Abstract
In this article, we present a method to delete genes in filamentous fungi that allows recycling of the selection marker and is efficient in a nonhomologous end-joining (NHEJ)-proficient strain. We exemplify the approach by deletion of the gene encoding the transcriptional regulator XlnR in the fungus Aspergillus niger. To show the efficiency and advantages of the method, we deleted 8 other genes and constructed a double mutant in this species. Moreover, we showed that the same principle also functions in a different genus of filamentous fungus (Talaromyces versatilis, basionym Penicillium funiculosum). This technique will increase the versatility of the toolboxes for genome manipulation of model and industrially relevant fungi.
INTRODUCTION
Genetic studies in filamentous fungi can be limited by the tools available for genome manipulation. For example, gene replacement in Aspergillus niger, a filamentous fungus used in many industrial processes (1–3), is typically based on transformation with a linear DNA fragment of selection marker flanked by target gene flanking regions. This is limited by the number of selection markers available and the requirement for fungal strains with multiple auxotrophies. Moreover, DNA integration through homologous recombination is inefficient. Most integration occurs ectopically through nonhomologous end joining (NHEJ) (4, 5). To circumvent this, NHEJ-inactivated strains are used (4–7), although such strains may show genomic instability (5, 7).
We present a method for gene deletion in filamentous fungi based on recombination between a plasmid and the chromosome initially developed for Saccharomyces cerevisiae (8). We use pyrG, encoding orotidine-5-phosphate decarboxylase, as a marker that can be selected and counterselected, allowing recycling of selection markers, hence leaving no mark on the genome (9). Indeed cells lacking this enzyme cannot grow without exogenous uridine/uracil, while these cells are resistant to the toxicity of 5-fluoro-orotic acid. To demonstrate the advantages and efficiency of this method in filamentous fungi, we deleted the xlnR gene coding for the xylanolytic transcriptional regulator in two species of filamentous fungi from different genera (A. niger and Talaromyces versatilis). Furthermore, 8 other genes were deleted from A. niger, and two genes coding for hydrophobins were deleted from the same strain. All of these gene deletions, in both strains, were performed in a strain proficient in NHEJ.
MATERIALS AND METHODS
Strains and growth media.
Escherichia coli strain XL1-Blue MRF′ (Stratagene) was grown according to the supplier's instructions, in medium supplemented with ampicillin (100 μg/ml), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-thiogalactopyranoside) (30 μg/ml), and IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mg/ml) (Sigma), where required, and was used for all DNA manipulation steps. The A. niger strain used was AB4.1 (ΔpyrG) (10). A. niger culture and medium composition were described in reference 11. The T. versatilis (ΔpyrG) strain used in this study originated from the industrial strain IMI378536, an Adisseo France S.A.S. property strain (18). This strain was maintained on potato dextrose agar (PDA) supplemented with 10 mM uridine. T. versatilis ΔpyrG cultures were grown in 200 ml of MN-Uri medium (NaNO3, 150 g/liter; KCl, 13 g/liter; KH2PO4, 38 g/liter; ZnSO4 · 7H2O, 22 g/liter; H3BO3, 11 g/liter; MnCl2 · 4H2O, 5 g/liter; FeSO4 · 7H2O, 5 g/liter; CoCl2 · 5H2O, 1.7 g/liter; CuSO4 · 5H2O, 1.6 g/liter; Na2MoO4 · 2H2O, 1.5 g/liter; EDTA Na2, 50 g/liter; glucose monohydrate, 15 g/liter; Casamino Acids, 1.25 g/liter; uridine, 2.43 g/liter; MgSO4, 0.24 g/liter).
Construction of plasmids.
Molecular cloning techniques were performed using standard procedures (12). DNA polymerase was Phusion (NEB). Primers (see the supplemental material) were designed using the A. niger CBS 513.88 (13) and T. versatilis sequences (unpublished data).
Transformation procedures.
