Abstract
Aerobic metabolism of vertebrates is linked to membrane fatty acid (FA) composition. Although the membrane pacemaker hypothesis posits that desaturation of FAs accounts for variation in resting or basal metabolic rate (BMR), little is known about the FA profiles that underpin variation in maximal metabolic rate (MMR). We examined membrane FA composition of liver and skeletal muscle in mice after seven generations of selection for increased MMR. In both liver and skeletal muscle, unsaturation index did not differ between control and high-MMR mice. We also examined membrane FA composition at the individual-level of variation. In liver, 18:0, 20:3 n-6, 20:4 n-6, and 22:6 n-3 FAs were significant predictors of MMR. In gastrocnemius muscle, 18:2 n-6, 20:4 n-6, and 22:6 n-3 FAs were significant predictors of MMR. In addition, muscle 16:1 n-7, 18:1 n-9, and 22:5 n-3 FAs were significant predictors of BMR, whereas no liver FAs were significant predictors of BMR. Our findings indicate that (i) individual variation in MMR and BMR appear to be linked to membrane FA composition in the skeletal muscle and liver, and (ii) FAs that differ between selected and control lines are involved in pathways that can affect MMR or BMR.
Keywords: Aerobic capacity, Artificial selection, Basal metabolic rate, Evolutionary physiology, Fatty acid, Metabolism, Metabolic rate, Network analysis
1. Introduction
Biological membranes profoundly influence the physiology of organisms ranging from vertebrates to eubacteria. Membrane phospholipid saturation/unsaturation (i.e., the fatty acid composition of membranes) has a marked effect on membrane fluidity, which can greatly affect cellular function, and hence the physiology and ecology of organisms (Sinensky 1974; Kogteva and Bezugov 1998; Hochachka and Somero 2002; Hoffman et al. 2009). Indeed, membrane fluidity is kept relatively constant respective to an animal’s body temperature, and it reflects an animal’s thermal environment (Cossins 1977; Hazel 1995; Hochachka and Somero 2002). For example, membrane fluidity of the brain is linked to the prevailing temperature in fish and other vertebrates (Hochachka and Somero 2002). In general, membrane fluidity increases as the fatty acid composition becomes more unsaturated (i.e., the number of double bond fatty acids increases). One of the pivotal physiological processes linked to membrane fatty acid composition is aerobic metabolism (Hulbert 2007).
Maximal metabolic rate (MMR) during exercise (also known as maximal aerobic capacity) and resting metabolic rate (RMR), vary substantially both within and among various species of vertebrates. For example, ectothermic vertebrates have lower metabolic rates than endothermic vertebrates. Maximal and minimal rates of metabolism are associated with many other biological attributes (e.g., predation risks and reproduction), so energy metabolism is profoundly important to a vertebrate’s biology and ecology (Brown et al. 2004; McNab 2006; Trebaticka et al. 2007; Speakman 2008). A number of hypotheses have attempted to explain the mechanistic bases for variation of energy metabolism among different species of vertebrates. While many observational studies have addressed these hypotheses, few manipulative experimental studies have tested them, so what accounts for variation in metabolism among vertebrates remains unclear.
Biologists are keenly interested in understanding why metabolic rates differ between ectotherms and endotherms, within various clades and within species (Bennett and Ruben 1979; Konarzewski and Diamond 1995; Lovegrove 2000, 2003; Rezende et al. 2004; Anderson and Jetz 2005; Clarke and Portner 2010). Nonetheless, a full understanding of the mechanistic and evolutionary factors responsible for variation in metabolic rate remains elusive. The membrane pacemaker model is a prominent mechanistic model that might help explain differences in metabolic rates between ectotherms and endotherms (Hulbert 2007). The membrane pacemaker hypothesis suggests that the differences in fatty acid desaturation of membrane phospholipids are the primary reason for the difference in RMR between ectotherms and endotherms (Hulbert and Else 1999, 2000). The model predicts that the membrane fatty acid unsaturation index (UI) is correlated positively with their RMR or basal metabolic rate (BMR). Whether or not the membrane pacemaker model extends to other levels of organization (e.g., within mammals or squamates, or intra-specifically) is an important question. Several recent studies suggest that the model might not apply to intra-specific differences in BMR (Brzęk et al. 2007; Haggerty et al. 2008), but we think additional studies are still worthwhile.
Why membrane fatty acid composition and metabolic rate vary among vertebrates is unclear (Brzęk et al. 2007; Haggerty et al. 2008; Konarzewski and Książek 2012), but the proximate effects of membrane fatty acid composition changes are easily studied. Environmental factors are generally considered to be the most important elements affecting muscle membrane fatty acid variation. These environmental factors include diet and exercise training. The fatty acid composition of muscle membranes reflects an individual’s diet (Ayre et al. 1996; Ranallo and Rhodes 1998; Andersson et al. 2002; Guglielmo et al. 2002; Guglielmo 2010), which can affect an individual’s aerobic performance (Guglielmo et al. 2002; Nagahuedi et al. 2009). To our knowledge, no one has looked at membrane fatty acid composition associated with MMR, which is surprising given that exercise training is linked with membrane fatty acid changes (Andersson et al. 1998; Hegle et al. 1999, 2001; Nikolaidis et al.2004; Petridou et al. 2005). In humans, research in how diet and exercise training affects membrane fatty acid composition has been intense because membrane fatty acid desaturation is linked to insulin sensitivity in the muscle (Storlien et al. 1991, 1996; Borkman et al. 1993; Vessby et al. 1994; Manco et al. 2000; Bouzakri et al. 2005). We used an experimental mouse model derived via artificial selection to quantify associations between fatty acid composition and metabolic rates. We posited that selection for high MMR would lead to a correlated increase in BMR if the assumptions underlying the aerobic capacity model for the evolution of endothermy are correct (Bennett and Ruben 1979; Hayes and Garland 1995; Wone et al. 2009). Accordingly, our selection experiment enabled us to test for effects at two levels: (1) effects of artificial selection on metabolism and (2) effects of individual variation of mice within selection treatments. More specifically, we artificially selected on mass-independent MMR (i.e., on residuals from regressions of MMR on body mass high-MMR mice) and tested whether this selection altered membrane fatty acid desaturation. We studied membrane fatty acid composition in a muscle, the gastrocnemius and a visceral organ, the liver. The gastrocnemius muscle was chosen because it is one of the main contributors to MMR (i.e., one of the main locomotor skeletal muscles involved in plantar flexion during walking or running), whereas the liver was chosen because it is one of the main contributors to BMR (Krebs 1950; Martin and Fuhrman 1955; Dann et al. 1990; Konarzewski and Diamond 1995; Weibel et al. 2004; Weibel and Hoppeler 2005; Wang et al. 2012).
2. Materials and Methods
2.1 Study organism and metabolic rates measurements
As described previously (Wone et al. 2011), we studied mice derived from an artificial selection experiment on aerobic metabolism (Wone et al. 2009). The base population for that selection experiment was HS/IBG mice (Heterogeneous Stock/Institute of Behavioral Genetics, University of Colorado, Boulder, CO, USA). The treatments were (1) control and (2) directional selection for increased mass-independent MMR (high-MMR). Each treatment was replicated four times (i.e., there were four blocks) such that there were eight lines of mice altogether (four control lines and four high-MMR). The mice we studied were offspring resulting from seven generations of selection (high MMR) or from seven generations of random breeding (controls). Our mice were not exercise trained, and standard laboratory rodent chow was available ad libitum. We measured both MMR and BMR. The metabolic rate measurements have been described previously in detail (Wone et al. 2009).
In brief, as we have described previously in Wone et al. (2011), MMR was measured once using an incremental step test during forced exercise on a motorized treadmill contained within a flow-through respirometry chamber. For the incremental step test, the treadmill rate was increased 8 m min−1 every 2 min. A shocker grid at the rear of the treadmill was used to motivate the mouse to run. When the mouse did not move off the grid, this was an indication that the mouse was exhausted and the trial was ended.
Likewise as we have described previously in Wone et al. (2011), BMR was measured at least two days after MMR with a flow-through respirometry system with 16 chambers. Twelve chambers were used to measure individual BMR and four chambers were used to record baseline concentrations of oxygen in ambient air. Mice were monitored during a 6 h period consisting of 6 cycles of 1 h each. Each mouse was monitored for 16 min out of every hour with an ambient oxygen baseline taken immediately before and immediately after each mouse measurement. BMR measurements were completed under post-absorptive conditions and at 32 °C which is within the animal’s thermal neutral zone. All mouse procedures and experimental protocols were approved by the University of Nevada, Reno Institutional Animal Care and Use Committee.
