Summary
ERBB receptors were linked to human cancer pathogenesis approximately three decades ago. Biomedical investigators have since developed substantial understanding of the biology underlying the dependence of cancers on aberrant ERBB receptor signaling. An array of cancer-associated genetic alterations in ERBB receptors has also been identified. These findings have led to the discovery and development of mechanism-based therapies targeting ERBB receptors that have improved outcome for many cancer patients. In this Perspective, we discuss current paradigms of targeting ERBB receptors with cancer therapeutics and our understanding of mechanisms of action and resistance to these drugs. As current strategies still have limitations, we also discuss challenges and opportunities that lie ahead as basic scientists and clinical investigators work toward more breakthroughs.
ERBB family: EGFR, HER2, HER3, HER4
The ERBB family of transmembrane receptor tyrosine kinases (RTKs) consists of the epidermal growth factor receptor EGFR (ERBB1), HER2 (ERBB2), HER3 (ERBB3) and HER4 (ERBB4). Binding of ligands to the extracellular domain of EGFR, HER3, and HER4 induces the formation of kinase active hetero-oligomers (Yarden and Sliwkowski, 2001). HER2 does not bind any of the ERBB ligands directly but it is in a conformation that resembles a ligand-activated state and favors dimerization (Cho et al., 2003; Garrett et al., 2003). Activation of HER2 and EGFR induces trans-phosphorylation of the ERBB dimer partner and stimulates intracellular pathways such as RAS/RAF/MEK/ERK, PI3K/AKT/TOR, Src kinases, and STAT transcription factors [reviewed in (Yarden and Pines, 2012)]. Although HER3 can bind ATP and catalyze autophosphorylation, it has a weak kinase activity compared to that of its ERBB co-receptors (Shi et al., 2010). However, upon trans-phosphorylation by another ERBB family member, HER3 serves as an efficient phosphotyrosine scaffold, leading to potent activation of downstream signaling. The specificity and potency of intracellular signaling cascades are determined by the expression of positive and negative regulators, the specific composition of activating ligand(s), receptor dimer constituents and the array of proteins that associate with the tyrosine phosphorylated C-terminal domain of the ERBB receptors (Avraham and Yarden, 2011).
Over the past several years, it has become evident the ERBB family members have a prominent role in the initiation and maintenance of several solid tumors. This has led to the development and widespread implementation of specific ERBB inhibitors as cancer therapies. In this Perspective, we will focus on the therapeutic approaches for targeting ERBB family members in cancer, with a particular emphasis on HER2 amplified breast cancer and EGFR mutant lung cancer.
LINKS TO CANCER
HER2
The first evidence for a role of ERBB2 or HER2 (for human EGFR2) in cancer was inferred from the connection to its rat ortholog, Neu, a mutant cDNA isolated from carcinogen-induced neuroblastomas (Schechter et al., 1984). (Please note that in this Perspective, ERBB2 and HER2 will be used when discussing mouse and human ERBB2, respectively.) Although rodent Neu is mutated, human HER2 is typically amplified in human cancers such as breast, gastric and esophageal (Table 1). Overexpression of either rat or human wild-type ERBB2 was shown to transform diploid cells. Consistent with its oncogenic activity, overexpression of wild-type Neu or HER2 under the control of a mammary-specific promoter leads to metastatic mammary tumors in transgenic mice (Andrechek et al., 2000; Finkle et al., 2004). In a seminal study, Slamon et al. found that HER2 is amplified in about 20% of breast cancers (Slamon et al., 1987). This was the first report of an oncogenic alteration associated with poor outcome in cancer patients, suggesting a causal relationship to cancer virulence. Further evidence linking HER2 with cancer progression is the improvement in survival of patients with HER2 amplified early-stage breast cancer treated with the HER2 antibody trastuzumab. More recent studies using next-generation sequencing have identified less frequent activating mutations in HER2 in several cancer types without HER2 gene amplification (discussed below).
Table 1.
Alterations of ERBB receptors and ligands in human cancer
| Molecule | Alteration | Cancer types | Notes | References |
|---|---|---|---|---|
| EGFR | mutation, insertion (L858R, etc.) | NSCLC (adenocarcinoma) | (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004) | |
| EGFR | vIII | glioma | deletion of exons 2–7 in the ectodomain | (Sugawa et al., 1990) |
| EGFR | amplification | NSCLC (squamous), head & neck, glioma, esophageal, colorectal, anal (?) | (Yarden et al., 2012) | |
| HER2 | amplification | breast, gastric, esophageal | (Koboldt et al., 2012) | |
| HER2 | mutation | breast (lobular), lung, gastric, bladder, endometrial | unclear if all those reported are activating or gain-of-function | (Kobodlt et al., 2012) |
| HER3 | mutation | breast, gastric | (Jaiswal et al., 2013) | |
| HER4 | mutation | melanoma, NSCLC, medulloblastoma | (Gilbertson et al., 2001; Prickett et al., 2009) | |
| TGFα | overexpression | prostate, lung, pancreas, ovary, colon, head & neck | androgen-independent prostate cancer; poor prognosis when associated with high EGFR | (Grandis et al., 1998; Yarden and Sliwkowski, 2001) |
| Neuregulin-1 | Overexpression | colorectal, head & neck | associated with drug sensitivity or resistance | (Wilson et al., 2011; Yonesaka et al., 2011) |
A recent study of >500 breast tumors by The Cancer Genome Atlas (TCGA) Network has shed light into the biological heterogeneity of clinical HER2 overexpressing cancers (HER2+ as defined by gene amplification) by further parsing into HER2-enriched (HER2E) and luminal subtypes as defined by gene expression (Koboldt et al., 2012). HER2E-HER2+ tumors had higher frequencies of aneuploidy, somatic mutation, and TP53 mutation, as well as amplification of FGFRs, EGFR, CDK4, and cyclin D1. Luminal-HER2+ breast cancers showed higher expression of a luminal gene cluster including GATA3, BCL2, and ESR1 and harbored a higher rate of GATA3 mutations. It is anticipated that because of these molecular differences, the clinical management of HER2E and luminal subtypes of HER2+ breast cancers will also be different. Finally, not all tumors of the HER2E gene expression subtype were HER2 amplified. One implication of these data is that some breast cancers with a single copy of HER2 harbor an expression signature of HER2 dependence and, as such, may benefit from anti-HER2 therapy. Consistent with this speculation are the results of the NSABP B-31 adjuvant trastuzumab trial, in which 9.7% of patients that did not meet criteria for HER2 overexpression by FISH or IHC also benefitted from adjuvant trastuzumab (Paik et al., 2008).
Somatic mutations in HER2 have been reported in several human cancers (Table 1). Most are missense mutations in the tyrosine kinase and extracellular domains or duplications/insertions in a small stretch within exon 20. HER2 mutations are almost exclusively observed in cancers without HER2 gene amplification. Several of these mutants have increased signaling activity, and are most commonly associated with lung adenocarcinoma, lobular breast, bladder, gastric, and endometrial cancers (Koboldt et al., 2012).
EGFR
The EGF receptor was originally identified as an oncogene because of its homology to v-ERBB, a retroviral protein that enables the avian erythroblastosis virus to transform chicken cells (Downward et al., 1984). Subsequently, EGFR overexpression was shown to be transforming in laboratory models, and EGFR gene amplification was reported in a wide range of carcinomas. Early studies by Mendelsohn and colleagues demonstrated that antibodies directed against EGFR block growth of A431 cells, demonstrating that EGFR signaling could drive cancer cell growth and setting the stage for clinical use of EGFR inhibitors (Kawamoto et al., 1983).
An oncogenic mutation that deletes exons 2–7 in the receptor ectodomain, denoted EGFRvIII, is found in about 40% of high-grade gliomas with wild-type EGFR amplification (Sugawa et al., 1990). EGFRvIII exhibits constitutive dimerization, impaired downregulation, and aberrant tyrosine kinase activity, all resulting in enhanced tumorigenicity (Nishikawa et al., 1994). In addition to glioblastoma multiforme (GBM), EGFRvIII has been found in a fraction of breast, lung, head and neck, ovarian, and prostate cancers (Moscatello et al., 1995). Because its expression is restricted to tumor tissues, EGFRvIII has been therapeutically targeted with specific antibodies and vaccines. There is clinical evidence suggesting that the presence of EGFRvIII can predict clinical responses of GBMs to the EGFR TKIs gefitinib and erlotinib (Haas-Kogan et al., 2005; Mellinghoff et al., 2005). The second most common EGFR variant in GBM is EGFRc958, observed in about 20% of tumors with wild-type EGFR amplification. EGFRc958 lacks amino acids 521–603 and displays increased, ligand-dependent kinase activity (Frederick et al., 2000).
The causal role of EGFR in tumorigenesis was further solidified in 2004 when somatic, activating mutations in EGFR were discovered in a subset of non-small cell lung cancers (NSCLC) (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004) (Table 1). The discovery was spurred by efforts to understand why occasional NSCLCs were highly sensitive to small molecule EGFR tyrosine kinase inhibitors (TKIs). It is now well established that lung cancers harboring these EGFR mutations are highly responsive to single-agent EGFR inhibitors with RECIST response rates of ~55%–75% (Mok et al., 2009; Rosell et al., 2012; Sequist et al., 2013b). EGFR mutations are primarily localized within two hot spots of the kinase domains, a series of overlapping deletions in exon 19 and a leucine to arginine substitution at amino acid position 858 (L858R) [reviewed in (Pao and Chmielecki, 2010)]. In addition, mutations are also rarely observed elsewhere in the kinase domain, including insertions in exon 20 (Yasuda et al., 2013). The prevalence of the mutations differs among distinct human populations. They are found in ~8%–10% of Caucasians, but in a higher proportion East Asians. Lung cancers with EGFR mutations are most highly associated with adenocarcinoma histology and in patients with a minimal smoking history. Of note, cancers with EGFR mutations often have amplification of the mutant EGFR allele as well (Cappuzzo et al., 2005). Cell culture and transgenic mouse model studies have shown that mutant EGFR has transforming activity (Greulich et al., 2005; Ji et al., 2006; Politi et al., 2006).
EGFR is important for the growth of some colorectal cancers (CRC) and head and neck cancers. In these cancers, genetic alterations in EGFR have not been consistently identified. However, the efficacy of the EGFR antibody cetuximab demonstrates the importance of EGFR signaling in these tumors. Although some reports suggest that EGFR amplification correlates with response to cetuximab (Moroni et al., 2005), this alteration is not currently used as a predictive biomarker. Importantly, cetuximab provides clinical benefit primarily in colorectal cancers that do not harbor KRAS mutations (Cunningham et al., 2004) and in those with high expression of EGFR ligands amphiregulin and epiregulin (Khambata-Ford et al., 2007). Presumably, cetuximab is effective in sensitive cancers because it blocks ligand-dependent activation of EGFR and down-regulates the receptor from the cell surface (Fan et al., 1994). Thus, in these colorectal cancers, we suspect that ligand-dependent activation of EGFR drives progression of these cancers. Currently, cetuximab is most often administered with chemotherapy in KRAS wild-type colorectal cancers. Similarly, in head and neck cancers, cetuximab is primarily used in conjunction with chemotherapy (Vermorken et al., 2008) as well as radiotherapy (Bonner et al., 2006). Despite conflicting reports on the utility of EGFR expression by IHC for patient selection in head and neck cancers (and CRC), there currently are no validated predictive biomarkers of response to EGFR inhibitors in head and neck cancers (Burtness et al., 2005; Cunningham et al., 2004; Licitra et al., 2011; Licitra et al., 2013; Vermorken et al., 2008). It is notable that cetuximab appears to be more effective than EGFR TKIs in cancers with ligand-dependent activation of EGFR, whereas TKIs are more effective in cancers with EGFR mutations. We speculate that this is so because mutant EGFR activation is not ligand-dependent and TKIs have higher affinity for mutant EGFR than wild-type EGFR, thus leading to a significant therapeutic window. In contrast, antibodies such as cetuximab are more effective in EGFR wild-type cancers because they are highly effective at blocking ligand-dependent activation of EGFR and are pharmacologically stable.
ERBB3 and ERBB4
ERBB3 has been linked to cancer, primarily due to its mechanistic role in promoting signaling from oncogenic HER2 and EGFR (discussed below). However, somatic mutations scattered throughout the ERBB3 gene were recently identified in subsets of breast and gastric cancers (Table 1). Many of the mutations were located in the extracellular domain, and they appear to have oncogenic potential, function in a ligand-independent manner, and require heterodimerization with HER2 for transforming activity (Jaiswal et al., 2013). Future studies are needed to determine if cancers with ERBB3 mutations are particularly sensitive to ERBB3 and/or HER2-targeted drugs. Similarly, mutations in ERBB4 were identified in cancer, particularly melanoma (Prickett et al., 2009), lung adenocarcinoma (Ding et al., 2008), and medulloblastoma (Gilbertson et al., 2001). Although laboratory studies demonstrated that melanoma cell lines harboring ERBB4 mutations were sensitive to lapatinib, it remains unknown if targeting ERBB4, or any other ERBB family member, will have therapeutic value in these cancers.
ERBB Ligands
Overproduction of ligands is one mechanism by which cancers aberrantly activate ERBB receptors. The source of these can be tumor cells or the tumor stroma. There are three groups of ligands. One group specifically binds EGFR and includes EGF, TGFα, amphiregulin (AR), and epigen (EPG). A second group binds both EGFR and HER4 and includes betacellulin (BTC), HB-EGF, and epiregulin (EPR). The third group includes all the neuregulins (NRG) 1–4, of which NRG1 and NRG2 bind HER3 and HER4, whereas NRG3 and NRG4 only bind HER4 (Hynes and MacDonald, 2009).
In transgenic mouse studies, mice that co-express TGFα and Neu in mammary epithelium developed multifocal mammary cancers that arose after a significantly shorter latency than those expressing either gene alone (Muller et al., 1996). TGFα is also co-overexpressed with EGFR in lung, colorectal, ovary and head and neck squamous cancers, where it is associated with poor patient prognosis (Grandis et al., 1998; Yarden and Sliwkowski, 2001) (Table 1). Recent reports suggest that in addition to overexpression, mistrafficking, and/or ‘extracrine’ (exosomal targeting receptor activation) signaling by ERBB ligands may also contribute to epithelial cell transformation (Singh and Coffey, 2014). For example, altered trafficking of EREG to the apical cell surface leads to prolonged EGFR phosphorylation and more proliferative and invasive tumors (Singh et al., 2013). Further, significantly enhanced levels of invasiveness are observed when breast cancer cells are incubated with exosomes containing high levels of AREG compared to incubation of cells with exosomes containing low levels of AREG or recombinant EGFR ligands (Higginbotham et al., 2011), suggesting a gain-of-function mode of EGFR signaling that might act in more distant environments. Other roles of ligand-dependent activation of EGFR were discussed above.