Polyethylene glycol (PEG)-mediated transformation of protoplasts was used with both fungal species using minor modifications to standard procedures (14). For both fungi, 25 mg/ml of lysing enzymes from Trichoderma harzianum (Sigma) were used per gram of wet mycelia. For A. niger, 4 mg/ml chitinase (Sigma) and 100 mg/ml bovine serum albumin (BSA) were added per gram of wet mycelium. For transformation of A. niger, 1.5 × 106 protoplasts were transformed with 3 μg of plasmid DNA. The transformed cell pellet was then resuspended in 300 μl STC (1.2 M sorbitol, 10 mM Tris base, 50 mM CaCl2 [pH 7.5]), and transformed mycelia were regenerated on osmotically stabilized medium containing 1.2 M sorbitol. For transformation of T. versatilis, 107 protoplasts were transformed with 10 μg plasmid DNA, and mycelia were regenerated using procedures described elsewhere (15).
Electroporation was used to transform E. coli.
RESULTS AND DISCUSSION
Construction of pC3 and pC7 integrative plasmids.
A 2,865-bp HindIII/EcoRI digest DNA fragment of plasmid pAo4-13 (16), containing the pyrG locus of Aspergillus oryzae, which include the gene and its native promoter, was blunt ended and cloned into the backbone of the plasmid pBluescript SK− (Stratagene) at the PsiI site, in both orientations, generating plasmids pC3 and pC7 (Fig. 1A). We constructed plasmids with both orientations of pyrG in case expression of the gene interfered with insertion or excision of the plasmid during the deletion steps. The plasmids retain the blue/white screening capability for cloning and 4 unique restriction sites (EcoRI, NotI, SpeI, and XhoI) in the multiple-cloning site (MCS) to clone the flanking sequences of the locus to be deleted. These plasmids are nonreplicative in filamentous fungi as they lack an origin of replication.
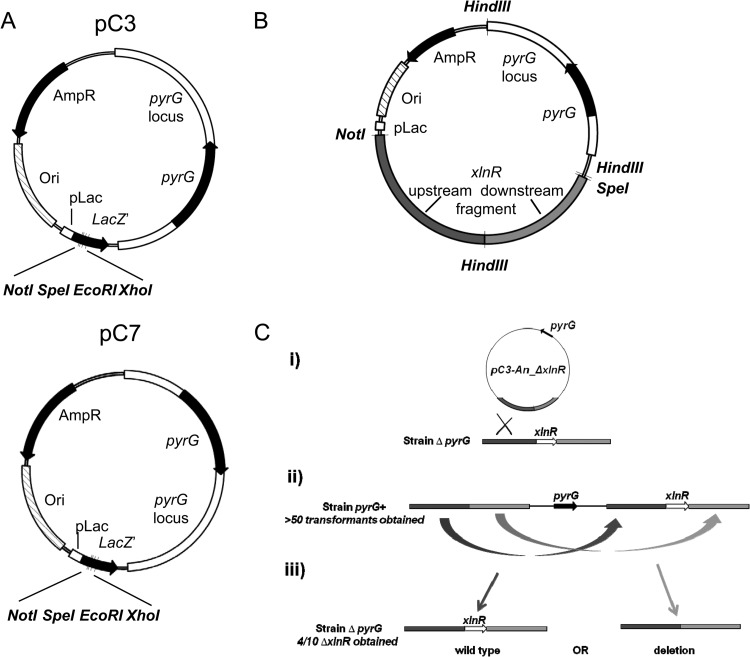
FIG 1.
Plasmid vectors pC3 and PC7 and the xlnR deletion strategy in Aspergillus niger. (A) Deletion vectors pC3 and pC7. Restriction enzyme sites available in the multiple-cloning site and component parts of the vectors are indicated. The white box represents the A. oryzae pyrG locus obtained from plasmid pAo4-13, and the black arrow inside the pyrG open reading frame. (B) pC3-An_ΔxlnR plasmid. Flanking regions of xlnR are shaded: upstream in dark gray and downstream in light gray. (C) Scheme for deletion of xlnR in A. niger by the intermediate of pC3-An_ΔxlnR plasmid. (i) Recombination between the plasmid and the chromosome using the homology of the flanking region of xlnR present on the chromosome and the plasmid lead to the integration of the plasmid on the chromosome. Transformants that have integrated the plasmid are pyrG+ and can grow on medium lacking uridine. Transformation with pC3-An_ΔxlnR gave 60 to 100 pyrG+ transformants. (ii) Releasing of the selective pressure for plasmid integration by growing cells on a medium containing uridine allows the excision of the plasmid. When the plasmid is excised and cured, the cells are ΔpyrG and can be selected by plating on a medium containing uridine and 5-fluoro-orotic acid. (iii) Excision using the homology of the flanking region can lead either to reversion to the wild-type copy of the gene or to deletion. For the xlnR gene, 4 out of 10 clones tested by PCR were shown to be deleted.