2.2 Tissue collection and phospholipid extraction/separation
We studied gastrocnemius muscle and liver membrane fatty acid composition from 80 mice (mean age = 117 days, 3.8 SD). Gastrocnemius muscle and liver tissues were collected from 10 male mice of each line, four control and four selected (high-MMR) lines. We did not sample female mice because of potential confounding fatty acid profiles resulting from pre- and post-breeding conditions, as well as, the effects of the estrus cycle. Mice were injected subcutaneously with a 0.3-ml mixture of Dormitor (10%; medetomidine hydrochloride; Orion Corp, Espoo, Finland), Ketaset (10%; ketamine hydrochloride; Fort Dodge Animal Health, Fort Dodge, Iowa, USA), and sterile water (80%), and tissue collection was performed after cervical dislocation. The gastrocnemius muscle and liver were rapidly dissected (< 90 s post-mortem time before freezing), snap frozen in liquid nitrogen, and stored at −80 °C until extraction.
Approximately 50 mg of tissue was pulverized with a BioPulverizer (BioSpec Products Inc., Bartlesville, OK, USA) under dry ice and liquid nitrogen. Lipids were then extracted from tissues using methanol-chloroform (Le Belle et al. 2002; Atherton et al. 2006). Ice-cold methanol-chloroform (2:1, 600 µL) was added and tissue samples were placed in a sonicating bath for 15 min. After sonication of the tissue samples, chloroform-water (1:1) was added (198 µL of each). Tissue samples were centrifuged at 13,500 g for 20 min. The aqueous layer was discarded, and the organic layers (i.e., extracted lipids) were stored at −80 °C until phospholipids (PL) and neutral lipids (i.e., triacylglycerides or TGA) were separated.
From the organic layer, 150 µL were transferred to activated silica Sep-Pak cartridges (Waters Corp, Milford, MA, USA) to separate into phospholipids and triacylglycerides. Sep-Pak cartridges were activated with 1 ml of chloroform with 0.01% butylated hydroxtoulene (BHT). Phospholipids were eluted with 2 × 1 ml of LC-MS grade methanol with 0.01% BHT under gentle pressure. Phospholipids extracts were stored at −80 °C or dried under a stream of nitrogen and derivatized. For derivation, samples were reconstituted with 750 µL of chloroform-methanol (1:1 vol/vol). We converted the phospholipid extracts to fatty acid methyl esters (FAMEs) by incubating with 150 µL of BF3-methanol at 80 °C for 90 min. Samples were room cooled and a methylated C19 internal standard (25 mg/l) dissolved in chloroform was added. We then added 300 µL of LC-MS grade water and 600 µL of hexane (1:2 ratio) to the samples. Samples were vortex mixed for 1 min and allowed to separate over night. The organic layer was transferred into a 2 ml auto-sampler vial and condensed under a stream of nitrogen. Samples were reconstituted with 150 µL of hexane with 0.01% BHT and transferred to auto-sampler vials with 1.5 ml glass inserts for gas chromatography mass spectrometry (GC-MS) analysis.
One microliter of the FAME was injected into the Thermo Finnigan GC equipped with a HP INNOWAX column (60 m × 0.25 mm-internal diameter column, part number 1909IN-136; Agilent Technologies, Santa Clara, CA, USA). The initial column temperature was 200 °C. Column temperature was increased 5 °C/min to 240 °C, and then the final temperature of 240 °C was held for 30 min. All column effluents were introduced into a Polaris Q trap mass spectrometer (Thermo Scientific, Waltham, MA, USA) for mass analysis.
GC-MS chromatograms were analyzed using Xcalibur v.1.3 (Thermo Scientific, Waltham, MA, USA). An individual FAME peak was identified in the chromatogram by comparing mass spectra to the National Institute of Standards and Technology (NIST) Mass Spectral Library (NIST 08) (Gaithersburg, MD, USA), the Golm Metabolite Database (Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany), and the University of Nevada, Reno standards database. We used MET-IDEA for semi-quantitation of each peak in the GC-MS chromatograms (Broeckling et al. 2006). Deconvolution and semi-quantitation of peaks and overlapping peaks was achieved by directed extraction of ion intensity values based on quantifier ion-retention time for the metabolite (Broeckling et al. 2006). A 0.1-min threshold window was used for the deviation of peaks away from the predicted retention time across the data set. The fatty acid results are presented in percentages of fatty acid detected. We used the molar percentage of individual fatty acids in all percentage-based analyses. Besides presenting percentage data for fatty acid, we calculated the following indices: saturated fatty acids (SFA), monounsaturated fatty acids (MUFA), polyunsaturated fatty acids (PUFA), n-6 fatty acids, n-3 fatty acids, and unsaturation index (the sum of the percentage of unsaturated fatty acids multiplied by the number of double bonds of each unsaturated fatty acid; Hulbert et al. 2006).
2.3 Data analysis
We analyzed the metabolic rates using a mixed-model approach. Selection treatment (selection for high residual MMR versus control) was fitted as a fixed effect, and block and line nested within treatment were fitted as random effects. Because metabolic rates correlate strongly with body mass, all analyses included body mass as a covariate because doing so adjusts (partials out) the effects of size on metabolic rate, so that the statistics are mass-independent or mass-adjusted. This approach is similar to the use of an ANCOVA with body mass as the covariate and selection or no selection (control) as the treatment, except that the mixed-model also includes random effects that are necessary to appropriately account for the experimental design of the selection experiment. Because selection experiments are conducted on lines of mice, not on individuals, the error degrees of freedom in the mixed-model analysis depends on the number of lines, not on the number of mice, so the error degrees of freedom are much lower as a result (Henderson 1989). For MMR analysis, age, treadmill (two treadmills were used to make the measurements), and observer (the person conducting the metabolic measurement, who determined when to terminate the measurement) were included as additional fixed effects. For BMR analysis, age and metabolism chamber (12 chambers were used to make the measurements) were included as additional fixed effects. For body mass analyses, we also used a mixed-model approach in which treatment and age (when MMR was measured, when BMR was measured, or when tissues were collected) were fitted as fixed effects. Percentages (i.e., relative to the sum of fatty acids in each phospholipid) of fatty acid were compared between control and high-MMR mice. The percentages were compared using separate one-way mixed model analysis of variance (ANOVA), where the block and line nested within treatment were fitted as random effects in the comparison. To account for multiple ANOVA comparisons, we estimated the false discovery rate as the maximum q value (Storey 2002). Besides analyzing the data from the perspective of the responses of the two treatments (control compared with selected), we also analyzed the individual-level of variation because much of the variation in metabolic rate, fatty acid composition, and unsaturation index was within treatments. These analyses were conducted by including all the percentages of individual fatty acids entered simultaneously as covariates in the mixed-model analyses of metabolic rates. Block and line nested within treatment were fitted as random effects in the mixed-models. For the MMR analysis, selection (effects of control compared with high-MMR mice), body mass at MMR, and age at MMR were included as fixed effects. For the BMR analysis, selection (effects of control compared with high-MMR mice), body mass at BMR, and age at BMR were included as fixed effects. Analyses were also done as above using MMR/BMR and MMR-BMR as dependent variables. However, none of the fatty acids were predictors based on these analyses, so these models were not reported. Unlike linear univariate models, linear mixed-models do not compute the model R2 statistic, nor the model fixed effects partial R2 statistics. Hence, we followed the method of Edwards et al. (2008) in computing the partial R2 statistic for all significant individual fatty acids. All mixed-model statistical analyses were performed using SAS, v. 9.3 (SAS Institute, Cary, NC, USA).
Because membrane fatty acids are correlated with each other (Haggerty et al. 2008) in that they can affect each other functionally and can be biosynthesized from the other (Brenner 1974; Wakil et al. 1983; Graber et al. 1993), we conducted a network analysis to examine their associations. We mapped the detected fatty acid species onto general biochemical pathways according to the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/; Kanehisa 2006) using Metscape v. 2.3.1 (Karnovsky et al. 2012), a plugin for Cytoscape v.2.7 (Shannon et al. 2003). This was done to obtain an overview of all the known components of the fatty acid network. Such a network map provides a complete list of KEGG metabolic pathways and genes related to the fatty acids studied.
