Abstract
A massive central Australian dust storm in September 2009 was associated with abundant fungal spores (150,000/m3) and hyphae in coastal waters between Brisbane (27°S) and Sydney (34°S). These spores were successfully germinated from formalin-preserved samples, and using molecular sequencing of three different genes (the large subunit rRNA gene [LSU], internal transcribed spacer [ITS[, and beta-tubulin gene), they were conclusively identified as Aspergillus sydowii, an organism circumstantially associated with gorgonian coral fan disease in the Caribbean. Surprisingly, no human health or marine ecosystem impacts were associated with this Australian dust storm event. Australian fungal cultures were nontoxic to fish gills and caused a minor reduction in the motility of Alexandrium or Chattonella algal cultures but had their greatest impacts on Symbiodinium dinoflagellate coral symbiont motility, with hyphae being more detrimental than spores. While we have not yet seen any soft coral disease outbreaks on the Australian Great Barrier Reef similar to those observed in the Caribbean and while this particular fungal population was non- or weakly pathogenic, our observations raise the possibility of future marine ecosystem pathogen impacts from similar dust storms harboring more pathogenic strains.
INTRODUCTION
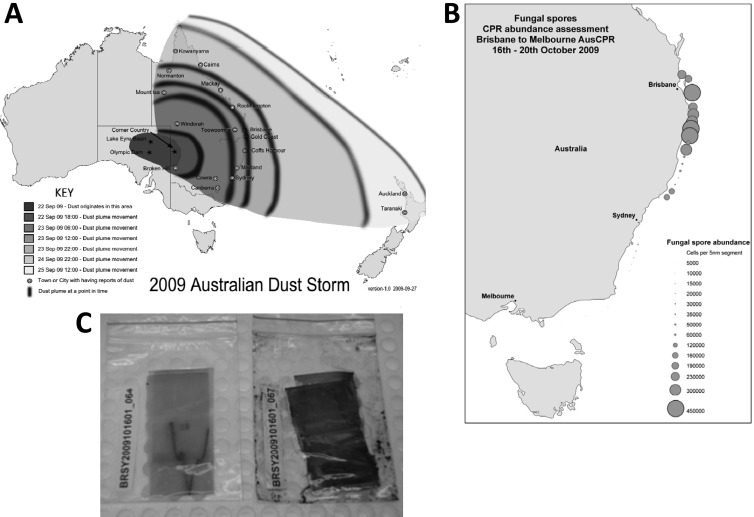
Fungi are rarely described for marine environments and are mainly considered terrestrial life forms. This may reflect a bias by fungal researchers but more likely relates to fungal cell biology. Fungi thrive in nutritionally rich environments, such as on plant or animal hosts and in soils, sediments, and detritus, where their enzymes break down complex biological polymers, allowing osmotrophic uptake of nutrients (1). During the 3 days following 22 September 2009, after a severe 10-year drought, a massive dust plume originating in the Australian Lake Eyre Basin swept eastward, reaching a width of 500 km and a length of 5,500 km before it traversed the Tasman Sea to reach New Zealand (Fig. 1A). Airports on the east coast of Australia were temporarily closed, and visibility was limited in some areas to <50 m. About 16 Tg of dust was stripped from central Australia, with 75,000 tons/h estimated to have crossed the New South Wales coast (2). Dust that originates in deserts is well known to be a vehicle for the spread of microbial communities and a concern for accidental spread of contaminants and diseases (3). The supply of dust-derived nutrients to the global ocean is also postulated to be a key control on marine primary production (4). In the wake of this dust event, we screened Continuous Plankton Recorder (CPR) samples for potential impacts on coastal phytoplankton communities. Unexpectedly, we discovered massive concentrations of 4- or 5-μm-diameter black spinose fungal spores and hyphae in coastal samples collected between Brisbane and Sydney from 16 to 20 Oct 2009 (up to 150,000 spores per m3). The formalin-preserved plankton recorder silks, which were returned to the lab within 10 days after towing, appeared black, as if they were covered in oil (Fig. 1C). Here we report on the conclusive morphological and molecular identification of the fungus and its cultivation and examine possible implications of this and future dust storm events for local marine ecosystems.
FIG 1.
