Abstract
Acetogenic bacteria use CO and/or CO2 plus H2 as their sole carbon and energy sources. Fermentation processes with these organisms hold promise for producing chemicals and biofuels from abundant waste gas feedstocks while simultaneously reducing industrial greenhouse gas emissions. The acetogen Clostridium autoethanogenum is known to synthesize the pyruvate-derived metabolites lactate and 2,3-butanediol during gas fermentation. Industrially, 2,3-butanediol is valuable for chemical production. Here we identify and characterize the C. autoethanogenum enzymes for lactate and 2,3-butanediol biosynthesis. The putative C. autoethanogenum lactate dehydrogenase was active when expressed in Escherichia coli. The 2,3-butanediol pathway was reconstituted in E. coli by cloning and expressing the candidate genes for acetolactate synthase, acetolactate decarboxylase, and 2,3-butanediol dehydrogenase. Under anaerobic conditions, the resulting E. coli strain produced 1.1 ± 0.2 mM 2R,3R-butanediol (23 μM h−1 optical density unit−1), which is comparable to the level produced by C. autoethanogenum during growth on CO-containing waste gases. In addition to the 2,3-butanediol dehydrogenase, we identified a strictly NADPH-dependent primary-secondary alcohol dehydrogenase (CaADH) that could reduce acetoin to 2,3-butanediol. Detailed kinetic analysis revealed that CaADH accepts a range of 2-, 3-, and 4-carbon substrates, including the nonphysiological ketones acetone and butanone. The high activity of CaADH toward acetone led us to predict, and confirm experimentally, that C. autoethanogenum can act as a whole-cell biocatalyst for converting exogenous acetone to isopropanol. Together, our results functionally validate the 2,3-butanediol pathway from C. autoethanogenum, identify CaADH as a target for further engineering, and demonstrate the potential of C. autoethanogenum as a platform for sustainable chemical production.
INTRODUCTION
Increasing awareness of the impact of rising CO2 levels in the atmosphere (1) is driving research into technologies that allow the production of fuels and chemicals in more environmentally sustainable processes that use fewer fossil carbon-intensive resources. While the transportation sector remains the single largest user of oil-derived products, the global chemical industry is also dependent on oil and natural gas feedstocks for the production of chemicals, including solvents, synthetic rubbers, fertilizers, and the intermediates for manufacturing plastics. Four-carbon (C4) petrochemicals are among those that are projected to become particularly scarce due to feedstock switching in the refining industry, with a global trend away from petroleum-derived naphtha and toward lower-cost natural gas (2).
Gas fermentation by acetogenic bacteria (acetogens) is a promising alternative route for producing chemicals and biofuels (3–5). Acetogens are obligate anaerobes that use the Wood-Ljungdahl pathway to synthesize acetyl-coenzyme A (acetyl-CoA) by reducing CO and/or CO2 plus H2 (6, 7). Not only do these gases comprise the sole source of carbon and energy for product synthesis, but also they can be sourced in large volumes from a range of waste and renewable resources, including industrial waste gases and synthesis gas (syngas) that is produced by gasification of biomass, coal, or municipal solid waste. Compared with traditional chemical syntheses from syngas, bacterial fermentation processes are less sensitive to variations in the composition of the gaseous substrate and more robust in the presence of contaminants, and they can lead to greater product specificity (3, 8). Gas fermentation also offers the opportunity to reduce industrial greenhouse gas emissions while producing chemicals and biofuels from feedstocks that are noncellulosic, and therefore do not consume food resources or reduce the availability of arable land for food cultivation.
