Abstract
Frataxin is a conserved mitochondrial protein deficient in patients with Friedreich’s ataxia. Frataxin has been implicated in control of iron homoeostasis and Fe–S cluster assembly. In yeast or human mitochondria, frataxin interacts with components of the Fe–S cluster synthesis machinery, including the cysteine desulfurase Nfs1, accessory protein Isd11 and scaffold protein Isu. In the present paper, we report that a single amino acid substitution (methionine to isoleucine) at position 107 in the mature form of Isu1 restored many deficient functions in Δyfh1 or frataxin-depleted yeast cells. Iron homoeostasis was improved such that soluble/usable mitochondrial iron was increased and accumulation of insoluble/non-usable iron within mitochondria was largely prevented. Cytochromes were returned to normal and haem synthesis was restored. In mitochondria carrying the mutant Isu1 and no frataxin, Fe–S cluster enzyme activities were improved. The efficiency of newFe–S cluster synthesis in isolated mitochondria was markedly increased compared with frataxin-negative cells, although the response to added iron was minimal. The M107I substitution in the highly conserved Isu scaffold protein is typically found in bacterial orthologues, suggesting that a unique feature of the bacterial Fe–S cluster machinery may be involved. The mechanism by which the mutant Isu bypasses the absence of frataxin remains to be determined, but could be related to direct effects on Fe–S cluster assembly and/or indirect effects on mitochondrial iron availability.
Keywords: frataxin, haem, iron, iron–sulfur cluster (Fe–S cluster), mitochondrion
INTRODUCTION
Iron is an essential nutrient for virtually all organisms because many important proteins require iron as a constituent of haem and Fe–S cofactors. Iron is toxic when present in excess, capable of reacting with oxygen to generate superoxide or hydroxyl radicals that damage proteins, lipids and DNA [1]. Therefore cells have evolved mechanisms for regulating the uptake, distribution and utilization of iron. In eukaryotic cells, mitochondria play a special role in iron homoeostasis [2]. Mitochondria contain large numbers of haem and Fe–S proteins in membrane-bound complexes that mediate cellular respiration or in matrix proteins of the tricarboxylic acid cycle (e.g. aconitase). Furthermore, synthesis of these cofactors takes place in mitochondria. Haem is made by insertion of iron into protoporphyrin IX by the enzyme ferrochelatase, which is found exclusively in mitochondria. Fe–S clusters are made in mitochondria by a catalysed process involving more than 20 proteins in mitochondria [3]. Iron uptake in mitochondria is controlled in the physiological setting such that iron is assimilated for biogenesis of iron proteins and excess iron does not accumulate. In some pathological settings, this homoeostasis is perturbed. For example, in cells lacking the mitochondrial protein frataxin, Fe–S cluster proteins are deficient and iron accumulates in mitochondria, producing cellular and organellar damage [2].
Frataxin is a conserved mitochondrial protein originally identified by its connection to Friedreich’s ataxia, an inherited degenerative disease affecting particular types of neurons, cardiac myocytes and pancreatic islet cells [4]. The affected tissues in individuals with this disease exhibit frataxin deficiency. The cellular phenotypes of the disease include iron homoeostatic abnormalities characterized by iron accumulation in mitochondria, Fe–S cluster deficiencies and haem protein deficiencies [5]. The principal role of frataxin has been a topic of extensive study and controversy, and primary and secondary effects of frataxin deficiency have been difficult to distinguish [2]. Recent evidence supports a critical role of frataxin in Fe–S cluster biogenesis in mitochondria. Fe–S cluster biogenesis can be broadly understood in terms of two phases: the formation of Fe–S cluster intermediates on the scaffold protein Isu, and the transfer of the intermediates to apoprotein recipients [6]. The formation of the intermediates requires a source of sulfur, which is provided by the cysteine desulfurase Nfs1. In eukaryotes, Nfs1 interacts with an essential small accessory protein Isd11. The Nfs1–Isd11 enzyme acts on the amino acid substrate cysteine, generating a persulfide and transferring sulfane sulfur to a critical cysteine residue of Isu. Iron must be provided at this stage, and frataxin may play a crucial role in this process. Frataxin appears to interact with iron [7] and also exhibits iron-dependent protein interactions with other components of the Fe–S cluster assembly machinery [8]. Electrons are also required, and these are provided by Arh1 and Yah1, a reductase and ferredoxin couple. For the transfer phase, a specialized Hsp70 (heat-shock protein 70) chaperone Ssq1 and co-chaperone Jac1 are involved, and they interact directly with the client protein Isu, thereby facilitating transfer of the Fe–S cluster intermediate to apoprotein recipients. Other components such as a glutaredoxin, Grx5, are also involved [9].
The role of frataxin in the first phase of Fe–S cluster assembly (formation of the Fe–S cluster intermediate on Isu) was initially shown by iron-labelling experiments in yeast cells. Frataxin-depleted cells failed to form the Fe–S cluster intermediate on Isu [8]. More recently a multiprotein complex has been described, consisting of Nfs1, Isd11, frataxin and Isu [10–12]. When all four components were present together in the complex, a large enhancement of cysteine desulfurase activity was observed [11]. The association of cysteine desulfurase with frataxin and the scaffold Isu is consistent with the functions of these proteins during formation of Fe–S cluster intermediates on Isu. In bacteria such as Escherichia coli, the Fe–S cluster assembly components are conserved and the frataxin (CyaY) deletion mutant exhibits very minimal phenotypes, suggesting a lesser role of frataxin in this organism [13]. Furthermore, although the bacterial cysteine desulfurase, scaffold and frataxin homologue are associated in a protein complex, Isd11 is not present. Another difference is that, unlike the human complex, the bacterial complex appears to inhibit rather than promote cysteine desulfurase activity [14].
We previously described the occurrence of spontaneous suppressor mutants of frataxin-deleted (Δyfh1) yeast strains [15]. These mutants recoveredmany of the functions that were deficient in Δyfh1 cells. In the present paper, we report that a single amino acid substitution in the scaffold protein Isu1 conferred most of the frataxin-bypassing activity. The substitution altered a conserved amino acid in Isu1 to one that is found exclusively in bacterial scaffolds, suggesting that the amino acid change might be mimicking a feature of the bacterial Fe–S cluster machinery.
EXPERIMENTAL
Spontaneously occurring mutants of Δyfh1
The frataxin-deletion strain of Saccharomyces cerevisiae, 5Cyfh1 (MATα Δyfh1::HIS3 ade2-1 leu2-3, 112 trp1-63 ura3-52) (strain no. 52-18), was derived from a cross of the parents CM3260 (strain no. 41–46) and Δyfh1 [strain no. 50–67; a gift from Jerry Kaplan (Department of Pathology, University of Utah, Salt Lake City, UT, U.S.A.)]. 5Cyfh1 was grown in rich YPAD medium (1% yeast extract, 2% peptone, 2% d-glucose and 100 mg/l adenine), and 1 ml of stationary phase culture was poured on an agar plate containing rich glycerol-based medium (same composition as above except glycerol was used in place of glucose) and allowed to dry. After 1 week, papillae appeared on top of a lawn of non-growing cells, and independent colonies called Sup3, Sup4 and Sup10 were picked with a fine needle. The suppressors ‘bred true’ in that they grew more rapidly than the background cells if re-inoculated under similar growth conditions. In crossing with the deletion of the opposite mating type, 2Dyfh1 (MATa Δyfh1::HIS3 ade2-1 leu2-3, 112, trp1-63 FRE1-HIS3::URA3) (strain no. 52–24), the suppressor mutants behaved as genetic dominants. Following sporulation of the diploid and meiosis, the enhanced growth characteristics of the suppressor phenotype could be recovered in the tetrads in a 2+ :2− pattern, suggesting that a single gene was involved [15]. In other genetic backgrounds (W303 and YPH499), similar effects were discerned, i.e. slow growth and respiratory deficiency of the knockout and appearance of papillae on glycerol plates after prolonged incubation [15].
