Abstract
Seabird ticks are known reservoirs of bacterial pathogens of medical importance; however, ticks parasitizing tropical seabirds have received less attention than their counterparts from temperate and subpolar regions. Recently, Rickettsia africae was described to infect seabird ticks of the western Indian Ocean and New Caledonia, constituting the only available data on bacterial pathogens associated with tropical seabird tick species. Here, we combined a pyrosequencing-based approach with a classical molecular analysis targeting bacteria of potential medical importance in order to describe the bacterial community in two tropical seabird ticks, Amblyomma loculosum and Carios (Ornithodoros) capensis. We also investigated the patterns of prevalence and host specificity within the biogeographical context of the western Indian Ocean islands. The bacterial community of the two tick species was characterized by a strong dominance of Coxiella and Rickettsia. Our data support a strict Coxiella-host tick specificity, a pattern resembling the one found for Rickettsia spp. in the same two seabird tick species. Both the high prevalence and stringent host tick specificity suggest that these bacteria may be tick symbionts with probable vertical transmission. Detailed studies of the pathogenicity of these bacteria will now be required to determine whether horizontal transmission can occur and to clarify their status as potential human pathogens. More generally, our results show that the combination of next generation sequencing with targeted detection/genotyping approaches proves to be efficient in poorly investigated fields where research can be considered to be starting from scratch.
INTRODUCTION
Ticks are considered vectors of major medical and veterinary importance. Their peculiar ecology, including their extended life spans and ability to transfer pathogens horizontally via cofeeding (1), gives these arthropod vectors the additional and important role of pathogen reservoirs (2). Among ticks, those parasitizing wild birds have been proposed to play an additional epidemiological role, given the potential of some bird species to play a part in the long-distance dispersal of ticks and tick-borne pathogens (2, 3).
Seabird ticks have received little attention, despite the fact that ticks in sometimes uninhabited and geographically remote areas have been shown to harbor human pathogens (4–6). The high mobility of seabirds and their wide geographical distribution, together with their gregarious breeding behaviors, may result in these hosts and their ticks playing an important epidemiological role (5). Many attempts to characterize pathogen circulation in seabird ticks have been restricted to viruses (5, 7), while bacteria of epidemiological importance, such as the Lyme disease bacterium Borrelia burgdorferi sensu lato, have been described in the seabird tick Ixodes uriae worldwide (4, 8–11). Despite these data on some important bacterial pathogens, to date, little is known about the broader array of bacterial communities in seabird ticks under natural conditions.
Seabirds represent the most abundant and widespread avifauna of the western Indian Ocean (WIO) (12). Located at the intersection of major intercontinental and transoceanic migratory routes between Europe, Africa, Asia, Oceania, and the subantarctic islands (13–15), the WIO, which encompasses distant and mostly uninhabited islands, is prone to pathogen introduction and emergence (16). Recent disease outbreaks related to birds (e.g., West Nile virus in Madagascar, Sindbis virus in Mauritius [17]) support this idea. Two distinct tick species have been documented from seabirds breeding on islands of the WIO: a hard tick species (Amblyomma loculosum) and a soft tick species (Carios [Ornithodoros] capensis). These species exhibit significant ecological differences, which may entail differences in infection patterns: C. capensis ticks are typically nest-associated ticks with a strong specificity for seabirds, while A. loculosum ticks are generalist ectoparasites able to feed on reptiles, birds, and mammals (18, 19). While a recent study showed the circulation of Rickettsia pathogens in these two tick species in the WIO (20), information pertaining to the epidemiological role of tropical seabird ticks in broader bacterial circulation is scarce.
In this study, we evaluated the potential of a metagenomic approach to describe the bacterial community in seabird ticks from islands of the western Indian Ocean and to detect potential tick-borne bacterial pathogens of epidemiological importance in this region. We evaluated the role of the tick species and distribution in structuring bacterial communities by comparing bacterial diversity between A. loculosum and C. capensis ticks within the same island and in C. capensis ticks collected from different islands, respectively, and investigated the prevalence of Coxiella bacteria in soft and hard ticks of the region.
MATERIALS AND METHODS
Tick collection.
Ticks were collected from different seabird species in 2011 and 2012 on six islands of the WIO: La Réunion Island, Bird and Aride Islands (the Seychelles), and Europa, Juan de Nova, and Tromelin Islands (the Eparses Islands). Ticks were collected either directly from adults and chicks or in the nest environment, as follows: soft ticks were sampled on red-tailed tropicbirds (Phaeton rubricauda) on Europa Island, on red-footed booby (Sula sula) specimens and nests on Tromelin Island, within reproductive colony boundaries of sooty terns (Onychoprion fuscatus) on Juan de Nova Island, and in wedge-tailed shearwater (Puffinus pacificus) nests on Petite Ile (La Réunion Island); hard ticks were collected on the ground of Petite Ile (La Réunion Island). Both soft and hard ticks were collected from a sooty tern colony at Bird Island. On Aride Island, ticks were collected from sooty terns (Onychoprion fuscata), tropical shearwaters (Pufinus bailloni), brown noddies (Anous stolidus), lesser noddies (Anous tenuirostris), white-tailed tropicbirds (Phaeton lepturus), and wedge-tailed shearwaters (Pufinus pacificus). All ticks were morphologically identified as A. loculosum or C. capensis using standard taxonomic keys (21, 22). Depending on the access to liquid nitrogen tanks, ticks were either conserved by dry flash freezing or by placing them in 70% ethanol until the samples were brought back to the laboratory and frozen at −80°C before analyses.