3. Results
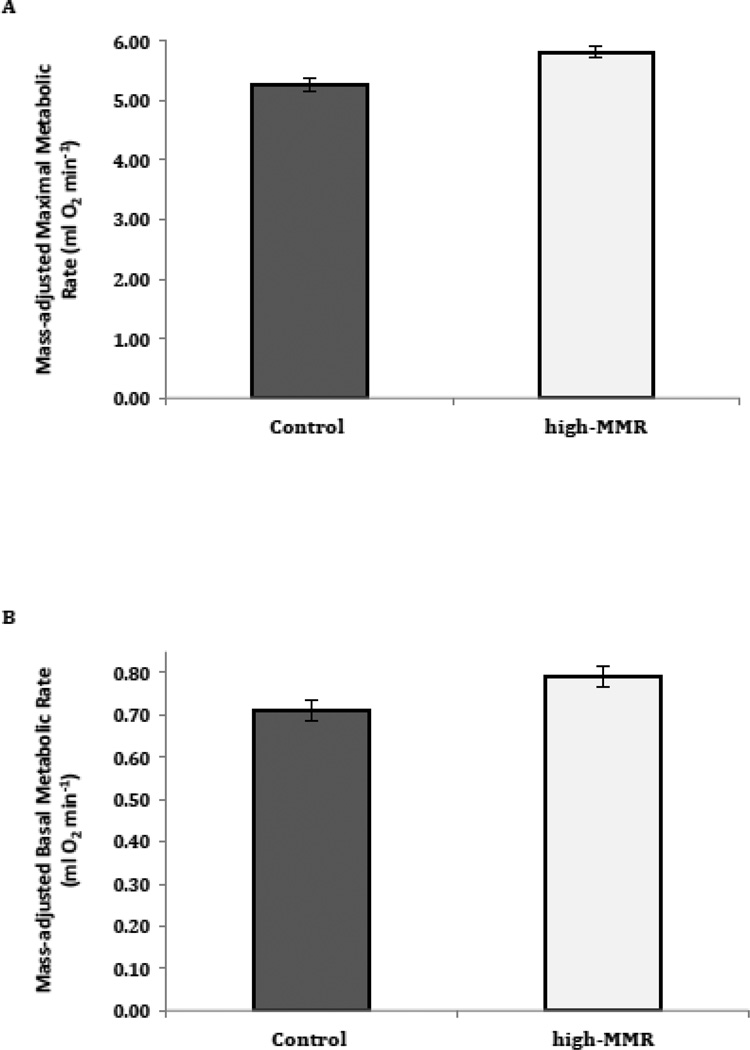
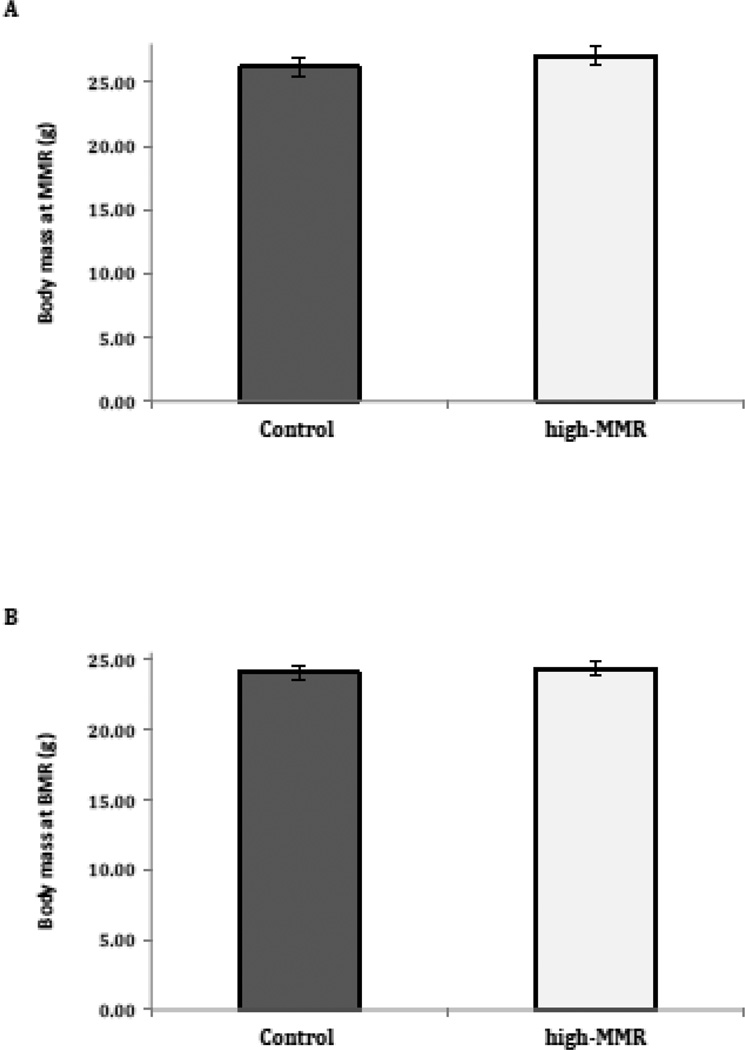
The mean mass-adjusted MMR was 10.2% higher in high-MMR selected mice than in control mice (Fig. 1A; n = 76, F1,3 = 16.4, p = 0.03). Mean body mass at the time MMR was measured in lines of high-MMR mice was 27.1 g (0.71 SE), whereas mean body mass at the time MMR was measured in lines of control mice was 26.2 g (0.72 SE), but this difference was not statistically significant (Fig. 2A; n = 76, F1,3 = 1.1, p = 0.38). Mass-adjusted BMR did not differ significantly between high-MMR and control mice (Fig. 1B, n = 72, F1,3 = 7.7, p = 0.07). Mean body mass at the time BMR was measured in lines of high-MMR mice was 24.3 g (0.53 SE), whereas mean body mass at the time BMR was measured in lines of control mice was 24.1 g (0.53 SE), but this difference was not statistically significant (Fig. 2B; n = 72, F1,3 = 0.1, p = 0.76). Metabolic rates were not available for all mice hence in some case n is less than 80 (Fig 1). Mean body mass at the time tissues were collected in lines of high-MMR mice was 30.1 g (0.72 SE), whereas mean body mass at the time tissues were collected in lines of control mice was 28.9 g (0.72 SE), but this difference was not statistically significant (n = 80, F1,3 = 3.2, p = 0.17).
Fig 1.
Metabolic rates of mice selected for 7 generations for high mass-independent MMR versus controls. Data are mass adjusted by inclusion of body mass as covariate in the mixed-model. Each mass-adjusted mean reflects data from 4 lines of mice per selection treatment (i.e., selection versus control). A. Mass-adjusted MMR (n = 40 for high-MMR mice, and n = 36 for control mice). B. Mass-adjusted BMR (n = 35 for high-MMR mice, and n = 37 for control mice). Values presented are mean (± sem). *Indicates p < 0.05.
Fig 2.
Body mass of mice selected for 7 generations for high mass-independent MMR versus controls. A. Mass at the time MMR was measured. B. Mass at the time BMR was measured. Values presented are mean (± sem).
The increase in MMR did not lead to changes in percentages of fatty acids or the unsaturation index (Table 1). In the liver, the most abundant unsaturated fatty acid species was 18:2 n-6, and the most abundant saturated fatty acid species was 16:0. Similarly, in gastrocnemius muscle the most abundant unsaturated fatty acid species was 18:2 n-6, and the most abundant saturated fatty acid species was 16:0.
Table 1.
Molar percentage distribution of individual fatty acids in gastrocnemius muscle and liver phospholipids in control and high-MMR mice (n = 40; mean ± SD).