Plasmid construction
Centromere-based TRP1-marked plasmids were constructed in the YCplac22 backbone as follows: YCplac22-YFH1 was made by introducing the YFH1 HindIII genomic fragment into the corresponding vector sites. YCplac22-ISU1 contains the 700 bp upstream sequence of ISU1 introduced into EagI/NdeI sites, followed by the open reading frame between NdeI and XhoI, followed by a stop codon and 200 base pairs of 3′ untranslated sequence between BamHI and SacI. Amino acid 107 of the mature protein was changed from methionine to isoleucine by site-directed mutagenesis in this plasmid to generate YCplac22-ISU1-M107I. Other point mutations were introduced in this plasmid, changing Met107 to valine, threonine, proline, histidine or glycine. The amino acid Asp37 was changed to alanine by site-directed mutagenesis, generating the plasmid YCplac22-ISU1-D37A.
Strains and media
A shuffle strain was constructed in the YPH499 background in which YFH1 was deleted from the genome and covered by a YFH1-expressing plasmid [MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 Δyfh1::HIS3 (pGal-Yfh1, URA3)] (no. 70-31). This strain was transformed with various TRP1-marked plasmids, and the covering plasmid was removed by counter selection in the presence of 5-fluoro-orotic acid under anaerobic conditions. Growth characteristics of the transformants were then evaluated aerobically. A promoter-swap strain was constructed in which the coding region of the YFH1 gene in YPH499 was placed under control of GAL1, using the HIS3MX6-PGAL1 cassette [16], creating strain Gal-Yfh1 (strain no. 99-51). This strain was transformed with YCplac22, YCplac22-ISU1-M107I or YCplac22-YFH1, and, after selection for tryptophan prototrophy in the presence of galactose, the promoter was turned off by growth in raffinose-based medium (6.7 g/l yeast nitrogen base without amino acids, 2 g/l complete medium supplements minus tryptophan, and 2% raffinose). As described elsewhere [17], iron-buffered medium was used to evaluate the growth of some strains. The composition was 50 mM Mes buffer, pH 6.5, standard defined medium, 1 mM ferrous iron chelator ferrozine and various concentrations of ferrous ammonium sulfate (0, 25, 100 and 350 µM). The unopposed activity of the chelator produces iron starvation, whereas iron addition to the chelator results in various amounts of soluble iron. Addition of 350 µM ferrous iron completely reverses the chelator effect.
Candidate gene sequencing
The open reading frames of NFS1, ISD11, ISU1 and ISU2 were amplified by yeast colony PCR from fresh patches of different yeast strains using a 1:1 mixture of Pfu and Taq polymerases with addition of 2.5 mM MgCl2. The products were subcloned using the StrataClone PCR cloning kit (Agilent Technologies), and two clones of each were subjected to DNA sequencing.
Biochemical assays
In order to measure new haem synthesis in isolated mitochondria, radioactive iron (1 µM [55Fe]ferrous ascorbate, 100 mCi/mg; CNL Scientific) and PPO (protoporphyrinogen, 1 µM final concentration) were added. The samples were incubated at 30°C for 5 min, acidified with 0.2 M hydrochloric acid, and extracted with methyl ethyl ketone followed by counting of the organic phase. PPO was prepared from protoporphyrin IX by reduction with sodium amalgam [18]. Low-temperature spectra (−191°C) of whole cells were recorded [19]. For assessment of new Fe–S cluster synthesis, mitochondria were incubated with [35S]cysteine and nucleotides, with or without 50 µM ferrous ascorbate. The radiolabelled Fe–S cluster of endogenous aconitase was visualized by native gel electrophoresis and autoradiography [17].
Iron labelling and cell fractionation
Yeast cells were radiolabelled during growth by addition of trace amounts of radioactive iron (20 or 50 nM [55Fe]ferrous ascorbate) to the cultures. Unincorporated iron was removed by washing, and total cellular iron was measured by scintillation counting of the entire lysate following spheroplasting and douncing. The PMS (post-mitochondrial supernatant) fraction was the supernatant remaining following differential centrifugation to obtain the mitochondrial pellet. The mitochondrial pellet was washed in isotonic buffer (50 mM Tris/HCl, pH 7.5, 0.6 M sorbitol and 5 mMEDTA) prior to scintillation counting. Purified mitochondria were subjected to sonication in hypotonic buffer (50 mM Tris/HCl, pH 7.5) followed by centrifugation (15000 g for 15 min) to separate the supernatant (‘soluble mito’) and pellet (‘insoluble mito’). Iron in the fractions was quantified by scintillation counting.
RESULTS
Characterization of spontaneously arising suppressors of a Δyfh1 yeast strain
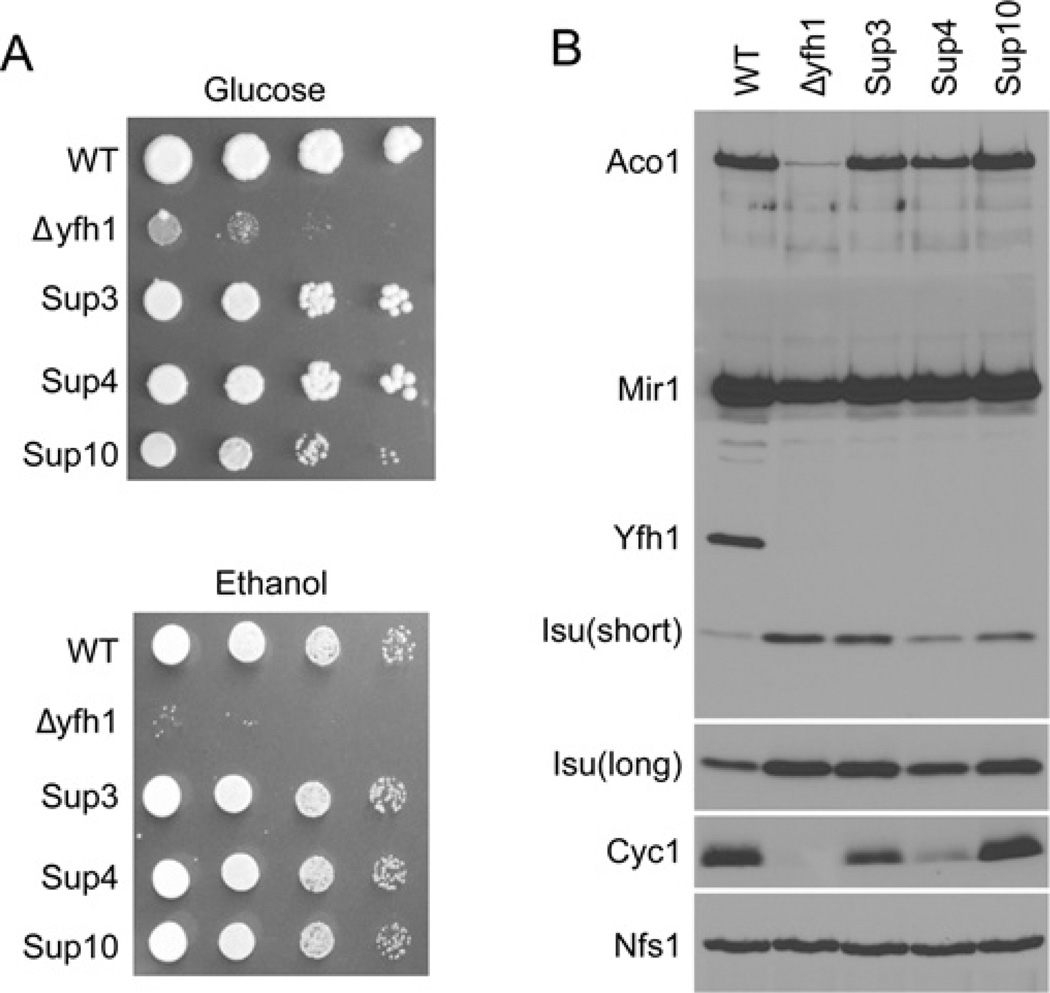
The parental yeast strain, Δyfh1, and Sup3, Sup4 and Sup10 were evaluated for their growth characteristics. The deletion mutant grew very poorly on glucose plates and virtually not at all on ethanol plates, consistent with the known respiratory deficiency of Δyfh1 cells. The suppressors recovered normal growth on glucose or ethanol plates (Figure 1A). Mitochondrial protein composition was examined by immunoblotting. Frataxin protein (Yfh1) was present only in the wild-type parental cells as expected. Aconitase protein was present in the wild-type, virtually absent from Δyfh1 mitochondria consistent with instability of the apoprotein, and recovered in the suppressors. Similarly, aconitase activity was present in the parent, virtually absent from Δyfh1 (2%of normal) and partially recovered in the suppressors (20%, 30% and 100% for Sup3, Sup4 and Sup10 respectively). Cytochrome c was present in the wild-type, not detected in Δyfh1, and recovered partially or completely in the suppressors. Isu was increased in Δyfh1 compared with the wild-type parent as described previously [20]. In the suppressors, Isu abundance was still increased compared with the parent, although to various extents (Figure 1B).