(A) Extent of the September/October 2009 Australian dust storm covering an area equivalent to 25 × surface of England (map adapted from Wikipedia [http://en.wikipedia.org/wiki/File:2009_Dust_Storm_-_Australia_and_New_Zealand_Map.png] [published under a Creative Commons license]; panel A appeared previously as Fig. 1a in reference 11). (B) Abundance of fungal spores in Brisbane-Sydney coastal waters on 16 to 20 October 2009, collected on CPR silks (panel B appeared previously as Fig. 1c in reference 11). (C) Photographs of CPR silks, with the one on the right derived from the “fungal bloom.”
MATERIALS AND METHODS
CPR.
The Continuous Plankton Recorder (CPR) collects plankton continuously from a standard depth of 6 to 10 m. Water enters the instrument through a square aperture (1.61 cm2) and flows down an expanding tunnel to reduce water pressure to minimize damage to the captured plankton before it exits through the rear of the device. The movement of the water turns an external impeller at the rear of the device that operates a drive shaft and gear system, which constantly advances a band of mesh silk onto which plankton are filtered. The filtering mesh (270 μm; 60% porosity) meets a second band of covering silk, effectively sandwiching the plankton, which is then wound onto a storage spool in a tank containing formaldehyde (4% final concentration [by volume]) buffered with sodium tetraborate at pH 7.4 to 7.5. The silk is cut into sections representing 5 nautical miles of tow. For phytoplankton species analysis, 10 fields on each of two diagonals (20 fields in total) are counted using a 50× objective and 12.5× ocular. The device captures phytoplankton down to ∼4 μm in what can be considered semiquantitative estimates (5).
Fungal cultures and bioassays.
Sporulating fungal cultures were established on potato dextrose agar plates kept in a dark 20°C culture cabinet. Aqueous cultures were established in GSe marine (28 ppt salinity) and MBL freshwater algal culture media (6). Different amounts of fungal spores (3-, 6-, and 9-mm2 portions) and/or mycelium (9, 18, and 27 mm3) were tested in incubation experiments (1, 2, and 4 days) with microalgal cultures of the dinoflagellate Alexandrium catenella (strain ACCH05), Symbiodinium sp. (CS-7317), raphidophyte flagellate Chattonella marina (CMPL01), and rainbow trout gill epithelium cells (RTW1). Cell motility and shape were monitored for the microalgal cultures by light microscopy, while vital staining was used to define fish gill impact (7).
DNA extraction.
CPR samples were stored at room temperature in formalin. Two small pellets of spores (∼10 μl) were produced by centrifugation at 10,000 rpm for 30 s in 1.5-ml microcentrifuge tubes of a gentle scraping of cells from the CPR silks. Pelleted spores were either resuspended and soaked in 1 ml of RNAlater (Ambion) for 5 days at 4°C or rinsed in RNAlater without soaking. Spores were repelleted, and the soaked sample was resuspended in 100 μl lysis buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.5% Igepal CA-630, 0.5% Tween 20, and 0.1 mg of proteinase K/ml) (modified from reference 8), while the rinsed sample was resuspended in 100 μl lysis buffer supplied in the MoBio microbial kit. No lysis of spores was observed after resuspension in either buffer.
Both samples were sonicated for 1 min using an MSE ultrasonic disintegrator (Measuring & Scientific Equipment Ltd., London, England) with a probe diameter of 2.5 mm, at an amplitude level of 9 μm peak to peak for 10 s at 100 W. An estimated 60 to 80% lysis was observed. DNA was purified from both samples following the manufacturer's instructions (MoBio microbial kit) and eluted in 40 μl of the supplied elution buffer. Purified DNA was quantified using the NanoDrop 8000 instrument (Thermo Fisher Scientific, Wilmington, DE, USA), and ∼12 ng was used in subsequent PCR. A low yield (2 to 4 ng/liter and 80 to 160 ng total) of good-quality DNA was obtained.
PCR and phylogenetic analysis.