More than 100 acetogenic species have been identified; however, over 90% of these produce acetate as the sole fermentation product (8). In addition to acetate, some acetogens are able to synthesize products, including ethanol, butanol, and butyrate (3), all of which are derived directly from acetyl-CoA. Until recently, it was unknown whether pyruvate-derived metabolites could be synthesized in significant quantities by acetogenic bacteria. We discovered that three acetogenic Clostridium species—Clostridium autoethanogenum, C. ljungdahlii, and C. ragsdalei — are able to produce the pyruvate-derived metabolites lactate and 2,3-butanediol from CO-containing steel mill waste gases (9). The identification of 2,3-butanediol as a native fermentation product was particularly significant because this C4 molecule is a precursor in the manufacture of a range of chemical products, including solvents, such as methyl ethyl ketone, and also 1,3-butadiene, which is an intermediate in the manufacture of nylon and the monomer used for producing synthetic rubber. Commercially, the key downstream products of 2,3-butanediol have a global market of ∼32 million tonnes per annum, valued at ∼$43 billion (10–12). Previously, 2,3-butanediol production has been shown only in sugar- or citrate-fermenting bacteria (the majority of which are risk group 2 organisms) (10, 12–18) or in engineered strains of C. acetobutylicum (19, 20), Escherichia coli (21, 22), or Synechococcus elongatus (22).
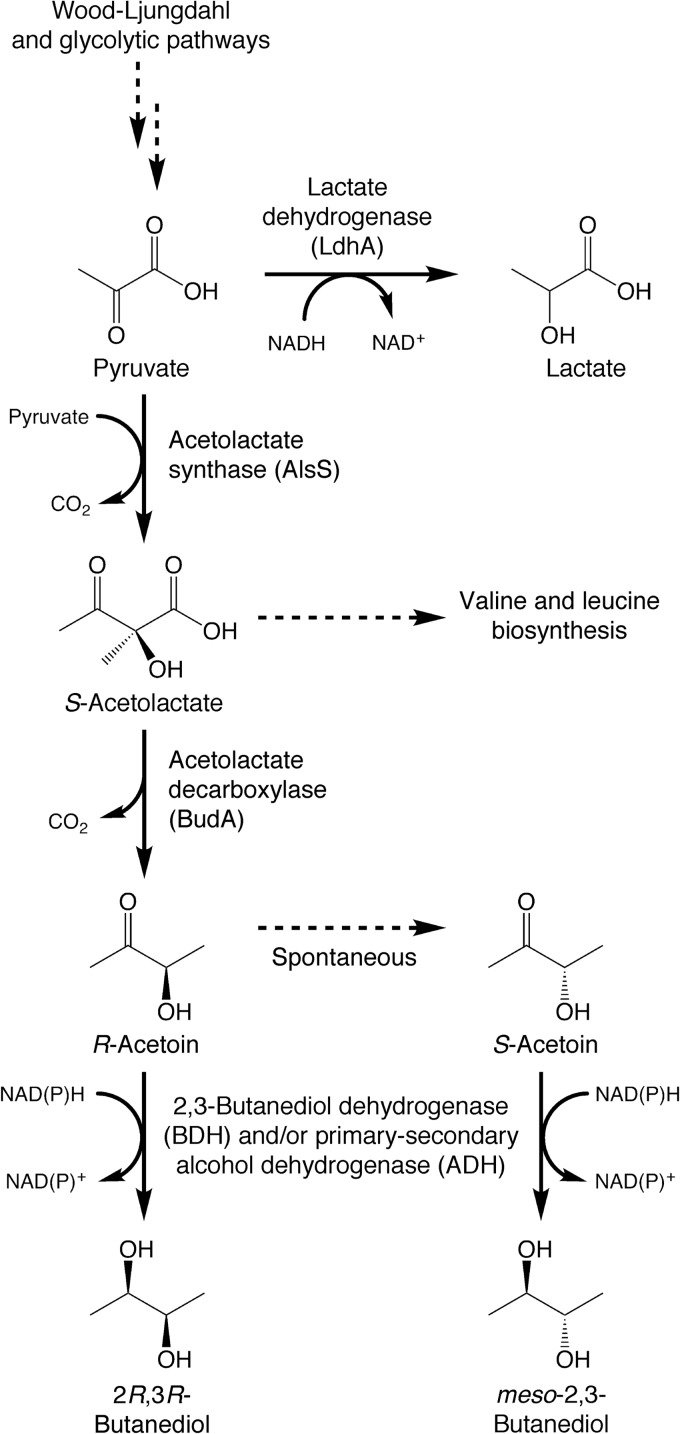
Our previous work highlighted the biotechnological potential of acetogens that produce 2,3-butanediol natively (9). However, in order to optimize an industrially relevant fermentation process, it is necessary to identify and characterize the enzymes that catalyze the synthesis of relevant metabolites, including 2,3-butanediol and lactate. We identified candidate genes for these enzymes through a combination of in silico sequence analysis and quantitative PCR to relate gene expression with chemical production (9). The proposed pathways for pyruvate utilization are shown in Fig. 1. In this study, we undertook direct functional validations of our previous predictions by using the relevant C. autoethanogenum genes to reconstruct the acetogenic 2,3-butanediol and lactate pathways in E. coli.