Figure 1. Extragenic suppressors of Δyfh1.
(A) Growth phenotypes. Wild-type (WT; CM3260), Δyfh1 (5Cyfh1), Sup3, Sup4 and Sup10 were serially diluted and spotted on to agar plates containing rich medium with glucose or ethanol. The plates were photographed after 3 days. (B) Protein expression. Mitochondria from the above strains were analysed by Western blotting for aconitase (Aco1), phosphate carrier (Mir1), frataxin (Yfh1), scaffolds (Isu1/2), cytochrome c (Cyc1) and cysteine desulfurase (Nfs1). Isu(short) refers to a short film exposure (15 s) and Isu(long) refers to a long film exposure (2 min). Cells were grown in rich raffinose-based medium until a D600 (attenuance at 600 nm) of 2.0 was reached, and mitochondria were isolated.
Identification of a common extragenic point mutation in Δyfh1 suppressors
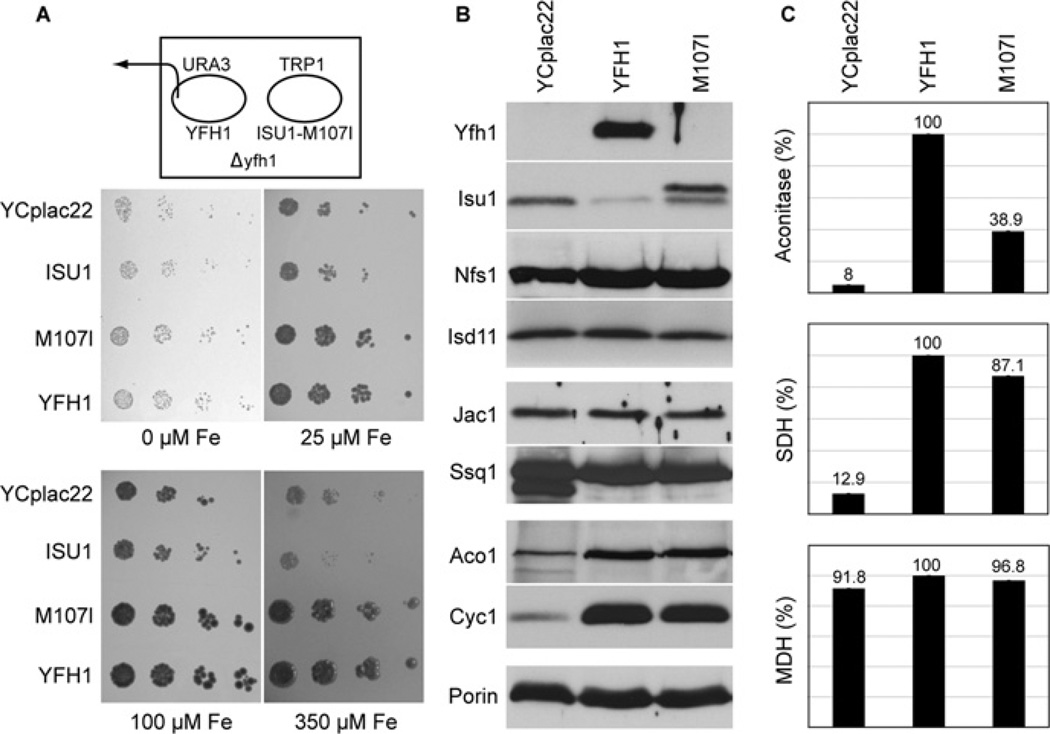
In view of the identification of a protein complex containing Yfh1, Nfs1, Isd11 and Isu [10–12], we considered that the improved phenotypes of the suppressor strains might arise from alterations in Yfh1-interacting proteins. Therefore the corresponding candidate genes were rescued from genomic DNA by PCR and subjected to sequencing. In the suppressor strains, NFS1 exhibited multiple changes compared with the database sequence, but these changes were also present in the wild-type parent and thus were likely to represent polymorphisms. No changes in ISD11 were found. Two redundant genes, ISU1 and ISU2, encode Isu, the scaffold protein for Fe–S cluster assembly in yeast. The ISU2 genes had no sequence alterations. Remarkably, however, the ISU1 genes from Sup3, Sup4 or Sup10 exhibited a single G to A transversion of nucleotide 423 of the full-length open reading frame, resulting in a change of amino acid 141 (amino acid 107 of the mature protein) from methionine to isoleucine. As expected, this ISU1 mutation was not found in the parental strain or in the Δyfh1 strain. In addition, the highly homologous ISU2 gene did not exhibit any mutations in the parental, Δyfh1 or suppressor strains. In order to demonstrate that the ISU1 mutation was responsible for the phenotypic changes observed in the suppressors, the mutation was generated by site-directed mutagenesis in a low-copy-number plasmid containing the complete ISU1 gene. The plasmid was introduced into a Δyfh1 shuffle strain constructed in the YPH499 yeast genetic background, distinct from the genetic background utilized for the original suppressor isolation. The covering YFH1 plasmid was removed from the shuffle strain by 5-fluoro-orotic acid treatment under anaerobic conditions, and the transformants were evaluated further. Markedly enhanced growth of the transformant carrying the mutated ISU1 was observed, compared with Δyfh1 carrying an empty plasmid. Iron-dependent growth was evaluated for Δyfh1 transformed with plasmids carrying no insert, YFH1, unmutated ISU1 or mutant ISU1-M107I (Figure 2A). No growth differences were observed under severe iron starvation (0 µM added iron in the presence of 1 mM ferrozine chelator). Under less-severe iron starvation or iron-replete conditions, the Δyfh1 mutant (YCplac22 transformant) grew slowly, and the unmutated ISU1 afforded no growth enhancement. In contrast, the suppressor (ISU1-M107I transformant) grew like the wild-type (YFH1 transformant) (Figure 2A). The results show that expression of ISU1-M107I from a plasmid in a different Δyfh1 strain and strain background recapitulated the improved growth observed for the original suppressor isolate.
