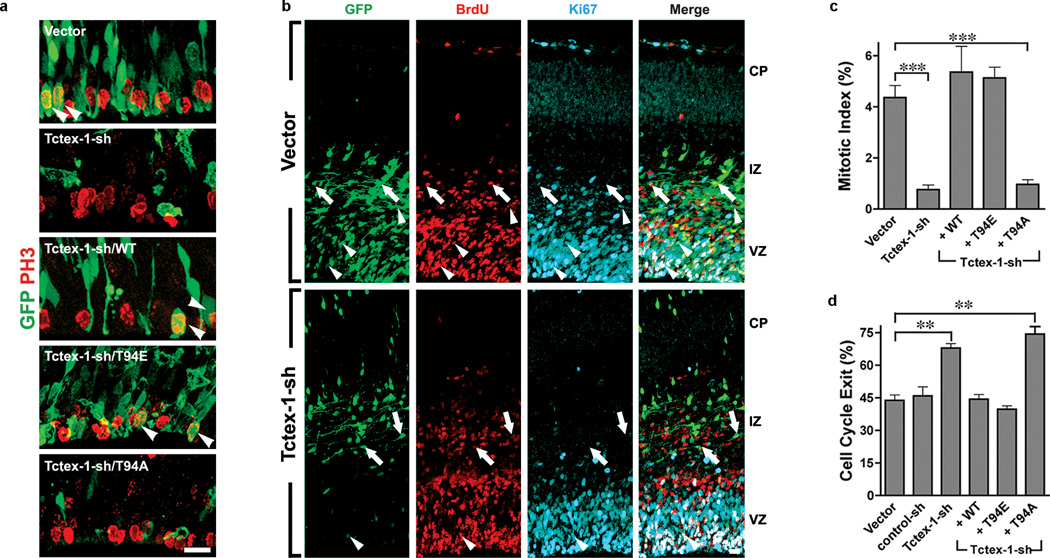
We consistently observed that the total numbers of GFP+ cells in Tctex-1-sh transfected brains were fewer than those of the control brains (Fig. 5a). This discrepancy in cell number was not due to low transfection efficiency (Supplementary Information, Fig. S5a, b) or apoptotic cell death (Supplementary Information, Fig. S5c). Thus, the most likely explanation to this is that the Tctex-1-sh transfected RG underwent a premature switch from proliferation to differentiation mode. Early depletion of the fast-proliferating progenitors could result in a detectably lower number of total GFP+ cells because the doubling times for E13-E14 RG are rather short (i.e., 11.4–15.1 hr) 3. Indeed, compared to control cells, a significantly reduced fraction of Tctex-1-sh transfected cells exhibited mitotic marker phosphohistone 3 (P-H3) (Fig. 6a, c). The low mitotic index also precluded the possibility that Tctex-1-depletion caused mitotic arrest. Furthermore, as expected 23,24, the large majority of ventricular zone mitotic cells transfected with vector alone had a vertical cleavage plane (i.e., DNAs in ana/metaphase cells are oriented 60°–90° relative to the axis of ventricle surfaces; Supplementary Information, Fig. S5d). Among the few Tctex-1-sh transfected mitotic cells, most of them also exhibited vertical cleavage. The difference in the fraction of cells displaying intermediate vs. horizontal cleavages between Tctex-1-suppressed and control cells was also negligible.
Figure 6.
Phospho-Tctex-1 is required for the cell cycling of RG. (a) Representative ventricular zone images of transfected brains immunolabeled with P-H3. Arrowheads indicate cells that were double positive for both GFP and P-H3. (b) Representative confocal images of transfected cortical slices subjected to cell cycle exit analysis: immunofluoresence of GFP (green), 24-hr BrdU (red), Ki67 (cyan) are shown. Arrows point to the cells that were double positive for GFP and BrdU but negative for Ki67. Arrowheads point to the cells that were triple positive for GFP, BrdU, and Ki67. (c) Fractions of GFP, P-H3-double positive cells out of total GFP+ cells (or mitotic index) are shown as means ±s.e.m.; n= 3, 6, 3, 5, 6 experiments for vector, Tctex-sh, Tctex-1-sh/WT, Tctex-1-sh/T94E and Tctex-1-sh/T94A, respectively; ***p<0.001; one-way ANOVA. (d) Fractions of GFP+, BrdU+, Ki67− cells out of total GFP+, BrdU+ cells (or cell cycle exit index) are listed as means ± s.e.m.; n=3 experiments; total 100 cells were scored; **p<0.01; one-way ANOVA.