| Gastrocnemius muscle | Liver | |||
|---|---|---|---|---|
| Fatty acid | Control | high-MMR | Control | high-MMR |
| 14:0 | 0.5 ± 0.40 | 0.5 ± 0.32 | 0.1 ± 0.04 | 0.1 ± 0.06 |
| 16:0 | 28.4 ± 8.49 | 25.9 ± 9.59 | 24.5 ± 3.47 | 24.5 ± 3.48 |
| 16:1n-7 | 0.1 ± 0.10 | 0.1 ± 0.17 | <0.1 ± 0.08 | <0.1 ± 0.02 |
| 18:0 | 18.4 ± 7.13 | 20.6 ± 7.35 | 23.0 ± 3.23 | 23.2 ± 2.60 |
| 18:1n-9 | 8.0 ± 7.00 | 9.3 ± 7.05 | 4.6 ± 1.75 | 4.5 ± 1.50 |
| 18:1n-7 | 2.3 ± 1.19 | 2.9 ± 1.73 | 2.4 ± 1.63 | 2.1 ± 0.83 |
| 18:2n-6 | 13.0 ± 6.08 | 11.7 ± 4.58 | 18.7 ± 2.79 | 19.1 ± 2.60 |
| 20:2n-6 | 1.3 ± 1.39 | 1.6 ± 1.99 | 0.8 ± 0.97 | 0.6 ± 0.75 |
| 20:3n-6 | 1.3 ± 0.63 | 1.4 ± 1.08 | 2.3 ± 0.76 | 2.1 ± 0.32 |
| 20:4n-6 | 7.1 ± 2.71 | 7.2 ± 3.06 | 15.1 ± 2.63 | 15.8 ± 4.69 |
| 20:5n-3 | 0.7 ± 0.86 | 0.9 ± 1.26 | 0.5 ± 0.72 | 0.4 ± 0.25 |
| 22:5n-3 | 3.9 ± 2.07 | 5.2 ± 5.49 | 0.5 ± 0.53 | 0.5 ± 0.41 |
| 22:6n-3 | 12.6 ± 7.84 | 12.9 ± 8.64 | 7.5 ± 3.31 | 7.1 ± 3.71 |
| Sum | 100.00 | 100.00 | 100.00 | 100.00 |
| Indices | ||||
| SFA (%) | 47.4 ± 11.48 | 47.0 ± 13.18 | 47.7 ± 6.28 | 47.9 ± 5.27 |
| MUFA (%) | 10.3 ± 7.45 | 12.2 ± 7.70 | 7.0 ± 2.79 | 6.6 ± 1.95 |
| PUFA (%) | 39.8 ± 8.36 | 40.9 ± 9.65 | 45.3 ± 5.05 | 45.6 ± 4.85 |
| n-6 PUFA (%) | 22.7 ± 5.89 | 21.9 ± 5.07 | 36.8 ± 4.57 | 37.6 ± 5.48 |
| n-3 PUFA(%) | 17.2 ± 8.47 | 19.0 ± 10.85 | 8.6 ± 3.45 | 8.0 ± 3.61 |
| UI | 174.2 ± 41.08 | 179.6 ± 59.66 | 166.2 ± 22.79 | 162.5 ± 21.88 |
SFA - saturated fatty acids
MUFA – monounsaturated fatty acids
PUFA – polyunsaturated fatty acids
UI – unsaturation index
Individual variation in the percentages of some fatty acids correlated with metabolic rates (Table. 2). For the liver, percentages of 18:0, 20:3 n-6, 20:4 n-6, and 22:6 n-3 fatty acids were significant predictors of MMR. For the gastrocnemius muscle, percentages of 18:2 n-6, 20:4 n-6, and 22:6 n-3 fatty acids were significant predictors of MMR. In contrast, in the gastrocnemius muscle, percentages of 16:1 n-7, 18:1 n-9, and 22:5 n-3 fatty acids were significant predictors of BMR. Interestingly in the liver, no fatty acids were predictors of BMR. Individual variation in unsaturation index was not significantly associated with either MMR or BMR.
Table 2.
Variables included in the individual level analyses, their associated p-values (bold indicates significance), direction, and partialR2* of prediction. The partial R2 were calculated for the analyses with significant effects only (e.g., for 16:1 n-7 in gastrocnemius the partial R2 is for the analysis of BMR and for 18:2 n-6 in gastrocnemius the partial R2 is for the analysis of MMR). See materials and methods for description of the variables.
| (Fatty Acid in molar %) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gastrocnemius | Liver | |||||||
| Effect | BMR | MMR | Direction | Partial R2 |
BMR | MMR | Direction | Partial R2 |
| Treatment | 0.10 | 0.02 | 0.43 | 0.04 | ||||
| Mass | 0.003 | <0.0001 | 0.009 | <0.0001 | ||||
| Age | 0.13 | 0.79 | 0.38 | 0.84 | ||||
| 14:0 | 0.11 | 0.21 | 0.20 | 0.19 | ||||
| 16:0 | 0.22 | 0.29 | 0.62 | 0.36 | ||||
| 16:1 n-7 | 0.006 | 0.91 | + | 0.14 | 0.48 | 0.92 | ||
| 18:0 | 0.82 | 0.90 | 0.12 | <0.05 | + | 0.08 | ||
| 18:1 n-9 | 0.0003 | 0.90 | − | 0.24 | 0.51 | 0.84 | ||
| 18:1 n-7 | 0.40 | 0.32 | 0.31 | 0.80 | ||||
| 18:2 n-6 | 0.13 | 0.03 | + | 0.09 | 0.37 | 0.10 | ||
| 20:2 n-6 | 0.20 | 0.23 | 0.09 | 0.07 | ||||
| 20:3 n-6 | 0.74 | 0.85 | 0.11 | 0.01 | − | 0.12 | ||
| 20:4 n-6 | 0.20 | 0.03 | + | 0.10 | 0.18 | 0.003 | + | 0.16 |
| 20:5 n-3 | 0.33 | 0.43 | 0.40 | 0.52 | ||||
| 22:5 n-3 | 0.04 | 0.89 | − | 0.09 | 0.07 | 0.62 | ||
| 22:6 n-3 | 0.53 | 0.03 | + | 0.09 | 0.68 | 0.03 | + | 0.08 |
| UI | 0.58 | 0.29 | 0.74 | 0.41 | ||||
partial R2 computed based on the method of Edwards et al. 2008
UI – unsaturation index
BMR – basal metabolic rate
MMR – maximal metabolic rate
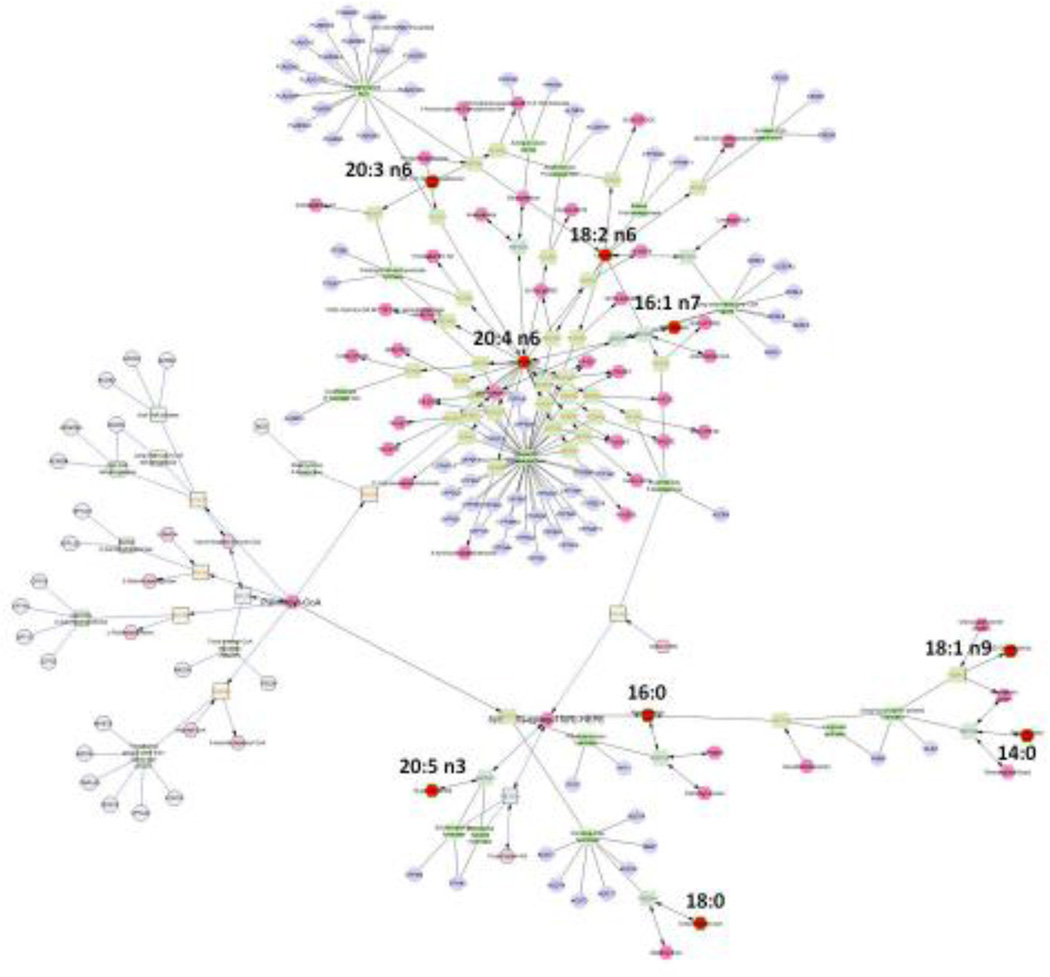
From the fatty acids that our analyses detected, we created a compound-reaction-enzyme-gene (CREG) network. Vaccenic (18:1 n-7) and docosapentaenoic (22:5 n3) acids were not available on KEGG, therefore they were not included in the network analysis. The resulting network consisted of three components: (i) a small anti-inflammatory subnetwork, (ii) a medium saturated fatty acids biosynthesis and beta-oxidation subnetwork, and (iii) a large subnetwork that contained the rest of the experimental fatty acids. We were able to connect the three components with two expand operations by adding the nodes related to the compounds of subnetwork (i) (i.e., 5(S),6(S)-epoxy-15(R)-HEPE - CE7109) and subnetwork (ii) (i.e., palmitoleoyl-CoA - CE0852) (Fig. 3). This expanded network provided a complete list of metabolic pathways (Table S1) and genes related to experimental fatty acids. To examine this expanded network further, we built subnetworks combining all fatty acids that were significant predictors of individual variation in MMR and/or BMR. The significant predictors of MMR in gastrocnemius muscle subnetwork included the pathways of arachidonic acid metabolism, di-unsaturated fatty acid beta-oxidation, leukotriene metabolism, linoleate metabolism, omega-6 fatty acid metabolism, and prostaglandin formation from arachidonate. Somewhat differently, the significant predictors of MMR in liver subnetwork included the pathways arachidonic acid metabolism, de novo fatty acid biosynthesis, leukotriene metabolism, omega-3 fatty acid metabolism, omega-6 fatty acid metabolism, propanoate metabolism, prostaglandin formation from arachidonate, and prostaglandin formation from dihomo gama-linoleic acid. We were unable to build a subnetwork for significant predictors of BMR in gastrocnemius muscle because 16:1 n7 fatty acid was the only one described or available on KEGG (Table 3)
Fig 3.