Figure 2. Effects of expression of mutant Isu1 (YCplac22-ISU1-M107I) on frataxin-deleted or frataxin-depleted yeast.
(A) Growth. The YFH1 shuffle strain was transformed with the TRP1 plasmids YCplac22, YCplac22-ISU1, YCplac22-ISU1-M107I or YCplac22-YFH1. The YFH1 covering plasmid carrying URA3 was removed by counterselection on 5-fluoro-orotic acid plates. Transformants were serially diluted and spotted on chelator plates containing 1 mM ferrozine and different amounts of ferrous ammonium sulfate (0 µM, 25 µM, 100 µM and 350 µM). (B) Mitochondrial proteins. Gal-Yfh1 transformants with plasmids YCplac22, YCplac22-YFH1 or YCplac22-ISU1-M107I were cultured in defined raffinose medium lacking tryptophan to repress expression of the chromosomal copy of YFH1. Mitochondria were isolated and immunoblotted with the indicated antibodies. (C) Enzyme activities. Mitochondria were analysed for aconitase, succinate dehydrogenase (SDH) or malate dehydrogenase (MDH) activities. Results shown are means ± S.D. for triplicate assays.
Mitochondrial protein profiles and enzyme activities
Owing to the extreme slow growth and genetic instability of the Δyfh1 mutant, subsequent experiments were performed with a Gal-Yfh1 strain in which the YFH1 gene was placed under the control of the regulated GAL1 promoter. Even under non-inducing conditions, this strain did not exhibit genetic instability, and the slow growth of the deletion strain was mitigated, perhaps due to leaky expression from the GAL1 promoter. The Gal-Yfh1 strain was transformed with several plasmids, and the transformants were cultured in raffinose-based medium to repress genomic Gal-Yfh1 expression. Yfh1 protein was detected only in the ‘wild-type’ (YFH1 transformant) (Figure 2B). Aconitase and cytochrome c, the most abundant Fe–S and haem proteins of mitochondria respectively, were decreased in the Yfh1-depleted mitochondria (YCplac22 transformant) compared with the ‘wild-type’ (YFH1 transformant), probably due to instability of the apoproteins. The levels were completely restored in the suppressor (ISU1-M107I transformant) (Figure 2B). Increased Isu levels have been described in Yfh1 mutants due to both increased transcription and decreased protein turnover [20]. Interestingly, in the suppressor (ISU1-M107I transformant), Isu levels were still abnormal, in contrast with aconitase and cytochrome c. Our antibody does not distinguish between Isu1 and Isu2, and so one or both may be increased. A slower-migrating band reacting with anti-Isu1 antibody was also increased in the ISU1-M107I transformant. This is probably attributable to the plasmid-borne ISU1-M107I, which carries three extra amino acids introduced at the C-terminus for ease of cloning. Activities of the Fe–S cluster proteins aconitase and succinate dehydrogenase were markedly reduced in Yfh1-depleted cells (YCplac22 transformant), and these were partially or completely restored in the suppressor (M107I transformant). Malate dehydrogenase does not contain any Fe–S clusters, and its activity was unaffected in all cases (Figure 2C).
Cytochromes and haem synthesis
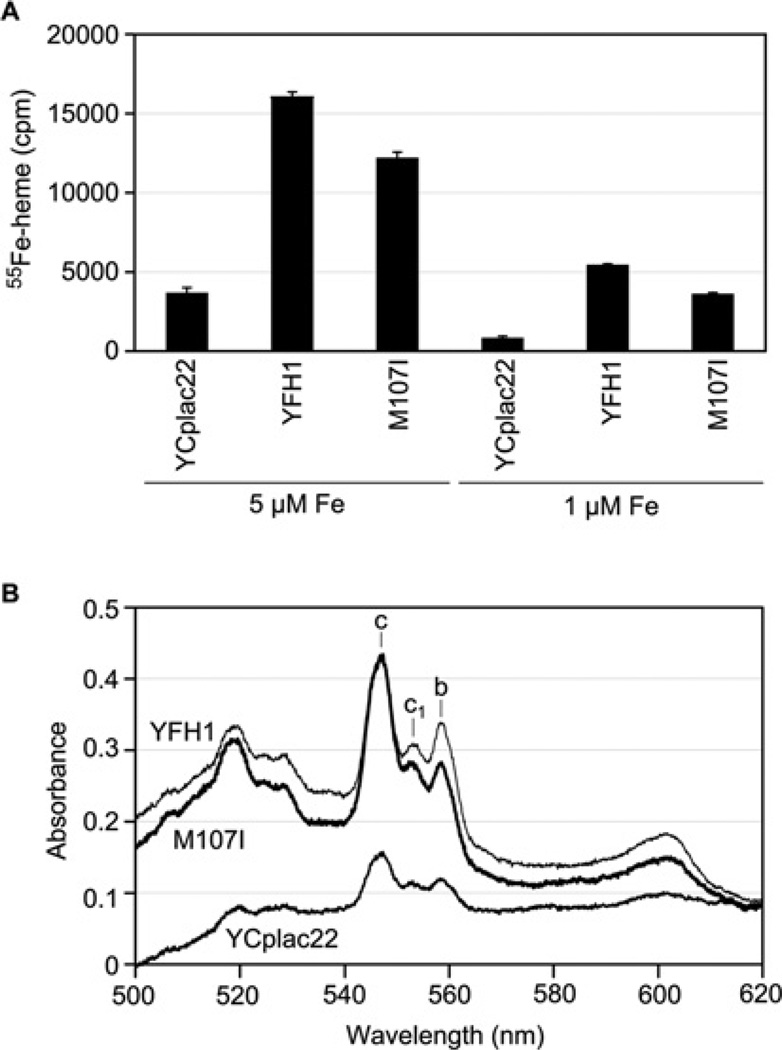
New haem synthesis was assessed in isolated mitochondria supplemented with the porphyrin precursor PPO and radioactive iron (1 or 5 µM [55Fe]ferrous ascorbate). The Yfh1-depleted mitochondria (YCplac22 transformant) synthesized haem slowly, and the ‘wild-type’ (YFH1 transformant) made 4-fold more haem during 5 min. The suppressor (ISU1-M107I transformant) mitochondria also made new radiolabelled haem, with 80% of the wild-type efficiency (Figure 3A). Steady-state levels of haem proteins were evaluated. Low-temperature spectra of whole cells showed deficient cytochromes in the YCplac22 transformant and equivalently restored cytochrome c in the YFH1 and ISU1-M107I transformants, with only slightly smaller c1 and b peaks in the latter (Figure 3B).
Figure 3. Effects of expression of the mutant Isu1 (YCpla22-ISU1-M107I) on haem synthesis and cytochromes in the absence of frataxin.
(A) Haem synthesis. Mitochondria (200 µg of proteins) from Gal-Yfh1 transformants with YCplac22, YCplac22-YFH1 or YCPlac22-ISU1-M107I were evaluated. Haem synthesis was measured by addition of 1 µM PPO and 1 µM or 5 µM [55Fe]ferrous ascorbate, followed by organic extraction of the haem and scintillation counting. Results shown are means ± S.D. for triplicate assays. (B) Cytochrome spectra. Gal-Yfh1 transformants as in (A) were grown in defined raffinose medium lacking tryptophan and shifted to rich raffinose medium for 12 h prior to measurement of low-temperature spectra of whole cells.