To test how Tctex-1 knockdown affects cell cycling, we performed a cell cycle exit analysis in situ 25. A single-pulse of BrdU was given to mice 24 hr after IUE, and the brain slices harvested 24 hr afterwards were immunolabeled for BrdU and Ki67. The fraction of GFP+BrdU+ cells that were Ki67− (i.e., cell-cycle-exit index) was found to be greater in Tctex-1-sh transfected cells than in control cells (Fig. 6b, d).
All phenotypes elicited by Tctex-1 silencing — the distribution and number of transfected cells (Fig. 5a), the neuron-to-progenitor ratio (Fig. 5d), the mitotic indices (Fig. 6 c), and the cell-cycle-exit indices (Fig. 6d) — were successfully restored by both WT and T94E Tctex-1, but not by T94A. Taken together, these results suggest that phospho(T94)Tctex-1 is the active form of Tctex-1 in RG.
Phospho-mimic Tctex-1 accelerates S-phase entry and increases the proliferating progenitor population
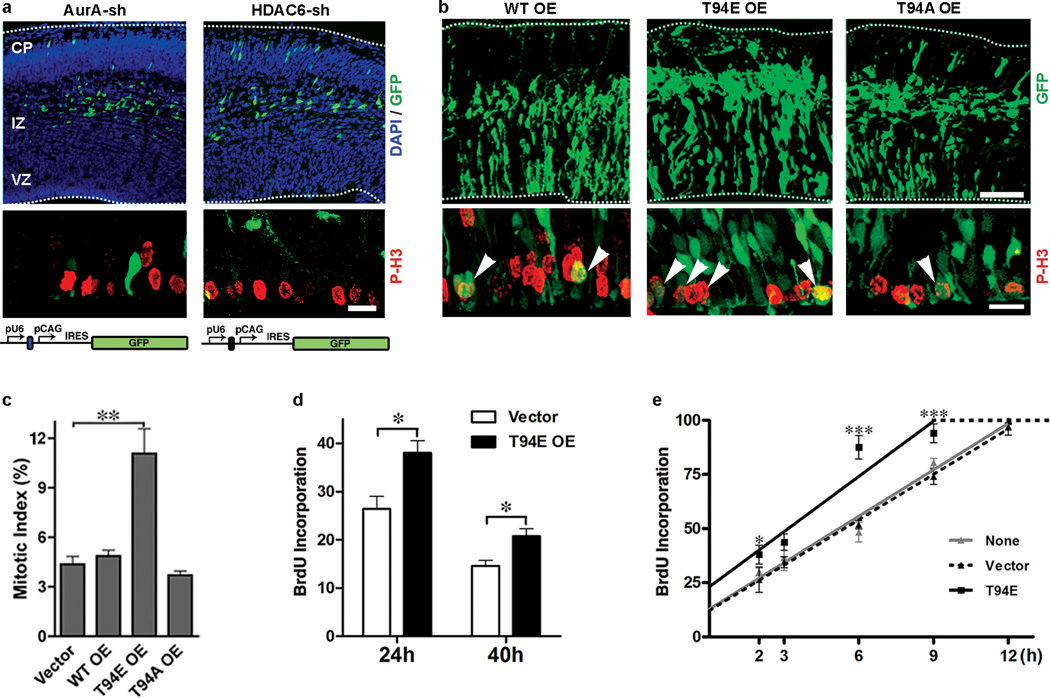
If impaired ciliary disassembly caused by Tctex-1 silencing underlies the S-entry block of RG, the perturbation of a known inhibitor of cilium resorption should mimic the phenotypes exerted by Tctex-1-sh. Indeed, a large majority of AurA-sh or HDAC6-sh transfected cells developed into intermediate zone-residing neurons 40-hr after electroporation (Fig. 7a), and displayed significantly lower mitotic indices (mitotic indices of AurA-sh and HDAC-6-sh transfected cells were 1% and 0.5%, respectively, vs. 4.4% of control cells).
Figure 7.