A fully connected network of fatty acids detected from the selection experiment on maximal metabolic rate. Fatty acids with experimental data are shown as red-filled hexagons. Blue-filled circles represent genes. Light-green filled squares represent enzymes. Pink-filled hexagons represent compounds or metabolites. Grey-filled squares represent reactions.
Table 3.
List of pathways involving the detected fatty acids mapped from KEGG.
| Common Name | Carbon Skeleton |
Metabolic pathway from KEGG |
|---|---|---|
| Myristic acid | 14:0 | De novo fatty acid biosynthesis |
| Palmitic acid | 16:0 | De novo fatty acid biosynthesis, Saturated fatty acids beta-oxidation |
| Palmitoleic acid* | 16:1 n7 | Mono-unsaturated fatty acid beta-oxidation |
| Stearic acid# | 18:0 | De novo fatty acid biosynthesis |
| Oleic acid* | 18:1 n9 | none described |
| Vaccenic acid | 18:1 n7 | not found |
| Linoleic acid+ | 18:2 n6 | Di-unsaturated fatty acid beta-oxidation, Linoleate metabolism |
| Eicosadienoic acid | 20:2 n6 | none described |
| Dihomo gamma linolenic acid# | 20:3 n6 | Prostaglandin formation from dihomo gama-linoleic acid |
| Eicosapentaenoic acid | 20:5 n3 | Putative anti-Inflammatory metabolites formation from EPA |
| Arachidonic acid+# | 20:4 n6 | Arachidonic acid metabolism, Leukotriene metabolism, Omega-6 fatty acid metabolism, Prostaglandin formation from arachidonate |
| Docosapentaenoic acid* | 22:5 n3 | not found |
| Docosahexaenoic acid+# | 22:6 n3 | none described |
significant predictors of BMR in the gastrocnemius muscle
significant predictors of MMR in the gastrocnemius muscle
significant predictors of MMR in the liver
4. Discussion
The metabolic data presented in this manuscript are for a subsample of mice from a larger selection experiment. These data are reported here because they represent the metabolic rates of the specific mice for which we conducted the fatty acid analyses. Directional selection for increased mass-independent MMR (i.e., residuals from analyses including mass as a covariate) resulted in significant increases in MMR but did not result in a correlated increase in mass-independent BMR in this subset of mice. Our previous report on a larger sample of mice from this same generation of the selection experiment yielded qualitatively similar results (Wone et al. 2011). The lack of correlated response in BMR is similar to another selection experiment where high voluntary wheel running was the selection criterion. High voluntary wheel running mice in this selection experiment had a significant increase in MMR, but no correlated response in RMR (Dlugosz et al. 2009).
Overall, the liver membrane fatty acid profiles in our mice were similar to profiles reported previously for mice and rats (Couture and Hulbert 1995; Hulbert et al. 2006; Brzęk et al. 2007; Haggerty et al. 2008). In agreement with previous studies, the most abundant saturated fatty acids in the liver of our mice were 16:0 and 18:0 fatty acids. Recall that unsaturation index is an index of the sum of the percentages of unsaturated fatty acids multiplied by the number of double bonds of each unsaturated fatty acid. The more unsaturated the membrane fatty acids of phospholipids, the higher the index value is expected to be (Hulbert et al. 2006; Hulbert 2007). The liver unsaturation index for our mice fell between the values of 230 and 166 reported previously for laboratory mice (Brzęk et al. 2007; Haggerty et al. 2008).
Selection for increased MMR did not alter membrane fatty acid desaturation in high-MMR mice. One likely explanation for this result is that our mice were not exercise trained prior to measuring MMR on the treadmill. Although increases in MMR are linked with exercise training (Brooks and White 1978; De Angelis et al. 2004) and exercise training is known to modulate muscle membrane fatty acid desaturation (Andersson et al. 1998; Helge et al. 1999, 2001; Mitchell et al. 2004; Nikolaidis et al. 2004; Petridou et al. 2005), our results suggest that exercise training is required to alter muscle membrane fatty acid desaturation. Another possible explanation for the lack of correlated response in membrane fatty acid desaturation is that perhaps selection for high MMR did not alter membrane fatty acid desaturation because selection resulted primarily in changes in motivation rather than in physiology (Rhodes et al. 2005; Waters et al. 2008; Mathes et al. 2010). Unlike MMR, elevated BMR has been extensively studied and is thought to be linked with membrane fatty acid desaturation (Hulbert 2003; Hulbert and Else 1999, 2000, 2005; Brzęk et al. 2007; Hulbert 2007; Haggerty et al. 2008). Indeed, this linkage led to the membrane pacemaker hypothesis of metabolism (Hulbert 2003; 2007). The hypothesis predicts that the membrane fatty acid unsaturation index is correlated positively with RMR or BMR.
The current study is not the first to be inconsistent with the membrane pacemaker hypothesis. For example, Brzęk et al. (2007) selected for low-BMR and high-BMR, and selection resulted in a change in membrane polyunsaturated fatty acid, but not in the direction predicted by the membrane pacemaker model. Brzęk et al. (2007) reported that the unsaturation index of liver and kidney membrane fatty acids was significantly higher in low-BMR mice, but if the membrane pacemaker hypothesis was correct, then one would predict that the unsaturation index would be lower in mice with low BMR. Likewise, Haggerty et al. (2008) reported no association of RMR and of mass-adjusted RMR with polyunsaturated fatty acid in an outbred strain (MF1) of mice. Valencak and Ruf (2007) have suggested that the link between membrane fatty acid desaturation and BMR is overstated. Their phylogenetic analysis, corrected for body mass, of 42 mammalian species did not support the predictions of the model (Valencak and Ruf 2007). However, Valencak and Ruf (2007) analyzed fatty acid profiles of muscle membrane, not fatty acid profiles of liver. The liver might be more important than skeletal muscle in determining BMR, and the possible link between liver membrane fatty acid desaturation and BMR remains unresolved.
Our individual-level analyses indicate that variation in MMR is linked to changes in the fatty acid composition of gastrocnemius muscle and liver membranes and that the fatty acids that differed between selected and control lines are involved in pathways that can affect BMR or MMR. Keep in mind that individual-level analyses are not analyses of percentages of fatty acid per se, but analyses of those variables as covariates. Interestingly, together with the individual-level analyses and the CREG network, we identified potential underlying molecular mechanistic explanations for the individual variation in MMR of the skeletal muscle and liver. These associations appear to reflect complex alterations and provide evidence for the involvement of such processes as fatty acid catabolism (e.g., beta oxidation), fatty acid anabolism (e.g., de novo fatty acid biosynthesis), and stress response (e.g., prostaglandin formation). This would suggest that the variation in MMR might be linked to the amounts of specific fatty acid species. Indeed, membrane fatty acids have a central role in determining membrane properties, cell signaling, and gene expression in the skeletal muscle and other tissues such as liver (Graber et al. 1993; Kogteva and Bezugov 1998; Ehrenborg and Krook 2009).
Our individual-level and CREG network analyses suggest possible functional implications of membrane fatty acids on metabolic rates. For example, our model predicts that as the percentages of 18:2 n-6, 20:4 n-6, and 22:6 n-3 fatty acids in muscle membranes increase, individual MMR is expected to increase. This increase in MMR appears to be linked to increases in fatty acid oxidation metabolism in the muscles. Indeed, 18:2 n-6, 20:4 n-6, and 22:6 n-3 fatty acids can increase fatty acid oxidation metabolism and affect the inflammatory response (Table 3; Ikeda et al. 1994, 1998; Grimsgaard et al. 1997). Mechanistically, elevated amounts of 18:2 n-6, 20:4 n-6, and 22:6 n-3 fatty acids in muscle membranes might have modulated the decreased amounts of free fatty acids and intra-muscular triacylglycerol fatty acids in high-MMR mice (Wone et al. 2011). Likewise, a gene expression study of rat heart muscle from Koch and Britton’s (2001) divergent selection experiment on exercise capacity showed that high-capacity runners switched toward fatty acid oxidation metabolism, whereas low-capacity runners switched toward glucose-based metabolism (Bye et al. 2008). Hence, one might expect that those fatty acids that are involved in fatty acid metabolism to be present in greater percentages in membranes of cardiac tissues of high-capacity runners compared with low-capacity runners. Lastly, a fatty acid metabolism study of high voluntary wheel running mice from Garland’s (Swallow et al. 1998) directional selection experiment on increased voluntary wheel running showed that high runners switched toward fatty acid metabolism (Templeman et al. 2012).