Construction of the Aspergillus niger xlnR deletion mutant.
To delete the xylanolytic transcriptional regulator-encoding gene xlnR in A. niger, 2,490-bp upstream and 1,878-bp downstream DNA fragments were amplified by PCR from A. niger N402 genomic DNA. Primers were designed so the upstream and downstream fragments contained a common HindIII restriction site to ligate them together and NotI and SpeI restriction sites for cloning the joined fragments into the plasmid MCS of pC3 and pC7, to create the pC3-An_ΔxlnR and pC7-An_ΔxlnR integrative plasmids (Fig. 1B). Transformations of the A. niger AB4.1 (ΔpyrG) strain were performed using pC3-An_ΔxlnR or pC7-An_ΔxlnR integrative plasmids. Transformations, done in triplicate, produced between 60 and 100 recombinants each, when plated on medium lacking uridine, to select for the integration of the plasmid carrying pyrG on the chromosome (Fig. 1C). No difference in efficiencies of transformation was observed between plasmids, indicating that there was no interference by the orientation of pyrG in the integration at the xlnR locus. Transformants were purified by propagating them twice successively on the same transformation medium but lacking sorbitol. Transformants were then propagated twice on PDA medium containing 10 mM uridine to release the selective pressure on the integrated plasmid. To select for clones that had excised the plasmid (ΔpyrG), spores were then resuspended in 0.001% (vol/vol) Tween 80 and spread on Aspergillus minimal medium containing 1% (wt/vol) glucose, 1.6 mM uridine, and 750 μg/ml 5-fluoro-orotic acid. The frequency of 5-fluoro-orotic acid-resistant strain was 8 × 10−4.
Excision can lead to reversal to the wild-type (WT) locus or deletion of the target locus (Fig. 1C). Ten candidates were screened, using PCR with a primer chosen to amplify the xlnR region. Four clones showed a product size expected for xlnR deletion (1,020 bp for the deletion mutant compared to 3,760 bp for the wild type) (see the supplemental material). Deletion was confirmed by sequencing the PCR product and the absence of product in another PCR with primers internal to the xlnR gene (see the supplemental material). All of these results confirmed that the xlnR gene was deleted in the A. niger AB4.1 strain. To check if extra copies of the plasmid remained integrated into the genome of A. niger due to multiple insertion of the plasmid in the chromosome of A. niger after the transformation, a Southern analysis of the genomic DNA restricted with BglII from the wild-type strain and the xlnR-deleted strain of A. niger was performed using pC3-An_ΔxlnR as a probe. The results showed no other bands than the expected 7-kbp band for the wild type and the 4.1-kbp band for the xlnR deletion mutant, excluding the possibility of the presence of supplementary copies of the plasmid in the genome of the xlnR-deleted strain (see the supplemental material).
Construction of A. niger single and double mutants.
To show the general applicability of the method, we deleted 8 other A. niger genes coding for various type of proteins; transcription factor, membrane protein, plant cell wall-degrading enzymes, and small hydrophobic proteins (Table 1). All of these deletions have been confirmed by two independent PCRs, one using primers external to the gene of interest (and sequencing of the PCR products) and a second PCR using primers internal to the gene. It is of note that while one of the flanking sequences of xlnR cloned into the integrating plasmid was over 2 kbp (2,490 bp for the upstream fragment), successful gene deletions in A. niger were obtained with shorter flanking regions of the targeted gene, which can facilitate construction of the plasmid. For example, we used about 2 kbp upstream and 1.5 kbp downstream of the hfbD gene and about 1.8 kbp upstream and 1.6 kbp downstream of the hsbA gene to obtain deletion mutants (Table 1). Finally, as this approach allows for recycling of the selection marker (pyrG), we also created a strain carrying deletion of both hydrophobin-encoding genes hfbD and hyp1. The double mutant strain obtained by this approach was still ΔpyrG, so further gene deletions are still possible in this strain, allowing for dissection of complex genetic networks.