Phenotypic characterization of loss-of-function of AurA and HDAC6, and gain-of-function of Tctex-1 in developing neocortex. (a) Cortical slices transfected with AurA-sh or HDAC6-sh plasmid (top panels) and their immunolabeling of P-H3 (red; middle panel). The schematic diagrams of these plasmids were shown in the bottom panel. (b) Cortical slices overexpressing the indicated Tctex-1 variants. Arrowheads point to P-H3-labeled, mitotic GFP+ cells at the ventricular zone. Bars=100 µm (top panel); 20 µm (bottom panel). (c) Mitotic indices of GFP transfected cells are shown as means ± s.e.m.; n=3 experiments; total 1,200 cells were scored; ** p < 0.01, one-way ANOVA. (d) The fractions of 2-hr BrdU incorporated, GFP+ cells out of total GFP+ cells in transfected brains, 24 hr and 40 hr post-electroporation (mean ± s.e.m.; n=4 experiments; total 700 cells were scored; *p < 0.05, t-Test). (e) Cumulative BrdU labeling curves of non-transfected, vector alone and T94E/GFP transfected cells (means ± s.e.m.; n=3 experiments; *p < 0.05, *** p < 0.001, one-way ANOVA). OE: overexpression.
To carry out gain-of-function analysis, bicistronic vectors expressing both Tctex-1 (or its variants) and GFP were employed to perform IUE. We first showed that overexpression of T94E, but not WT or T94A, almost doubled the mitotic index in transfected neocortices (Fig. 7 b, c). Furthermore, cell-cycle entry analysis, in which a single pulse of BrdU was given to the mice 2 hr prior to harvest, was carried out. The fraction of GFP+ cells that were also BrdU+ was significantly higher in T94E transfected cells than in WT or T94A-transfected cells (Fig. 7d), suggesting that S-phase entry is accelerated in T94E overexpressed cells. Finally, we employed a cumulative BrdU labeling experiment to determine the cell cycle length of the RG 3. The length of the G2/M phase of both T94E-, GFP- and non-transfected progenitors was the same (i.e., 2 hr). However, the G1 length of T94E-transfected progenitors (6.8 hr) was significantly shorter than that of untransfected or GFP-transfected progenitors (10 hr) (Fig. 7e). These results collectively suggest that forced expression of phospho-mimic Tctex-1 expands the proliferating progenitor pool by shortening the G1 phase and accelerating S-phase re-entry.
DISCUSSION
Human genetic studies have suggested a causal relationship between several ciliary/centrosomal proteins and the cellular over-proliferation phenotype seen in cilium-related disorders (e.g., cystic kidney disease, and certain brain development diseases) 26. However, the relationships between cilia, centrosomes, and cell division is not well understood. Complete ciliary resorption prior to mitosis is thought to be necessary for the release of centrioles from basal bodies to form mitotic poles 18. Several ciliated proteins have indeed been linked to the G2/M transition and/or to cytokinesis 27,28. The significance of ciliary shortening at late-G1 phase and its relevance to the subsequent S-phase entry, however, is unknown. Here we present evidence that phospho(T94)Tctex-1 is required for cilium resorption and, hence, S-phase entry.
Our data supports a model in which cilia provide a brake in the cell cycle to retain cells in G0/G1. Extracellular cues that activate the signaling pathways involved may determine the time required for this cellular event to take place and the duration of G0/G1. Cilia-mediated quiescence control may be absent in some non-ciliated cancerous cells 29. These cells are thus likely to be insensitive to the environmental cues that time normal cells for division.
An understanding of the molecular switch between self-renewal and neuronal differentiation in cortical progenitors is critical in elucidating mechanisms that control brain size 30–32. We speculate that RG receive ventricular cues via the cilium, allowing for subsequent recruitment of phospho(T94)Tctex-1 to guide timely ciliary resorption and proliferation. Blocking the activation of phospho(T94)Tctex-1 induces neuronal differentiation at the expense of proliferating progenitors, mimicking the developmental program of late progenitors. Interestingly, several protein mutations found in human diseases associated with small brain size are localized to the centrosomes 33. Although both previous 4,5,19,20 and our studies suggest G1 length controls the cell fate determination of RG during corticogenesis, direct visualization and comparison of life cycle of RG with varying G1 lengths remains to be shown.
The primary cilium is vital for the proper development of multiple brain regions. Sonic hedgehog signaling pathway transduced through cilia delivers a proliferative signal, which is required for the establishment and maintenance of neural progenitors of the cerebellum and hippocampus 34–36 37. Thus, it appears that while the cilia are required to sense “proliferation” signals, they are reabsorbed in a timely manner to permit the cells to re-enter the cell cycle. These two seemingly opposing roles of cilia could, in fact, represent two temporally related events, working in conjunction to regulate cell division.