An autocrine loop has been described in ovarian cancer cells and tumors that overexpress NRG1 and HER3, where suppression of HER3 with RNAi or with a neutralizing HER3 antibody suppressed ovarian cancer growth in laboratory models (Sheng et al., 2010) (Table 1). A NRG1-mediated autocrine loop inducing HER3 activation was also discovered in head and neck cancer cells without HER2 amplification. These cells were particularly sensitive to the EGFR/HER2 TKI lapatinib (Wilson et al., 2011), suggesting NRG1-driven tumors depend on HER3 activated by HER2 and/or EGFR. Finally, Hegde et al. found high levels of NRG1 and its receptor HER4 in NSCLC residual tumor cells that remained after cytotoxic chemotherapy. Inhibition of HER3/4 signaling with a NRG1-blocking antibody increased the magnitude and duration of response to chemotherapy in these in vivo models (Hegde et al., 2013). This causal association of ERBB ligand overexpression and drug resistance is not limited to NRG1 or to chemotherapy. For example HGF has been found to confer resistance to the BRAF inhibitor vemurafenib in BRAF-mutant melanoma cells (Wilson et al., 2012).
DOWNSTREAM SIGNALING
Oncogenic addiction to EGFR and HER2 are intimately linked to regulation of downstream signaling. In cancers highly sensitive to inhibition of EGFR or HER2 inhibitors, EGFR or HER2 is the main driver of downstream signaling, particularly via the PI3K/AKT and MEK/ERK pathways. Thus, in cancers addicted to EGFR or HER2, inhibition of the respective RTK leads to concomitant loss of flux through these pathways. Loss of these signaling events leads to growth arrest and converges on the BCL-2 family of proteins to promote apoptosis [reviewed in (Niederst and Engelman, 2013)].
In EGFR and HER2 driven cancers, HER3 is an important heterodimer partner because it potently activates the phosphatidylinositide-3 kinase (PI3K)/AKT survival pathway via its six docking sites for the p85 regulatory subunit of PI3K. Although HER2 potently activates ERK signaling, it does not bind p85 nor directly activate PI3K/AKT. Thus, HER2-mediated activation of HER3 is essential for stimulation of the PI3K/AKT pathway. In transgenic mice, genetic ablation of ERBB3 in the mammary gland via Cre-mediated recombination abrogates ERBB2-driven mammary hyperplasias, DCIS, invasive cancers and metastases (Vaught et al., 2012). Similarly, shRNA-mediated knockdown of HER3 but not EGFR inhibits viability of HER2-overexpressing breast cancer cells. Further, HER3 but not EGFR is always phosphorylated in human HER2-amplified breast cancers (Lee-Hoeflich et al., 2008), suggesting it is an obligatory co-biomarker of aberrant HER2 activity and dependence. More recently, an inducible HER3 shRNA (Lee-Hoeflich et al., 2008) and a HER3 neutralizing antibody (Garrett et al., 2013b) were shown to inhibit growth of established HER2-amplified xenografts, further suggesting HER3 is essential for survival of HER2-dependent tumors. Analogous to HER2-induced signal transduction, mutant EGFR often activates PI3K via HER3 (Engelman et al., 2005), and maintenance of HER3 signaling can promote resistance to EGFR inhibitors (Engelman et al., 2007; Schoeberl et al., 2010). However, unlike HER2, EGFR is also able to signal to PI3K via GAB1 in a HER3-independent manner (Mattoon et al., 2004; Turke et al., 2010), suggesting that EGFR mutant cancers may be better equipped than HER2 amplified cancers to adapt to the loss of HER3 function.
HER2 amplified tumors have a strong dependence on PI3K/AKT signaling, as sustained blockade of this pathway appears to be required for the antitumor effect of HER2 antagonists (Chakrabarty et al., 2013; Yakes et al., 2002). Comprehensive cancer cell line panels screened for sensitivity to pan-PI3K, p110α-specific, and AKT inhibitors have consistently shown preferential activity of these drugs against HER2-amplified breast cancer lines (Heiser et al., 2012; O’Brien et al., 2010). Further, genetic ablation of p110α has been shown to abrogate ERBB2-induced mammary tumor formation in transgenic mice (Utermark et al., 2012). Preclinical studies have shown that, compared to HER2 amplified cancers, EGFR mutant cancers are less sensitive to single-agent PI3K/AKT inhibitors. Rather, inhibition of the PI3K and MEK pathways is necessary in order to induce apoptosis and cause tumor regressions (Faber et al., 2010). Importantly, mechanisms of de novo and acquired resistance to HER2 and EGFR directed therapies involve persistence or reactivation of PI3K/AKT signaling via alternate amplified RTKs and/or mutations in the PI3K pathway (Rexer and Arteaga, 2013).
Other downstream signaling pathways such as Src kinases, JAK/STAT and WNT are also activated by ERBB receptors (Yarden and Sliwkowski, 2001). Examples below suggest they are involved and/or mediate resistance to ERBB receptor-targeted therapies. However, evidence that ERBB receptors depend on Src, JAK/STAT or WNT for their effects on transformation and cancer progression is less clear and will not be discussed further.
Feedback activation of ERBB signaling promoting resistance to inhibition of alternative kinases
More recently, EGFR and HER3 activation have been observed as important cellular adaptations to inhibitors of downstream signaling. For example, in BRAF mutant CRC, BRAF inhibitors fail to inhibit ERK signaling in sustained fashion due to activation of EGFR which, in turn, re-activates ERK in the presence of the BRAF inhibitor (Corcoran et al., 2012; Prahallad et al., 2012). However, combined inhibition of EGFR and BRAF blocks re-activation of ERK and leads to regressions of BRAF mutant CRC in vivo. This combination is now being actively developed in clinic for this subset of CRCs. Similarly, inhibition of the MEK pathway in many cancers, including KRAS mutant cancers, activates ERBB signaling by releasing a negative feedback on ERBB dimerization (Turke et al., 2012). This further suggests the ERBB activation could mitigate the responsiveness of other cancers to MEK inhibition.
Analogous to the effects of inhibiting the MEK pathway, inhibition of the PI3K pathway leads to potent activation of HER3-dependent signaling in HER2-amplified breast cancers (Chakrabarty et al., 2012; Chandarlapaty et al., 2011). In these cancers, co-inhibition of HER3 and PI3K provided substantially greater anti-tumor efficacy. In other examples, EGFR activation has been observed as a resistance mechanism to small molecules targeting other tyrosine kinases. For example, EGFR activation is a resistance mechanism to ALK and MET inhibitors in ALK-positive lung and MET-amplified gastric cancers, respectively. Inhibition of EGFR re-sensitizes the resistant cancers to their respective TKI (Katayama et al., 2012; McDermott et al., 2010; Qi et al., 2011; Sasaki et al., 2011). Thus, activation of ERBB family members has emerged as a common mechanism of adaptation upon inhibition of downstream signaling, and inhibition of ERBB family members may be used to augment the efficacy of other pathway inhibitors.
MECHANISMS OF ACTION OF EGFR AND HER2 INHIBITORS
HER2
Trastuzumab is a humanized IgG1 antibody that binds to an epitope in juxtamembrane region IV of the HER2 receptor. It inhibits cleavage of the HER2 ectodomain, uncouples ligand-independent HER2-containing dimers leading to partial inhibition of downstream signaling, and triggers antibody-dependent, cell-mediated cytotoxicity (ADCC) (Clynes et al., 2000; Ghosh et al., 2011; Junttila et al., 2009; Molina et al., 2001; Yakes et al., 2002) (Table 2). This last mechanism cooperates with the recruitment of a T-cell population mediating an adaptive immune (memory) response that enhances tumor eradication (Park et al., 2010; Stagg et al., 2011). The importance of the immune response is underscored by the finding that the therapeutic effect of trastuzumab was markedly diminished in mice that were engineered to be deficient in NK cells and macrophages capable of binding the Fc region of trastuzumab (Clynes et al., 2000). Pertuzumab is a monoclonal antibody that recognizes an epitope in heterodimerization domain II of HER2, thus blocking ligand-induced HER2-HER3 dimerization, resulting in partial inhibition of PI3K/AKT signaling (Agus et al., 2002). Because pertuzumab and trastuzumab bind to different epitopes in the HER2 ectodomain (Franklin et al., 2004), hence their complementary abilities to disrupt HER2-containing dimers, the combination of pertuzumab and trastuzumab has shown synergy in preclinical studies (Scheuer et al., 2009) and clinical trials (Baselga et al., 2012b; Gianni et al., 2012) and is now approved for treatment of patients with HER2+ breast cancer. Trastuzumab-derivative of maytansine 1 (T-DM1 or trastuzumab emtansine) is an antibody-drug conjugate in which one molecule of trastuzumab is covalently bonded via a non-cleavable linker to 3.5 molecules of a maytansinoid that inhibits microtubule polymerization (DM1). After binding to the receptor, the T-DM1/HER2 complex is internalized followed by degradation in lysosomes, release of DM1 and subsequent cell lysis (Lewis Phillips et al., 2008). T-DM1 binds to HER2 with similar affinity as trastuzumab, thus retaining the ability of the naked antibody to inhibit ligand-independent HER2-containing dimers and signal transduction as well as to mediate ADCC (Junttila et al., 2011).
Table 2.
ERBB receptor inhibitors: Mechanisms of action and key clinical trials
| Drug | Type of molecule | Mechanism of action | FDA approval | Key clinical trial(s) |
|---|---|---|---|---|
| Trastuzumab | Humanized IgG1, binds juxtamembrane domain IV | Inhibits ectodomain cleavage and ligand-independent HER2-containing dimers ADCC and adaptive immunity to HER2 |
1998 (metastatic breast) 2006 (adjuvant early breast) 2010 (advanced gastric) |
(Slamon et al., 2001) (Piccart-Gebhart et al., 2005; Robert et al., 2006; Romond et al., 2005) (Bang et al., 2010) |
| Pertuzumab | Humanized IgG1, binds heterodimerization domain II | Inhibits ligand-induced HER2-containing dimers | 2012 (metastatic breast) 2013 (neoadjuvant breast) |
(Baselga et al., 2012b; Gianni et al., 2012; Schneeweiss et al., 2013) |
| Lapatinib | Small molecule | Reversible, ATP-competitive TKI | 2006 (advanced breast) | (Geyer et al., 2006) |
| Trastuzumab emtansine (T-DM1) | Antibody-drug conjugate | Same as trastuzumab plus inhibition of microtubules and cell lysis (DM-1) | 2013 (advanced breast) | (Verma et al., 2012) |
| Erlotinib | Small molecule | Reversible, ATP-competitive TKI | 2004 (3rd line advanced NSCLC) 2005 (pancreas cancer) 2013 (1st line EGFR mutant NSCLC) |
(Mok et al., 2009; Moore et al., 2007; Shepherd et al., 2005) |
| Afatinib | Small molecule | Irreversible ATP-competitive TKI | 2013 (metastatic EGFR mutant NSCLC) | (Sequist et al., 2013b) |
| Neratinib | Small molecule | Irreversible ATP-competitive TKI | N/A | Trials in patients with HER2 mutant tumors in progress |
| Cetuximab | Human-murine chimeric IgG2, binds ligand-binding domain | Inhibits ligand-dependent activation of EGFR | 2004 (originally for late line EGFR+ CRC but now only used in earlier line wild-type KRAS CRC) 2006 (head & neck with radiotherapy or chemotherapy) |
(Van Cutsem et al., 2009; Vermorken et al., 2008) (Bonner et al., 2006) |
| Panitumumab | Human IgG1, binds ligand-binding domain | Inhibits ligand-dependent activation of EGFR | 2006 (originally for late line EGFR+ CRC but now only used in earlier line wild-type KRAS CRC) | (Van Cutsem et al., 2007) |
| AZD9291 | Small molecule | Irreversible ATP-competitive TKI (3rd generation) | N/A | Trials in EGFR mutant lung cancer in progress |
| CLO-1686 | Small molecule | Irreversible ATP-competitive TKI (3rd generation) | N/A | Trials in EGFR mutant lung cancer in progress |
Lapatinib is an ATP-competitive, reversible small molecule inhibitor of the HER2 and EGFR tyrosine kinases (Konecny et al., 2006). In HER2+ breast cancers, lapatinib quickly disables HER2 signaling, resulting in inhibition of the PI3K/AKT and MAPK pathways, and it has shown clinical activity in HER2+ breast cancers that have progressed on trastuzumab (Geyer et al., 2006). Lapatinib also binds the inactive conformation of EGFR (Wood et al., 2004) but has not been active against cancers for which EGFR antibodies or TKIs are approved. Afatinib (Minkovsky and Berezov, 2008) and neratinib (Burstein et al., 2010) are irreversible, covalent HER2/EGFR TKIs with activity against HER2, HER4, EGFR, and some HER2 insertion mutants (Bose et al., 2013). Of note, the clinical efficacy of all therapeutic inhibitors of HER2 has been predominantly limited to breast cancers that overexpress HER2 as measured by intense membrane staining in the majority of tumor cells with HER2 antibodies [3+ by immunohistochemistry (IHC)] or excess copies of the HER2 gene determined by fluorescent in situ hybridization (FISH).
EGFR
Gefitinib and erlotinib are ATP-competitive EGFR TKIs (Table 2). Biochemical and crystallography analyses demonstrate that the mutants possess a higher affinity for the first-generation EGFR inhibitors gefitinib and erlotinib compared to the wild-type receptor (Carey et al., 2006; Yun et al., 2007). Thus, the mutant enzymes are inhibited at lower concentrations of drug, which leads to a favorable therapeutic index. As will be discussed in greater detail below, EGFR mutant lung cancers often develop a second mutation in the gatekeeper residue, T790M, as they become resistant to gefitinib or erlotinib. Thus, there have been intense efforts to develop a drug that can inhibit T790M EGFR to overcome resistance. One such effort was the development of second-generation EGFR inhibitors, such as afatinib and dacomitinib. These drugs are irreversible ATP-competitors that form covalent links with the Cys773 residue of EGFR. Although these second-generation drugs have the capacity to inhibit the EGFR T790M, they do so at concentrations that also inhibit wild-type EGFR. Thus, there is not a favorable therapeutic index, and dose-limiting toxicities due to inhibition of wild-type EGFR (such as rash and diarrhea) prevent increasing doses high enough to fully suppress T790M. Thus, they have been largely ineffective at overcoming T790M-mediated resistance in the clinic. Pao et al. found that mouse lung transgenic tumors expressing T790M EGFR are sensitive to the combination of afatinib and cetuximab (Regales et al., 2009). This combination has progressed to the clinic where it has demonstrated significant clinical activity against T790M EGFR lung cancers, although it is also associated with significant toxicity (Janjigian et al., 2011).