Sampling in La Réunion Island and in the Eparses Islands (Europa, Juan de Nova, and Tromelin Islands) was conducted under the approval of the Direction de l'Environnement, de l'Aménagement et du Logement and the Terres Australes and Antarctiques Françaises. Sample collection at Aride and Bird Islands and sample export to La Réunion Island were performed with the approval of the Seychelles Bureau of Standards and the Ministry of Environment.
Bacterial metabarcoding design.
Ticks analyzed through metabarcoding were randomly selected from samples of 3 distinct and geographically distant sites, i.e., Juan de Nova, Tromelin, and La Réunion Islands. Pooled tick samples consisted of 5 adult ticks for both C. capensis and A. loculosum. For each pool, ticks were placed in a single 1.5-ml Eppendorf tube and crushed for 2 min at 25 Hz using two 3-mm tungsten beads in a TissueLyser apparatus (Qiagen, Valencia, CA). Total nucleic acids were extracted using a V2 minikit (Qiagen, Valencia, CA), and each pool was prepared for bacterial 16S rRNA metabarcoding (23) with the use of the GsFLX technology (Genoscreen, Lille, France). Briefly, 16S rRNA V3 and V4 variable regions were amplified via specific PCR primers targeting the upstream and downstream regions of the V3–V4 segment: the 3′ ends of forward (TACGGRAGGCAGCAG) and reverse (GGACTACCAGGGTATCTAAT) bacterium-specific primers were associated at the 5′ end with multiplex identifier (MID) tags, a GsFLX key, and GsFLX adapters. Each pool was independently amplified twice with distinct MID tags, allowing the individual identification of each pool, as well as each of the two PCR duplicates. The quantity of each PCR product was then determined with a PicoGreen kit, and all products were mixed together in equimolar concentrations prior to 454 GsFLX sequencing.
Sequence analyses and species identification.
All 454 GsFLX sequences were separated into their respective pools by MID identification, performed using the Geneious software package (24). Those sequences that were less than 250 bp in length or that contained sequence ambiguities were discarded. Grouped sequences were exported in the FASTA format, and a locally executed nucleotide BLAST search was performed using the BLAST+ program, downloaded from the NCBI website (25). The reference 16S rRNA database used was also obtained from the NCBI file listings. One hundred alignments and 10 descriptions were exported for each BLAST result; all other search parameters were retained in their default settings. The MEGAN (v5) program was used to assign the BLAST results to bacterial taxa and to visualize the data (26). Normalized data sets were compared in MEGAN (v5), and unassigned reads were excluded from the analyses. The new minimum taxon cover functionality of MEGAN (v5) was exploited to provide accurate BLAST hit assignments within taxonomic trees, and the new core biome and shared biome functions were used to generate representations of the major bacterial groups present in each pooled sample. Grouped sequence similarities were compared using the principle component analysis (PCoA) methodology, and Shannon and Simpson indices were calculated from the leaves of a fully expanded taxonomic tree, irrespective of the taxonomic level of classification.
Coxiella PCR detection and genotyping.
Due to the high prevalence of Coxiella detection in all tick DNA pools (see Results), the 16S rRNA gene was amplified using endpoint PCR, as previously described (27). Positivity was assessed on agarose gels, and randomly selected PCR products were sequenced for each tick species and for samples from each island. Sequences were aligned with those of known Coxiella symbionts as well as with the sequence of Coxiella burnetii using the ClustalW program and subsequently trimmed, and the alignment was eventually refined over 100 iterations using the MUSCLE algorithm implemented in the Geneious Pro (v5.6.0) software package (24). A maximum likelihood phylogenetic tree with 1,000 bootstrap iterations was generated using the PhyML plugin from Geneious Pro (v5.6.0) and the optimal substitution model (GTR+I+G), as selected using the jModelTest (v2.1.2) tool (28). The appearance of the tree was edited in the FigTree (v1.4) program.
Nucleotide sequence accession numbers.
Partial 16S rRNA gene sequences have been submitted to GenBank and may be found under accession numbers KF913893 to KF913928.
RESULTS
Bacterial identification by pyrosequencing.
A total of 125,880 sequences were obtained from the four tick pools. The lengths of the sequences ranged from 30 to 568 bp, and 12,851 (10%) were shorter than 250 bp. The average and median read lengths were 401 and 442 bp, respectively, and sequence reads were evenly distributed across each multiplexed pool. Sequences were filtered using quality criteria to exclude reads containing ambiguities and/or sequences with length of less than 250 bp.