FIG 1.
Pathways for production of 2,3-butanediol and lactate from pyruvate in C. autoethanogenum.
An unanswered question from our previous study centered on the identity of the enzyme that reduces acetoin to 2,3-butanediol (Fig. 1). We identified a putative 2,3-butanediol dehydrogenase which shares 78% amino acid identity with an acetoin reductase from C. beijerinckii (20) and which appeared to be NADH and zinc dependent (9). However, the gene for this enzyme was expressed constitutively in C. autoethanogenum, at a high normalized mRNA level (9). This was in sharp contrast to all the other genes of the 2,3-butanediol and lactate pathways, which were highly upregulated (∼15-fold) only during stationary phase (which is when the cells were producing 2,3-butanediol and lactate). Furthermore, a recent analysis of C. autoethanogenum cell extracts detected acetoin reductase activities that were both NADH and NADPH dependent (23). These observations led us to hypothesize that a second dehydrogenase may also be important for the production of 2,3-butanediol in C. autoethanogenum. In this work, we identified this dehydrogenase and characterized its kinetic properties in vitro.
MATERIALS AND METHODS
Materials.
All molecular biology enzymes were purchased from New England BioLabs (Ipswich, MA). Oligonucleotides were from Integrated DNA Technologies (Coralville, IA). l-Arabinose was from Gold Biotechnology (St. Louis, MO). Dithiothreitol (DTT) was from Melford Laboratories (Ipswich, United Kingdom). Benzonase nuclease and reduced β-NAD (NADH) were from Merck Millipore (Billerica, MA). Talon metal affinity resin was from Clontech (Mountain View, CA). Reduced and oxidized forms of β-NAD 2′-phosphate (NADPH and NADP+, respectively), protease inhibitor cocktail, chicken egg white lysozyme, and the assayed substrates (acetaldehyde, RS-acetoin, 2R,3R-butanediol, meso-2,3-butanediol, 2S,3S-butanediol, acetone, and butanone) were from Sigma Chemical Co. (St. Louis, MO).
Bacterial strains.
Acetobacterium woodii WB1 (DSM 1030), C. aceticum (DSM 1496), C. autoethanogenum JAI-1 (DSM 10061), C. carboxidivorans P7 (DSM 15243), C. ljungdahlii PETC (DSM 13528), and C. beijerinckii NRRL-B593 (DSM 6423) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ), Braunschweig, Germany. C. ragsdalei P11 (ATCC BAA-622), C. beijerinckii NCIMB 8052 (ATCC 51743), and C. acetobutylicum (ATCC 824) were from the American Type Culture Collection (ATCC), Manassas, VA. E. coli strains DH5α and LMG194 were from Invitrogen (Carlsbad, CA). E. coli strain JW1375 was from the Coli Genetic Stock Center (CGSC), New Haven, CT. Cloning was carried out in E. coli strain XL1-Blue MRF′ (Stratagene, La Jolla, CA).
Reconstruction of the C. autoethanogenum 2,3-butanediol and lactate pathways in E. coli.
PCR amplification was performed using iProof High Fidelity DNA polymerase (Bio-Rad, Hercules, CA), and the primers used are listed in Table 1. Genomic DNA was extracted from C. autoethanogenum strain DSM 10061 as described previously (24). The complete sequence of this genome is available from GenBank (accession number CP006763) and has been described in detail elsewhere (25).
TABLE 1.
Oligonucleotides used in this study

Restriction sites are underlined.