More recently, third-generation EGFR inhibitors have been developed. The first of such compounds, WZ-4002, was designed to be much more potent against the resistant T790M mutation than the wild-type receptor, thus restoring a favorable therapeutic index in which the drugs can be dosed high enough to inhibit T790M without inducing toxicity from inhibiting wild-type EGFR (Walter et al., 2013; Zhou et al., 2009). Of note, this drug is not a quinazoline derivative like the first- and second-generation EGFR inhibitors. WZ-4002 has not been developed clinically, whereas two drugs with similar properties, AZD9291 and CLO-1686, have been (Walter et al., 2013). Clinical data are emerging for these compounds, and the high rate of clinical responses, with minimal toxicity, are increasing enthusiasm for the class of drugs (Ranson et al., 2013; Sequist et al., 2013a; Soria et al., 2013).
In contrast to the EGFR TKIs, the EGFR neutralizing antibody cetuximab blocks ligand binding to the EGFR. Thus, it is most effective in cancers that harbor ligand-activated, wild-type EGFR. In colorectal cancers with wild-type KRAS, inhibition of EGFR leads mainly to loss of downstream ERK signaling. However, since mutant KRAS directly activates ERK, cetuximab fails to suppress ERK in these cancers, likely explaining the lack of clinical activity (Ebi et al., 2011). As a result, cetuximab is now used primarily in cancers with wild-type KRAS. Panitumumab is another EGFR-targeted antibody that has activity in wild-type KRAS CRC. Unlike cetuximab, it is an IgG2, and is predicted not to engage immune effector cells to mediate ADCC. Despite this difference, phase III studies have demonstrated clinical efficacy similar to that of cetuximab (Douillard et al., 2013; Jonker et al., 2007; Van Cutsem et al., 2009; Van Cutsem et al., 2007). Thus, it seems plausible that the primary mechanism of action of cetuximab and panitumumab is due to its inhibition of EGFR signaling and not engagement of ADCC.
HER3 inhibitors
Several HER3 neutralizing antibodies are in clinical development. MM-121 and U3-1287 (formerly AMG-888) bind the extracellular domain of HER3, block heregulin-induced phosphorylation and reduce expression of HER3 at the cell surface (Garrett et al., 2011; Schoeberl et al., 2010). MM-121 (IgG2) is most effective against tumors with ligand-dependent activation of HER3 (Sheng et al., 2010). U3-1287 synergizes with trastuzumab and lapatinib to suppress the growth of HER2-amplified xenografts (Garrett et al., 2013a) and has single-agent activity against transgenic mouse mammary cancers induced by Polyomavirus middle T antigen (Muraoka-Cook et al., 2011). RG7116 is an IgG1 that selectively binds domain 1 of human HER3. It blocks ligand binding and down-regulates HER3 from the cell surface. Through glycoengineering of its Fc moiety, RG7116 mediates enhanced ADCC that correlates with HER3 receptor density (Mirschberger et al., 2013). At this time, these antibodies have completed phase I safety and dose-finding trials but their clinical efficacy remains to be shown.
LJM716 is a novel anti-HER3 antibody that binds an epitope within domains 2 and 4 in the receptor’s extracellular domain, thus trapping HER3 in an inactive conformation. In contrast to the other anti-HER3 antibodies, it blocks both ligand-induced and ligand-independent HER3 dimerization and activation (Garner et al., 2013). This property may be particularly advantageous in HER2-amplified breast cancers, in which HER2 appears to activate HER3 in a ligand-independent manner. Accordingly, in laboratory studies, LJM716 reduced growth of established HER2-amplified xenografts when given as a single agent and synergized with PI3K inhibitors to suppress growth of HER2-amplified/PIK3CA mutant tumors (Garrett et al., 2013b).
More recently, bi-specific antibodies targeting HER3 have been introduced. MM-111 is an antibody that docks onto HER2 and subsequently binds HER3, thus blocking ligand-dependent activation of HER2/HER3 dimers (McDonagh et al., 2012). Finally, MEHD7945A is a two-in-one IgG1 generated by phage display engineering that specifically binds HER3 and EGFR with high affinity, thus blocking TGFα- and HRG-induced activation of both receptors and downstream PI3K/AKT and ERK signaling. MEHD7945A mediates ADCC in vivo and demonstrated superior antitumor activity against multiple tumor models compared to monospecific antibodies (Schaefer et al., 2011). Currently HER3 inhibitors are being developed in combination with trastuzumab, EGFR antibodies and TKIs, PI3K inhibitors and cytotoxic chemotherapy. In addition to HER2 amplification and EGFR mutation, high heregulin expression and HER3 mutations are being explored as predictive biomarkers of response in clinical trials.
MECHANISMS OF RESISTANCE TO ERBB INHIBITORS
Although ERBB targeted therapies have provided substantial benefit to patients with advanced cancer, cancers ultimately have developed resistance to the current approaches. In this Perspective, we will discuss both de novo and acquired resistance. The distinction is primarily a clinical one: de novo or intrinsic resistance refers to cancers that do not exhibit an initial response, whereas acquired resistance develops following an initial, often marked and durable, clinical response. It is important to appreciate that the same molecular mechanism may cause both types of resistance, underscoring the robustness of the biological principles underlying how cancers evade these therapies.
Mechanisms of resistance have been discovered by several approaches, including the maintenance of cell lines and xenografts in the presence of drug until resistance emerges, or infecting sensitive cell lines with open reading frame (ORF) or shRNA libraries to identify genes whose expression or loss leads to resistance. These efforts have also been coupled to biopsy programs, in which cancers are systematically biopsied upon the development of resistance to interrogate acquired molecular changes upon treatment pressures (Sequist et al., 2011; Yano et al., 2011; Yu et al., 2013). However, there are significant limitations with many of the laboratory studies. Although EGFR TKIs are primarily used as single-agents for EGFR mutant lung cancers, HER2-directed therapies and EGFR antibodies are generally used in combination with chemotherapy in the clinic. However, most laboratory studies have modeled resistance to these agents as single therapies, thus not recapitulating the selective pressure of combination therapies applied in the clinic. Other data about potential resistance mechanisms have been derived from correlative clinical trials where patients have been treated with anti-HER2 drug(s) in combination with chemotherapy, a variable not always considered in the interpretation of the studies of drug resistance. Finally, even though combinations of HER2 antagonists are increasingly used in the clinic, resistance to these combinations has yet to be modeled widely in the laboratory.
Intrinsic HER2 alterations
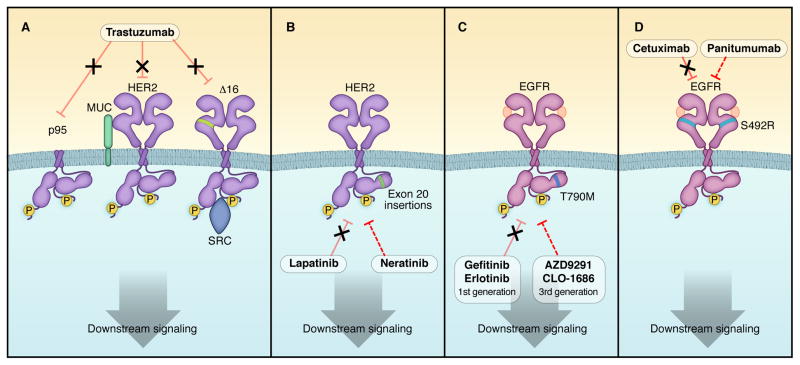
Some resistance mechanisms affect the capacity for HER2 inhibitors to directly engage HER2. Anido et al. described p95-HER2, a truncated form of HER2, lacking the trastuzumab binding region, which may arise from alternate transcription initiation sites in HER2 (Anido et al., 2006) (Figure 1A). Patients with metastatic breast cancer harboring cytosolic expression of p95-HER2 exhibit a very low response rate to trastuzumab compared to those patients without p95-HER2 in their tumors (Scaltriti et al., 2007). This form of HER2 retains kinase activity, and tumors with p95-HER2 may still be susceptible to kinase inhibition with a TKI, as suggested by the observation that p95-HER2 tumors exhibit a similar response rate to the combination of capecitabine and lapatinib compared to breast cancers expressing full-length HER2 (Scaltriti et al., 2010).
Figure 1. Schema depicting intragenic alterations leading to resistance to HER2 and EGFR inhibitors.
A) HER2 truncations (p95) and splice variants (Δ16) are not inhibited by trastuzumab. In addition, expression of specific mucin isoforms can prevent trastuzumab from binding HER2 (Price-Schiavi et al., 2002). Not shown in the figure, pertuzumab and T-DM1 cannot recognize p95 either. B) HER2 harboring exon 20 insertions are not inhibited by lapatinib, but may be sensitive to irreversible HER2 inhibitors afatinib and neratinib. They are also resistant to trastuzumab. C) The EGFR T790M gatekeeper mutation leads to acquired resistance to first generation EGFR inhibitors, but is effectively inhibited by third generation EGFR inhibitors. D) An EGFR mutation in the extracellular domain is associated with acquired resistance to cetuximab, but may still be sensitive to another anti-EGFR antibody, panitumumab. Dashed lines indicate inhibition via alternative antibodies and inhibitors.
A splice variant that eliminates exon 16 in the extracellular domain of the HER2 receptor has also been identified in HER2+ primary breast cancers and cell lines (Kwong and Hung, 1998) (Figure 1A). This variant does not eliminate the trastuzumab epitope on HER2, but stabilizes HER2 homodimers and prevents their disruption upon binding by the antibody, resulting in trastuzumab resistance in cell lines. The Δ16 isoform was found to interact directly with Src, and treatment with the Src inhibitor dasatinib overcame the resistance to the antibody conferred by the alternative splicing variant (Mitra et al., 2009). However, clinical evidence of an association between HER2-Δ16 and resistance to trastuzumab has not been shown.
HER2 mutations have been found in a small proportion of lung, gastric, colorectal, breast and head and neck cancers (Lee et al., 2006; Ross et al., 2013; Stephens et al., 2004; Willmore-Payne et al., 2006). These mutants of HER2 are resistant to lapatinib and trastuzumab (Figure 1B) but sensitive to the covalent HER2 TKI neratinib (Bose et al., 2013; Wang et al., 2006). To the best of our knowledge, HER2 mutations in HER2 gene-amplified breast tumors are very rare. As such, they have not been identified as a resistance mechanism to trastuzumab. One possible reason is that these mutations may comprise only a portion of the amplified HER2 alleles and, therefore, exist below the limits of sensitivity of traditional DNA sequencing methods (Zito et al., 2008). It is possible that cancer cells harboring these mutations will be selected, or acquired, after the selective pressure of anti-HER2 treatment. If so, they may only be detected in tumors that are progressing after primary HER2-targeted therapy. However, comprehensive studies profiling HER2+ tumors that have progressed on primary anti-HER2 therapies have not been reported.
Intrinsic EGFR alterations
In EGFR mutant lung cancer, the most common mechanism of acquired resistance to EGFR inhibitors is the development of a mutation in the gatekeeper residue of EGFR, T790M (Kobayashi et al., 2005; Pao et al., 2005). T790M abrogates the inhibitor effects of gefitinib and erlotinib by increasing the affinity of the receptor for ATP (Yun et al., 2008), thereby lessening the potency of first-generation EGRF inhibitors (Figure 1C). At least 50% of biopsies from patients with acquired resistance harbor the T790M mutation. Recent studies suggest that highly sensitive methods can detect the T790M mutation in ~35% of pre-treatment biopsies. This suggests, but does not prove, that it pre-exists in a small fraction of cells, and that those cells are selected for during the course of treatment (Maheswaran et al., 2008; Rosell et al., 2011). Currently, the third-generation EGFR inhibitors (discussed above) are in early clinical trials to overcome this resistance.
An analogous finding has been observed in wild-type KRAS CRCs that develop resistance to cetuximab. A small study reported the development of a S492R mutation in the extracellular domain of EGFR that interferes with cetuximab binding, but does not interfere with ligand-dependent activation nor abrogates receptor engagement by panitumumab (Montagut et al., 2012) (Figure 1D).
BYPASS TRACK RESISTANCE
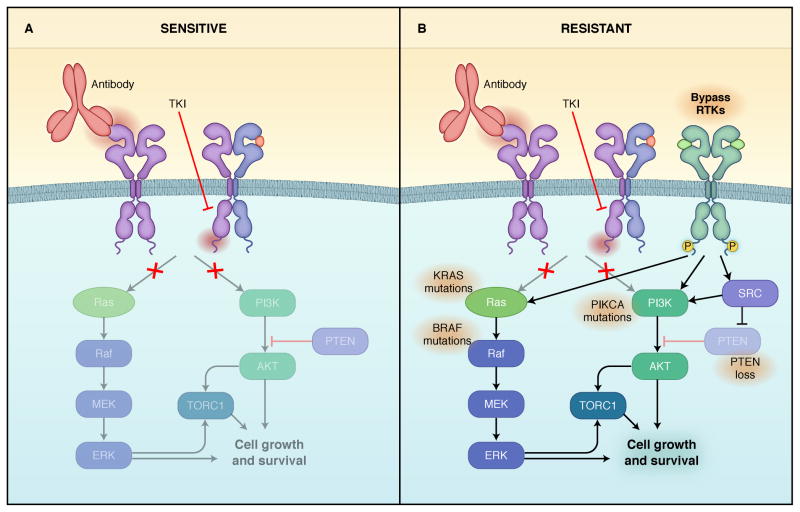
Other than the immune effects of ERBB antibodies, it is believed that most of activity of these drugs is due to suppression of downstream signaling, particularly PI3K/AKT and MEK/ERK. Thus, many cancers are resistant to single-agent ERBB inhibitors because at least one of these critical downstream pathways is maintained despite inhibition of the targeted receptor. This type of resistance, also termed “bypass track” resistance, is often used to describe resistance resulting from maintenance of these key downstream signaling pathways despite adequate inhibition of the respective RTK [reviewed in (Niederst and Engelman, 2013) and Figure 2].
Figure 2. Schematic depicting resistance to EGFR and HER2 inhibitors due to activation of bypass track signaling.