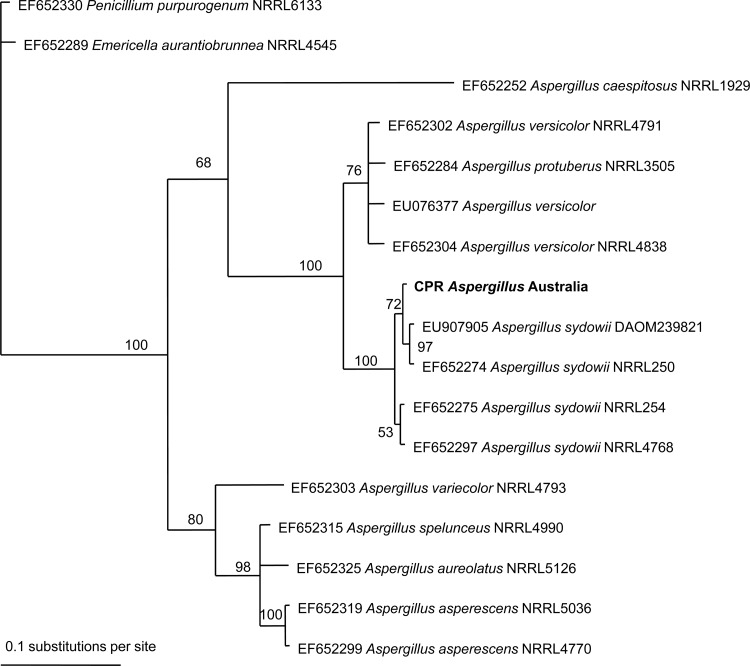
Three genes were amplified by PCR: the large subunit rRNA gene (LSU), internal transcribed spacer (ITS), and beta-tubulin (BT2) gene. PCR primers (Table 1) were used at a 1 μM final concentration using GoTaq Green master mix (Promega, Madison, WI). The BT2 gene and ITS were amplified using the following cycling conditions: 1 cycle of 94°C for 1 min, followed by 35 cycles at 94°C for 1 min, 58°C for 30 s, and 72°C for 30 s, and one final cycle of 72°C for 10 min. The LSU gene was amplified using the following cycling conditions: one cycle of 94°C for 2 min, followed by 35 cycles at 94°C for 1 min, 55°C for 30 s, and 72°C for 1.5 min, and 1 final cycle of 72°C for 10 min. PCR products were purified using a QIAquick PCR purification kit (Qiagen, Doncaster, Australia), and DNA sequencing was performed by the Australian Genome Research Facility, Brisbane, Australia. Electropherograms were visually inspected, and sequence results were of high quality. Phylogenetic relationships were inferred for the BT2 gene using the computer program MrBayes v3.1.2 (9). The analysis was run for 1 million generations, and trees were sampled every 1,000 generations. Of 1,000 trees saved, the last 500 were used to construct a 50% majority-rule consensus tree. The model used was a general time-reversible evolutionary model with gamma-distributed among-site rate variation. The analysis included seven species of Aspergillus, including four strains of A. sydowii. Two outgroup species, Penicillium purpurogenum (GenBank accession number EF652330) and Emericella aurantiobrunnea (GenBank accession number EF652289), were included.
TABLE 1.
Primers used for PCR and DNA sequencing
| Gene | Primer name | Sequence (5′–3′) |
|---|---|---|
| LSU | D1R-F | ACCCGCTGAATTTAAGCATA |
| 28-1483 | GCTACTACCACCAAGATCTGC | |
| BT2 | Bt2a | GGTAACCAAATCGGTGCTGCTTTC |
| Bt2b | ACCCTCAGTGTAGTGACCCTTGGC | |
| ITS | f-ITS1 | TCCGTAGGTGAACCTGCGG |
| f-ITS4 | TCCTCCGCTTATTGATATGC |
Nucleotide sequence accession numbers.
GenBank accession numbers for the Australian strain of Aspergillus sydowii ASAU-1, maintained at the Institute for Marine and Antarctic Studies of the University of Tasmania, have been lodged as KJ524908 (for LSU), KJ524907 (for ITS), and KJ524906 (for the BT2 gene).