A link between the spindle orientation and the fate determination of mouse RG has been previously proposed 38, but remains controversial 23,24. We did not observe any significant change in spindle orientation in Tctex-1 suppressed cells, suggesting that altering the cleavage plane was not a major contributor to the neural fate adoption of these cells. This finding differed from a previous report by Gauthier-Fisher et al., 39 where it was suggested that suppression of Tctex-1 expression has an impact on the spindle orientation of RG. Differences were noted between these two studies. First, different Tctex-1 sh-plasmids were used; we were able to rule out any off-target effect by performing in vivo rescue experiments. Second, we considered cells with cleavage plane at an angle greater than or equal to 30° oblique to the vertical plane to be anomalous 23,24,38; Gauthier-Fisher et al. considered 15° to be their benchmark 39.
A recent report suggested that actin dynamics is involved in ciliogenesis 40. Our data show that phospho(T94)Tctex-1-regulated ciliary resorption also occurs through a process involving actin cytoskeleton rearrangement. We envision that at the base of the clilium, Tctex-1 is dissociated from the dynein complex, via phosphorylation at Thr94. Phospho-Tctex-1 activates local F-actin polymerization, which may trigger a cascade of cellular events that coordinately reabsorb the cilia and modify the basal bodies. The latter may transmit signals to program the cells for S-phase entry, a mechanism which agrees with the finding that the centrosome/basal body is a central hub for almost all cell cycle-regulating proteins 41.
METHODS
Cell cultures, transfection, and cilium assembly/disassembly assay
IFT20KD-RPE-1 was a gift from Dr. Greg Pazour 13. Immortalized WT MEF and Ift88 mutant MEF cells were gifts from Dr. Aimin Liu 14. MEF, RPE-1, and 3T3 cells were transfected using nucleofection (Amaxa, Gaithersburg, MD). HeLa cells were transfected using Lipofectamine 2000 (Invitrogen). COS-7 and HEK cells were transfected using the polyethylenimine method 44. Protein expression levels were estimated by immunoblotting and quantified by Odyssey Infrared scanner (LI-COR, Lincoln, NB). For most immunolabeling, cultured cells were fixed in 4% paraformaldehyde for 10 min and then submerged in cold methanol for 5 min prior to the blocking and Ab incubation using standard procedures.
A cilium assembly/disassembly assay was performed as previously described 11. Specifically, cells were starved in serum-free medium for 48 hr to induce cilium formation. Serum was then added back to the medium to stimulate cilium resorption and cell cycle re-entry. Cells were harvested at various time points for immunolabeling assays. All plasmids used for overexpression experiments in cultures were under the CMV promoter. Cells receiving more than one plasmid were routinely immunolabeled to confirm high (>90%) double or triple transfection efficiency.
Purified recombinant proteins (e.g., GFP-9R, T94E-9R and T94A-9R; 1 µM) were added into cilium-preformed cells in the absence of serum for 30 min 18. Cells were then gently rinsed and transferred to peptide-free, serum-free medium different time periods before fixation. In some experiments, CytoD (0.5 µM, Sigma) was included in the medium 1 hr prior to the peptide addition and thereafter.
BrdU incorporation index
For the experiments carried out in synchronized cells, 12-hr post-transfected cells were induced to growth arrest by 48 h serum starvation. Cells were then cultured in regular medium for 12 hr, pulse labeled with 1 hr BrdU (10 µM), and followed by a 12 hr chase. These cells were subsequently treated with acid followed by BrdU labeling. The fractions of GFP+ transfected cells with BrdU incorporated were counted, and the BrdU incorporation indices were shown by considering the control as 100%. For the experiments carried out in unsynchronized cells, cells 48 hr post-transfection wee treated with BrdU for 12 hr, followed by GFP and BrdU labeling. For the protein transduction experiments, BrdU was added into serum-free medium for 12hr immediately after the 30-min incubation of peptides.