TABLE 1.
List of selected genes deleted from A. niger in this studya
| Gene identification no. (CBS 513.88) | CADREb annotation | Sizes of flanking regions cloned (bp) |
|---|---|---|
| An04g08600 | Transcriptional activator XlnR | 2,490 and 1,878 |
| An02g03830 | Carbon catabolite repressor CreA | 2,517 and 2,000 |
| An08g09880 | Hydrophobin HfbD | 1,937 and 1,476 |
| An07g03340 | Hydrophobin Hyp1 | 1,931 and 1,608 |
| An09g00840 | Similarity to HsbA (Aspergillus oryzae) | 1,770 and 1,561 |
| An18g02730 | Similarity to PTH11 (Magnaporthe grisea) | 2,036 and 1,556 |
| An07g09330 | 1,4-β-d-Glucan cellobiohydrolase CbhA | 1,908 and 1,727 |
| An01g11660 | 1,4-β-d-Glucan cellobiohydrolase CbhB | 1,960 and 1,519 |
| An12g04610 | Copper-dependent lytic polysaccharide monooxygenase | 2,070 and 1,519 |
Other deletions have also been made, including double deletions (e.g., hfbD hyp1).
Central Aspergillus Data REpository.
Gene deletion in T. versatilis.
To check if the method could be extended to other fungi, deletion of the xlnR gene of T. versatilis using the pC3 integrative plasmid was performed. T. versatilis is an important industrial fungal species used, for example, to produce a mixture of glycosyl hydrolase enzymes. To delete the T. versatilis xlnR gene, 1,791-bp upstream and 1,613-bp downstream DNA fragments of the gene were amplified from genomic DNA, ligated, and cloned in pC3 to give pC3-Tv_ΔxlnR. Transformation of this plasmid into a pyrG-deleted T. versatilis strain was done in duplicate, and plating on a medium lacking uracil gave 20 recombinants and 25 recombinants. This indicates that there is a functional complementation of the T. versatilis pyrG-deleted strain by the A. oryzae pyrG gene. After release of the selective pressure for plasmid integration and selection for excision of the plasmid, 9 colonies were screened for the deletion of xlnR by PCR using primers external to the gene. Five clones showed the expected 673-bp band corresponding to the deletion of the locus, compared to 3,470 bp of the wild-type locus (see the supplemental material). This result indicates that the method can be extended to other genera of fungi.
Conclusions.
Genetic analysis in filamentous fungi can be impaired by the lack of selection markers for gene deletion and the low efficiency of homologous recombination compared to ectopic integration of linear DNA fragments. In this study, we developed a method for gene deletion that worked in two filamentous fungal species, A. niger and T. versatilis. This method should also be effective with other species and genera of filamentous fungi, providing the availability of a pyrG deletion strain and a functional complementation of the strain by the pyrG present on the plasmid. This approach is effective in more genetically stable NHEJ-proficient strains, and as it is based on recycling of the selection marker, it allows the construction of strains with multiple gene deletions. We did not examine any potential pitfalls of repeated use of selection based on the use of 5-fluoro-orotic acid, but we are aware that repeated use of 5-fluoro-orotic acid may cause chromosome alterations (16). Although it has not been directly examined here, the approach could be made more amenable for high-throughput approaches, such as the Clontech In-Fusion cloning method or, with modifications, the Life Technologies Gateway system. The method described will increase the toolbox for genome manipulation in these important and industrially relevant organisms and adds to those transformation procedures already described (2, 17).
Supplementary Material
ACKNOWLEDGMENTS
The research reported here was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) Sustainable Bioenergy Centre (BSBEC), under the program for “Lignocellulosic Conversion to Ethanol” (LACE) (grant no. BB/G01616X/1). A.L. and J.-L.P. were supported by CIFRE (grant no. 1558/2010).
Footnotes
Published ahead of print 28 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00625-14.