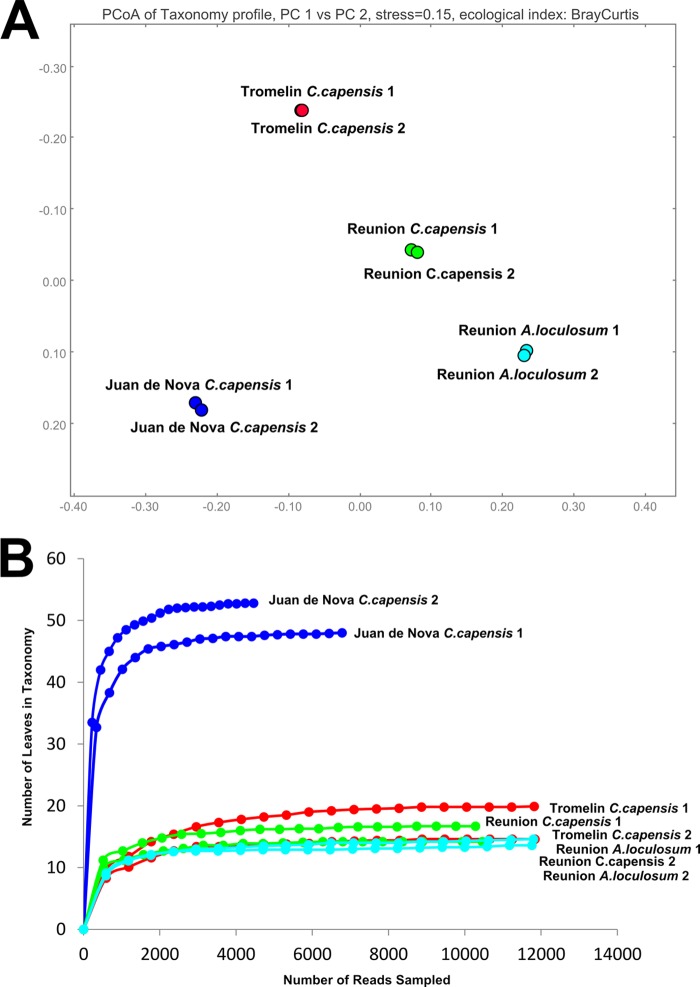
Independent sequencing repeats generated highly similar taxonomic representations, whereas samples from different islands or tick species generated characteristic diversity profiles (Fig. 1A). The overall levels of bacterial diversity measured in samples from A. loculosum from La Réunion Island and C. capensis from La Réunion and Tromelin Islands were comparable, whereas a greater level of diversity was obtained from C. capensis ticks from Juan de Nova Island (Fig. 1B and Table 1). However, rarefaction curves based on the leaves of a fully expanded taxonomic tree in MEGAN (v5) plateaued for each of the obtained samples, suggesting that the biodiversity of each sample had been exhaustively categorized.
FIG 1.
Pyrosequencing data. (A) Similarity testing by principle component analysis on the basis of the Bray-Curtis ecological index of fully expanded comparative taxonomic trees in the MEGAN (v5) program. PC1 is presented on the horizontal axis, and PC2 is presented on the vertical axis. (B) Rarefaction curves generated from data assigned to the leaves of fully expanded comparative taxonomic trees.
TABLE 1.
Summary of pyrosequencing data
| Sample origin | Tick species | No. of ticks | Replicate no. | MID used | No. of raw reads | No. of reads after quality control | Shannon index | Simpson index |
|---|---|---|---|---|---|---|---|---|
| Tromelin Island | Carios capensis | 5 | 1 | MID1 | 14,743 | 13,652 | 1.63 | 2.48 |
| 2 | MID2 | 15,738 | 14,243 | 1.61 | 2.46 | |||
| Juan de Nova Island | Carios capensis | 5 | 1 | MID3 | 15,115 | 13,423 | 3.71 | 6.88 |
| 2 | MID7 | 17,259 | 15,292 | 4.23 | 11.07 | |||
| La Réunion Island | Carios capensis | 5 | 1 | MID9 | 13,691 | 12,417 | 1.79 | 2.02 |
| 2 | MID10 | 16,395 | 14,838 | 1.63 | 1.88 | |||
| La Réunion Island | Amblyomma loculosum | 5 | 1 | MID11 | 17,336 | 15,746 | 1.47 | 2.27 |
| 2 | MID12 | 15,065 | 13,418 | 1.54 | 2.35 |
Of the sequence reads assigned with MEGAN to a minimum taxonomic level of order, the bacterial orders Legionales, Rickettsiales, Bacillales, Actinomycetales, and Rhodobacterales were the most prominently represented across the studied tick samples (Fig. 1C).
Core and shared biomes and genera of possible medical importance.