RESULTS
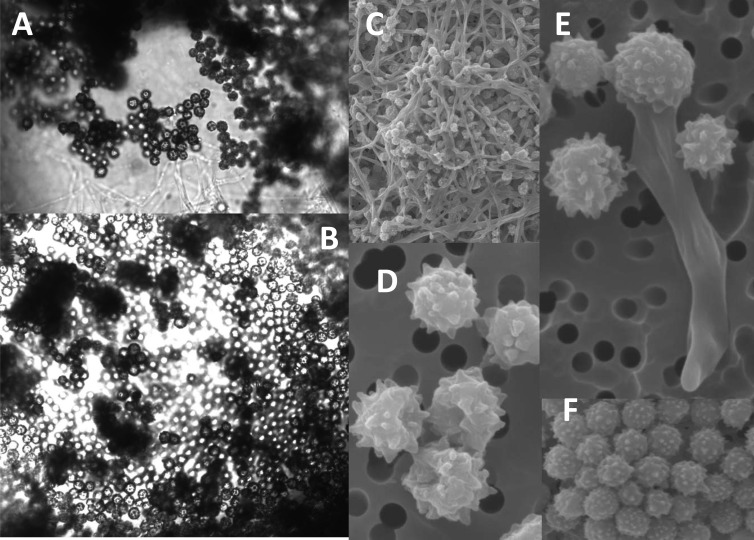
The abundance of fungal spores on CPR silks from Brisbane-Sydney coastal waters (up to 150,000/m3 seawater) (Fig. 1B) on 16 to 20 October 2009 was closely associated with the wake of the September/October 2009 Australian dust storm, covering an area equivalent to 25 times the surface of England. We succeeded in establishing viable sporulating cultures from formalin-preserved material and even from samples kept at −20°C for several months. In aqueous medium, fungal cultures grew better in freshwater (0 ppt) than in salt water (28 ppt salinity) but with spore formation taking place exclusively at the air-water interface and only hyphae proliferating submerged in the liquid culture medium. The conidia were echinulate, spherical to subspherical, and 2.5 to 4.0 μm in diameter (Fig. 2). Conidial heads were radiate, and conidiophore stipes measured up to 500 μm in length and were hyaline and smooth walled (see Fig. 4a and c). Colonies on potato dextrose agar were blue-green in color (Fig. 4b). Fungal spores have not been detected in similar CPR tows collected in the same coastal waters using the identical instrument before or after the dust storm (3 tows were collected before the dust storm and since the survey started in Jun 2009, and 13 tows have been collected afterwards, up to Dec 2011, equivalent to 7,200 nautical miles of sampling).
FIG 2.
Aspergillus sydowii conidiospores and hyphae from Australian coastal waters. (A and B) Light micrographs of spore masses from CPR silks. (C to F) Scanning micrographs of conidiospores and hyphae from CPR silks (panels C to F appeared previously as Fig. 1b in reference 11).
FIG 4.
Fungal cultures established from CPR silks and dust samples. (a and c) Light microscopy (LM) of Aspergillus sydowii fungal cultures obtained from Australian CPR silks (September 2009). (b) Aspergillus colony (1-cm diameter) on potato dextrose agar. (d and e) LM of unidentified fungal culture obtained from North Atlantic CPR silk (256M-10; June 2000). (f to h) LM (f and g) or scanning electron microscopy (SEM) (h) of unidentified fungal culture obtained from 2009 Brisbane dust storm sample.
Using molecular sequencing of three different genes (the conservative LSU, ITS, and variable beta-tubulin genes) (10), we unambiguously identified this fungus as Aspergillus sydowii (99 to 100% sequence match). The beta-tubulin (BT2) gene sequences showed a 99% match (391/392 bp). A similar level of variation (number of single nucleotide polymorphisms [SNPs]) for BT2 gene sequences for other A. sydowii strains is available in GenBank. The phylogram (50% majority-rule consensus tree) in Fig. 3 showed that in all trees sampled during the analysis, the BT2 gene sequence in this study grouped with A. sydowii (clade support value, 100%). No other species, out of sequences available in GenBank, fell into this group. Relationships between the four A. sydowii strains and the CPR spore sample were poorly resolved (clade support values of 53 and 72). The more conserved ITS gene showed a 100% match to Aspergillus versicolor, Aspergillus carneus, and A. sydowii.
FIG 3.
Bayesian phylogenetic analysis of the beta-tubulin gene for Aspergillus strains based on nucleotide sequences available in GenBank. The CPR fungal spore sequence for this study is denoted in bold type. Clade credibility values are shown at each node as a percentage.
Australian fungal cultures were not toxic to fish gill cells, caused a minor reduction in motility for Alexandrium (from 74% down to 53% swimming cells after 2 days), or altered morphology of Chattonella algal cultures (incidence of 1 to 2% cells with aberrant elongated morphology) but had greatest impacts on Symbiodinium dinoflagellate coral symbiont motility, with hyphae (from 48% down to 0 to 5% motility after 1 to 2 days) being more detrimental than spores (from 64% down to 9 to 14% motility after 1 day). Symbiodinium cells did recover after 4 days, however (Table 2).