Fe–S cluster synthesis
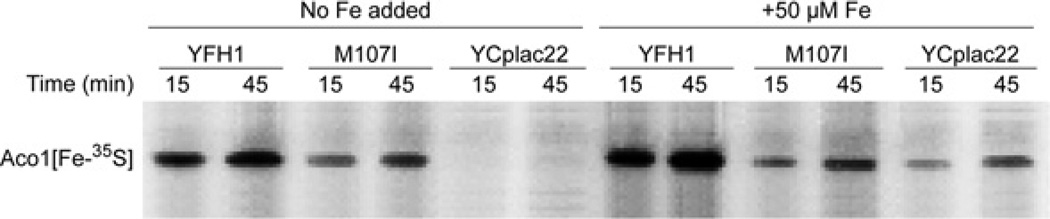
An assay has been developed for examining the synthesis of new Fe–S clusters in isolated mitochondria [17]. If intact mitochondria are incubated with [35S]cysteine, the radiolabelled amino acid is processed by the endogenous Fe–S cluster assembly machinery, and the radiolabelled sulfur is incorporated into newly formed aconitase clusters that can be detected on a native gel. Using this assay, in the frataxin-depleted (YCplac22) mitochondria, no new Fe–S clusters were detected on aconitase. In contrast, in the suppressor (ISU1-M107I transformant), significant restoration of new Fe–S cluster synthesis was detected, consistent with the presence of aconitase activity in these mitochondria. However, the efficiency and rate of formation of new Fe–S clusters were compromised compared with ‘wild-type’ mitochondria (YFH1 transformant). A parallel set of experiments was performed in the presence of iron added during the assay (50 µM iron). Surprisingly, iron addition to the isolated mitochondria resulted in detectable and markedly improved Fe–S cluster synthesis in the frataxin-depleted mitochondria. In contrast, Fe–S cluster synthesis in the mitochondria from the ISU1-M107I transformant was marginally if at all improved by iron addition. The ‘wild-type’ (YFH1 transformant) showed iron-enhanced labelling of aconitase at both the 15 and 45 min time points (Figure 4). In summary, the expression of the mutant ISU1 conferred improved Fe–S cluster assembly to frataxin-depleted mitochondria. The effect was mimicked to some extent by the addition of iron during the assay.
Figure 4. Fe–S cluster synthesis in isolated mitochondria.
Mitochondria from cells grown as described in the legend to Figure 2(B) were incubated with [35S]cysteine for different time periods (15 and 45 min) and new Fe–35S clusters on aconitase were visualized by native gel electrophoresis. In a parallel set of samples, 50 µM ferrous ascorbate was added.
Iron homoeostasis
One of the salient features of the frataxin mutant is its abnormal iron homoeostasis [21]. The mutant phenotype is characterized by constitutively induced cellular iron uptake and diversion of cellular iron to mitochondria [21]. Within mitochondria, iron accumulates as insoluble ferric phosphate nanoparticles [15]. The Gal-Yfh1 transformants were evaluated by addition of a trace amount of radioactive iron to the growth medium followed by cell fractionation and scintillation counting of the fractions. Mitochondrial iron was further assessed by sonication and centrifugation to separate soluble from insoluble iron species. Results confirmed that total cellular and mitochondrial iron were markedly increased in the frataxin-depleted cells (Table 1, compare YCplac22 with YFH1 in two separate experiments). The ratio of soluble iron to insoluble iron was also decreased in these frataxin-depleted cells, reflecting iron aggregation/precipitation in the mutant mitochondria (Table 1). The effect of the ISU1-M107I expression was to reverse to a large extent the features of the frataxin mutant phenotype in relation to iron homoeostasis. The whole-cell iron, mitochondrial iron and mitochondrial iron solubility were all markedly improved in the suppressor (ISU1-M107I transformant) (Table 1, compare M107I with YCplac22 and YFH1).
Table 1. Iron uptake and cellular distribution.
Gal-Yfh1 was transformed with plasmids YCplac22, YCplac22-YFH1 or YCplac22-ISU1-M107I. The transformants were grown in defined raffinose medium lacking tryptophan to maintain plasmid selection while repressing expression of the genomic YFH1. In experiment 1, cells were cultured for 14 h in low-copper medium containing 50 nM [55Fe]ferrous ascorbate and 10 µM unlabelled ferrous ascorbate. Copper sulfate (1 µM) was then added for 3 h. In experiment 2, cells were grown in standard defined medium supplemented with 20 nM [55Fe]ferrous ascorbate and 10 µM ferrous ascorbate for 14 h. Radiolabelled cells were washed and subjected to cell fractionation to obtain PMS and mitochondria (mito) fractions. Mitochondria (mito) were sonicated and centrifuged to separate the supernatant (‘soluble mito’) from the pellet (‘insoluble mito’). Iron in the various fractions was assessed by scintillation counting of the radiolabel. S/I represents the ratio of soluble to insoluble iron in mitochondria.
| (a) Fe55 in cellular fractions (c.p.m. ×103 per 100 D600 units or 2×109 cells) | ||||||
|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | |||||
| Plasmid | YCplac22 | M107I | YFH1 | YCplac22 | M107I | YFH1 |
| Total cell | 640 | 215 | 87 | 324 | 111 | 68 |
| PMS | 173 | 128 | 56 | 219 | 93 | 58 |
| Mito | 194 | 37 | 26 | 114 | 47 | 31 |
| (b) Fe55 in mitochondrial fractions (c.p.m. ×103 per mg of mitochondrial proteins) | ||||||
|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | |||||
| Plasmid | YCplac22 | M107I | YFH1 | YCplac22 | M107I | YFH1 |
| Total mito | 278 | 47 | 31 | 131 | 35 | 25 |
| Soluble mito | 105 | 32 | 23 | 39 | 20 | 15 |
| Insoluble mito | 221 | 20 | 10 | 105 | 18 | 10 |
| Ratio (S/I) | 0.48 | 1.60 | 2.30 | 0.37 | 1.11 | 1.50 |
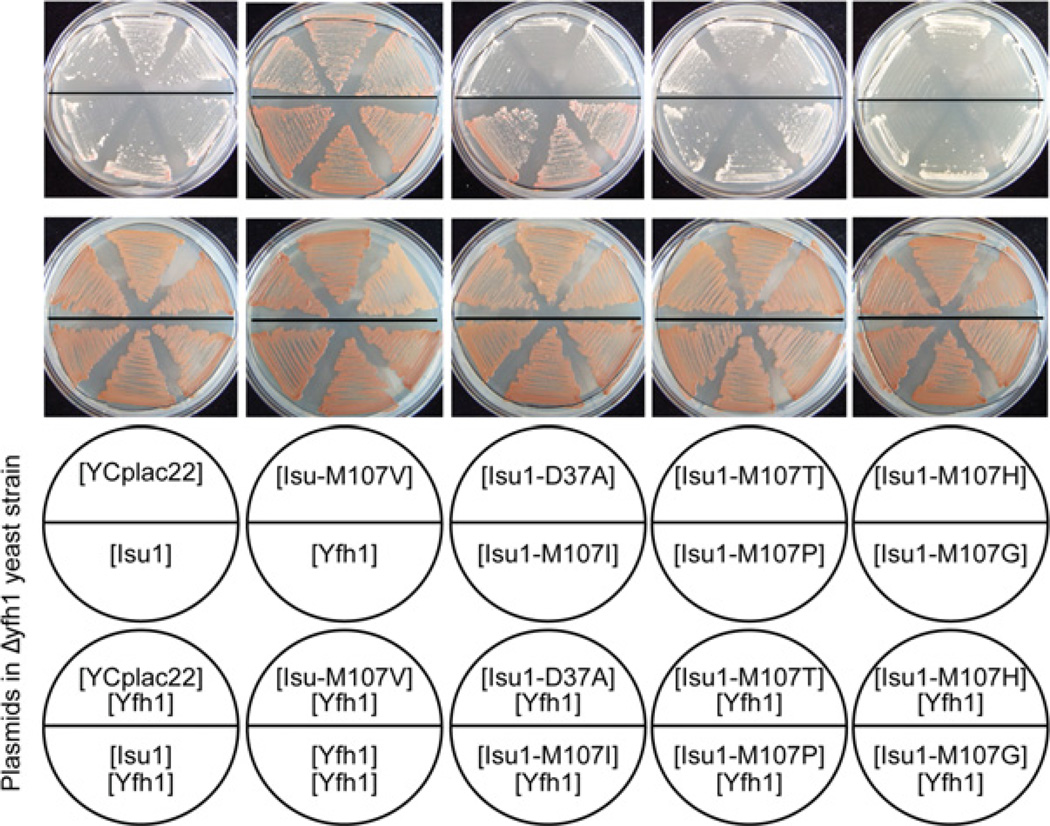
Other substitutions of amino acid 107 of Isu1