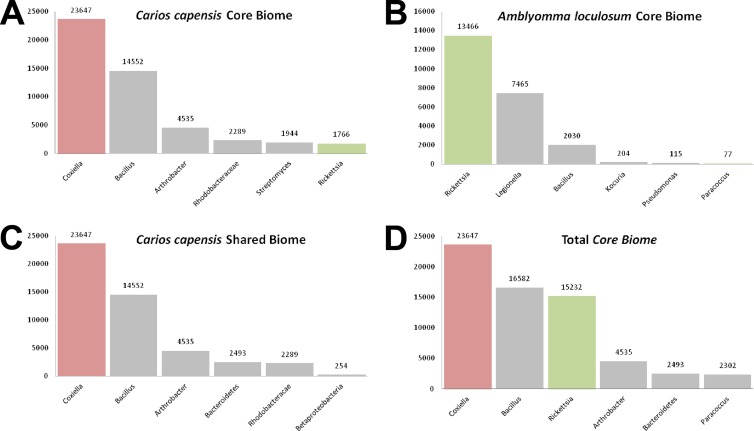
Characterization of the core bacterial biome constituted by combining data from all pooled samples identified Coxiella, Bacillus, and Rickettsia as the three most prominent bacterial genera present in the tested ticks of the region (Fig. 2D).
FIG 2.
Bacteriome data from ticks of the WIO. Bars indicate the number of raw sequence reads assigned to named bacterial taxa. The numbers of sequences assigned to the named taxa are indicated above the bars. Bars corresponding to Coxiella spp. and Rickettsia spp. are colored red and green, respectively.
Interestingly, two of the most abundant genera identified in the tested ticks included known human pathogens: Rickettsia, the causative agent of rickettsioses such as African tick bite fever, has previously been identified to be abundant in ticks of the region (20) and was observed to be prevalent within samples of A. loculosum from La Réunion Island. Rickettsia bacteria were also present but were less well represented in some samples of C. capensis. In contrast, Coxiella bacteria were present in large quantities in samples of C. capensis but at low levels in A. loculosum. Coxiella sequences exhibited between 98.4 and 98.7% genetic identities with Coxiella burnetii, the causative agent of Q fever (29). Interestingly Borrelia spp., the causative agents of Lyme disease and relapsing fever, were also identified in C. capensis from Tromelin Island; however, they were present only in trace quantities (13 of 24,797 reads). Additionally, Kocuria bacteria, which are increasingly associated with human pathologies (30), were identified in A. loculosum.
Coxiella prevalence in seabird ticks.
Although Coxiella spp. were found to be ubiquitous from 454 pyrosequencing, precise species identification and measures of population prevalence were impossible to obtain from these data. Thus, we set out to characterize the regional Coxiella prevalence by targeted PCR and classical Sanger sequencing using Coxiella-specific primer pairs that target the 16S rRNA gene (31).
Targeted PCR was performed on the full bank of available individual tick samples, which consisted of C. capensis and A. loculosum specimens collected from seabirds and their nests in Juan de Nova, Tromelin, Europa, Bird, Aride, and La Réunion Islands (Fig. 3). The Coxiella detection prevalence ranged from 46% to 100% (Table 2). Of all C. capensis ticks tested, 65.7% were positive for Coxiella, whereas this figure was 71.2% for A. loculosum. The prevalence of Coxiella bacteria was significantly lower in C. capensis ticks from La Réunion Island than in C. capensis ticks from the other locations (P < 0.001 by Fisher's exact test in all pairwise comparisons).
FIG 3.
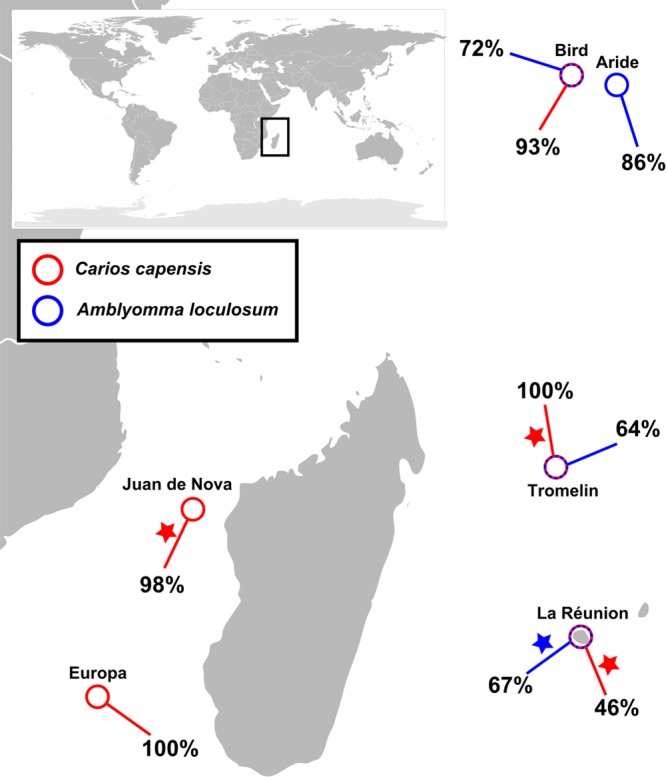
Coxiella in ticks of the southwestern Indian Ocean (SWIO) islands. The prevalence of Coxiella spp. detected in tick samples from different locations of the SWIO is shown. Stars, samples that were included in the 16S rRNA pyrosequencing analyses.