As previously mentioned, the fatty acid composition of muscle membranes can change due to exercise training, and some of our individual-level analysis predictions match those reported for exercise training. In particular, of the three fatty acids that were related significantly to MMR in muscle membranes, exercise training also increased two fatty acids significantly. Notably, the fatty acid 18:2 n-6 was significantly increased after 8 weeks of wheel running in muscle membranes of Wistar rats (Petridou et al. 2005), and the fatty acid 22:6 n-3 was significantly increased after 8 weeks of aerobic training in muscle membranes of humans (Andersson et al. 2000; Hegle et al. 2001). Together, these similar changes suggest that alterations in membrane fatty acid composition, whether due to exercise training or selection for increased MMR, reflect the same physiological adaptations to enhance aerobic performance (Guglielmo et al. 2002; Nagahuedi et al. 2009).
Our individual-level analyses also indicate that BMR is linked to the fatty acid composition of gastrocnemius muscle membranes, but not to the fatty acid composition of liver membranes or to the unsaturation index in either organ. BMR and MMR are often correlated at the inter-specific level, but whether or not there is a functional mechanism responsible for this correlation remains unclear. The basis of a functional link between BMR and MMR is difficult to predict a priori, because the main contributors to BMR are thought to be visceral organs, such the liver (Dann et al. 1990; Konarzewski and Diamond 1995; Konarzewski and Książek 2012), whereas the main contributor to MMR is the musculature (Weibel et al. 2004; Weibel and Hoppeler 2005). However, recent analyses of mammals suggest that our notions about which organs are responsible for BMR might need to be revisited because these analyses indicate that BMR correlates strongly with variation in muscle mass (Raichlen et al. 2010). If the musculature is indeed one of the contributors to BMR in addition to the visceral organs, then the elevated metabolic rate of muscles might be the mechanistic connection that accounts for the correlation between BMR and MMR. In the current study, 16:1 n-7, 18:1 n-9, and 22:5 n-3 fatty acids in muscle membranes were significant predictors of BMR. Interestingly, increased amounts of 16:1 n-7 and 18:1 n-9 fatty acids in tissues and serum have been implicated in the up-regulation of glucose uptake and lipid catabolism (Table 3; Kien et al. 2005; Dimopoulos et al. 2006). Mechanistically, it might be that elevated BMR in our high-MMR mice is due to changes in the percentages of certain fatty acid species in the muscle membrane (Wone et al. 2011; this study).
In summary, selection for increased MMR did not result in correlated responses in fatty acid desaturation of membrane phospholipids from the liver and gastrocnemius muscle of mice. Our findings did not support the prediction that the unsaturation index is correlated with BMR, but more broadly MMR and BMR were linked to membrane fatty acid composition changes in the skeletal muscle and liver. Our findings indicate that the type of fatty acid of membrane phospholipids partially accounts for the variation in intra-specific MMR and BMR. Lastly, given that aerobic training increases fatty acid oxidation (Talanian et al. 2007), and that muscle membrane fatty acids can affect aerobic performance (Guglielmo et al. 2002; Nagahuedi et al. 2009), our expanded CREG network analyses provide a springboard to generate hypotheses regarding the functional associations of membrane fatty acids and the individual variation in MMR and/or BMR.
Understanding the functional associations of membrane fatty acids and the variation in MMR will be an important goal for future studies. This area of research will not only advance basic knowledge of mammalian physiology, but also might reveal how alterations in muscle membrane fatty acids due to diet and exercise training can enhance aerobic performance.
Supplementary Material
Acknowledgments
This study was funded by NSF IOS 0344994 to J. P. Hayes and University of Nevada, Reno Graduate Student Association Research Grants to B. Wone. We thank Beate Wone for help with the tissue extractions. We thank Dave Shintani for the use of his lab facilities. We thank Angelica Adrian, Cynthia Downs, Amber Huleva, Andy Hicks, Else Huerta, Marta Labocha, Kim Mclean, Mike Sears, and Alexandra Watson for help with the selection experiment. We thank Lloyd Edwards for help with the partial R2 statistic calculation. We thank the anonymous reviewers for helpful suggestions to improve the manuscript. The UNR Proteomics Center acknowledges support from NIH Grant P20 RR-016464 from the INBRE Program of the National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures. No conflicts of interest, financial or otherwise, are declared by the authors.
References
- Andersson A, Sjödin A, Olsson R, Vessby B. Effects of physical exercise on phospholipid fatty acid composition in skeletal muscle. Am J Physiol Endocrinol Metab. 1998;274:E432–E438. doi: 10.1152/ajpendo.1998.274.3.E432. [DOI] [PubMed] [Google Scholar]
- Andersson A, Sjödin A, Hedman A, Olsson R, Vessby B. Fatty acid profile of skeletal muscle phospholipids in trained and untrained young men. Am. J. Physiol. 2000;279:744–751. doi: 10.1152/ajpendo.2000.279.4.E744. [DOI] [PubMed] [Google Scholar]
- Andersson A, Nalsen C, Tengblad S, Vessby B. Fatty acid composition of skeletal muscle reflects dietary fat composition in humans. Am J Clin Nutr. 2002;76:1222–1229. doi: 10.1093/ajcn/76.6.1222. [DOI] [PubMed] [Google Scholar]
- Anderson KJ, Jetz W. The broad-scale ecology of energy expenditure of endotherms. Ecology Letters. 2005;8:310–318. [Google Scholar]
- Atherton HJ, Bailey NJ, Zhang W, Taylor J, Major H, Shockcor J, Clarke K, Griffin JL. A combined 1H-NMR spectroscopy- and mass spectrometry-based metabolomic study of the PPAR-α null mutant mouse defines profound systemic changes in metabolism linked to the metabolic syndrome. Physiol Genomics. 2006;27:178–186. doi: 10.1152/physiolgenomics.00060.2006. [DOI] [PubMed] [Google Scholar]
- Ayre KJ, Hulbert AJ. Effects of changes in dietary fatty acids on isolated skeletal muscle functions in rats. J Appl Physiol. 1996;80:464–471. doi: 10.1152/jappl.1996.80.2.464. [DOI] [PubMed] [Google Scholar]
- Bennett AF, Ruben JA. Endothermy and activity in vertebrates. Science. 1979;206:649–654. doi: 10.1126/science.493968. [DOI] [PubMed] [Google Scholar]
- Borkman M, Storlien LH, Pan DA, Jenkins AB, Chisholm DJ, Campbell LV. The relation between insulin sensitivity and the fatty-acid composition of skeletal-muscle phospholipids. N Engl J Med. 1993;28:238–244. doi: 10.1056/NEJM199301283280404. [DOI] [PubMed] [Google Scholar]
- Bouzakri K, Koistinen HA, Zierath JR. Molecular mechanisms of skeletal muscle insulin resistance in type 2 diabetes. Curr Diabetes Rev. 2005;1:167–174. doi: 10.2174/1573399054022785. [DOI] [PubMed] [Google Scholar]
- Brenner RR. The oxidative desaturation of unsaturated fatty acids in animals. Mol Cell Biochem. 1974;3:41–52. doi: 10.1007/BF01660076. [DOI] [PubMed] [Google Scholar]
- Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW. MET-IDEA: a data extraction tool for mass spectrometry-based metabolomics. Anal Chem. 2006;78:4334–4431. doi: 10.1021/ac0521596. [DOI] [PubMed] [Google Scholar]
- Brooks GA, White TP. Determination of metabolic and heart rate responses of rats to treadmill exercise. J Appl Physiol. 1978;45:1009–1015. doi: 10.1152/jappl.1978.45.6.1009. [DOI] [PubMed] [Google Scholar]
- Brown JH, Gillooly JF, Allen AP, Savage VM, West GB. Toward a metabolic theory of ecology. Ecology. 2004;85:1771–1789. [Google Scholar]
- Brzęk P, Bielawska K, Ksiazek A, Konarzewski M. Anatomic and molecular correlates of divergent selection for basal metabolic rate in laboratory mice. Physiol Biochem Zool. 2007;80:491–499. doi: 10.1086/520617. [DOI] [PubMed] [Google Scholar]