TABLE 2.
Bioassays with Australian Aspergillus sydowii spores and/or hyphaea
| Test organism | Spores and/or hyphae | Parameter | Value (mean ± SD) with exposure time (days) |
|||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 4 | |||
| Rainbow trout fish gill cells | Spores | % viability (alamarBlue staining) | 100 ± 3 | 95 ± 16*** | NT | NT |
| Raphidophyte Chattonella marina | Spores | % elongated cells | 0 | 2.2 ± 1.7** | 1.2 ± 1.2** | NT |
| Dinoflagellate Alexandrium catenella | Spores | % swimming cells | 74 ± 7.2 | 68 ± 4.2*** | 53 ± 3.3*** | NT |
| Dinoflagellate Symbiodinium sp. | Spores | % swimming cells | 64 ± 5; 64 ± 3.2 | 14 ± 15***; 9 ± 6.2** | 52 ± 3.8***; 49 ± 4.6** | 51 ± 4.4***; 49 ± 4.4** |
| Dinoflagellate Symbiodinium sp. | Spores + hyphae | % swimming cells | 48 ± 6 | 0 ± 0*** | 5 ± 0 *** | 41 ± 18**; 11 ± 8.3*** |
Dosing for spores: *, 3 mm2; **, 6 mm2; ***, 9 mm2. Dosing for spores plus hyphae: *, 9 mm3; **, 18 mm3; ***, 27 mm3. NT, not tested.
DISCUSSION
The present report of fungal spore masses in Australian coastal waters following a dust storm is unprecedented. An early account of our finding appeared in the AusCPR newsletter, April 2011 (11). There has not been a similar event reported by the North Atlantic CPR survey (20 to 60°N) over the past 70 years, the U.S. CPR survey in the Northwest Atlantic (over the past 30 years), the Southern Ocean survey over the past 20 years, or the North Pacific survey over the past decade. However, we have occasionally observed low concentrations of viable fungal spores in other CPR samples (e.g., from the North Atlantic; sample code 256M-10, 57.72°N, 0.48°E [Fig. 4d and e]), and branched, strand-like fibers suspected to be fungal hyphae were detected in August 2012 on one of the mid-North Atlantic subtropical routes (12). Further, molecular analyses of North Atlantic CPR silks have revealed a substantial amount of Ascomycetes fungi (13), even though plankton microscopists are not trained to recognize 4- to 5-μm fungal spores in seawater samples. Similarly, Australian phytoplankton workers in Sydney coastal waters failed to detect the September-October 2009 fungal event reported here. This can be partially explained by the much larger sample size afforded by the CPR, that is, each 5 cm of silk sample represents 1.5 m3 of seawater. Spore concentrations in Sydney coastal waters were estimated at 55,000 cells/m3, which equates to detecting 3 spores in a 200-ml routine plankton count subsample. We also had to consider the possibility of a sampling artifact, whereby low concentrations of spores collected on the CPR silks succeeded in growing on the formalin-drenched silks before they were returned to the lab. Fungal spores were never detected in similar CPR tows using the identical instrument in the months immediately before or after the dust storm. Further, as is common protocol, extra formalin was added to the silk roll immediately on completion of the tow. Samples were returned to the Brisbane laboratory within 10 days of towing, smelling of formalin, and the shear abundance of fungal spores on the silks would rule out this possibility. On arrival in the laboratory, it was obvious to the plankton analysts that the silk had been “blackened” even before any microscopic analysis, and this has never been seen before or since (Fig. 1). In simulated laboratory experiments, we succeeded in growing the fungus on CPR silks inoculated on potato dextrose agar plates but never on formalin-drenched silks.
Aspergillus sydowii is believed to essentially be a terrestrial fungus, but salt tolerant and capable of growing in the sea. We did not detect via genetic sequencing any other fungal contaminants in the black spore masses harvested by the Continuous Plankton Recorder, which we attribute to A. sydowii being uniquely equipped to tolerate and grow in seawater. The Australian marine isolate showed a perfect (100%) match to a Malaysian isolate (accession number KC795920) derived from sandy beach soil from Malaysia (14). This species has previously been cultured from Caribbean air samples and used to inoculate Gorgonia sea fan corals, where it caused a disease termed “aspergillosis” (15, 16). This initial work claimed A. sydowii to be the main causative agent of a widespread outbreak of sea fan disease in the Caribbean, and increased influx of African dust was proposed as the source of the coral mortality (17). Subsequent physiological and toxicological work suggested that the strains in African dust and as sea fan pathogens were distinct, hence raising the possibility that African dust was not the source of the pathogen, although the dust still could have played a nutrient enrichment role to allow fungal pathogen proliferation (18, 19). In the past 5 years, Caribbean sea fans have developed increased resistance to this disease. The changing host/pathogen dynamics and the precise role of aeolian dust therefore are still unclear. While on the Australian Great Barrier Reef we have not yet seen any soft coral disease outbreaks similar to those observed in the Caribbean, this significant 2009 Australian Aspergillus fungal bloom raises similar questions.