IUE, immunohistochemistry of mouse brain slices, and quantitative analyses
IUE procedures were performed on E13.5 CD1 mouse brains using plasmids driven by either CAG or U6 promoter, as described 45. At specific time points, electroporated brains were harvested, fixed with 4% paraformaldehyde overnight at 4°C, embedded in low melting agarose, and sectioned by vibratome. Sectioned brain slices were subsequently subjected to immunostaining, and all immunolabeled sections were analyzed by Leica TCS SP2 spectral confocal system (Nussloch, Germany), as previously described 10. In some experiments, electroporated brains were dissociated with papain (Worthington) and dispersed on poly-L-lysine coated coverslips. After a 2-hr 37°C incubation, cells were fixed with 4% paraformaldehyde followed by immunostaining. All animal manipulations were performed in accordance with the guidelines for animal experiments at Weill Medical Cornell IACUC.
Mitotic index, cell-cycle exit and cell-cycling analyses in brain slices
The mitotic index of treated brain slices was determined as the ratios of GFP and P-H3-double positive cells to the total GFP+ transfected cells in the ventricular zone/subventricular zone. Cell cycle analysis was performed as described 46. Briefly, pregnant mice were injected with BrdU (50 mg/kg body weight) 24 hr after IUE, and fetal brains were harvested 24 hr after BrdU treatment. Brain slices were then immunolabeled for GFP, BrdU and Ki67. The cell-cycle-exit index of GFP-transfected cells was determined as a ratio of the GFP-labeled cells that exited the cell cycle (GFP+, BrdU+ and Ki67−) to total GFP-labeled cells with BrdU incorporation (GFP+ and BrdU+). For S-phase entry analysis, IUE treated animals were treated with BrdU (50 mg/kg body weight) for 2 hr prior to harvest. In all cases, similar areas in the transfected neocortices were selected for analysis. Calculation of cell cycle phases was carried out using cumulative BrdU incorporation, as described 3. Briefly, BrdU injection started at E14.5 (24 hr post-electroporation) and continued at 3-hr intervals. Mice were sacrificed 2, 3, 6, 9, or 12 hr after the first BrdU injection. 100% BrdU incorporation refers to when all ventricular zone cells are labeled with BrdU. The length of G2/M was taken to be the time required to label all mitotic cells, based on chromatin condensation. The length of G1 was estimated as described 3.
Production and specificity verification of phospho-specific antibodies for Tctex-1
Rabbits were injected with a peptide (TDGSC(pT)VRWEN) corresponding to amino acid positions 89–99 of human Tctex-1 with chemical phosphorylation on the Thr94 residue. The resulting serum was affinity purified with a phospho-peptide column. The affinity purified Ab specifically detected the phospho-peptide but not the unphosphorylated peptide in ELISA assays.
Reagents
Two human Tctex-1-siRNA oligonucleotides (targeting sequence: 5’-GAGGCTAUAGAAAGCGCAATT-3’ and 5’-TACATCGTGACCTGTGTAATT-3’) and a mouse Tctex-1-siRNA (targeting sequence: 5'-GTCAACCAGTGGACCACT-3') were synthesized by Thermo Fisher (Waltham, MA). Both control-siRNA and AurA-si RNA oligonucleotide were purchased from Santa Cruz Biotech (Santa Cruz, CA). To generate short-hairpin encoding plasmid, the annealed targeting-oligos were first inserted into pU6-promoter driven vector (gift of Dr. Yang Shi), as suggested47. The entire shRNA expression cassette was then transferred into pCAG-IRES-GFP vector (gift of Dr. Connie Cepko); the resulting construct thus also encoded GFP under the CAG promoter. The targeting sequences for mouse and human Tctex-1-sh were 5’-GGGTTACACTCCGCAAGTTCC-3’ and 5’-GGGACAGCTCTACTGACGGGA-3’, respectively. The targeting sequence for (mouse and human) HDAC6-sh was 5’-GGGTTATGCCCACCTCACCCAC-3’. Three control shRNA plasmids were used; one was purchased from Applied Biosystem (Austin, TX) and two were synthesized (the targeting sequences were 5’-GGGATCGTAGGTTCTCGAAA-3’ and 5’-GGGCTATACCAAGAGACATGC-3’). For ectopic expression in vivo, cDNA fragments encoding Tctex-1 (e.g., WT, T94E, or T94A 9) were cloned into either pCAG-IRES-GFP or Tctex-1-sh/GFP plasmid. Other vectors (e.g., pEGFPC1, pRK5) were used to generate various constructs for in vitro expression. Detailed procedures are available upon request. GFP-9R and Tctex-1-9R were generated in pET21a vector (Novagen, Madison, WI), and recombinant proteins were expressed in BL21 (DE3) cells and purified by Ni-NTA agarose column (Invitrogen, San Diego, CA) 18.