- Bye A, Langaas M, Höydal MA, Kemi OJ, Heinrich G, Koch LG, Britton SL, Najjar SM, Ellingsen Ø, Wisløff U. Aerobic capacity-dependent differences in cardiac gene expression. Physiol Genomics. 2008;33:100–109. doi: 10.1152/physiolgenomics.00269.2007. [DOI] [PubMed] [Google Scholar]
- Clarke A, Portner HO. Temperature, metabolic power and the evolution of endothermy. Biol Revs. 2010;85:703–727. doi: 10.1111/j.1469-185X.2010.00122.x. [DOI] [PubMed] [Google Scholar]
- Cossins AR. Adaptation of biological-membranes to temperature - effect of temperature-acclimation of goldfish upon viscosity of synaptosomal membranes. Biochim Biophys Acta - Biomembranes. 1977;470:395–411. doi: 10.1016/0005-2736(77)90131-6. [DOI] [PubMed] [Google Scholar]
- Couture P, Hulbert AJ. Membrane fatty acid composition of tissues is related to body mass of mammals. J Membr Biol. 1995;148:27–39. doi: 10.1007/BF00234153. [DOI] [PubMed] [Google Scholar]
- Daan S, Masman D, Groenewold A. Avian basal metabolic rates: their association with body composition and energy expenditure. Am J Physiol. 1990;259:R333–R340. doi: 10.1152/ajpregu.1990.259.2.R333. [DOI] [PubMed] [Google Scholar]
- De Angelis K, Wichi RB, Jesus WRA, Moreira ED, Morris M, Krieger EM, Irigoyen MC. Exercise training changes autonomic cardiovascular balance in mice. J Appl Physiol. 2004;96:2174–2178. doi: 10.1152/japplphysiol.00870.2003. [DOI] [PubMed] [Google Scholar]
- Dimopoulos N, Watson M, Sakamoto K, Hundal HS. Differential effects of palmitate and palmitoleate on insulin action and glucose utilization in rat L6 skeletal muscle cells. Biochem J. 2006;399:473–481. doi: 10.1042/BJ20060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosz EM, Chappell MA, McGillivray DG, Syme DA, Garland T., Jr Locomotor trade-offs in mice selectively bred for high voluntary wheel running. J Exp Biol. 2009;212:2612–2618. doi: 10.1242/jeb.029058. [DOI] [PubMed] [Google Scholar]
- Edwards LJ, Muller KE, Wolfinger RD, Qaqish BF, Schabenberger O. An R2 statistic for fixed effects in the linear mixed model. Stat Med. 2008;27:6137–6157. doi: 10.1002/sim.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenborg E, Krook A. Regulation of skeletal muscle physiology and metabolism by peroxisome proliferator-activated receptor delta. Pharmacol Revs. 2009;61:373–393. doi: 10.1124/pr.109.001560. [DOI] [PubMed] [Google Scholar]
- Graber R, Sumida C, Nunez EA. Fatty acids and cell signal transduction. J Lipid Mediat Cell Signal. 1994;2:91–116. [PubMed] [Google Scholar]
- Grimsgaard S, Bønaa KH, Hansen JB, Nordøy A. Highly purified eicosapentaenoic acid and docosahexaenoic acid in humans have similar triacylglycerol-lowering effects but divergent effects on serum fatty acids. Am J Clin Nutr. 1997;66:649–659. doi: 10.1093/ajcn/66.3.649. [DOI] [PubMed] [Google Scholar]
- Guglielmo CG, Williams TD, Zwingelstein G, Brichon G, Weber JM. Plasma and muscle phospholipids are involved in the metabolic response to long-distance migration in a shorebird. J Comp Physiol B. 2002;172:409–417. doi: 10.1007/s00360-002-0266-z. [DOI] [PubMed] [Google Scholar]
- Guglielmo CG. Move that fatty acid: Fuel selection and transport in migratory birds and bats. Integr Comp Biol. 2010;50:336–345. doi: 10.1093/icb/icq097. [DOI] [PubMed] [Google Scholar]
- Haggerty C, Hoggard N, Brown DS, Clapham JC, Speakman JR. Intra-specific variation in resting metabolic rate in MF1 mice is not associated with membrane lipid desaturation in the liver. Mech Ageing Dev. 2008;129:129–137. doi: 10.1016/j.mad.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Hayes JP, Garland T. The evolution of endothermy: testing the aerobic capacity model. Evolution. 1995;49:836–847. doi: 10.1111/j.1558-5646.1995.tb02320.x. [DOI] [PubMed] [Google Scholar]
- Hazel JR. Thermal adaptation in biological-membranes - is homeoviscous adaptation the explanation. Annu Rev Physiol. 1995;57:19–42. doi: 10.1146/annurev.ph.57.030195.000315. [DOI] [PubMed] [Google Scholar]
- Helge JW, Ayre KJ, Hulbert AJ, Kiens B, Storlien LH. Regular exercise modulates membrane fatty acid phospholipid composition in rats. J Nutr. 1999;129:1636–1642. doi: 10.1093/jn/129.9.1636. [DOI] [PubMed] [Google Scholar]
- Helge JW, Wu BJ, Willer M, Daugaard JR, Storlien LH, Kiens B. Training affects muscle phospholipid fatty acid composition in humans. J Appl Physiol. 2001;90:670–677. doi: 10.1152/jappl.2001.90.2.670. [DOI] [PubMed] [Google Scholar]
- Henderson ND. Interpreting studies that compare high- and low-selected lines on new characters. Behav Genet. 1989;19:473–502. doi: 10.1007/BF01066250. [DOI] [PubMed] [Google Scholar]
- Hochachka PW, Somero GN. Biochemical Adaptation: Mechanism and Process in Physiological Evolution. Oxford: Oxford University Press; 2002. p. 466. [Google Scholar]
- Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol Revs. 2009;89:193–277. doi: 10.1152/physrev.00037.2007. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ. Life, death and membrane bilayers. J Exp Biol. 2003;206:2303–2311. doi: 10.1242/jeb.00399. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ. Membrane fatty acids as pacemakers of animal metabolism. Lipids. 2007;42:811–819. doi: 10.1007/s11745-007-3058-0. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ, Else PL. Membranes as possible pacemakers of metabolism. J Theor Biol. 1999;199:257–274. doi: 10.1006/jtbi.1999.0955. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ, Else PL. Mechanisms underlying the cost of living in animals. Annu Rev Physiol. 2000;62:207–235. doi: 10.1146/annurev.physiol.62.1.207. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ, Else PL. Membranes and the setting of energy demand. J Exp Biol. 2005;208:1593–1599. doi: 10.1242/jeb.01482. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ, Turner N, Hinde J, Else P, Guderley H. How might you compare mitochondria from different tissues and different species? J Comp Physiol. 2006;176:93–105. doi: 10.1007/s00360-005-0025-z. [DOI] [PubMed] [Google Scholar]
- Ikeda I, Wakamatsu K, Inayoshi A, Imaizumi K, Sugano M, Yazawa K. alpha-Linolenic, eicosapentaenoic and docosahexaenoic acids affect lipid metabolism differently in rats. J Nutr. 1994;124:1898–1906. doi: 10.1093/jn/124.10.1898. [DOI] [PubMed] [Google Scholar]
- Ikeda I, Cha JY, Yanagita T, Nakatani N, Oogami K, Imaizumi K, Yazawa K. Effects of dietary alpha-linolenic, eicosapentaenoic and docosahexaenoic acid on hepatic lipogenesis and beta-oxidation in rats. Biosci Biotechnol Biochem. 1998;62:675–680. doi: 10.1271/bbb.62.675. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh1 M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, Sartor MA, Stringer KA, Jagadish HV, Burant C, Athey B, Omenn GS. Metscape 2 Bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kien CL, Bunn JY, Ugrasbul F. Increasing dietary palmitic acid decreases fat oxidation and daily energy expenditure. Am J Clin Nutr. 2005;82:320–326. doi: 10.1093/ajcn.82.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch LG, Britton SL. Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol Genomics. 2001;5:45–52. doi: 10.1152/physiolgenomics.2001.5.1.45. [DOI] [PubMed] [Google Scholar]
- Kogteva GS, Bezuglov VV. Unsaturated fatty acids as endogenous bioregulators. Biochemistry-Moscow. 1998;63:4–12. [PubMed] [Google Scholar]
- Konarzewski M, Diamond J. Evolution of basal metabolic rate and organ masses in laboratory mice. Evolution. 1995;49:1239–1248. doi: 10.1111/j.1558-5646.1995.tb04450.x. [DOI] [PubMed] [Google Scholar]
- Konarzewski M, Książek A. Determinants of intra-specific variation in basal metabolic rate. J Comp Physiol B. 2012;183:27–41. doi: 10.1007/s00360-012-0698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs H. Body size and tissue respiration. Biochim Biophys Acta. 1950;4:249–269. doi: 10.1016/0006-3002(50)90032-1. [DOI] [PubMed] [Google Scholar]