Left) Model of a sensitive EGFR or HER2-addicted cancer treated with an ERBB small molecule inhibitor or antibody resulting in suppression of downstream signaling. EGFR or HER2 homodimers and heterodimers are shown. Right) Model of a EGFR mutant or HER2-amplified cancer with resistance due to maintenance of downstream signaling in the presence of the EGFR or HER2 inhibitors. Activation of signaling can be caused by activation of other RTKs or mutational activation of downstream signaling.
Ligand- and RTK-mediated resistance
One of the earliest bona fide observations that RTK bypass signaling induces resistance to ERBB inhibitors was in EGFR-mutant NSCLCs. Amplification of the MET gene was found in EGFR mutant cancers with acquired resistance to EGFR TKIs but not in pre-treatment biopsies (Bean et al., 2007; Engelman et al., 2007). In EGFR-mutant NSCLC cells with acquired resistance, MET reactivates both PI3K/AKT and MEK/ERK signaling despite the inhibition of EGFR. The combination of MET and EGFR inhibitors was sufficient to block downstream signaling and induce marked tumor regressions (Engelman et al., 2007; Turke et al., 2010). Activation of MET by its ligand hepatic growth factor (HGF) was also sufficient to promote resistance through activation of downstream signaling (Yano et al., 2008). MET has also been implicated in trastuzumab resistance. HGF-induced signaling through MET was shown to abrogate the action of trastuzumab (Shattuck et al., 2008; Turke et al., 2010). Further, gene amplification of MET and HGF was reported in a cohort of HER2+ patients who did not respond to trastuzumab and chemotherapy (Minuti et al., 2012). Thus, MET activation by either gene amplification or ligand stimulation can cause bypass resistance to EGFR and HER2 inhibitors.
Re-activation of EGFR and HER3 can also serve as a mechanism of resistance to ERBB inhibitors. In laboratory models of HER2-amplified breast cancer treated with trastuzumab, increased levels of EGFR and ERBB ligands led to an increase in active EGFR/HER3 and EGFR/HER2 dimers to promote resistance (Ritter et al., 2007). This is consistent with data showing that trastuzumab is unable to block ligand-induced HER2-containing heterodimers (Agus et al., 2002). Similarly, activation of TGFβ receptors can increase ERBB ligand production and cleavage, particularly TGFα, amphiregulin and heregulin, via activation of the TACE/ADAM17 sheddase; this results in activation of HER3 and PI3K and promotes drug resistance (Wang et al., 2008). Further, a gene signature of TGFβ activity was developed and shown to correlate with resistance to trastuzumab and poor clinical outcome in patients (Wang et al., 2008).
Similarly, in EGFR mutant cancers, MET amplification leads to resistance to EGFR TKIs through re-activation of HER3 (Engelman et al., 2007). In a subset of EGFR mutant lung cancers, amplification of HER2, presumably involving HER3 reactivation, was also identified as a resistance mechanism to EGFR TKIs (Takezawa et al., 2012). Consistent with these data, blockade of HER3 with the neutralizing antibody MM-121 increases the efficacy of cetuximab in a mouse model of EGFR mutant lung cancer (Schoeberl et al., 2010). Along those lines, a selective ADAM inhibitor, INCB3619, which prevents the processing and activation of multiple ERBB ligands including heregulin, inhibits HER3 signaling and enhances gefitinib-mediated inhibition of EGFR in NSCLC (Zhou et al., 2006). Further supporting a role of amplified HER2-HER3 signaling in resistance to EGFR-targeted therapies, colorectal cancer patients with de novo or acquired resistance to cetuximab-based therapy exhibit HER2 amplification in their tumor or high levels of circulating heregulin (Yonesaka et al., 2011). Finally, using patient-derived colon cancer xenografts, Bertotti et al. identified HER2 gene amplification as a predictor of resistance to cetuximab among KRAS wild-type tumors (Bertotti et al., 2011).
IGF-I receptors have also been implicated in driving resistance to both EGFR and HER2 inhibitors. Overexpression of IGF-1R or an increase in levels of IGF-1R/HER2 heterodimers can potently activate PI3K/AKT signaling and confer resistance to trastuzumab in laboratory studies (Huang et al., 2010). Inhibition of IGF-1R with a neutralizing antibody or a small molecule TKI, or targeting the HER2 kinase with lapatinib was found to overcome IGF-1R-mediated resistance to trastuzumab (Nahta et al., 2007). In a neoadjuvant trial of chemotherapy plus trastuzumab, a high level of IGF-1R expression measured by IHC correlated with a poor clinical response (Harris et al., 2007). Similarly, activation of IGFIR, via loss of expression of IGFBP3 and IGFBP4, which encode insulin-like growth factor binding proteins 3 and 4, respectively, maintains PI3K/AKT signaling despite blockade of EGFR and promotes resistance to EGFR inhibitors in multiple cell lines (Guix et al., 2008). In these cases, inhibition of IGF1R re-sensitized to EGFR inhibition. In addition, inhibition of IGF-IR also suppressed the development of “persistor cells,” the small population of PC9 EGFR mutant cells that survives the initial inhibition of EGFR, described by Settleman and colleagues (Sharma et al., 2010).
In addition to the bypass pathways mentioned above, numerous other RTK-mediated resistance mechanisms have been observed. The EphA2 receptor has been shown to confer resistance to trastuzumab in cell lines, and EphA2 expression was shown to predict poor outcome patients with HER2+ breast cancer. (Zhuang et al.). Most recently, the erythropoietin (Epo) receptor was found to be co-expressed in cell lines and primary tumors that overexpress HER2. In these cell lines, concurrent treatment with recombinant erythropoietin conferred trastuzumab resistance. In patients with HER2+ breast cancer, the concurrent administration of erythropoietin and trastuzumab correlated with a shorter progression-free and overall survival compared to patients not receiving erythropoietin (Liang et al., 2010). Finally, in erlotinib-resistant EGFR mutant lung cancer cells and lapatinib-resistant HER2-amplified breast cancer cells, levels of the AXL RTK were markedly increased (Liu et al., 2009; Zhang et al., 2012). Targeting AXL was able to re-sensitize some of these resistant cancers to the original TKI.
We should note that most of these RTK-mediated mechanisms do not necessarily involve genetic activation of the RTK, as mainly protein assays (i.e., IHC for IGF-IR, AXL, EphA2, etc.) have been employed to measure their levels in tumor tissues. Such correlations do not prove that the putative bypass RTK is causal to drug resistance in the clinic or in a particular patient. Ultimately, clinical efficacy using specific drugs that target the bypass RTK will be needed for true validation.
Intracellular kinases
Molecules in the pathways downstream of RTKs can be aberrantly activated as a result of genetic alterations, also resulting in drug resistance (Figure 2). Somatic alterations in the PI3K/AKT pathway are the most frequent in breast cancer, occurring in approximately 30% of HER2+ tumors. These include mutation and/or amplification of the genes encoding the PI3K catalytic subunits p110α (PIK3CA) and p110β (PIK3CB), the PI3K regulatory subunit p85α (PIK3R1), the PI3K effectors AKT1, AKT2, and PDK1, and loss of the lipid phosphatases PTEN and INPP4B [reviewed in (Engelman, 2009)]. It is generally accepted that the antitumor activity of HER2 inhibitors depends on inhibition of PI3K-AKT downstream of HER2. Thus, one would expect that activating mutations in the PI3K pathway would confer resistance to HER2 inhibitors.
Constitutive activation of PI3K, via overexpression of PIK3CA mutants, conferred resistance to the antibody in laboratory studies (Chakrabarty et al., 2010; Eichhorn et al., 2008; Serra et al., 2008). Patients with ‘hot spot’ PIK3CA mutations and undetectable or low PTEN measured by IHC, exhibited a poorer outcome after treatment with chemotherapy and trastuzumab compared to patients without those alterations (Berns et al., 2007; Dave et al., 2011; Esteva et al., 2010). In the EMILIA and Neo-ALTTO randomized trials in HER2+ breast cancer, patients with PIK3CA mutant tumors did not benefit from lapatinib and capecitabine (Baselga, 2013b) and from lapatinib and trastuzumab (Baselga, 2013a), respectively. It remains to be determined whether T-DM1, because of its ability to deliver high levels of cytotoxic chemotherapy to HER2-overexpressing cells, trumps this mechanism of resistance.
One of the first discoveries linking constitutive activation of PI3K signaling and resistance to HER2 inhibitors was accomplished by Berns et al. Using a large-scale siRNA genetic screen, they identified PTEN as the only gene whose knockdown conferred trastuzumab resistance (Berns et al., 2007). However, the association of PTEN loss with drug resistance in the clinic is less clear. In one early study in patients with metastatic breast cancer, loss or low levels of PTEN correlated with a lower response to trastuzumab (Nagata et al., 2004). This correlation was not found in patients with early breast cancer treated with adjuvant trastuzumab (Perez et al., 2013). We speculate this was because of the concomitant administration of chemotherapy in an adjuvant setting (Rexer et al., 2013).
Similar findings have been observed in cancers with acquired resistance to EGFR inhibitors. Introduction of PIK3CA mutations into EGFR mutant lung cancer cell lines is sufficient to maintain PI3K signaling and promote resistance (Engelman et al., 2006). Accordingly, PIK3CA mutations have been identified in biopsies of EGFR mutant cancers with acquired resistance to EGFR inhibitors (Sequist et al., 2011). Similarly, a report found that PTEN loss may be associated with resistance to EGFR inhibitors (Sos et al., 2009). In addition to reactivation of PI3K, re-activation of ERK signaling can promote resistance to EGFR inhibitors as evidenced by the finding of a BRAF mutation in an EGFR mutant lung cancer with acquired resistance (Ohashi et al., 2012). In a second example, an EGFR mutant cell line made resistant to third-generation EGFR inhibitors developed amplification of ERK and was re-sensitized upon inhibition of MEK (Ercan et al., 2012).
A compelling recent discovery underlying this type of resistance mechanism was the study of KRAS wild-type colorectal cancers that had developed resistance to cetuximab. By performing repeat biopsies and evaluating circulating tumor DNA, investigators observed the emergence of KRAS mutations as a resistance mechanism (Diaz et al., 2012; Misale et al., 2012). From a signaling perspective, one would expect that the KRAS mutant clones fail to down-regulate the ERK pathway in response to cetuximab, underlying the resistance. As the presence of KRAS mutations predicts for lack of initial response to cetuximab, these findings underscore the convergence of intrinsic and acquired resistance mechanisms.
Src family kinase (SFK) signaling has been implicated by several studies in promoting resistance to HER2 inhibitors. In HER2+ breast cancer cells with acquired resistance to lapatinib, up-regulation of SFK activity, particularly Yes, was observed in several resistant cell lines. Resistance was associated with recovery of PI3K/AKT signaling despite inhibition of HER2. Addition of a Src TKI partially blocked PI3K/AKT and restored sensitivity to lapatinib (Rexer et al., 2011). In another study, the authors suggest that PTEN was no longer capable of dephosphorylating and suppressing Src in trastuzumab-resistant HER2+ cells, and the addition of a Src kinase inhibitor overcame trastuzumab resistance (Zhang et al., 2011). Src activity is also involved in the resistance conferred by the Δ16 HER2 isoform and the EpoR (Mitra et al., 2009). Src is thought to mediate resistance in part via phosphorylation and inhibition of PTEN, leading to constitutive PI3K signaling (Liang et al., 2010).
Defects in apoptosis and cell cycle control
Inhibition of a driver oncogene such as EGFR and HER2 results in proliferation arrest and apoptosis. Therefore, alterations in the normal apoptotic machinery can also induce resistance to EGFR- and HER2-targeted therapies. Indeed, we observed that levels of the pro-apoptotic BH3-only Bcl2 family member, BIM, are predictive of response to targeted therapy in EGFR mutant lung cancers, HER2-amplified breast cancers and PIK3CA mutant cancers (Faber et al., 2011). BIM protein normally is induced following inhibition of EGFR and HER2 in these cancers. In this study, although erlotinib or lapatinib inhibited EGFR and HER2 and downstream signaling in EGFR mutant and HER2-amplified cancers, respectively, only those cell lines with high levels of BIM underwent marked apoptosis. This suggests that BIM levels are a biomarker predictive of response to a TKI in an oncogene-addicted cancer. Other groups have reported similar results in EGFR mutant lung cancers (Ng et al., 2012) and HER2 gene-amplified breast cell lines with and without activating mutations (Tanizaki et al., 2011). In cancers with concurrent PIK3CA mutant cells, however, both the growth inhibitory effect and induction of BIM following treatment with lapatinib were blunted (Tanizaki et al., 2011).
Survivin, a member of the inhibitor of apoptosis (IAP) protein family that inhibits the activity of caspases, has been shown to be a point of convergence of several pathways that can lead to resistance to HER2 inhibitors. In HER2+ breast cancer cells, inhibition of HER2-PI3K reduces survivin expression resulting in apoptosis. HER2-amplified breast cancer cells with acquired resistance to lapatinib up-regulate ERα which, in turn, induces FoxO3a-mediated transcription of survivin (Xia et al., 2006). In turn, high survivin levels allow for escape from lapatinib. Accordingly, elevated levels of survivin and MCL-1 have been found in trastuzumab-resistant cells (Chakrabarty et al., 2013).
Altered control of progression through the cell cycle in response to HER2 inhibition also plays a role in resistance. Cell lines made resistant to trastuzumab by chronic exposure exhibited focal amplification of cyclin E. CDK2 inhibitors reduced growth of these trastuzumab-resistant xenografts (Scaltriti et al., 2011). Further, in a cohort of patients with HER2+ breast cancers treated with trastuzumab, amplification of cyclin E was associated with a diminished clinical response. Downregulation of the Cdk inhibitor p27KIP1 and a resulting increase in Cdk activity has also been associated with trastuzumab resistance (Nahta et al., 2004; Yakes et al., 2002). Indeed, modulation of levels of p27KIP1 appears to be a common endpoint for several of the resistance pathways noted above, including signaling from IGF-1R and MET (Nahta et al., 2005; Shattuck et al., 2008).