TABLE 2.
Rate of detection of Coxiella spp. in ticks from WIO region
| Tick species | Origin | No. of ticks: |
Prevalence (%) of Coxiella detection | |
|---|---|---|---|---|
| Positive | Tested | |||
| C. capensis | Tromelin Island | 15 | 15 | 100 |
| Juan de Nova Island | 42 | 43 | 98 | |
| Europa Island | 41 | 41 | 100 | |
| La Réunion Island | 87 | 189 | 46 | |
| Bird Island | 14 | 15 | 93 | |
| A. loculosum | Aride Island | 25 | 29 | 86 |
| Tromelin Island | 9 | 14 | 64 | |
| La Réunion Island | 37 | 55 | 67 | |
| Bird Island | 28 | 39 | 72 | |
Coxiella diversity shows a strict tick-host specificity.
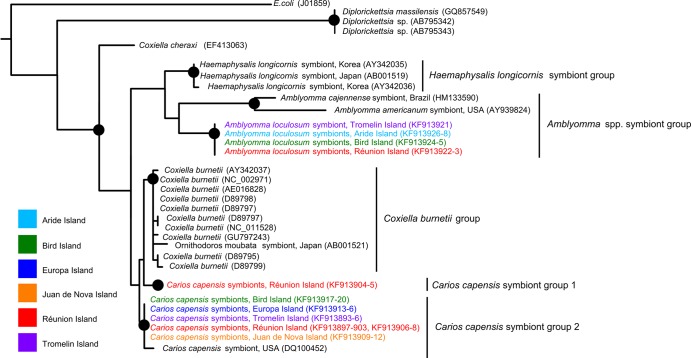
Phylogenetic analyses demonstrated that all Coxiella strains originating from the WIO were closely related to Coxiella burnetii and demonstrated nearly perfect species specificity for their tick hosts (Fig. 4). Two well-supported, separate bacterial clades were identified in hard ticks (e.g., A. loculosum, Amblyomma americanum, Amblyomma cajennense, and Haemaphysalis longicornis) and soft ticks (C. capensis and Ornithodoros moubata). Additionally, distinct Coxiella clades were identifiable for C. capensis and A. loculosum hosts. Furthermore, two distinct Coxiella haplotypes were identified in C. capensis ticks: one in a single P. pacificus nest in Petite Ile and the other in all other sequenced samples from the WIO. Genetic distances revealed that Coxiella strains from C. capensis were the most closely related to Coxiella burnetii. The mean ± standard deviation genetic identities to the C. burnetii group of the phylogenetic groups presented in Fig. 4 were as follows: 98.7% ± 0.19% for C. capensis symbiont group 1, 98.9% ± 0.31% for C. capensis symbiont group 2, 96.4% ± 0.21% for the Amblyomma species symbiont group, and 96.2% ± 0.20% for the Hemaphysalis longicornis symbiont group.
FIG 4.
Phylogenetic relationships between the strains of Coxiella detected. The maximum likelihood phylogenetic tree was calculated with 1,000 bootstrap replicates of Coxiella MUSCLE-aligned partial 16S rRNA sequences from ticks, using selected sequences from GenBank (accession numbers are indicated in parentheses) as reference strains for C. burnetii and C. cheraxi. Escherichia coli (outgroup) and Diplorickettsia spp. (family Coxiellacae) were also included in the analysis. Dots indicate nodes with bootstrap support greater than 90%. The major phylogenetic clades are indicated on the right. Sequences generated as part of this study are indicated with a color code corresponding to the sampled island.
DISCUSSION
The present investigation shows that a combination of nontargeted 454 pyrosequencing with a subsequent specific investigation of a taxon of interest quickly provides a powerful set of data for poorly investigated biological material, such as tropical seabird ticks. Products from two independent PCRs carried out on DNA from four independent pooled tick samples were sequenced, and sequencing indicated little intrasample variability, which may have occurred due to the experimental variability in deep sequencing and/or PCRs. This suggests that increasing the number of samples may be more relevant than duplicating PCRs and sequencing when designing such approaches.