First, the promoter region of the phosphotransacetylase-acetate kinase operon (Ppta-ack) was amplified from C. autoethanogenum genomic DNA by using primers Ppta-ack-NotI-F and Ppta-ack-NdeI-R. The amplified product was cloned into vectors pMTL85141 and pMTL85241 by using NotI and NdeI restriction sites, generating the plasmids pMTL85141-P and pMTL85241-P, respectively. The pMTL system of shuttle vectors has been described elsewhere (26); it is a modular system that comprises different components with standard numbering systems, as described at http://clostron.com/pMTL80000.php. Plasmids pMTL85141 and pMTL85241 each contain the Gram-positive replication origin from pIM13, the Gram-negative origin from ColE1, and a multiple-cloning site. Plasmid pMTL85141 contains a chloramphenicol resistance marker, whereas pMTL85241 confers erythromycin resistance.
To reconstruct the 2,3-butanediol pathway, the candidate C. autoethanogenum alsS gene (proposed to encode a catabolic acetolactate synthase; locus tag CAETHG_1740) was amplified using primers als-NdeI-F and als-EcoRI-R, and the resulting amplicon was cloned into pMTL85141-P by using NdeI and EcoRI restriction sites. The putative budA gene (encoding an acetolactate decarboxylase; locus tag CAETHG_2932) was then added to this plasmid by amplification using primers ALDC-EcoRI-F and ALDC-SacI-R, with subsequent cloning using EcoRI and SacI restriction sites, generating the plasmid pMTL85141-P-alsS-budA. Finally, primers BDH-SacI-F and BDH-KpnI-R were used to amplify bdh (encoding the putative 2,3-butanediol dehydrogenase; locus tag CAETHG_0385) from C. autoethanogenum genomic DNA, and the resulting amplicon was cloned using SacI and KpnI restriction sites, generating the plasmid pMTL85141-P-alsS-budA-bdh.
The genes for a primary-secondary alcohol dehydrogenase from C. autoethanogenum (adh) and a 2,3-butanediol dehydrogenase from Klebsiella pneumoniae (27) were also each cloned into plasmid pMTL85141-P-alsS-budA, to test their activities in reducing acetoin to 2,3-butanediol and to confirm the stereospecificity of the enzymes. The C. autoethanogenum adh gene (locus tag CAETHG_0553) was amplified from genomic DNA by using primers LZAdh469-SalI-F and LZAdh469-XhoI-R, while the K. pneumoniae 2,3-butanediol dehydrogenase (encoded by the budC gene; accession number AF098800) was synthesized by GeneArt (Regensburg, Germany). The genes were cloned using SalI and XhoI restriction sites, generating the plasmids pMTL85141-P-alsS-budA-adh and pMTL85141-P-alsS-budA-budC, respectively.
To reconstruct the pathway for lactate production, the putative lactate dehydrogenase gene, ldhA (locus tag CAETHG_1147), was amplified from C. autoethanogenum genomic DNA by using primers Ldh-BamHI-F and Ldh-SalI-R and then cloned into pMTL85241-P, between BamHI and SalI restriction sites.
The plasmids described above and the control plasmids pMTL85141-P and pMTL85241-P were used to transform either E. coli XL1-Blue or E. coli JW1375, which is a lactate dehydrogenase-negative strain from the Keio collection (28). Transformants were verified by plasmid purification and restriction digestion. Single colonies were picked to inoculate overnight cultures into LB medium. Inoculants were concentrated by pelleting cells, removing 80% of the original volume from the supernatant, and then resuspending the pellet to generate a 5× concentrated inoculum. Anaerobic 125-ml serum bottles (Bellco Glass Inc., Vineland, NJ) containing 50 ml M9 medium with 2.5 g liter−1 glucose and either 25 mg liter−1 chloramphenicol (for maintenance of pMTL85141 derivatives) or 250 mg liter−1 erythromycin (for pMTL85241-derived plasmids) were inoculated with 0.5-ml aliquots of the resuspended cells. The cultures were shaken at 37°C and analyzed for production of acetoin and 2,3-butanediol, or lactate, by high-pressure liquid chromatography (HPLC). Growth was monitored by measuring the optical density at 600 nm (OD600).
Analytics.