Amino acid 107 of Isu1 was changed to various other amino acids by site-directed mutagenesis of YCplac22-ISU1, a centromere-based plasmid carrying the genomic ISU1. These mutated plasmids were reintroduced into the Δyfh1 shuffle strain and treated with 5-fluoro-orotic acid to remove the covering YFH1 plasmid (Figure 5). The red colour from adenine pigment deposition indicates robust growth in this assay. The YCplac22 vector and unmutated Isu1 showed no growth-promoting effect. The plasmids carrying isoleucine and valine substitutions of Isu1 at position 107 were effective in restoring growth and red colour to the Δyfh1 patches, similar to the YFH1-containing plasmid. On the other hand, proline, glycine, threonine or histidine substitutions at position 107 of Isu1 were inactive. The D37A substitution of Isu1 further worsened the defective growth phenotype of Δyfh1. This Isu1 allele has been shown to impede Fe–S cluster synthesis by blocking cluster transfer from Isu scaffolds [22].
Figure 5. Amino acid substitutions of Isu1.
ISU1 on a TRP1-marked plasmid (YCplac22-ISU1) was mutated, changing amino acid 107 of the coding region to valine (Isu1-M107V), isoleucine (Isu1-M107I), threonine (Isu1-M107T), proline (Isu1-M107P), histidine (Isu1-M107H) or glycine (Isu1-M107G). On the same plasmid, amino acid 37 was changed from aspartic acid to alanine (Isu1-D37A). YCplac22 alone and YCplac22-YFH1 were included as controls. All of the above plasmids were transformed into the YFH1 shuffle strain (Δyfh1 covered by a YFH1-URA3 plasmid), and three transformants of each were reselected and streaked on a fresh agar plate (bottom row of plates). Following counterselection against the covering YFH1-URA3 plasmid by growth on 5-fluoro-orotic-acid-containing plates, the transformants were again selected for tryptophan auxotrophy (top row of plates). The red colour is caused by accumulated adenine precursor and reflects robust growth. The results show growth complementation of Δyfh1 by plasmids carrying Yfh1, Isu1-M107V or Isu1-M07I.
DISCUSSION
Frataxin, a conserved mitochondrial protein, has been implicated in control of iron homoeostasis, Fe–S cluster assembly and other iron-related processes. The striking phenotype of frataxin mutant cells is characterized by mitochondrial iron accumulation and deficient iron proteins, both haem and Fe–S [21]. In the present paper we report that in yeast cells lacking frataxin (Δyfh1 or Yfh1-depleted), a single amino acid substitution in the Fe–S cluster assembly scaffold protein Isu1 effectively reversed many of the mutant phenotypes. The cells with the mutated Isu1 and no frataxin were able to grow well, with little apparent difference from the wild-type. The iron-homoeostatic defects of the frataxin-depleted or Δyfh1 cells were mitigated or reversed by the mutant Isu1. Some cellular iron accumulation still occurred, but this was much less than in the original Δyfh1, and diversion of iron to mitochondria was minimal. Iron solubility and bioavailability were improved.
The fact that a single amino acid change in the Fe–S scaffold protein Isu1 was responsible for these changes in the phenotype of the Δyfh1 frataxin-depleted cells suggests that the effect is mediated via changes in Fe–S cluster assembly. Consistent with this idea, in assays with isolated mitochondria, significant Fe–S cluster synthesis was restored in the suppressor (M107I-ISU1 expressing and frataxin-depleted) compared with frataxin-depleted mitochondria alone. The restored activity was still abnormal, however, and exhibited slower kinetics, lower efficiency and less iron responsiveness than in the wild-type. A conundrum that is difficult to resolve is whether the restored Fe–S cluster assembly activity is responsible for the improved iron homoeostasis or vice versa. It is probable that both are true to some extent. Iron overload and mitochondrial iron accumulation have been shown to worsen the Fe–S cluster defect inΔyfh1 cells. Conversely, mitochondrial iron overload itself is thought to arise as a consequence of regulatory signals initiated by deficient Fe–S cluster proteins [23]. The problem with Fe–S cluster assembly in Δyfh1 cells is unlikely to be entirely due to iron toxicity, as shown by experiments in which Mrs3 and Mrs4 iron transporters were deleted. In these experiments, mitochondrial iron accumulation was abrogated, and yet the Fe–S cluster assembly defect persisted [24]. Similarly, in a time course of frataxin depletion in genetically engineered mice, Fe–S cluster deficiency was found to develop before mitochondrial iron accumulation, suggesting an upstream or causal role for Fe–S cluster deficiency [25].
In mammalian cells, knockout or knockdown of frataxin leads to inviability, consistent with the key roles of this protein in iron metabolism and Fe–S cluster assembly [26]. In yeast, the frataxin deletion is viable. However,Δyfh1 strains in various yeast genetic backgrounds are extremely sick and slow growing, exhibiting severe defects in iron metabolism and Fe–S cluster assembly [4,21]. Some strains have been noted to grow significantly better and to harbour Fe–S cluster enzyme activities, but these may have been selected for suppressor mutation(s) by prior growth on non-fermentable substrates [27,28]. The implication of the results of the present study is not that frataxin is superfluous for Fe–S cluster assembly, but that a genetic bypass of frataxin deficiency can occur, mediated by a small alteration in Isu1, the Fe–S cluster scaffold protein.