Primary Ab used were acetylated α-tubulin mouse IgG2b (1:400, Sigma, St. Louis, MO), Arl13b rabbit Ab (1:500, Gift from Kathyrn Anderson 42), α-tubulin mouse Ab (1:1,000, GE Health, Piscataway, NJ), brain lipid binding protein rabbit Ab (1:2,500, Gift from Nathaniel Heintz 48), BrdU rat Ab (1:200, Harlan, Loughborough, England), active caspase 3 rabbit Ab (1:200, Abcam, Cambridge, MA), dynein intermediate chain mouse Ab (1:10,000, Millipore, Temecula, CA), FLAG mouse and rabbit Abs (1:500, Sigma), GFP rabbit Ab (1:1,000, Invitrogen), GFP chicken Ab (1:2,000, Abcam), GFP mouse Ab (1: 1,000, Millipore), γ-tubulin mouse Ab (clone GTU-88, IgG1, 1: 2,000, Sigma), Ki67 rabbit Ab (1:500, Novocastra, Newcastle, UK), Nestin mouse Ab (clone 401, 1: 200, Developmental Studies Hybridoma Bank, IA), P-H3 rabbit Ab (1:200, Millipore), Tbr2 rabbit Ab (1:1,000, Gift from Robert Hevner 49), protein A-purified Tctex-1 mouse Ab (1:1,000, This paper), affinity-purified Tctex-1 rabbit Ab 50 (1:100), Tuj1 mouse Ab (1:500, Covance, Princeton, NJ), ZO-1 mouse Ab (1:500, Invitrogen), phospho-Rb (Ser795), phospho-Rb(Ser807/811), and phospho-cdc2 (Tyr15) rabbit Abs (1:1,000, Cell Signaling Technology, Beverly, MA). Alexa-conjugated streptavidin or secondary Abs (anti-mouse, anti-rabbit, anti-rat, anti-IgG2b; 1:400) were purchased from Invitrogen; biotinylated anti-IgG1 and Cy5-conjugated streptavidin were purchased from Jackson Lab (used as 1:200; West Grove, PA). In this study, negative controls involved omission of the primary Ab and use of Ab that had been pre-absorbed with antigen. Co-labeling of acetylated α-tubulin (mouse IgG2b) and γ-tubulin (mouse IgG1) in this study involved using isotype-specific mouse IgGs.
Microscopic and statistical analyses
All the quantification studies were carried out in transfected cells localized within the dorsolateral neocortex to avoid possible variation within particular brain regions. At least 3 independent brains were analyzed for each DNA construct. Analysis of the immunostained sections was carried out on a Leica TCS SP2 spectral confocal system. In order to improve observer objectivity, image capture and analysis were done at separate times in a double-blind fashion. To score images, the GFP channel was first judged independently, followed by judgments of the other markers. For Ki67 and BrdU-nuclear labeling, small puncta or signals that were not compliant with the DAPI nuclear labeling were ignored.
Statistical analysis was performed with GraphPad software (GraphPad Prism v4.0, GraphPad Software Inc., San Diego, CA, USA). Data are presented as the mean ± SEM from at least three representative independent experiments. t-test was designed for the comparison of two groups. One-way analysis of variance (ANOVA) was applied for the comparisons in which only one independent variable was being analyzed. The Dunnet test (as a post hoc test) was used to compare data samples versus control. Statistical significance was defined as p < 0.05, 0.01 or 0.001.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to the following grant support: Tri-Institutional Starr Foundation, NYSTEM, NIH (EY11307, EY016805), RPB (to CHS), Tohoku University (to MS), New Energy and Industrial Technology Development Organization, and Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan (to KT). We thank Drs. Greg Pazour, Kathryn Anderson, Robert Hevner, Yang Shi, Nathaniel Heintz, Connie Cepko, Aiming Liu, and Steven Doxsey for reagents, and Drs. Stewart Anderson, M. Elizabeth Ross, David Cobrinik, and Bryan Tsou for discussion.
Footnotes
AUTHOR CONTRIBUTIONS
A.L., J.-Z., C., and C.-H. Sung designed the overall studies. A.L. and C.D. performed IUE experiments and phenotype characterization. J.-Z. C. generated all constructs. A. L., Y.-Y. T, and M.S. performed cell culture studies. K.T. generated the anti-phospho(T94)Tctex-1 Ab. K. T., and T. K. generated 9R peptides. A. L., J.-Z. C., and C.-H. S. wrote the paper.