We speculate that A. sydowii spores originated in terrestrial Lake Eyre habitats and were deposited in the form of nutrient-rich floating dust slicks in Australian coastal waters, which allowed rapid fungal growth and sporulation. In laboratory experiments, sporulation occurred only at the air-water interface. While we have not yet succeeded in detecting A. sydowii from an admittedly limited number of Sydney and Brisbane dust samples, a range of other fungal cultures could readily be established from this source (Fig. 4f to h). Critically, CPR samples have proved to be suitable for interrogation by light and scanning electron microscopy, molecular probes, and also cultivation of fungal spores to provide an early warning of future events and address potential marine ecosystem impacts from fungal-spore-laden dust storms. Aerosol fungal impacts have been well documented in terms of human health, agriculture, ice nucleation, cloud formation, and atmospheric chemistry (20), but broader impacts on marine ecosystems remain largely undocumented. Ongoing studies seek to characterize the bioactive molecules of marine-derived fungi (21). We also note that some mycotoxins, such as those produced by Aspergillus fumigatus, can bioaccumulate in shellfish and are thus potentially of human health risk (22). Importantly, southern and central Australia are projected to become drier under most climate change scenarios, and so severe droughts are likely to increase in frequency (23). While this particular fungal population was non- or weakly virulent, our observations raise the possibility of future marine ecosystem pathogen impacts from similar dust storms harboring more pathogenic strains.
ACKNOWLEDGMENTS
Grant McTainsh, Griffiths University, provided us with Brisbane DustWatch samples, and John Kirkwood, Team Leader, Air Quality Monitoring Unit, Department of Environment and Climate Change, Lidcombe, NSW, Australia, kindly provided us with 2009 dust storm samples intercepted in the Sydney region. Helen Bond (University of Tasmania) assisted with fungal culturing, and Karsten Goemann helped with scanning electron microscopy.
AusCPR is funded by the Integrated Marine Observing System (IMOS), which is funded by the Australian Government through the National Collaborative Research Infrastructure Strategy and the Super Science Initiative. This work was funded by Australia Research Council grant DP130102725.
Footnotes
Published ahead of print 21 March 2014
REFERENCES
- 1.Richards TA, Jones MDM, Leonard G, Bass D. 2012. Marine fungi: their ecology and molecular diversity. Annu. Rev. Mar. Sci. 4:495–522. 10.1146/annurev-marine-120710-100802 [DOI] [PubMed] [Google Scholar]
- 2.Jayaratne R, Johnson GR, McGarry PD, Cheung HC, Morawska L. 2011. Characteristics of airborne ultrafine and coarse particles during the Australian dust storm of 23 September 2009. Atmos. Environ. 45:3996–4001. 10.1016/j.atmosenv.2011.04.059 [DOI] [Google Scholar]
- 3.Gorbushina AA, Kort R, Schulte A, Lazarus D, Schnetger B, Brumsack H-J, Broughton FJ. 2007. Life in Darwin's dust: intercontinental transport and survival of microbes in the nineteenth century. Environ. Microbiol. 9:2911–2922. 10.1111/j.1462-2920.2007.01461.x [DOI] [PubMed] [Google Scholar]
- 4.Cropp R, Gabric AJ, Levasseur M, McTainsh GH, Bowie A, Hassler CS, Law CS, McGowan H, Tindale N, Viscarra Rossel R. 2013. The likelihood of observing dust-stimulated phytoplankton growth in waters proximal to the Australian continent. J. Mar. Syst. 117-118:43–52. 10.1016/j.jmarsys.2013.02.013 [DOI] [Google Scholar]