Tumor host factors
Host factors that affect the immunomodulatory function of trastuzumab can also contribute to trastuzumab resistance. In mice lacking FcγRIII and, thus, deficient in NK cells and macrophages capable of binding the Fc region of trastuzumab, the therapeutic effect of trastuzumab was markedly diminished (Clynes et al., 2000). Polymorphisms in the gene encoding FcγRIII in humans were associated with resistance to trastuzumab in patients with metastatic HER2+ breast cancer (Musolino et al., 2008). In the same study, PBMCs from patients with FCGR3 polymorphisms associated with an improved outcome after trastuzumab induce a stronger trastuzumab-mediated ADCC in vitro. A follow-up study found that the quantity and lytic efficiency of CD16(+) lymphocytes are the major factors affecting the level of ADCC induced by trastuzumab. This, in turn, correlates with tumor response (Gennari et al., 2004). We should note, however, that a large trial of trastuzumab-based adjuvant chemotherapy in patients with early HER2+ breast cancer did not show an association between FCGR3A and FCGR2A polymorphisms with patient outcome (Hurvitz et al., 2012)
STRATEGIES TO OVERCOME RESISTANCE: COMBINATION THERAPIES
Clinical experience has validated EGFR and HER2 as effective drug targets. However, in the metastatic setting, these inhibitors do not lead to cures, and cancers ultimately develop resistance. Thus, there is a great need to identify therapeutic strategies that will improve upon the current approaches. We believe that one strategy will include maximal blockade of the oncogene target itself as well as inhibition of the key bypass tracks that promote resistance. The growing number of escape routes will likely necessitate combinations of multiple agents, whose delivery will require innovative dosing and scheduling.
HER2
All currently available HER2 inhibitors target or exploit mechanisms of HER2 function. As single drugs, however, they do not potently suppress HER2 signaling. This may explain the generally short-lived responses of metastatic HER2+ breast cancers to single-agent HER2 inhibitors. Trastuzumab and pertuzumab, in particular, are each weak signaling inhibitors, possibly because they incompletely block HER2-containing dimers (Junttila et al., 2009). Treatment with lapatinib (and likely other HER2 TKIs) leads to an increase in HER2 and HER2-containing dimers at the plasma membrane, and fails to completely and persistently inhibit the HER2 kinase (Garrett et al., 2011; Scaltriti et al., 2009). This may be explained by both the narrow therapeutic index of current HER2 TKIs and the challenge of complete and sustained inhibition of an amplified drug target, i.e., HER2, with small molecules. Moreover, inhibition of the HER2 kinase leads to an initial reduction of PI3K/AKT signaling, which releases negative feedbacks resulting in up-regulation of HER3 and other RTKs as wells as survival factors such as BCL2 and ERα, thereby mitigating the efficacy of HER2 inhibitors(Chakrabarty et al., 2012; Chandarlapaty et al., 2011; Garrett et al., 2011; Muranen et al., 2012; Xia et al., 2006). We also recognize that HER2-amplified breast cancers vary with respect to their addiction to HER2 signaling, although they are grouped together using clinical criteria (FISH, IHC) for HER2 overexpression. Thus, we speculate many HER2 gene amplified cancers are not truly ‘addicted’ to HER2 signaling and, as such, are not sensitive to HER2 TKIs. Finally, resistance to T-DM1 may be due to several reasons, including the sparing of HER2-negative tumor cells within heterogeneous cancers containing HER2+ and HER-negative tumor cells, a scenario not uncommon in clinical practice. Finally, T-DM1, trastuzumab and pertuzumab cannot bind p95-HER2 and, thus, would be inactive against tumor cells with an abundance of cytosolic fragments of HER2.
One strategy to address the limitations of anti-HER2 drugs as single agents has been to combine multiple HER2 antagonists that have different but complementary mechanisms of action. Clinical experience had suggested that trastuzumab-refractory tumors remained dependent on HER2 as continuing trastuzumab in new treatment regimens beyond progression to trastuzumab demonstrated clinical benefit (von Minckwitz et al., 2009). Currently, dual blockade of HER2 is well-entrenched in the clinic. For example, the combination of trastuzumab and lapatinib is superior to each agent alone in both the metastatic (Blackwell et al., 2010) and neoadjuvant settings (Baselga et al., 2012a). Similarly, the combination of trastuzumab and pertuzumab was shown to be superior to each antibody alone in both neoadjuvant trials in patients with early disease (Gianni et al., 2012) and in patients with advanced disease, as assessed by progression-free survival (Baselga et al., 2012b). The combination of T-DM1 and pertuzumab is also in progress. This novel approach would incorporate dual receptor blockade with two HER2 antibodies (trastuzumab and pertuzumab) plus the delivery of a potent cytotoxic (DM1) to HER2 amplified cells while mostly sparing host tissues (Phillips et al., 2013).
At the time of writing this Perspective, several novel anti-HER2 combinations are being tested in clinical trials. Some of these include a third drug targeted to the HER2 network (Table 3). It is anticipated that for a cohort of HER2+ breast cancers that escape anti-HER2 dual therapy, a third drug targeted against a signaling hub in the receptor network might be necessary. Supporting this possibility, a recent study showed that transgenic mammary tumors expressing HER2 and PIK3CAH1047R were completely resistant to the combinations of trastuzumab plus pertuzumab and trastuzumab plus lapatinib. Addition of the pan-PI3K inhibitor BKM120 to each combination resulted in inhibition of tumor growth, but only partially and temporarily (Hanker et al., 2013). Currently, a main clinical focus is the addition of PI3K inhibitors and/or HER3 neutralizing antibodies (Garrett et al., 2013b) to the established combinations of anti-HER2 therapies (Table 3). More recent data suggest that blockade of mTOR downstream HER2 with the TORC1 inhibitor everolimus, while maintaining trastuzumab therapy, can induce clinical responses in HER2+ cancers that have progressed on trastuzumab (Hurvitz et al., 2013; Morrow et al., 2011; O’Regan, 2013). Along the same lines, neratinib in combination with the TORC1 inhibitor temsirolimus recently demonstrated clinical activity in HER2 mutant lung cancers (Gandhi et al., 2014). Finally, one proposed novel strategy is the combination of trastuzumab with anti-PD1 and anti-CD37 monoclonal antibodies. In this case, anti-PD1 would inhibit IFNγ-activated T cells and anti-CD37 would block CD8+ T cells; both of these T-cell subtypes are required for the adaptive immune response triggered by trastuzumab (Stagg et al., 2011).
Table 3.
Anti-ERBB combinations
| Combination | Mechanism(s) of action | Relevant clinical trials |
|---|---|---|
| Trastuzumab + lapatinib (or neratinib, afatinib) | ADCC, partial disruption of HER2-HER3 dimers, inhibition of HER2 and EGFR tyrosine kinases | (Baselga et al., 2012a; Blackwell et al., 2010) |
| Trastuzumab + pertuzumab (only approved combination) | More complete inhibition of ligand-induced and ligand-independent HER2-containing heterodimers, ADCC, downregulation of HER2 from cell surface | (Baselga et al., 2012b; Gianni et al., 2012; Schneeweiss et al., 2013) |
| T-DM1 + pertuzumab | Same as above plus inhibition of polymerization of microtubules with DM1 | MARIANNE (NCT01120184) |
| Trastuzumab + everolimus | ADCC, disruption of ligand-independent HER2-HER3 dimers, inhibition of TORC1 | BOLERO-3 (NCT01007942) |
| Trastuzumab + pertuzumab + PI3K inhibitor | Inhibition of ligand-induced and ligand-independent HER2-containing heterodimers, ADCC, ATP-competitive inhibition of catalytic activity of p110 | Pending |
| Trastuzumab + HER3 neutralizing antibody | ADCC, partial disruption of HER2-HER3 dimers, inhibition of heregulin binding, downregulation of HER3 and/or HER3 dimerization | Pending |
| Trastuzumab + HER3 antibody + PI3K inhibitor | Same as above plus direct inhibition of p110 | Pending |
| T-DM1 + PI3K inhibitor | ADCC, partial disruption of HER2-HER3 dimers, inhibition of polymerization of microtubules, direct inhibition of p110 | Pending |
| Afatinib + cetuximab | Combined targeting of EGFR T790M to compensate for complete inhibition of target by either approach alone. Afatinib may also target resistance due to HER2 activation | Pending |
| EGFR + PI3K inhibitor | Block resistance due to re-activation of PI3K signaling | Pending |
| Erlotinib + MET inhibitor | Block MET dependent resistance to EGFR inhibitors | MetLung Trial and others |
| EGFR inhibitor + IGF-IR antibody | Block IGF-IR dependent resistance to EGFR inhibitors | Pending |
| Erlotinib + Hydroxychloroquine | Effort to block the survival of “drug tolerant” cells following treatment with EGFR TKIs (Sharma et al, Cell, 2010) | (Goldberg et al., 2012) |
| Irreversible EGFR inhibitor + MET inhibitor | Overcome both T790M and MET mediated resistance | Pending |
EGFR
One major mechanism of resistance to EGFR inhibitors in EGFR mutant cancers is the development of the T790M gatekeeper residue. The newer third generation EGFR TKIs that suppress T790M are showing remarkable progress in this subset of cancers (Ranson et al., 2013; Sequist et al., 2013a; Soria et al., 2013). Ultimately, clinical trials will be needed to determine if third generation EGFR inhibitors become first-line therapy for EGFR mutant lung cancers. We anticipate that metastatic EGFR mutant lung cancers will likely become resistant to drugs that target T790M since there are several additional potential resistance mechanisms. Thus, it will likely be necessary to combine inhibitors of bypass tracks with T790M-specific inhibitors to provide greater durations of remission and prolongation of patient survival.
In general, combinations that overcome resistance to EGFR inhibitors have generally required continued inhibition of EGFR combined with a drug that blocks the bypass track [reviewed in (Niederst and Engelman, 2013)]. For example, in EGFR mutant lung cancers that are resistant via MET amplification, combined EGFR and MET inhibition is required to suppress downstream PI3K/AKT and MEK/ERK and induce tumor regressions in vivo (Turke et al., 2010). In similar examples of resistance mediated by IGF-IR and AXL, inhibition of the bypass RTK in combination with the EGFR is needed to overcome resistance (Cortot et al., 2013; Guix et al., 2008; Zhang et al., 2012). Thus, one central strategy involving combinations centers on maintaining potent inhibition mutant EGFR while adding different inhibitors to these accessory pathways. This has been employed in early clinical trials that have combined EGFR inhibitors with MET inhibitors, PI3K inhibitors and IGF-IR inhibitors (Table 3). However, none of these trials utilized third generation EGFR inhibitors, which are the only drugs that appear capable of overcoming T790M. The advent of these third generation inhibitors may now unleash the potential of targeting bypass tracks once T790M is effectively inhibited.
In KRAS wild-type colorectal cancers, the recent finding that resistant cancers develop EGFR mutations that abrogate cetuximab binding or KRAS mutations suggests approaches analogous to those discussed above. Again, the development of point mutations in EGFR suggests that alternative approaches to suppress EGFR may be warranted. Preclinical studies suggest panitumumab may overcome this type of resistance (Montagut et al., 2012). In colorectal cancers that develop KRAS mutations upon development of resistance, current trials are examining the efficacy of EGFR inhibitors in combination with inhibitors to overcome resistance (Alberto Bardelli, personal communication).
Projections to the future: Novel approaches
Monitoring tumor evolution
One of the major challenges will be to determine the optimal combinations for individual patients. Some examples have clear biomarkers pointing to specific combinations, such as the use of combined HER2 and PI3K inhibitors for HER2-amplified breast cancers harboring PIK3CA mutations, as well as EGFR and MET inhibitors for EGFR mutant lung cancers harboring MET amplifications. However, several of the other combinations do not have straightforward biomarkers for patient selection. In the cases of IGF-IR and AXL, it is quite unlikely that expression of the RTK alone will accurately identify those cancers in which those proteins are driving resistance. Thus, more precise assessments of RTK and signaling activation via novel proteomic methods would be potentially valuable to identify the most appropriate combinations.
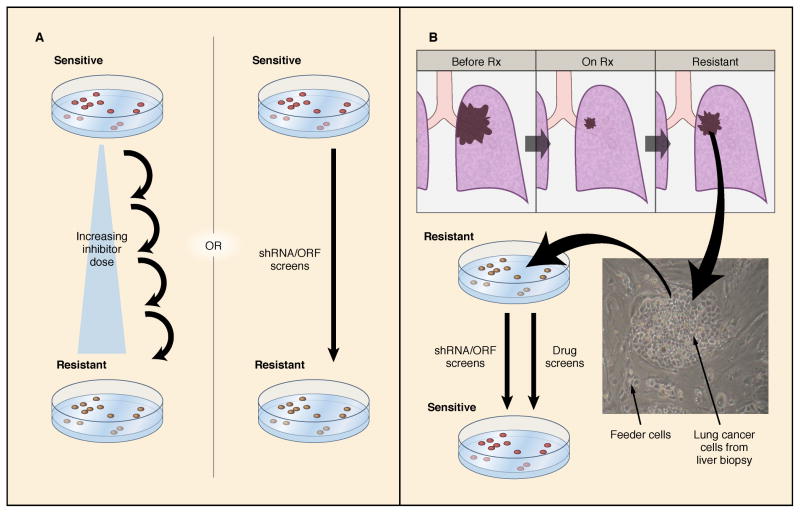
As the cancers progress through therapies, there will be a need to continually interrogate the cancer to understand how it has adapted to treatment pressures and become drug resistant. Many centers have utilized repeat biopsy programs to perform biopsies after the development of resistance to targeted therapies to determine how the cancer has evolved. Indeed, with the increasing use of next-generation sequencing approaches, it is likely the genetic landscape for resistance mechanisms will increase dramatically over the coming years. Even though this approach has great promise to discover resistance mechanism, it may also have potential limitations when the results are used to determine the next course of therapy for an individual patient as the (acquired) alterations identified in a single biopsy may not reflect all of the resistant clones in multiple metastatic sites in an individual patient (Bean et al., 2007; Engelman et al., 2007; Turke et al., 2010). Thus, non-invasive measures such as molecular interrogation of circulating tumor cells or plasma DNA may help capture the heterogeneity of resistance in patients, as was done to identify the development of KRAS mutations in colorectal cancers that acquire resistance to cetuximab (Diaz et al., 2012; Misale et al., 2012). As efforts to identify new therapeutic strategies to overcome resistance are intensified, the development of cell lines and patient-derived xenografts from resistant biopsies may facilitate the identification of new therapeutic strategies. Recent advances in technology may help bring these ‘live’ biopsies directly into the laboratory for interrogation (Liu et al., 2012). Such models could be interrogated by high-throughput shRNA and drug screens to identify novel therapeutic approaches to overcome resistance (Figure 3).
Figure 3. Developing laboratory models to discover mechanisms of resistance.