Eukaryotic compared with prokaryotic Isu scaffolds
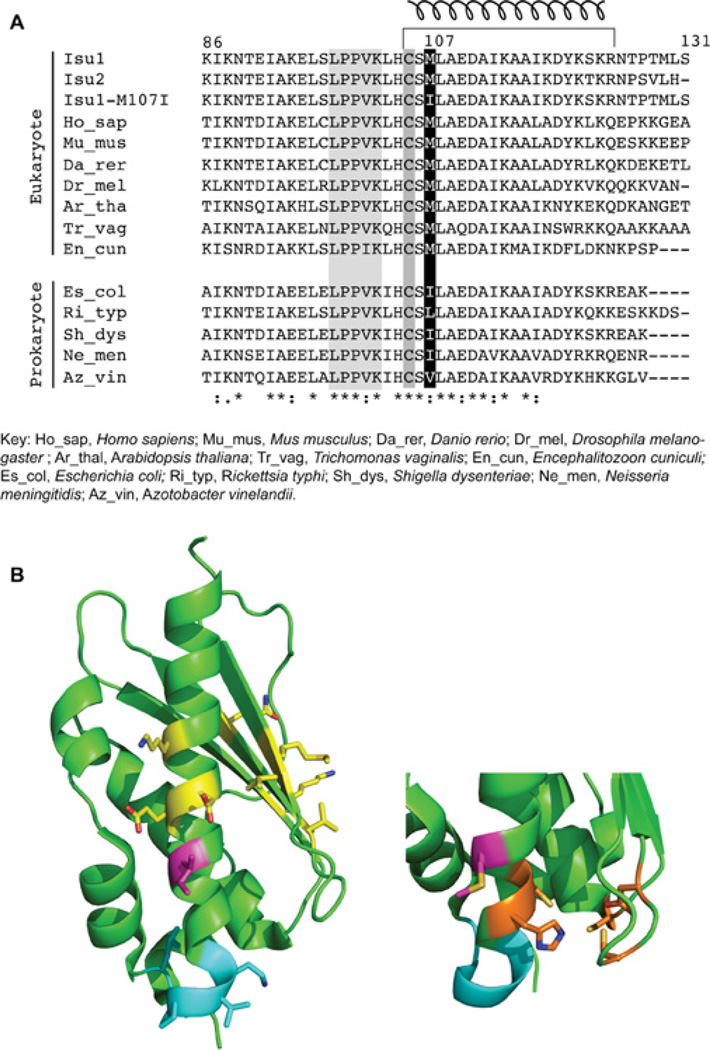
A large number of amino acid sequences for Isu scaffold are available in the databases, and the proteins are extremely similar in both prokaryotes and eukaryotes [29]. The change from methionine to isoleucine in the Isu1 suppressor appears in a highly conserved region of the protein (Figure 6A). Interestingly, the choice of conserved methionine at this position segregates with eukaryotes across different kingdoms and large evolutionary distances. Methionine occurs not only in budding yeast Isu1 and Isu2 paralogues, but also in the human scaffold orthologue ISCU2 at this location. Similarly, methionine occurs in vertebrates (mouse and zebrafish), insects (Drosophila), plants (Arabidopsis) and primitive protists (Trichomonas) [29]. In contrast, in prokaryotes, the amino acid found at this conserved location is almost always a branched-chain amino acid, such as isoleucine, valine or leucine. Examples include Gram-negative bacteria (E. coli), primitive intracellular pathogens (Rickettsia), and nitrogen-fixing bacteria (Azotobacter). Thus the amino acid change in the yeast Isu1 mutant with frataxin-bypassing activity apparently utilizes an amino acid preference of prokaryotic-type proteins (Figure 6A).
Figure 6. Isu orthologues.
(A) Alignment. Amino acid sequences from the C-terminal portions of Isu scaffold proteins from various species were aligned using ClustalW. The substituted amino acid 107 in the mature Isu1 is shaded in black; the cluster-liganding Cys105 and the Hsp70-binding LPPVK motif are shaded in grey. The C-terminal helix is indicated by a corkscrew symbol at the top. (B) Structure. Left-hand panel: the structure of the E. coli IscU [35] is shown with the isoleucine residue (magenta) as found in bacteria and in the yeast Isu1 suppressor. Also shown are the Hsp70-interaction site LPPVK (cyan) and HscB/Jac1-interaction sites (yellow) [32]. Right-hand panel: the bottom portion of the structure is rotated to the left so that the Fe–S cluster-liganding residues can be seen (orange). The bacterial isoleucine residue is replaced with methionine as found in eukaryotes (magenta).
Fe–S cluster assembly in mitochondria of eukaryotes such as yeast is similar to that in prokaryotes such as E. coli. However, there are additional eukaryote-specific features, such as the requirement for Isd11, an essential protein associated with Nfs1. In prokaryotes the frataxin-deletion phenotype is very mild, indicating that Fe–S cluster synthesis is less frataxin-dependent [13]. It is possible that the methionine-to-isoleucine substitution in yeast Isu1 alters Fe–S cluster synthesis to a more bacterial type, thereby relieving the dependence on frataxin.
Mechanism of frataxin bypass
Isu structures from different species are highly similar, exhibiting α + β features with cluster-liganding amino acids somewhat buried in the protein interior [30]. The amino acid substitution in the M107I Isu1 is predicted to be exposed on the surface of a prominent C-terminal α-helix (Figure 6B). How might the substitution in Isu1 enhance Fe–S cluster synthesis in the absence of frataxin? The methionine-to-isoleucine substitution in Isu1 is unlikely to produce a major conformational change in the protein. Interestingly, the position in question is separated by two amino acids in the primary sequence from a critical Fe–S cluster-liganding cysteine residue. Thus the altered Isu might facilitate increased exposure of this cysteine residue and improved sulfur transfer from Nfs1/Isd11 to Isu. In an alternative scenario, the iron-donation pathway could be targeted. The physiological iron source for forming the scaffold Fe–S may involve iron donation by frataxin [31], and in the absence of frataxin, the mutant form of Isu1 might recruit an alternative iron donor. Finally, the Isu1 amino acid substitution might work by enhancing transfer of the scaffold cluster intermediate to recipient apoproteins. Residues introduced at position 107 designed to disrupt helices (glycine or proline), thereby exposing the critical Cys105, had little effect. The lack of effect of threonine and the positive effect of the isosteric valine suggest that hydrophobicity is important for the suppression effect. The M107I (or M107V) substitution might enhance Isu1 interaction with Jac1 by improving a hydrophobic interface. A similar hydrophobic interface between HscB (heat-shock cognate B) and the IscU helix has been described [32]. The Isu interaction with Hsp70 has been mapped to a conserved peptide LPPVK [33], very close to the Isu1 mutation in the suppressor (Figure 6B, cyan loop in structure). Thus the footprint of the chaperone Hsp70 and co-chaperone Jac1 on Isu may overlap the area of the suppressor substitution (Figure 6B). Further work will be required to discriminate between these possible mechanisms.
Therapeutic potential
Frataxin deficiency in patients with Friedreich’s ataxia causes a progressive degenerative disease affecting dorsal root ganglia, heartmyocytes and pancreatic β -cells. Emergent therapies seek to increase frataxin levels or to decrease cellular toxicity by chelating iron or mitigating oxidative stress [34]. Data from the present study raise the possibility that the genetically dominant effect of the Isu suppressor mutation observed in yeast may provide a therapeutic avenue for treatment of the human disease. A drug that mimics the effects of the mutant Isu might enhance Fe–S cluster synthesis in the absence of frataxin.
ACKNOWLEDGEMENTS
We acknowledge Dr Robert Wilson (Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia, PA,U.S.A.),Dr Timothy Stemmler (Department of Biochemistry and Molecular Biology, Wayne State University School of Medicine, Detroit, MI, U.S.A.) and Dr David Barondeau (Department of Chemistry, Texas A&M University, College Station, TX, U.S.A.) for insightful conversations.
FUNDING
This work was supported by the National Institutes of Health [grant number R37DK053953 (to A.D.)]; the National Institute on Aging [grant number AG030504 (to D.P.)]; and the American Heart Association [grant number 09GRNT2260364 (to D.P.)].