The depth of the analysis provided by this next generation sequencing methodology was sufficient to identify bacterial species of Kocuria (an emerging human pathogen) and other genera that were present in minute trace quantities, such as Borrelia spp., which are known to be transmitted by seabird ticks (5) and which are the causative agents of Lyme disease and relapsing fever. Furthermore, high read numbers of Rickettsia and Coxiella spp. were obtained from the majority of tested tick samples, suggesting that ticks are a frequent and possibly amplifying reservoir host for these bacterial species. Our results corroborate recent work that has shown that these ticks are infected with the human pathogen Rickettsia africae (20). The investigation presented herein reveals massive infection of both hard and soft ticks with Coxiella-like organisms, with the bacteria infecting soft ticks presenting up to 99% identity with C. burnetii. The strict host specificity reported herein supports the vertical transmission of this bacterial species and the previously proposed long coevolution of both Coxiella lineages following an ancient infection in the ancestor of both main families of ticks: the Argasidae (soft ticks) and the Ixodidae (hard ticks) (31). We did not observe any bird species-associated specificity or any spatial structure, which suggests that the presented Coxiella spp. are likely transmitted among islands and bird species. However, the resolution of a 16S rRNA gene marker is not sufficient to completely address these points (O. Duron, personal communication), and the use of additional markers will be necessary to resolve the regional structure of this bacterial population.
Another important point raised by the 454 approach is that the results provide information not only on bacterial diversity but also on bacterial abundance, as previously discussed (32). Indeed, 29.5% of all bacterial 16S rRNA gene haplotypes were related to Coxiella, and another 19.0% were related to Rickettsia. Thus, although a quantitative PCR study of individual ticks will be required to determine bacterial copy numbers per host, our data show that these two bacterial genera are clearly prominent in C. capensis and A. loculosum.
The absence of some symbionts in the sample is also of interest. For instance, the absence of Wolbachia in these seabird ticks is coherent with the proposition of Plantard et al. that Wolbachia isolates that were previously detected in ticks had a parasitoid origin (33).
Two C. capensis ticks from the same nest of a wedge-tailed shearwater (P. pacificus) were infected with a clearly distinct Coxiella haplotype. Interestingly, the same two tick specimens were shown to be infected with two Rickettsia spp. substantially different from the other Rickettsia spp. infecting the WIO sample and were strongly related to Rickettsia belli, considered the earliest diverging Rickettsia species (34, 35). These two specimens may descend from a distinct tick population or lineage, as recently described for C. capensis (19), and thus be infected by distinct Rickettsia and Coxiella strains. Such a pattern would suggest that “a single bird with reservoir ticks may serve to seed a new geographic area,” as previously suggested (36). A population genetics investigation of these ticks at a global scale may allow testing of this tempting but still speculative hypothesis.
Coxiella burnetii is the causative agent of Q fever, which can cause illness in many domesticated mammals, birds, and humans (37). Coxiella-like bacteria have now been identified in a number of distinct tick species (31, 38–42) and are considered symbionts with probable vertical transmission. Whether Rickettsia may be primarily considered an invertebrate symbiont or a vertebrate pathogen is still an open question (34, 43) and can be subjectively related to the main scientific interest of each researcher. Given the growing number of Coxiella species described in several distinct tick species and the prominent prevalence of this taxon in the seabird ticks investigated herein, it seems that a proper assessment of the pathogenicity of these Coxiella species should today be considered an important issue.
ACKNOWLEDGMENTS
Licia Calabrese, Chris Feare, Christine Larose, Sébastien Lefort, Aurélien Prudor, Bernard Rota, and Gérard Rocamora are thanked for their help in tick collection. We thank the Island Conservation Society for permission and support to work on Aride Island. We also acknowledge Marie-France and Guy Savy for supporting the work performed on Bird Island. Sample collection at Aride and Bird Islands and export of the samples to La Réunion Island were performed with the approval of the Seychelles Bureau of Standards and the Ministry of Environment.
This work was supported by FEDER POCT Réunion (Pathogènes Associés à la Faune Sauvage Océan Indien no. 31189), by the Fédération de Recherche Environnement, Biodiversité, Santé, Université de La Réunion (Genoticks), and by the CNRS Institute of Ecologie and the Environment (INEE) (APP Iles Eparses, Program PathOrnithoTiques). The postdoctoral positions of David A. Wilkinson, Muriel Dietrich, Camille Lebarbenchon, and Audrey Jaeger are supported by the European Community FP7 Capacity RegPot Run-Emerge Program. Matthieu Bastien and Céline Le Rouzic were supported by a Fédération de Recherche Environnement, Biodiversité, Santé, training program fellowship.