REFERENCES
- 1.Pan J, Snell W. The primary cilium: keeper of the key to cell division. Cell. 2007;129:1255–1257. doi: 10.1016/j.cell.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi T, Nowakowski RS, Caviness VS., Jr The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15:6046–6057. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Pilaz LJ, et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci U S A. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calegari F, Huttner WB. An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J Cell Sci. 2003;116:4947–4955. doi: 10.1242/jcs.00825. [DOI] [PubMed] [Google Scholar]
- 7.Pfister KK, et al. Cytoplasmic dynein nomenclature. J Cell Biol. 2005;171:411–413. doi: 10.1083/jcb.200508078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King SM, et al. The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J. Biol. Chem. 1996;271:32281–32287. doi: 10.1074/jbc.271.50.32281. [DOI] [PubMed] [Google Scholar]
- 9.Chuang JZ, et al. The dynein light chain Tctex-1 has a dynein-independent role in actin remodeling during neurite outgrowth. Dev Cell. 2005;9:75–86. doi: 10.1016/j.devcel.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dedesma C, Chuang JZ, Alfinito PD, Sung CH. Dynein light chain Tctex-1 identifies neural progenitors in adult brain. J Comp Neurol. 2006;496:773–786. doi: 10.1002/cne.20958. [DOI] [PubMed] [Google Scholar]
- 11.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 13.Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia J, et al. Suppressor of Fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev Biol. 2009;330:452–460. doi: 10.1016/j.ydbio.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murcia NS, et al. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- 16.Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- 17.Schneider L, et al. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Matsushita M, et al. A high-efficiency protein transduction system demonstrating the role of PKA in long-lasting long-term potentiation. J Neurosci. 2001;21:6000–6007. doi: 10.1523/JNEUROSCI.21-16-06000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 20.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 21.Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Shu T, et al. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 23.Konno D, et al. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 24.Morin X, Jaouen F, Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat Neurosci. 2007;10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- 25.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 26.Davenport JR, Yoder BK. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am J Physiol Renal Physiol. 2005;289:F1159–F1169. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- 27.Mahjoub MR, Qasim Rasi M, Quarmby LM. A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol Biol Cell. 2004;15:5172–5186. doi: 10.1091/mbc.E04-07-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin H, Wang Z, Diener D, Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seeley ES, Carriere C, Goetze T, Longnecker DS, Korc M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Res. 2009;69:422–430. doi: 10.1158/0008-5472.CAN-08-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Zhong X, Pfeifer GP, Xu X. Microcephalin encodes a centrosomal protein. Cell Cycle. 2006;5:457–458. doi: 10.4161/cc.5.4.2481. [DOI] [PubMed] [Google Scholar]
- 33.Bond J, Woods CG. Cytoskeletal genes regulating brain size. Curr Opin Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Chizhikov VV, et al. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J Neurosci. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spassky N, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han YG, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 37.Lai K, Kaspar BK, Gage FH, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- 38.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 39.Gauthier-Fisher A, et al. Lfc and Tctex-1 regulate the genesis of neurons from cortical precursor cells. Nat Neurosci. 2009;12:735–744. doi: 10.1038/nn.2339. [DOI] [PubMed] [Google Scholar]
- 40.Kim J, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi A, O'Leary DD. Radial migration of superficial layer cortical neurons controlled by novel Ig cell adhesion molecule MDGA1. J Neurosci. 2006;26:4460–4464. doi: 10.1523/JNEUROSCI.4935-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas M, et al. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proc Natl Acad Sci U S A. 2005;102:5679–5684. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- 46.Sanada K, Tsai LH. G Protein betagamma Subunits and AGS3 Control Spindle Orientation and Asymmetric Cell Fate of Cerebral Cortical Progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Xia XG, et al. An enhanced U6 promoter for synthesis of short hairpin RNA. Nucleic Acids Res. 2003;31:e100. doi: 10.1093/nar/gng098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 49.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 50.Tai AW, Chuang J-Z, Sung C-H. Localization of Tctex-1, a cytoplasmic dynein light chain, to the Golgi apparatus and evidence for dynein complex heterogeneity. J. Biol. Chem. 1998;273:19639–19649. doi: 10.1074/jbc.273.31.19639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.