- 5.Richardson AJ, Walne AW, John AWG, Jonas TD, Lindley JA, Sims DW, Stevens D, Witt M. 2006. Using continuous plankton recorder data. Prog. Oceanogr. 68:27–74. 10.1016/j.pocean.2005.09.011 [DOI] [Google Scholar]
- 6.Andersen RA. (ed). 2005. Algal culturing techniques. Elsevier, New York, NY [Google Scholar]
- 7.Dorantes-Aranda JJ, Waite TD, Godrant A, Rose AL, Tovar CD, Woods GM, Hallegraeff GM. 2011. Novel application of a fish gill cell line assay to assess ichthyotoxicity of harmful marine microalgae. Harmful Algae 10:366–373. 10.1016/j.hal.2011.01.002 [DOI] [Google Scholar]
- 8.Galluzzi L, Penna A, Bertozzini E, Vila M, Garces E, Magnani M. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199–1206. 10.1128/AEM.70.2.1199-1206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 10.Peterson SW. 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226. 10.3852/mycologia.100.2.205 [DOI] [PubMed] [Google Scholar]
- 11.Integrated Marine Observing System. 2011. AusCPR Newsletter, no. 2, p 3–4 Integrated Marine Observing System, Hobart, Tasmania: http://imos.org.au/uploads/media/AusCPR_newsletter_No.2_April2011.pdf [Google Scholar]
- 12.Sir Alister Hardy Foundation for Ocean Science. 2012. 2012 SAHFOS annual report. Sir Alister Hardy Foundation for Ocean Science, Plymouth, United Kingdom [Google Scholar]
- 13.Sir Alister Hardy Foundation for Ocean Science. 2010. 2010 SAHFOS annual report. Sir Alister Hardy Foundation for Ocean Science, Plymouth, United Kingdom [Google Scholar]
- 14.Zakaria L, Yee TL, Zakaria M, Salleh B. 2011. Diversity of microfungi in sandy beach soil of Teluk Aling, Pulau Pinang. Trop. Life Sci. Res. 22:71–80 [PMC free article] [PubMed] [Google Scholar]
- 15.Geiser DM, Taylor JW, Ritchie KB, Smith GW. 1998. Cause of sea-fan death in the West Indies. Nature 394:137–138. 10.1038/280799671296 [DOI] [Google Scholar]
- 16.Smith GW, Ives LD, Nagelkerken IA, Ritchie KB. 1996. Caribbean sea-fan mortalities. Nature 383:487. 10.1038/383487a0 [DOI] [Google Scholar]
- 17.Weir-Brush JR, Garrison VH, Smith GW, Shinn EA. 2004. The relationship between gorgonian coral (Cnidaria: Gorgonacea) disease and African dust storms. Aerobiologia 20:119–126 10.1023/B:AERO.0000032949.14023.3a [DOI] [Google Scholar]
- 18.Bruno JF, Petes L, Harvell CD, Hettinger A. 2003. Nutrient enrichment can increase the severity of coral diseases. Ecol. Lett. 6:1056–1061. 10.1046/j.1461-0248.2003.00544.x [DOI] [Google Scholar]
- 19.Rypien KL, Andras JP, Harvell CD. 2008. Globally panmictic population structure in the opportunistic fungal pathogen Aspergillus sydowii. Mol. Ecol. 17:4068–4078. 10.1111/j.1365-294X.2008.03894.x [DOI] [PubMed] [Google Scholar]
- 20.Griffin DW. 2007. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin. Microbiol. Rev. 20:459–477. 10.1128/CMR.00039-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamasaki T, Sato Y, Hatsuda Y. 1975. Structure of sydowinin A, sydowinin B, and sydowinol, metabolites from Aspergillus sydowi. Agric. Biol. Chem. 39:2341–2345. 10.1271/bbb1961.39.2341 [DOI] [Google Scholar]
- 22.Grovel O, Pouchus YF, Verbist J-F. 2003. Accumulation of gliotoxin, a cytotoxic mycotoxin from Aspergillus fumigatus, in blue mussel (Mytilus edulis). Toxicon 42:297–300. 10.1016/S0041-0101(03)00146-6 [DOI] [PubMed] [Google Scholar]
- 23.Poloczanska ES, Hobday AJ, Richardson AJ. (ed). 2009. Report card of marine climate change for Australia: detailed scientific assessment. NCCARF publication 05/09. National Climate Change Adaptation Research Facility, Southport, Australia [Google Scholar]