REFERENCES
- 1.Lubertozzi D, Keasling JD. 2009. Developing Aspergillus as a host for heterologous expression. Biotechnol. Adv. 27:53–75. 10.1016/j.biotechadv.2008.09.001 [DOI] [PubMed] [Google Scholar]
- 2.Meyer V. 2008. Genetic engineering of filamentous fungi—progress, obstacles and future trends. Biotechnol. Adv. 26:177–185. 10.1016/j.biotechadv.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 3.de Vries R. 2003. Regulation of Aspergillus genes encoding plant cell wall polysaccharide-degrading enzymes; relevance for industrial production. Appl. Microbiol. Biotechnol. 61:10–20. 10.1007/s00253-002-1171-9 [DOI] [PubMed] [Google Scholar]
- 4.Honda Y, Kobayashi K, Kirimura K. 2011. Increases in gene-targeting frequencies due to disruption of kueA as a ku80 homolog in citric acid-producing Aspergillus niger. Biosci. Biotechnol. Biochem. 75:1594–1596. 10.1271/bbb.110015 [DOI] [PubMed] [Google Scholar]
- 5.Meyer V, Arentshorst M, El-Ghezal A, Drews A-C, Kooistra R, van den Hondel CAMJJ, Ram AFJ. 2007. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 128:770–775. 10.1016/j.jbiotec.2006.12.021 [DOI] [PubMed] [Google Scholar]
- 6.Carvalho NDSP, Arentshorst M, Kwon MJ, Meyer V, Ram AFJ. 2010. Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl. Microbiol. Biotechnol. 87:1463–1473. 10.1007/s00253-010-2588-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Mao Z, Xue W, Li Y, Tang G, Wang A, Zhang Y, Wang H. 2011. Ku80 gene is related to non-homologous end-joining and genome stability in Aspergillus niger. Curr. Microbiol. 62:1342–1346. 10.1007/s00284-010-9853-5 [DOI] [PubMed] [Google Scholar]
- 8.Scherer S, Davis RW. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. U. S. A. 76:4951–4955. 10.1073/pnas.76.10.4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeke JD, LaCroute F, Fink GR. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345–346. 10.1007/BF00330984 [DOI] [PubMed] [Google Scholar]
- 10.Van Hartingsveldt W, Mattern IE, van Zeijl CM, Pouwels PH, van den Hondel CA. 1987. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Gen. Genet. 206:71–75. 10.1007/BF00326538 [DOI] [PubMed] [Google Scholar]
- 11.Delmas S, Pullan ST, Gaddipati S, Kokolski M, Malla S, Blythe MJ, Ibbett R, Campbell M, Liddell S, Aboobaker A, Tucker GA, Archer DB. 2012. Uncovering the genome-wide transcriptional responses of the filamentous fungus Aspergillus niger to lignocellulose using RNA sequencing. PLoS Genet. 8:e1002875. 10.1371/journal.pgen.1002875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, van Dijck PWM, van Dijk A, Dijkhuizen L, Driessen AJM, d'Enfert C, Geysens S, Goosen C, Groot GSP, de Groot PWJ, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JPTW, van den Hondel CAMJJ, van der Heijden RTJM, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJEC, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NNME, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJJ, Wedler H, Wösten HAB, Zeng A-P, van Ooyen AJJ, Visser J, Stam H. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221–231. 10.1038/nbt1282 [DOI] [PubMed] [Google Scholar]
- 14.Ballance DJ, Turner G. 1985. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene 36:321–331. 10.1016/0378-1119(85)90187-8 [DOI] [PubMed] [Google Scholar]
- 15.de Ruiter-Jacobs YM, Broekhuijsen M, Unkles SE, Campbell EI, Kinghorn JR, Contreras R, Pouwels PH, van den Hondel CA. 1989. A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae. Curr. Genet. 16:159–163. 10.1007/BF00391472 [DOI] [PubMed] [Google Scholar]
- 16.Wellington M, Kabir MA, Rustchenko E. 2006. 5-Fluoro-orotic acid induces chromosomal alterations in genetically manipulated strains of Candida albicans. Mycologia 98:393–398. 10.3852/mycologia.98.3.393 [DOI] [PubMed] [Google Scholar]
- 17.Meyer V, Wu B, Ram AFJ. 2011. Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol. Lett. 33:469–476. 10.1007/s10529-010-0473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabatier A, Fish NM, Haigh NP. November 1999. Enzymes mixture obtained from Penicillium funiculosum. International patent no. WO99/57325
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.