Abbreviations used
- HscB
heat-shock cognate B
- Hsp70
heat-shock protein 70
- PMS
post-mitochondrial supernatant
- PPO
protoporphyrinogen
Footnotes
AUTHOR CONTRIBUTION
Heeyong Yoon and Ramesh Golla performed many of the experiments. Emmanuel Lesuisse isolated the original suppressors, recorded cytochrome spectra and contributed key ideas. Jayashree Pain performed the Fe–S cluster loading experiments. Jason Donald researched and analysed protein structural data. Elise Lyver sequenced the mutant gene(s) and constructed plasmids and strains. Debkumar Pain contributed ideas, designed key experiments, executed experiments and participated in writing the paper. Andrew Dancis isolated suppressors, supervised experiments and wrote the paper.
REFERENCES
- 1.Kaplan CD, Kaplan J. Iron acquisition and transcriptional regulation. Chem. Rev. 2009;109:4536–4552. doi: 10.1021/cr9001676. [DOI] [PubMed] [Google Scholar]
- 2.Richardson DR, Lane DJ, Becker EM, Huang ML, Whitnall M, Rahmanto YS, Sheftel AD, Ponka P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. U.SA. 2010;107:10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muhlenhoff U, Lill R. Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta. 2000;1459:370–382. doi: 10.1016/s0005-2728(00)00174-2. [DOI] [PubMed] [Google Scholar]
- 4.Wilson RB, Roof DM. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat. Genet. 1997;16:352–357. doi: 10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 5.Ye H, Rouault TA. Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry. 2010;49:4945–4956. doi: 10.1021/bi1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 7.He Y, Alam SL, Proteasa SV, Zhang Y, Lesuisse E, Dancis A, Stemmler TL. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43:16254–16262. doi: 10.1021/bi0488193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber J, Muhlenhoff U, Lill R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003;4:906–911. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Craig EA. Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu. J. Biol. Chem. 2008;283:12674–12679. doi: 10.1074/jbc.M800399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai CL, Barondeau DP. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry. 2010;49:9132–9139. doi: 10.1021/bi1013062. [DOI] [PubMed] [Google Scholar]
- 12.Schmucker S, Martelli A, Collin F, Page A, Wittenhofer-Donze M, Reutenauer L, Puccio H. Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS ONE. 2011;26:e16199. doi: 10.1371/journal.pone.0016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pohl T, Walter J, Stolpe S, Soufo JH, Grauman PL, Friedrich T. Effects of the deletion of the Escherichia coli frataxin homologue CyaY on the respiratory NADH:ubiquinone oxidoreductase. BMC Biochem. 2007;8:13. doi: 10.1186/1471-2091-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adinolfi S, Iannuzzi C, Prischi F, Pastore C, Iametti S, Martin SR, Bonomi F, Pastore A. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat. Struct. Mol. Biol. 2009;16:390–396. doi: 10.1038/nsmb.1579. [DOI] [PubMed] [Google Scholar]
- 15.Lesuisse E, Santos R, Matzanke BF, Knight SA, Camadro JM, Dancis A. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1) Hum. Mol. Genet. 2003;12:879–889. doi: 10.1093/hmg/ddg096. [DOI] [PubMed] [Google Scholar]
- 16.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Amutha B, Gordon DM, Dancis A, Pain D. Nucleotide-dependent iron-sulfur cluster biogenesis of endogenous and imported apoproteins in isolated intact mitochondria. Methods Enzymol. 2009;456:247–266. doi: 10.1016/S0076-6879(08)04414-5. [DOI] [PubMed] [Google Scholar]
- 18.Camadro JM, Chambon H, Jolles J, Labbe P. Purification and properties of coproporphyrinogen oxidase from the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 1986;156:579–587. doi: 10.1111/j.1432-1033.1986.tb09617.x. [DOI] [PubMed] [Google Scholar]
- 19.Lesuisse E, Labbe P. Reductive and non-reductive mechanisms of iron assimilation by the yeast Saccharomyces cerevisiae. J. Gen. Microbiol. 1989;135:257–263. doi: 10.1099/00221287-135-2-257. [DOI] [PubMed] [Google Scholar]
- 20.Andrew AJ, Song JY, Schilke B, Craig EA. Posttranslational regulation of the scaffold for Fe-S cluster biogenesis, Isu. Mol. Biol. Cell. 2008;19:5259–5266. doi: 10.1091/mbc.E08-06-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 22.Raulfs EC, O’Carroll IP, Dos Santos PC, Unciuleac MC, Dean DR. In vivo iron-sulfur cluster formation. Proc. Natl. Acad. Sci. U.SA. 2008;105:8591–8596. doi: 10.1073/pnas.0803173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen OS, Crisp RJ, Valachovic M, Bard M, Winge DR, Kaplan J. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron–sulfur cluster biosynthesis. J. Biol. Chem. 2004;279:29513–29518. doi: 10.1074/jbc.M403209200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Lyver ER, Knight SA, Pain D, Lesuisse E, Dancis A. Mrs3p, Mrs4p, and frataxin provide iron for Fe–S cluster synthesis in mitochondria. J. Biol. Chem. 2006;281:22493–22502. doi: 10.1074/jbc.M604246200. [DOI] [PubMed] [Google Scholar]
- 25.Puccio H, Simon D, Cossee M, Criqui-Filipe P, Tiziano F, Melki J, Hindelang C, Matyas R, Rustin P, Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 26.Calmels N, Schmucker S, Wattenhofer-Donze M, Martelli A, Vaucamps N, Reutenauer L, Messaddeq N, Bouton C, Koenig M, Puccio H. The first cellular models based on frataxin missense mutations that reproduce spontaneously the defects associated with Friedreich ataxia. PloS ONE. 2009;4:e6379. doi: 10.1371/journal.pone.0006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duby G, Foury F, Ramazzotti A, Herrmann J, Lutz T. A non-essential function for yeast frataxin in iron-sulfur cluster assembly. Hum. Mol. Genet. 2002;11:2635–2643. doi: 10.1093/hmg/11.21.2635. [DOI] [PubMed] [Google Scholar]
- 28.Lesuisse E, Knight SA, Courel M, Santos R, Camadro JM, Dancis A. Genome-wide screen for genes with effects on distinct iron uptake activities in Saccharomyces cerevisiae. Genetics. 2005;169:107–122. doi: 10.1534/genetics.104.035873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberg AV, Molik S, Tsaousis AD, Neumann K, Kuhnke G, Delbac F, Vivares CP, Hirt RP, Lill R, Embley TM. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–628. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- 30.Adinolfi S, Rizzo F, Masino L, Nair M, Martin SR, Pastore A, Temussi PA. Bacterial IscU is a well folded and functional single domain protein. Eur. J. Biochem. 2004;271:2093–2100. doi: 10.1111/j.1432-1033.2004.04112.x. [DOI] [PubMed] [Google Scholar]
- 31.Cook JD, Kondapalli KC, Rawat S, Childs WC, Murugesan Y, Dancis A, Stemmler TL. Molecular details of the yeast frataxin-Isu1 interaction during mitochondrial Fe–S cluster assembly. Biochemistry. 2010;49:8756–8765. doi: 10.1021/bi1008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JH, Fuzery AK, Tonelli M, Ta DT, Westler WM, Vickery LE, Markley JL. Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry. 2009;48:6062–6071. doi: 10.1021/bi9002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vickery LE, Cupp-Vickery JR. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol. 2007;42:95–111. doi: 10.1080/10409230701322298. [DOI] [PubMed] [Google Scholar]
- 34.Tsou AY, Friedman LS, Wilson RB, Lynch DR. Pharmacotherapy for Friedreich ataxia. CNS Drugs. 2009;23:213–223. doi: 10.2165/00023210-200923030-00003. [DOI] [PubMed] [Google Scholar]
- 35.Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang L, Trempe JF, Matte A, Armengod ME, Cygler M. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010;8:e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]