A) Resistance mechanisms can be discovered by culturing sensitive cell lines in the presence of a specific HER2 or EGFR inhibitor until resistance develops, or by introducing shRNA or ORF libraries to determine genes whose overexpression or suppression will lead to resistance. B) Alternatively, when resistance develops in the clinic, a cell line can be generated from a biopsy of the resistant lesion, and the resulting resistant line can be screened with drugs and/or shRNA libraries to determine strategies to re-sensitize them.
In addition to profiling of metastatic recurrences, the increasing use of neoadjuvant anti-HER2 therapy in patients with newly diagnosed HER2+ breast cancers provides a novel platform for discovery of mechanisms of resistance and tumor heterogeneity. At least two studies have shown that some patients with HER2+ tumors convert to HER2-negative following neoadjuvant trastuzumab and chemotherapy, and these patients exhibit a shorter relapse-free survival compared to those with residual tumors that remain HER2 amplified (Hurley et al., 2006; Mittendorf et al., 2009). These results suggest, first, that with heterogeneity in HER2 overexpression in the primary tumor, the anti-oncogene therapy eliminates the HER2-dependent compartment and enriches for HER2-negative clones. Second, patients with HER2+ tumors that change to HER2-negative upon primary anti-HER2 therapy are at a high risk of early recurrence. This neoadjuvant approach facilitates interrogation of the drug-resistant cancer and the identification of targetable mechanisms potentiallly driving subsequent metastatic recurrences (discussed below).
Innovative trial designs
For both HER2- and EGFR-driven cancers, it is becoming apparent that new treatment paradigms will be necessary to lead to durable remissions or even cures. Ultimately, we posit that combination therapies will be needed, and that it is more rational to consider a proactive regimen that employs alternating regimens of combinations that eliminate cancer cells before they adapt and become resistant rather than treating cancers after the development of clinically overt resistance. However, the large number of potential resistance mechanisms will likely necessitate the use of more drugs than will be tolerable if they are all delivered simultaneously, and each drug is dosed to achieve continual target suppression. When developing combinations, the use of mutant specific inhibitors will be highly attractive components because of their greater therapeutic windows. However, even with such an approach, given the large number of potential resistance mechanisms, it may become necessary to use even more creative approaches to proactively kill the various resistant clones as they emerge. In the future, we envision developing regimens that rotate and intercalate tolerable combinations to prevent or substantially delay the development of resistance. In particular, regimens that include immunotherapy and other disparate approaches may be needed.
In breast cancer, the increasing use of neoadjuvant therapy lends itself to some innovative possibilities to develop novel therapeutic regimens, accelerate drug approvals and discover mechanisms of drug resistance. Achievement of a pathological complete response (path CR) in the breast and axillary lymph nodes after neoadjuvant trastuzumab or chemotherapy has been associated with improved long-term outcome (Gianni et al., 2010; Liedtke et al., 2008). Because of this association, the FDA recently proposed that randomized neoadjuvant trials can be considered for accelerated drug approval using path CR as a surrogate that is ‘reasonably likely to predict longer term benefit,’ at least for some subtypes of breast cancer, particularly the HER2+ subtype (Prowell and Pazdur, 2012). Recently, the FDA approved the HER2 antibody pertuzumab as neoadjuvant treatment in patients with HER2+ early breast cancer (http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm370393.htm). This approval was based on the results of two neoadjuvant studies, NeoSphere and TRYPHAENA, where the combination of pertuzumab and trastuzumab was superior to trastuzumab alone. The potential impact of this recommendation is quite transformative, as it can accelerate the approval of novel and effective combinations. Further, the early delivery of these anti-HER2 combinations to patients with treatment-naive HER2+ tumors should at least partially trump acquired drug resistance. The use of the pre-operative therapy setting as a clinical research platform where novel combinations and regimens can be compared and triaged using path CR as a clinical endpoint predictive of long-term outcome has been discussed recently (Bardia and Baselga, 2013).
Another benefit of a pre-operative approach is that, except for patients who experience a complete response, tumor tissue is always available at the time of surgery. These ‘drug resistant’ residual cancers should harbor mechanisms and/or biomarkers of resistance to the primary therapy and, potentially, a similar molecular profile to that of drug-resistant micro-metastases that can be interrogated with massive parallel sequencing of DNA extracted from the mastectomy specimen (Balko et al., 2012; Balko et al., 2013). Thus, we propose ‘drug-resistant’ HER2+ residual cancers in the breast harbor targetable genomic alterations causally associated with resistance to neoadjuvant anti-HER2 therapy (Figure 4). Molecular profiling of these residual tumors should identify these alterations. In addition, patient-derived xenografts (PDXs) generated with these residual cancers can be used to test novel combinations with activity against these drug-resistant cancer that can be later applied to patients on an individual basis. Drugs that target novel mechanisms of resistance identified in the residual tumors can be examined in subsequent randomized neoadjuvant trials. In the future, we anticipate that tumor types other than HER2+ breast cancer could also effectively utilize neoadjuvant trials to accelerate drug development and discover mechanisms of resistance.
Figure 4. Targetable alterations in (residual) breast cancers after neoadjuvant anti-HER2 therapy may identify actionable mechanisms of drug resistance.
Systemic neoadjuvant anti-HER2 therapy reduces or eliminates the primary HER2+ tumor as well as micrometastases (top row). We propose that ‘drug-resistant’ residual cancers in the breast following neoadjuvant therapy harbor targetable genomic alterations causally associated with resistance to HER2 inhibitors. Molecular profiling of these residual tumors should identify these genomic alterations. Further, patient-derived xenografts (PDXs) generated with these residual cancers can be used to test novel combinations with activity against these drug-resistant cancers that can be later applied to patients on an individual basis. Drugs that target novel mechanisms of resistance identified in the residual tumors can be examined in subsequent neoadjuvant trials (bottom row).
Conclusions
Ultimately, to cure ERBB-dependent cancers, we will likely have to incorporate therapeutics that are toxic to cancer cells via mechanisms that are not solely based on suppressing ERBB signaling, the associated bypass tracks, and antibodies targeting ERBB receptors to induce ADCC. The timing of treatment may also make a difference. For example, deploying ERBB targeted combinations early in the natural history of these cancers, i.e., in the adjuvant setting to treat micro-metastatic subclinical disease, may yield better outcomes than treating patients with metastatic disease, where the effect will not be curative. We feel that optimizing the timing and intensity of this approach will provide substantial clinical benefit to patients, and will serve as the foundation for incorporating complementary, independent therapeutic strategies that may ultimately lead to highly durable responses and further cures.
Acknowledgments
This work was supported by NCI grant R01 CA80195 (CLA), R01 CA137008 (JAE), R01 CA164273 (JAE), R01 CA140954 (JAE), ACS Clinical Research Professorship Grant CRP-07-234 (CLA), Breast Cancer SPORE grant P50 CA98131, GI Cancer SPORE grant P50 CA127003, Department of Defense (JAE), and Vanderbilt-Ingram Cancer Center Support grant P30 CA68485
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agus DB, Akita RW, Fox WD, Lewis GD, Higgins B, Pisacane PI, Lofgren JA, Tindell C, Evans DP, Maiese K, et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer cell. 2002;2:127–137. doi: 10.1016/s1535-6108(02)00097-1. [DOI] [PubMed] [Google Scholar]
- Andrechek ER, Hardy WR, Siegel PM, Rudnicki MA, Cardiff RD, Muller WJ. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3444–3449. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anido J, Scaltriti M, Bech Serra JJ, Santiago Josefat B, Todo FR, Baselga J, Arribas J. Biosynthesis of tumorigenic HER2 C-terminal fragments by alternative initiation of translation. Embo J. 2006;25:3234–3244. doi: 10.1038/sj.emboj.7601191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nature reviews Molecular cell biology. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, Sanders ME, Granja-Ingram NM, Smith JJ, Meszoely IM, et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med. 2012;18:1052–1059. doi: 10.1038/nm.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko JM, Giltnane J, Wang K, Schwarz LJ, Young CD, Cook RS, Owens P, Sanders ME, Kuba MG, Sanchez V, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardia A, Baselga J. Neoadjuvant therapy as a platform for drug development and approval in breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:6360–6370. doi: 10.1158/1078-0432.CCR-13-0916. [DOI] [PubMed] [Google Scholar]
- Baselga J, Bradbury I, Eidtmann H, Di Cosimo S, de Azambuja E, Aura C, Gomez H, Dinh P, Fauria K, Van Dooren V, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012a;379:633–640. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Cortes J, Kim SB, Im SA, Hegg R, Im YH, Roman L, Pedrini JL, Pienkowski T, Knott A, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012b;366:109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Majewski I, Nuciforo P, et al. PIK3CA mutations and correlation with pCR in the NeoALTTO trial (BIG 01–06). European Cancer Congress; 2013.2013a. [Google Scholar]
- Baselga J, Verma S, Ro J, et al. Relationship between tumor biomarkers (BM) and efficacy in EMILIA, a phase III study of trastuzumab emtansine (T-DM1) in HER2-positive metastatic breast cancer (MBC) Cancer research. 2013b:73. doi: 10.1158/1078-0432.CCR-15-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Cora D, Di Nicolantonio F, Buscarino M, Petti C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- Blackwell KL, Burstein HJ, Storniolo AM, Rugo H, Sledge G, Koehler M, Ellis C, Casey M, Vukelja S, Bischoff J, et al. Randomized study of Lapatinib alone or in combination with trastuzumab in women with ErbB2-positive, trastuzumab-refractory metastatic breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:1124–1130. doi: 10.1200/JCO.2008.21.4437. [DOI] [PubMed] [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, Monsey J, Goel N, Aronson AB, Li S, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein HJ, Sun Y, Dirix LY, Jiang Z, Paridaens R, Tan AR, Awada A, Ranade A, Jiao S, Schwartz G, et al. Neratinib, an irreversible ErbB receptor tyrosine kinase inhibitor, in patients with advanced ErbB2-positive breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:1301–1307. doi: 10.1200/JCO.2009.25.8707. [DOI] [PubMed] [Google Scholar]
- Burtness B, Goldwasser MA, Flood W, Mattar B, Forastiere AA Eastern Cooperative Oncology G. Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:8646–8654. doi: 10.1200/JCO.2005.02.4646. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Varella-Garcia M, Shigematsu H, Domenichini I, Bartolini S, Ceresoli GL, Rossi E, Ludovini V, Gregorc V, Toschi L, et al. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor-positive non-small-cell lung cancer patients. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:5007–5018. doi: 10.1200/JCO.2005.09.111. [DOI] [PubMed] [Google Scholar]
- Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer research. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A, Bhola NE, Sutton C, Ghosh R, Kuba MG, Dave B, Chang JC, Arteaga CL. Trastuzumab-resistant cells rely on a HER2-PI3K-FoxO-survivin axis and are sensitive to PI3K inhibitors. Cancer research. 2013;73:1190–1200. doi: 10.1158/0008-5472.CAN-12-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A, Rexer BN, Wang SE, Cook RS, Engelman JA, Arteaga CL. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29:5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A, Sanchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, Brown RD, Della Pelle P, Dias-Santagata D, Hung KE, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortot AB, Repellin CE, Shimamura T, Capelletti M, Zejnullahu K, Ercan D, Christensen JG, Wong KK, Gray NS, Janne PA. Resistance to irreversible EGF receptor tyrosine kinase inhibitors through a multistep mechanism involving the IGF1R pathway. Cancer research. 2013;73:834–843. doi: 10.1158/0008-5472.CAN-12-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- Dave B, Migliaccio I, Gutierrez MC, Wu MF, Chamness GC, Wong H, Narasanna A, Chakrabarty A, Hilsenbeck SG, Huang J, et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard JY, Zemelka T, Fountzilas G, Barone C, Schlichting M, Heighway J, Eggleton SP, Srimuninnimit V. FOLFOX4 With Cetuximab vs. UFOX With Cetuximab as First-Line Therapy in Metastatic Colorectal Cancer: The Randomized Phase II FUTURE Study. Clin Colorectal Cancer. 2013 doi: 10.1016/j.clcc.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Downward J, Yarden Y, Mayes E, Scrace G, Totty N, Stockwell P, Ullrich A, Schlessinger J, Waterfield MD. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, Ryan DP, Meyerhardt JA, Benes C, Settleman J, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest. 2011;121:4311–4321. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn PJ, Gili M, Scaltriti M, Serra V, Guzman M, Nijkamp W, Beijersbergen RL, Valero V, Seoane J, Bernards R, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer research. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun J, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, Montero J, Butaney M, Shimamura T, Sholl L, Ivanova EV, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2012;2:934–947. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva FJ, Guo H, Zhang S, Santa-Maria C, Stone S, Lanchbury JS, Sahin AA, Hortobagyi GN, Yu D. PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol. 2010;177:1647–1656. doi: 10.2353/ajpath.2010.090885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AC, Wong KK, Engelman JA. Differences underlying EGFR and HER2 oncogene addiction. Cell Cycle. 2010;9:851–852. doi: 10.4161/cc.9.5.11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Lu Y, Wu X, Mendelsohn J. Antibody-induced epidermal growth factor receptor dimerization mediates inhibition of autocrine proliferation of A431 squamous carcinoma cells. The Journal of biological chemistry. 1994;269:27595–27602. [PubMed] [Google Scholar]
- Finkle D, Quan ZR, Asghari V, Kloss J, Ghaboosi N, Mai E, Wong WL, Hollingshead P, Schwall R, Koeppen H, et al. HER2-targeted therapy reduces incidence and progression of midlife mammary tumors in female murine mammary tumor virus huHER2-transgenic mice. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:2499–2511. doi: 10.1158/1078-0432.ccr-03-0448. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer cell. 2004;5:317–328. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer research. 2000;60:1383–1387. [PubMed] [Google Scholar]