- Le Belle JE, Harris NG, Williams SR, Bhakoo KK. A comparison of cell and tissue extraction techniques using high-resolution 1H-NMR spectroscopy. NMR Biomed. 2002;15:37–44. doi: 10.1002/nbm.740. [DOI] [PubMed] [Google Scholar]
- Lovegrove BG. The zoogeography of mammalian basal metabolic rate. Am Nat. 2000;156:201–219. doi: 10.1086/303383. [DOI] [PubMed] [Google Scholar]
- Lovegrove BG. The influence of climate on the basal metabolic rate of small mammals: A slow-fast metabolic continuum. J Comp Physiol B. 2003;173:87–112. doi: 10.1007/s00360-002-0309-5. [DOI] [PubMed] [Google Scholar]
- Martin AW, Fuhrman FA. The relationship between summated tissue respiration and metabolic rate in the mouse and dog. Physiol Zool. 1955;28:18–34. [Google Scholar]
- Mathes WF, Nehrenberg DL, Gordon R, Hua K, Garland T, Jr, Pomp D. Dopaminergic dysregulation in mice selectively bred for excessive exercise or obesity. Beh Brain Res. 2010;210:155–163. doi: 10.1016/j.bbr.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manco M, Mingrone G, Greco AV, Capristo E, Gniuli D, De Gaetano A, Gasbarrini G. Insulin resistance directly correlates with increased saturated fatty acids in skeletal muscle triglycerides. Metabolism. 2000;49:220–224. doi: 10.1016/s0026-0495(00)91377-5. [DOI] [PubMed] [Google Scholar]
- McNab BK. The energetics of reproduction in endotherms and its implication for their conservation. Integr Comp Biol. 2006;46:1159–1168. doi: 10.1093/icb/icl016. [DOI] [PubMed] [Google Scholar]
- Mitchell TW, Turner N, Hulbert AJ, Else PL, Hawley JA, Lee JS, Bruce CR, Blanksby SJ. Exercise alters the profile of phospholipid molecular species in rat skeletal muscle. J Appl Physiol. 2004;97:1823–1829. doi: 10.1152/japplphysiol.00344.2004. [DOI] [PubMed] [Google Scholar]
- Nagahuedi S, Popesku JT, Trudeau VL, Weber JM. Mimicking the natural doping of migrant sandpipers in sedentary quails: effects of dietary n-3 fatty acids on muscle membranes and PPAR expression. J Exp Biol. 2009;212:1106–1114. doi: 10.1242/jeb.027888. [DOI] [PubMed] [Google Scholar]
- Nikolaidis MG, Petridou A, Matsakas A, Schulz T, Michna H, Mougios V. Effect of chronic wheel running on the fatty acid composition of phospholipids and triacylglycerols in rat serum, skeletal muscle and heart. Acta Physiol Scand. 2004;181:199–208. doi: 10.1111/j.1365-201X.2004.01277.x. [DOI] [PubMed] [Google Scholar]
- Petridou A, Nikolaidis MG, Matsakas A, Schulz T, Michna H, Mougios V. Effect of exercise training on the fatty acid composition of lipid classes in rat liver, skeletal muscle, and adipose tissue. Eur J Appl Physiol. 2005;94:84–92. doi: 10.1007/s00421-004-1294-z. [DOI] [PubMed] [Google Scholar]
- Raichlen DA, Gordon AD, Muchlinski MN, Snodgrass JJ. Causes and significance of variation in mammalian basal metabolism. J Comp Physiol B. 2010;180:301–311. doi: 10.1007/s00360-009-0399-4. [DOI] [PubMed] [Google Scholar]
- Ranallo RF, Rhodes EC. Lipid metabolism during exercise. Sports Med. 1998;26:29–42. doi: 10.2165/00007256-199826010-00003. [DOI] [PubMed] [Google Scholar]
- Rezende EL, Bozinovic F, Garland T. Climatic adaptation and the evolution of basal and maximum rates of metabolism in rodents. Evolution. 2004;58:1361–1374. doi: 10.1111/j.0014-3820.2004.tb01714.x. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Gammie SC, Garland T., Jr Neurobiology of mice selected for high voluntary wheel-running activity. Integr Comp Biol. 2005;45:438–455. doi: 10.1093/icb/45.3.438. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape User Manual. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;11:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinensky M. Homeoviscous adaptation - homeostatic process that regulates viscosity of membrane lipids in Escherichia coli. Proc Natl Acad Sci USA. 1974;71:522–525. doi: 10.1073/pnas.71.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR. The physiological costs of reproduction in small mammals. Phil Trans R Soc B. 2008;363:375–398. doi: 10.1098/rstb.2007.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow JG, Carter PA, Garland T. Artificial selection for increased wheel-running behavior in house mice. Behav Genet. 1998;28:227–237. doi: 10.1023/a:1021479331779. [DOI] [PubMed] [Google Scholar]
- Swallow JG, Hayes JP, Koteja P, Garland T., Jr . Selection experiments and experimental evolution of performance and physiology. In: Garland T Jr, Rose MR, editors. Experimental evolution: concepts, methods, and applications of selection experiments. Berkeley, California: University of California Press; 2009. pp. 301–351. [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B. 2002;64:479–498. [Google Scholar]
- Storlien LH, Jenkins AB, Chisholm DJ, Pascoe WS, Khouri S, Kraegen KW. Influence of dietary fat composition on development of insulin resistance in rats. Relationship to muscle triglyceride and omega-3 fatty acids in muscle phospholipid. Diabetes. 1991;40:280–289. doi: 10.2337/diab.40.2.280. [DOI] [PubMed] [Google Scholar]
- Storlien LH, Pan DA, Kriketos AD. Skeletal muscle membrane lipids and insulin resistance. Lipids. 1996;31:S261–S265. doi: 10.1007/BF02637087. [DOI] [PubMed] [Google Scholar]
- Talanian JL, Galloway SDR, Heigenhauser GJF, Bonen A, Spriet LL. Two weeks of high-intensity aerobic interval training increases the capacity for fat oxidation during exercise in women. J Appl Physiol. 2007;102:1439–1447. doi: 10.1152/japplphysiol.01098.2006. [DOI] [PubMed] [Google Scholar]
- Templeman NM, Schutz H, Garland T, Jr, McClelland GB. Do mice bred selectively for high locomotor activity have a greater reliance on lipids to power submaximal aerobic exercise? Am J Physiol Reg Integr Comp Physiol. 2012;303:R101–R111. doi: 10.1152/ajpregu.00511.2011. [DOI] [PubMed] [Google Scholar]
- Trebaticka L, Ketola T, Klemme I, Eccard JA, Ylonen H. Is reproduction really costly? Energy metabolism of bank vole (Clethrionomys glareolus) females through the reproductive cycle. Ecoscience. 2007;14:306–313. [Google Scholar]
- Valencak TG, Ruf T. N-3 polyunsaturated fatty acids impair lifespan but have no role for metabolism. Aging Cell. 2007;6:15–25. doi: 10.1111/j.1474-9726.2006.00257.x. [DOI] [PubMed] [Google Scholar]
- Vessby B, Tengblad S, Lithell H. Insulin sensitivity is related to the fatty acid composition of serum lipids and skeletal muscle phospholipids in 70-year-old men. Diabetologia. 1994;37:1044–1050. doi: 10.1007/BF00400468. [DOI] [PubMed] [Google Scholar]
- Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ying Z, Bosy-Westphal A, Zhang J, Heller M, Later W, Heymsfield SB, Müller MJ. Evaluation of specific metabolic rates of major organs and tissues: comparison between nonobese and obese women. Obesity. 2012;1:95–100. doi: 10.1038/oby.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters RP, Renner KJ, Pringlea RB, Summers CH, Britton SL, Koch LG, Swallow JG. Selection for aerobic capacity affects corticosterone, monoamines and wheel-running activity. Physiol Behav. 2008;93:1044–1054. doi: 10.1016/j.physbeh.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel R, Hoppeler H. Exercise-induced maximal metabolic rate scales with muscle aerobic capacity. J Exp Biol. 2005;208:1635–1644. doi: 10.1242/jeb.01548. [DOI] [PubMed] [Google Scholar]
- Weibel ER, Bacigalupe LD, Schmitt B, Hoppeler H. Allometric scaling of maximal metabolic rate in mammals: muscle aerobic capacity as determinant factor. Resp Physiol Neurobiol. 2004;140:115–132. doi: 10.1016/j.resp.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Wone B, Sears MW, Labocha MK, Donovan ER, Hayes JP. Genetic variances and covariances of aerobic metabolic rates in laboratory mice. Proc Roy Soc B-Biol Sci. 2009;276:3695–3704. doi: 10.1098/rspb.2009.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wone B, Donovan ER, Hayes JP. Metabolomics of aerobic metabolism in mice selected for increased maximal metabolic rate. Comp Biochem Physiol Part-D. 2011;6:399–405. doi: 10.1016/j.cbd.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.