Footnotes
Published ahead of print 21 March 2014
REFERENCES
- 1.Jones LD, Davies CR, Steele GM, Nuttall PA. 1987. A novel mode of arbovirus transmission involving a nonviremic host. Science 237:775–777. 10.1126/science.3616608 [DOI] [PubMed] [Google Scholar]
- 2.Jongejan F, Uilenberg G. 2004. The global importance of ticks. Parasitology 129(Suppl):S3–S14. 10.1017/S0031182004005967 [DOI] [PubMed] [Google Scholar]
- 3.Hubálek Z. 2004. An annotated checklist of pathogenic microorganisms associated with migratory birds. J. Wildl. Dis. 40:639–659. 10.7589/0090-3558-40.4.639 [DOI] [PubMed] [Google Scholar]
- 4.Olsen B, Jaenson TGT, Noppa L, Bunikis J, Bergstrom S. 1993. A Lyme borreliosis cycle in seabirds and Ixodes uriae ticks. Nature 362:340–342. 10.1038/362340a0 [DOI] [PubMed] [Google Scholar]
- 5.Dietrich M, Gómez-Díaz E, McCoy KD. 2011. Worldwide distribution and diversity of seabird ticks: implications for the ecology and epidemiology of tick-borne pathogens. Vector Borne Zoonotic Dis. 11:453–470. 10.1089/vbz.2010.0009 [DOI] [PubMed] [Google Scholar]
- 6.Tsiodras S, Kelesidis T, Kelesidis I, Bauchinger U, Falagas ME. 2008. Human infections associated with wild birds. J. Infect. 56:83–98. 10.1016/j.jinf.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Major L, Linn ML, Slade RW, Schroder WA, Hyatt AD, Gardner J, Cowley J, Suhrbier A. 2009. Ticks associated with Macquarie Island penguins carry arboviruses from four genera. PLoS One 4:e4375. 10.1371/journal.pone.0004375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gylfe A, Yabuki M, Drotz M, Bergström S, Fukunaga M, Olsen B. 2001. Phylogeographic relationships of Ixodes uriae (Acari: Ixodidae) and their significance to transequatorial dispersal of Borrelia garinii. Hereditas 134:195–199. 10.1111/j.1601-5223.2001.00195.x [DOI] [PubMed] [Google Scholar]
- 9.Smith RP, Jr, Muzaffar SB, Lavers J, Lacombe EH, Cahill BK, Lubelczyk CB, Kinsler A, Mathers AJ, Rand PW. 2006. Borrelia garinii in seabird ticks (Ixodes uriae), Atlantic Coast, North America. Emerg. Infect. Dis. 12:1909–1912. 10.3201/eid1212.060448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duneau D, Boulinier T, Gómez-Díaz E, Petersen A, Tveraa T, Barrett RT, McCoy KD. 2008. Prevalence and diversity of Lyme borreliosis bacteria in marine birds. Infect. Genet. Evol. 8:352–359. 10.1016/j.meegid.2008.02.006 [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Diaz E, Boulinier T, Sertour N, Cornet M, Ferquel E, Mccoy KD. 2011. Genetic structure of marine Borrelia garinii and population admixture with the terrestrial cycle of Lyme borreliosis. Environ. Microbiol. 13:2453–2467. 10.1111/j.1462-2920.2011.02515.x [DOI] [PubMed] [Google Scholar]
- 12.Sinclair I, Langrand O. 2013. Birds of the Indian Ocean islands. Random House Struik, Cape Town, South Africa [Google Scholar]
- 13.Le Corre M, Salamolard M, Portier MC. 2003. Transoceanic dispersion of the red-tailed tropicbird in the Indian Ocean. Emu 103:183–184. 10.1071/MU02026 [DOI] [Google Scholar]
- 14.Weimerskirch H, Le Corre M, Marsac F, Barbraud C, Tostain O, Chastel O. 2006. Postbreeding movements of frigatebirds tracked with satellite telemetry. The Condor 108:220–225. 10.1650/0010-5422(2006)108[0220:PMOFTW]2.0.CO;2 [DOI] [Google Scholar]
- 15.Newton I. 2008. The migration ecology of birds. Academic Press, London, United Kingdom [Google Scholar]
- 16.Tortosa P, Pascalis H, Guernier V, Cardinale E, Le Corre M, Goodman SM, Dellagi K. 2012. Deciphering arboviral emergence within insular ecosystems. Infect. Genet. Evol. 12:1333–1339. 10.1016/j.meegid.2012.03.024 [DOI] [PubMed] [Google Scholar]
- 17.Lonchampt C, Migliani R, Ratsitorahina M, Rabarijaona LP, Ramarokoto CE, Rakoto Andrianarivelo M, Rousset D. 2003. Persistence of an endemic circulation of the West Nile virus in Madagascar. Arch. Inst. Pasteur Madagascar 69:33–36 (In French.) [PubMed] [Google Scholar]
- 18.Hoogstraal H, Wassef HY, Converse JD, Keirans JE, Clifford CM, Feare CJ. 1976. Amblyomma loculosum (Ixodoidea: Ixodidae): identity, marine bird and human hosts, virus infection, and distribution in the southern oceans. Ann. Entomol. Soc. Am. 69:3-14 [Google Scholar]
- 19.Gómez-Díaz E, Morris-Pocock JA, González-Solís J, McCoy KD. 2012. Trans-oceanic host dispersal explains high seabird tick diversity on Cape Verde Islands. Biol. Lett. 8:616–619. 10.1098/rsbl.2012.0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietrich M, Lebarbenchon C, Jaeger A, Le Rouzic C, McCoy K, Pascalis H, Lecorre M, Dellagi K, Tortosa P. Rickettsia species in seabird ticks of the western Indian Ocean: a wide distribution with strong host specificity. Emerg. Infect. Dis., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohls GM. 1957. Insects of Micronesia. Acarina: Ixodoidea. Bernice P. Bishop Museum, Honolulu, HI [Google Scholar]