- Gandhi L, Bahleda R, Tolaney SM, Kwak EL, Cleary JM, Pandya SS, Hollebecque A, Abbas R, Ananthakrishnan R, Berkenblit A, et al. Phase I study of neratinib in combination with temsirolimus in patients with human epidermal growth factor receptor 2-dependent and other solid tumors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2014;32:68–75. doi: 10.1200/JCO.2012.47.2787. [DOI] [PubMed] [Google Scholar]
- Garner AP, Bialucha CU, Sprague ER, Garrett JT, Sheng Q, Li S, Sineshchekova O, Saxena P, Sutton CR, Chen D, et al. An antibody that locks HER3 in the inactive conformation inhibits tumor growth driven by HER2 or neuregulin. Cancer research. 2013;73:6024–6035. doi: 10.1158/0008-5472.CAN-13-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, Dave B, Cook RS, Pao W, McKinely E, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett JT, Sutton CR, Kuba MG, Cook RS, Arteaga CL. Dual blockade of HER2 in HER2-overexpressing tumor cells does not completely eliminate HER3 function. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013a;19:610–619. doi: 10.1158/1078-0432.CCR-12-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett JT, Sutton CR, Kurupi R, Bialucha CU, Ettenberg SA, Collins SD, Sheng Q, Wallweber J, Defazio-Eli L, Arteaga CL. Combination of antibody that inhibits ligand-independent HER3 dimerization and a p110alpha inhibitor potently blocks PI3K signaling and growth of HER2+ breast cancers. Cancer research. 2013b;73:6013–6023. doi: 10.1158/0008-5472.CAN-13-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Molecular cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- Gennari R, Menard S, Fagnoni F, Ponchio L, Scelsi M, Tagliabue E, Castiglioni F, Villani L, Magalotti C, Gibelli N, et al. Pilot study of the mechanism of action of preoperative trastuzumab in patients with primary operable breast tumors overexpressing HER2. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:5650–5655. doi: 10.1158/1078-0432.CCR-04-0225. [DOI] [PubMed] [Google Scholar]
- Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A, Kaufman B, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Narasanna A, Wang SE, Liu S, Chakrabarty A, Balko JM, Gonzalez-Angulo AM, Mills GB, Penuel E, Winslow J, et al. Trastuzumab has preferential activity against breast cancers driven by HER2 homodimers. Cancer research. 2011;71:1871–1882. doi: 10.1158/0008-5472.CAN-10-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni L, Eiermann W, Semiglazov V, Manikhas A, Lluch A, Tjulandin S, Zambetti M, Vazquez F, Byakhow M, Lichinitser M, et al. Neoadjuvant chemotherapy with trastuzumab followed by adjuvant trastuzumab versus neoadjuvant chemotherapy alone, in patients with HER2-positive locally advanced breast cancer (the NOAH trial): a randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet. 2010;375:377–384. doi: 10.1016/S0140-6736(09)61964-4. [DOI] [PubMed] [Google Scholar]
- Gianni L, Pienkowski T, Im YH, Roman L, Tseng LM, Liu MC, Lluch A, Staroslawska E, de la Haba-Rodriguez J, Im SA, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- Gilbertson R, Hernan R, Pietsch T, Pinto L, Scotting P, Allibone R, Ellison D, Perry R, Pearson A, Lunec J. Novel ERBB4 juxtamembrane splice variants are frequently expressed in childhood medulloblastoma. Genes, chromosomes & cancer. 2001;31:288–294. doi: 10.1002/gcc.1146. [DOI] [PubMed] [Google Scholar]
- Grandis JR, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, Drenning SD, Tweardy DJ. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. Journal of the National Cancer Institute. 1998;90:824–832. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, Bulmer SE, Frank DA, Hahn WC, Sellers WR, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, Rinehart C, Seidel B, Yee D, Arteaga CL, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, Baumber R, Lamborn KR, Kapadia A, Malec M, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. Journal of the National Cancer Institute. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- Hanker AB, Pfefferle AD, Balko JM, Kuba MG, Young CD, Sanchez V, Sutton CR, Cheng H, Perou CM, Zhao JJ, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LN, You F, Schnitt SJ, Witkiewicz A, Lu X, Sgroi D, Ryan PD, Come SE, Burstein HJ, Lesnikoski BA, et al. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:1198–1207. doi: 10.1158/1078-0432.CCR-06-1304. [DOI] [PubMed] [Google Scholar]
- Hegde GV, de la Cruz CC, Chiu C, Alag N, Schaefer G, Crocker L, Ross S, Goldenberg D, Merchant M, Tien J, et al. Blocking NRG1 and other ligand-mediated Her4 signaling enhances the magnitude and duration of the chemotherapeutic response of non-small cell lung cancer. Science translational medicine. 2013;5:171ra118. doi: 10.1126/scitranslmed.3004438. [DOI] [PubMed] [Google Scholar]
- Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham JN, Demory Beckler M, Gephart JD, Franklin JL, Bogatcheva G, Kremers GJ, Piston DW, Ayers GD, McConnell RE, Tyska MJ, et al. Amphiregulin exosomes increase cancer cell invasion. Current biology: CB. 2011;21:779–786. doi: 10.1016/j.cub.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Gao L, Wang S, McManaman JL, Thor AD, Yang X, Esteva FJ, Liu B. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-i receptor in breast cancer cells resistant to herceptin. Cancer research. 2010;70:1204–1214. doi: 10.1158/0008-5472.CAN-09-3321. [DOI] [PubMed] [Google Scholar]
- Hurley J, Doliny P, Reis I, Silva O, Gomez-Fernandez C, Velez P, Pauletti G, Powell JE, Pegram MD, Slamon DJ. Docetaxel, cisplatin, and trastuzumab as primary systemic therapy for human epidermal growth factor receptor 2-positive locally advanced breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:1831–1838. doi: 10.1200/JCO.2005.02.8886. [DOI] [PubMed] [Google Scholar]
- Hurvitz SA, Betting DJ, Stern HM, Quinaux E, Stinson J, Seshagiri S, Zhao Y, Buyse M, Mackey J, Driga A, et al. Analysis of Fcgamma receptor IIIa and IIa polymorphisms: lack of correlation with outcome in trastuzumab-treated breast cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:3478–3486. doi: 10.1158/1078-0432.CCR-11-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurvitz SA, Dalenc F, Campone M, O’Regan RM, Tjan-Heijnen VC, Gligorov J, Llombart A, Jhangiani H, Mirshahidi HR, Tan-Chiu E, et al. A phase 2 study of everolimus combined with trastuzumab and paclitaxel in patients with HER2-overexpressing advanced breast cancer that progressed during prior trastuzumab and taxane therapy. Breast Cancer Res Treat. 2013;141:437–446. doi: 10.1007/s10549-013-2689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, Chaudhuri S, Pujara K, Guillory J, Edgar KA, et al. Oncogenic ERBB3 mutations in human cancers. Cancer cell. 2013;23:603–617. doi: 10.1016/j.ccr.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Janjigian YY, Groen HJ, Horn L, Smit EF, Fu Y, Wang F, Shahidi M, Denis LJ, Pao W, Miller VA. Activity and tolerability of afatinib (BIBW 2992) and cetuximab in NSCLC patients with acquired resistance to erlotinib or gefitinib. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011 suppl; abstr 7525. [Google Scholar]
- Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, Mahmood U, Mitchell A, Sun Y, Al-Hashem R, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, Sampath D, Sliwkowski MX. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Junttila TT, Li G, Parsons K, Phillips GL, Sliwkowski MX. Trastuzumab-DM1 (T-DM1) retains all the mechanisms of action of trastuzumab and efficiently inhibits growth of lapatinib insensitive breast cancer. Breast Cancer Res Treat. 2011;128:347–356. doi: 10.1007/s10549-010-1090-x. [DOI] [PubMed] [Google Scholar]
- Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, Jessop NA, Wain JC, Yeo AT, Benes C, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Science translational medicine. 2012;4:120ra117. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Sato JD, Le A, Polikoff J, Sato GH, Mendelsohn J. Growth stimulation of A431 cells by epidermal growth factor: identification of high-affinity receptors for epidermal growth factor by an anti-receptor monoclonal antibody. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:1337–1341. doi: 10.1073/pnas.80.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecny GE, Pegram MD, Venkatesan N, Finn R, Yang G, Rahmeh M, Untch M, Rusnak DW, Spehar G, Mullin RJ, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer research. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- Kwong KY, Hung MC. A novel splice variant of HER2 with increased transformation activity. Mol Carcinog. 1998;23:62–68. doi: 10.1002/(sici)1098-2744(199810)23:2<62::aid-mc2>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP, Sliwkowski MX, Stern HM. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer research. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Seo SH, Kim SY, Park CH, Wang YP, Park K, Nam SW, Park WS, Kim SH, et al. Somatic mutations of ERBB2 kinase domain in gastric, colorectal, and breast carcinomas. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:57–61. doi: 10.1158/1078-0432.CCR-05-0976. [DOI] [PubMed] [Google Scholar]
- Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, Blattler WA, Lambert JM, Chari RV, Lutz RJ, et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer research. 2008;68:9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- Liang K, Esteva FJ, Albarracin C, Stemke-Hale K, Lu Y, Bianchini G, Yang CY, Li Y, Li X, Chen CT, et al. Recombinant human erythropoietin antagonizes trastuzumab treatment of breast cancer cells via Jak2-mediated Src activation and PTEN inactivation. Cancer cell. 2010;18:423–435. doi: 10.1016/j.ccr.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra L, Mesia R, Rivera F, Remenar E, Hitt R, Erfan J, Rottey S, Kawecki A, Zabolotnyy D, Benasso M, et al. Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first-line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Ann Oncol. 2011;22:1078–1087. doi: 10.1093/annonc/mdq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra L, Storkel S, Kerr KM, Van Cutsem E, Pirker R, Hirsch FR, Vermorken JB, von Heydebreck A, Esser R, Celik I, et al. Predictive value of epidermal growth factor receptor expression for first-line chemotherapy plus cetuximab in patients with head and neck and colorectal cancer: analysis of data from the EXTREME and CRYSTAL studies. Eur J Cancer. 2013;49:1161–1168. doi: 10.1016/j.ejca.2012.11.018. [DOI] [PubMed] [Google Scholar]
- Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- Liu L, Greger J, Shi H, Liu Y, Greshock J, Annan R, Halsey W, Sathe GM, Martin AM, Gilmer TM. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer research. 2009;69:6871–6878. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]
- Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoon DR, Lamothe B, Lax I, Schlessinger J. The docking protein Gab1 is the primary mediator of EGF-stimulated activation of the PI-3K/Akt cell survival pathway. BMC Biol. 2004;2:24. doi: 10.1186/1741-7007-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer research. 2010;70:1625–1634. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh CF, Huhalov A, Harms BD, Adams S, Paragas V, Oyama S, Zhang B, Luus L, Overland R, Nguyen S, et al. Antitumor activity of a novel bispecific antibody that targets the ErbB2/ErbB3 oncogenic unit and inhibits heregulin-induced activation of ErbB3. Mol Cancer Ther. 2012;11:582–593. doi: 10.1158/1535-7163.MCT-11-0820. [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Minkovsky N, Berezov A. BIBW-2992, a dual receptor tyrosine kinase inhibitor for the treatment of solid tumors. Curr Opin Investig Drugs. 2008;9:1336–1346. [PubMed] [Google Scholar]
- Minuti G, Cappuzzo F, Duchnowska R, Jassem J, Fabi A, O’Brien T, Mendoza AD, Landi L, Biernat W, Czartoryska-Arlukowicz B, et al. Increased MET and HGF gene copy numbers are associated with trastuzumab failure in HER2-positive metastatic breast cancer. British journal of cancer. 2012;107:793–799. doi: 10.1038/bjc.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirschberger C, Schiller CB, Schraml M, Dimoudis N, Friess T, Gerdes CA, Reiff U, Lifke V, Hoelzlwimmer G, Kolm I, et al. RG7116, a therapeutic antibody that binds the inactive HER3 receptor and is optimized for immune effector activation. Cancer research. 2013;73:5183–5194. doi: 10.1158/0008-5472.CAN-13-0099. [DOI] [PubMed] [Google Scholar]
- Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D, Brumlik MJ, Okamgba SU, Zhu Y, Duplessis TT, Parvani JG, Lesko SM, Brogi E, Jones FE. An oncogenic isoform of HER2 associated with locally disseminated breast cancer and trastuzumab resistance. Mol Cancer Ther. 2009;8:2152–2162. doi: 10.1158/1535-7163.MCT-09-0295. [DOI] [PubMed] [Google Scholar]
- Mittendorf EA, Wu Y, Scaltriti M, Meric-Bernstam F, Hunt KK, Dawood S, Esteva FJ, Buzdar AU, Chen H, Eksambi S, et al. Loss of HER2 amplification following trastuzumab-based neoadjuvant systemic therapy and survival outcomes. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:7381–7388. doi: 10.1158/1078-0432.CCR-09-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Molina MA, Codony-Servat J, Albanell J, Rojo F, Arribas J, Baselga J. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer research. 2001;61:4744–4749. [PubMed] [Google Scholar]
- Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, Salido M, Gallen M, Marsters S, Tsai SP, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18:221–223. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- Morrow PK, Wulf GM, Ensor J, Booser DJ, Moore JA, Flores PR, Xiong Y, Zhang S, Krop IE, Winer EP, et al. Phase I/II study of trastuzumab in combination with everolimus (RAD001) in patients with HER2-overexpressing metastatic breast cancer who progressed on trastuzumab-based therapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:3126–3132. doi: 10.1200/JCO.2010.32.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, Zoltick PW, Biegel JA, Hayes RL, Wong AJ. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer research. 1995;55:5536–5539. [PubMed] [Google Scholar]
- Muller WJ, Arteaga CL, Muthuswamy SK, Siegel PM, Webster MA, Cardiff RD, Meise KS, Li F, Halter SA, Coffey RJ. Synergistic interaction of the Neu proto-oncogene product and transforming growth factor alpha in the mammary epithelium of transgenic mice. Mol Cell Biol. 1996;16:5726–5736. doi: 10.1128/mcb.16.10.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka-Cook RS, Garrett J, Sanchez VK, Stanford JC, Young C, Chakrabarty A, Rinehart C, Zhang Y, Wu Y, Greenberger LM, et al. ErbB3 ablation impairs phosphatidylinositol 3-kinase (PI3K)/AKT-dependent mammary tumorigenesis. Cancer research. 2011 doi: 10.1158/0008-5472.CAN-10-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, Laccabue D, Zerbini A, Camisa R, Bisagni G, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:1789–1796. doi: 10.1200/JCO.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Nahta R, Takahashi T, Ueno NT, Hung MC, Esteva FJ. P27(kip1) down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer research. 2004;64:3981–3986. doi: 10.1158/0008-5472.CAN-03-3900. [DOI] [PubMed] [Google Scholar]
- Nahta R, Yuan LX, Du Y, Esteva FJ. Lapatinib induces apoptosis in trastuzumab-resistant breast cancer cells: effects on insulin-like growth factor I signaling. Mol Cancer Ther. 2007;6:667–674. doi: 10.1158/1535-7163.MCT-06-0423. [DOI] [PubMed] [Google Scholar]
- Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer research. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, Ariyaratne PN, Takahashi N, Sawada K, Fei Y, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- Niederst MJ, Engelman JA. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci Signal. 2013;6:re6. doi: 10.1126/scisignal.2004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, Punnoose EA, Guan J, Berry L, Prior WW, Amler LC, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast cancer preclinical models. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:3670–3683. doi: 10.1158/1078-0432.CCR-09-2828. [DOI] [PubMed] [Google Scholar]
- O’Regan R, Ozguroglu M, Andre F, Toi M, Jerusalem G, Wilks S, Isaacs C, Xu B, Masuda N, Arena FP, Yardley DA, Yap YS, Mukhopadhyay P, Douma S, El-Hashimy M, Taran T, Sahmoud T, Lebwohl DE, Gianni L. Phase III randomized, double-blind, placebo-controlled, multicenter trial of daily everolimus plus weekly trastuzumab and vinorelbine in trastuzumab-resistant, advanced breast cancer (BOLERO-3) Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013:31. [Google Scholar]
- Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, Pan Y, Wang L, de Stanchina E, Shien K, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2127–2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358:1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Jiang Z, Mortenson ED, Deng L, Radkevich-Brown O, Yang X, Sattar H, Wang Y, Brown NK, Greene M, et al. The therapeutic effect of anti-HER2/neu antibody depends on both innate and adaptive immunity. Cancer cell. 2010;18:160–170. doi: 10.1016/j.ccr.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EA, Dueck AC, McCullough AE, Chen B, Geiger XJ, Jenkins RB, Lingle WL, Davidson NE, Martino S, Kaufman PA, et al. Impact of PTEN protein expression on benefit from adjuvant trastuzumab in early-stage human epidermal growth factor receptor 2-positive breast cancer in the North Central Cancer Treatment Group N9831 trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:2115–2122. doi: 10.1200/JCO.2012.42.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GD, Fields CT, Li G, Dowbenko D, Schaefer G, Miller K, Andre F, Burris HA, 3rd, Albain KS, Harbeck N, et al. Dual Targeting of HER2-Positive Cancer with Trastuzumab Emtansine and Pertuzumab: Critical Role for Neuregulin Blockade in Antitumor Response to Combination Therapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013 doi: 10.1158/1078-0432.CCR-13-0358. [DOI] [PubMed] [Google Scholar]
- Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Price-Schiavi SA, Jepson S, Li P, Arango M, Rudland PS, Yee L, Carraway KL. Rat Muc4 (sialomucin complex) reduces binding of anti-ErbB2 antibodies to tumor cell surfaces, a potential mechanism for herceptin resistance. Int J Cancer. 2002;99:783–791. doi: 10.1002/ijc.10410. [DOI] [PubMed] [Google Scholar]
- Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA, Samuels Y. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowell TM, Pazdur R. Pathological complete response and accelerated drug approval in early breast cancer. N Engl J Med. 2012;366:2438–2441. doi: 10.1056/NEJMp1205737. [DOI] [PubMed] [Google Scholar]
- Qi J, McTigue MA, Rogers A, Lifshits E, Christensen JG, Janne PA, Engelman JA. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer research. 2011;71:1081–1091. doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson M, Pao W, Kim DW, Kim SW, Ohe Y, Felip E, Planchard D, Ghiorghiu S, Cantarini M, Cross D, et al. AZD9291: AN IRREVERSIBLE, POTENT AND SELECTIVE TYROSINE KINASE INHIBITOR (TKI) OF ACTIVATING (EGFRM+) AND RESISTANCE (T790M) MUTATIONS IN ADVANCED NSCLC. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2013;8:s389. [Google Scholar]
- Regales L, Gong Y, Shen R, de Stanchina E, Vivanco I, Goel A, Koutcher JA, Spassova M, Ouerfelli O, Mellinghoff IK, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest. 2009;119:3000–3010. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexer BN, Arteaga CL. Optimal targeting of HER2-PI3K signaling in breast cancer: mechanistic insights and clinical implications. Cancer research. 2013;73:3817–3820. doi: 10.1158/0008-5472.CAN-13-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexer BN, Ham AJ, Rinehart C, Hill S, de Matos Granja-Ingram N, Gonzalez-Angulo AM, Mills GB, Dave B, Chang JC, Liebler DC, et al. Phosphoproteomic mass spectrometry profiling links Src family kinases to escape from HER2 tyrosine kinase inhibition. Oncogene. 2011 doi: 10.1038/onc.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexer BN, Shyr Y, Arteaga CL. Phosphatase and tensin homolog deficiency and resistance to trastuzumab and chemotherapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:2073–2075. doi: 10.1200/JCO.2012.48.5243. [DOI] [PubMed] [Google Scholar]
- Ritter CA, Perez-Torres M, Rinehart C, Guix M, Dugger T, Engelman JA, Arteaga CL. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- Rosell R, Molina MA, Costa C, Simonetti S, Gimenez-Capitan A, Bertran-Alamillo J, Mayo C, Moran T, Mendez P, Cardenal F, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:1160–1168. doi: 10.1158/1078-0432.CCR-10-2158. [DOI] [PubMed] [Google Scholar]
- Ross JS, Wang K, Sheehan CE, Boguniewicz AB, Otto G, Downing SR, Sun J, He J, Curran JA, Ali S, et al. Relapsed classic E-cadherin (CDH1)-mutated invasive lobular breast cancer shows a high frequency of HER2 (ERBB2) gene mutations. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:2668–2676. doi: 10.1158/1078-0432.CCR-13-0295. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, Lathan C, Marcoux JP, Du J, Okuda K, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer research. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti M, Chandarlapaty S, Prudkin L, Aura C, Jimenez J, Angelini PD, Sanchez G, Guzman M, Parra JL, Ellis C, et al. Clinical benefit of lapatinib-based therapy in patients with human epidermal growth factor receptor 2-positive breast tumors coexpressing the truncated p95HER2 receptor. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2688–2695. doi: 10.1158/1078-0432.CCR-09-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti M, Eichhorn PJ, Cortes J, Prudkin L, Aura C, Jimenez J, Chandarlapaty S, Serra V, Prat A, Ibrahim YH, et al. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3761–3766. doi: 10.1073/pnas.1014835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti M, Rojo F, Ocana A, Anido J, Guzman M, Cortes J, Di Cosimo S, Matias-Guiu X, Ramon y Cajal S, Arribas J, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. Journal of the National Cancer Institute. 2007;99:628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- Scaltriti M, Verma C, Guzman M, Jimenez J, Parra JL, Pedersen K, Smith DJ, Landolfi S, Ramon y Cajal S, Arribas J, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene. 2009;28:803–814. doi: 10.1038/onc.2008.432. [DOI] [PubMed] [Google Scholar]
- Schaefer G, Haber L, Crocker LM, Shia S, Shao L, Dowbenko D, Totpal K, Wong A, Lee CV, Stawicki S, et al. A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies. Cancer cell. 2011;20:472–486. doi: 10.1016/j.ccr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Schechter AL, Stern DF, Vaidyanathan L, Decker SJ, Drebin JA, Greene MI, Weinberg RA. The neu oncogene: an erb-B-related gene encoding a 185,000-Mr tumour antigen. Nature. 1984;312:513–516. doi: 10.1038/312513a0. [DOI] [PubMed] [Google Scholar]
- Scheuer W, Friess T, Burtscher H, Bossenmaier B, Endl J, Hasmann M. Strongly enhanced antitumor activity of trastuzumab and pertuzumab combination treatment on HER2-positive human xenograft tumor models. Cancer research. 2009;69:9330–9336. doi: 10.1158/0008-5472.CAN-08-4597. [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Faber AC, Li D, Liang MC, Crosby K, Onsum M, Burenkova O, Pace E, Walton Z, Nie L, et al. An ErbB3 antibody, MM-121, is active in cancers with ligand-dependent activation. Cancer research. 2010;70:2485–2494. doi: 10.1158/0008-5472.CAN-09-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Soria J-C, Gadgeel SM, Wakelee HA, Camidge DR, Varga A, Fidias P, Wozniak AJ, Neal JW, Doebele RC, et al. First-in-human evaluation of CO-1686, an irreversible, selective, and potent tyrosine kinase inhibitor of EGFR T790M. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013a:31. [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013b;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer research. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DL, Miller JK, Carraway KL, 3rd, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer research. 2008;68:1471–1477. doi: 10.1158/0008-5472.CAN-07-5962. [DOI] [PubMed] [Google Scholar]
- Sheng Q, Liu X, Fleming E, Yuan K, Piao H, Chen J, Moustafa Z, Thomas RK, Greulich H, Schinzel A, et al. An activated ErbB3/NRG1 autocrine loop supports in vivo proliferation in ovarian cancer cells. Cancer cell. 2010;17:298–310. doi: 10.1016/j.ccr.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Bogatcheva G, Washington MK, Coffey RJ. Transformation of polarized epithelial cells by apical mistrafficking of epiregulin. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8960–8965. doi: 10.1073/pnas.1305508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Coffey RJ. Trafficking of epidermal growth factor receptor ligands in polarized epithelial cells. Annual review of physiology. 2014;76:275–300. doi: 10.1146/annurev-physiol-021113-170406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Soria JC, Sequist LV, Gadgeel S, Goldman J, Wakelee H, Varga A, Fidias P, Wozniak AJ, Neal JW, Doebele RC, et al. FIRST-IN-HUMAN EVALUATION OF CO-1686, AN IRREVERSIBLE, HIGHLY, SELECTIVE TYROSINE KINASE INHIBITOR OF MUTATIONS OF EGFR (ACTIVATING AND T790M) Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2013;8:s141. [Google Scholar]
- Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, Seeger JM, Weiss J, Fischer F, Frommolt P, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer research. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg J, Loi S, Divisekera U, Ngiow SF, Duret H, Yagita H, Teng MW, Smyth MJ. Anti-ErbB-2 mAb therapy requires type I and II interferons and synergizes with anti-PD-1 or anti-CD137 mAb therapy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7142–7147. doi: 10.1073/pnas.1016569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, Stevens C, O’Meara S, Smith R, Parker A, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, Ohashi K, Janjigian YY, Spitzler PJ, Melnick MA, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizaki J, Okamoto I, Fumita S, Okamoto W, Nishio K, Nakagawa K. Roles of BIM induction and survivin downregulation in lapatinib-induced apoptosis in breast cancer cells with HER2 amplification. Oncogene. 2011;30:4097–4106. doi: 10.1038/onc.2011.111. [DOI] [PubMed] [Google Scholar]
- Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, Engelman JA. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer research. 2012;72:3228–3237. doi: 10.1158/0008-5472.CAN-11-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermark T, Rao T, Cheng H, Wang Q, Lee SH, Wang ZC, Iglehart JD, Roberts TM, Muller WJ, Zhao JJ. The p110alpha and p110beta isoforms of PI3K play divergent roles in mammary gland development and tumorigenesis. Genes Dev. 2012;26:1573–1586. doi: 10.1101/gad.191973.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pinter T, Lim R, Bodoky G, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- Vaught DB, Stanford JC, Young C, Hicks DJ, Wheeler F, Rinehart C, Sanchez V, Koland J, Muller WJ, Arteaga CL, et al. HER3 is required for HER2-induced preneoplastic changes to the breast epithelium and tumor formation. Cancer research. 2012;72:2672–2682. doi: 10.1158/0008-5472.CAN-11-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- von Minckwitz G, du Bois A, Schmidt M, Maass N, Cufer T, de Jongh FE, Maartense E, Zielinski C, Kaufmann M, Bauer W, et al. Trastuzumab beyond progression in human epidermal growth factor receptor 2-positive advanced breast cancer: a german breast group 26/breast international group 03–05 study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:1999–2006. doi: 10.1200/JCO.2008.19.6618. [DOI] [PubMed] [Google Scholar]
- Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, et al. Discovery of a Mutant-Selective Covalent Inhibitor of EGFR that Overcomes T790M-Mediated Resistance in NSCLC. Cancer Discov. 2013;3:1404–1415. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, Carpenter G, Gazdar AF, Muthuswamy SK, Arteaga CL. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Wang SE, Xiang B, Guix M, Olivares MG, Parker J, Chung CH, Pandiella A, Arteaga CL. Transforming growth factor beta engages TACE and ErbB3 to activate phosphatidylinositol-3 kinase/Akt in ErbB2-overexpressing breast cancer and desensitizes cells to trastuzumab. Mol Cell Biol. 2008;28:5605–5620. doi: 10.1128/MCB.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore-Payne C, Holden JA, Layfield LJ. Detection of EGFR- and HER2-activating mutations in squamous cell carcinoma involving the head and neck. Mod Pathol. 2006;19:634–640. doi: 10.1038/modpathol.3800552. [DOI] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TR, Lee DY, Berry L, Shames DS, Settleman J. Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer cell. 2011;20:158–172. doi: 10.1016/j.ccr.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer research. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- Xia W, Bacus S, Hegde P, Husain I, Strum J, Liu L, Paulazzo G, Lyass L, Trusk P, Hill J, et al. A model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7795–7800. doi: 10.1073/pnas.0602468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer research. 2002;62:4132–4141. [PubMed] [Google Scholar]
- Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, Ogino H, Kakiuchi S, Hanibuchi M, Nishioka Y, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer research. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- Yano S, Yamada T, Takeuchi S, Tachibana K, Minami Y, Yatabe Y, Mitsudomi T, Tanaka H, Kimura T, Kudoh S, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:2011–2017. doi: 10.1097/JTO.0b013e31823ab0dd. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature reviews Molecular cell biology. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, Huberman MS, Cohen DW, Nakayama S, Ishioka K, et al. Structural, Biochemical, and Clinical Characterization of Epidermal Growth Factor Receptor (EGFR) Exon 20 Insertion Mutations in Lung Cancer. Science translational medicine. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, Ercan D, Rogers A, Roncalli M, Takeda M, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Science translational medicine. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M, Riely GJ. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, Abdel-Rahman M, Wang X, Levine AD, Rho JK, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, Lo Y, Baribaud F, Mikami I, Reguart N, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang G, Brantley-Sieders DM, Vaught D, Yu J, Xie L, Wells S, Jackson D, Muraoka-Cook R, Arteaga C, Chen J. Elevation of receptor tyrosine kinase EphA2 mediates resistance to trastuzumab therapy. Cancer research. 70:299–308. doi: 10.1158/0008-5472.CAN-09-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito CI, Riches D, Kolmakova J, Simons J, Egholm M, Stern DF. Direct resequencing of the complete ERBB2 coding sequence reveals an absence of activating mutations in ERBB2 amplified breast cancer. Genes, chromosomes & cancer. 2008;47:633–638. doi: 10.1002/gcc.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]