- 22.Robinson LE. 1926. Ticks: the genus Amblyomma. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 23.Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. 2009. Metagenomic pyrosequencing and microbial identification. Clin. Chem. 55:856–866. 10.1373/clinchem.2008.107565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 26.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386. 10.1101/gr.5969107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeves WK, Loftis AD, Sanders F, Spinks MD, Wills W, Denison AM, Dasch GA. 2006. Borrelia, Coxiella, and Rickettsia in Carios capensis (Acari: Argasidae) from a brown pelican (Pelecanus occidentalis) rookery in South Carolina, USA. Exp. Appl. Acarol. 39:321–329. 10.1007/s10493-006-9012-7 [DOI] [PubMed] [Google Scholar]
- 28.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 29.Hechemy KE. 2012. History and prospects of Coxiella burnetii research. Adv. Exp. Med. Biol. 984:1–11. 10.1007/978-94-007-4315-1_1 [DOI] [PubMed] [Google Scholar]
- 30.Purty S, Saranathan R, Prashanth K, Narayanan K, Asir J, Sheela Devi C, Kumar Amarnath S. 2013. The expanding spectrum of human infections caused by Kocuria species: a case report and literature review. Emerg. Microbes Infect. 2:e71. 10.1038/emi.2013.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almeida AP, Marcili A, Leite RC, Nieri-Bastos FA, Domingues LN, Martins JR, Labruna MB. 2012. Coxiella symbiont in the tick Ornithodoros rostratus (Acari: Argasidae). Ticks Tick Borne Dis. 3:203–206. 10.1016/j.ttbdis.2012.02.003 [DOI] [PubMed] [Google Scholar]
- 32.Amend AS, Seifert KA, Bruns TD. 2010. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol. Ecol. 19:5555–5565. 10.1111/j.1365-294X.2010.04898.x [DOI] [PubMed] [Google Scholar]
- 33.Plantard O, Bouju-Albert A, Malard M-A, Hermouet A, Capron G, Verheyden H. 2012. Detection of Wolbachia in the tick Ixodes ricinus is due to the presence of the hymenoptera endoparasitoid Ixodiphagus hookeri. PLoS One 7:e30692. 10.1371/journal.pone.0030692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perlman SJ, Hunter MS, Zchori-Fein E. 2006. The emerging diversity of Rickettsia. Proc. Biol. Soc. 273:2097–2106. 10.1098/rspb.2006.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, Fournier P-E, Claverie J-M, Raoult D. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:e76. 10.1371/journal.pgen.0020076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoogstraal H. 1961. Migrating birds and their ectoparasites in relation to disease. East Afr. Med. J. 38:221–226 [PubMed] [Google Scholar]
- 37.Maurin M, Raoult D. 1999. Q fever. Clin. Microbiol. Rev. 12:518–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernasconi MV, Casati S, Péter O, Piffaretti J-C. 2002. Rhipicephalus ticks infected with Rickettsia and Coxiella in southern Switzerland (Canton Ticino). Infect. Genet. Evol. 2:111–120. 10.1016/S1567-1348(02)00092-8 [DOI] [PubMed] [Google Scholar]
- 39.Mediannikov O, Ivanov L, Nishikawa M, Saito R, Sidelnikov YN, Zdanovskaya NI, Tarasevich IV, Suzuki H. 2003. Molecular evidence of Coxiella-like microorganism harbored by Haemaphysalis concinnae ticks in the Russian Far East. Ann. N. Y. Acad. Sci. 990:226–228. 10.1111/j.1749-6632.2003.tb07367.x [DOI] [PubMed] [Google Scholar]
- 40.Machado-Ferreira E, Dietrich G, Hojgaard A, Levin M, Piesman J, Zeidner NS, Soares CAG. 2011. Coxiella symbionts in the Cayenne tick Amblyomma cajennense. Microb. Ecol. 62:134–142. 10.1007/s00248-011-9868-x [DOI] [PubMed] [Google Scholar]
- 41.Reeves WK. 2008. Molecular evidence for a novel Coxiella from Argas monolakensis (Acari: Argasidae) from Mono Lake, California, USA. Exp. Appl. Acarol. 44:57–60. 10.1007/s10493-008-9128-z [DOI] [PubMed] [Google Scholar]
- 42.Reeves WK, Loftis AD, Priestley RA, Wills W, Sanders F, Dasch GA. 2005. Molecular and biological characterization of a novel Coxiella-like agent from Carios capensis. Ann. N. Y. Acad. Sci. 1063:343–345. 10.1196/annals.1355.055 [DOI] [PubMed] [Google Scholar]
- 43.La Scola B, Raoult D. 1997. Laboratory diagnosis of rickettsioses: current approaches to diagnosis of old and new rickettsial diseases. J. Clin. Microbiol. 35:2715–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]