Acetoin, 2,3-butanediol, lactate, acetate, and ethanol concentrations were determined using an Agilent 1100 series HPLC system (Agilent Technologies, Santa Clara, CA) equipped with a refractive index detector operated at 35°C and an Alltech IOA-2000 organic acid column. The column was kept at 60°C. Slightly acidified water (0.005 M H2SO4) was used as the mobile phase, with a flow rate of 0.7 ml/min. To remove proteins and other cell residues, each 400-μl sample was mixed with 100 μl of 2% (wt/vol) 5-sulfosalicylic acid, and the samples were centrifuged at 14,000 × g for 3 min. The supernatant (10 μl) was then injected into the HPLC system for analysis. Reliable detection was achieved with ≥1 mg of each metabolite.
Acetone and isopropanol were measured using gas chromatography (GC) analysis, employing an Agilent 6890N headspace GC equipped with a Supelco polyethylene glycol (PEG) 60-μm solid-phase microextraction fiber, a Restek Rtx-1 (30 m × 0.32 μm × 5 μm) column, and a flame ionization detector (FID). Samples (4 ml) were transferred into a 20-ml headspace vial, upon which the fiber was incubated (exposed) for 10 min at 50°C. The sample was desorbed in the injector at 250°C for 9 min. Chromatography was performed with an oven program of 40°C (5-min hold) and 10°C/min to 200°C, followed by a 5-min hold at 220°C. The column flow rate was 1 ml/min, with hydrogen as the carrier gas. The FID was kept at 250°C, with hydrogen at 40 ml/min, air at 450 ml/min, and nitrogen at 15 ml/min as the makeup gas. The detection limit of the system was 1 mg of either acetone or isopropanol.
Optical activity was measured at room temperature on a polartronic NH8 instrument (Schmidt+Haensch, Berlin, Germany) at 589 nm and 20°C. Pure chiral standards (2R,3R-butanediol, 2S,3S-butanediol, and meso-2,3-butanediol) were obtained from Sigma Chemical Co. For identification of 2,3-butanediol stereoisomers, an Agilent GC-mass spectrometry (GC-MS) system with a quadrupole mass selective detector operated at 70 eV was used. A ZB-1701 column (30 m × 250 μm × 150-μm film thickness; Phenomenex, Torrance, CA) with a 5-m guard column was used for all analyses.
Expression and purification of C. autoethanogenum primary-secondary alcohol dehydrogenase (CaADH).
The C. autoethanogenum adh gene was amplified from the plasmid pMTL85141-P-alsS-budA-adh by using Phusion polymerase and the primers adh_KpnI.for and adh_HindIII.rev (Table 1). The product was cloned into the expression vector pBAD(KpnI) by using KpnI and HindIII. This vector had been constructed previously by modifying the multiple-cloning site of pBAD/Myc-His B (Invitrogen) to include a sequence that encoded an N-terminal His6 tag and a cleavage site for the tobacco etch virus (TEV) protease. The resulting plasmid, pBAD(KpnI)-CaADH, was used to transform the expression host, E. coli strain LMG194 (29).
Expression of CaADH was induced in mid-log-phase cultures (OD600 ≈ 0.5) by adding l-arabinose to a final concentration of 0.2% (wt/vol). The cultures were incubated at 28°C for an additional 5 h. Cells were harvested by centrifugation, and the pellets were stored at −80°C. Each pellet (∼3.0 g cell wet weight from a 300-ml culture) was resuspended in 10 ml of lysis buffer (50 mM potassium phosphate, 300 mM NaCl, pH 7.0). Protease inhibitor cocktail (150 μl), Benzonase nuclease (37.5 U), and lysozyme (final concentration, 0.2 mg ml−1) were added. After 20 min of incubation at 4°C, cells were lysed by sonication on ice, and the lysates were clarified by centrifugation (21,000 × g, 4°C, 30 min). The clarified lysate was mixed with 500 μl Talon metal affinity resin (50% [wt/vol] slurry), and the mixture was gently agitated at 4°C for 1 h to allow the His6-tagged CaADH protein to bind the resin. The resin was washed twice with 10 bed volumes of lysis buffer before being transferred to a gravity flow column. After two further washes with 10 bed volumes of lysis buffer, containing 5 mM imidazole and then 10 mM imidazole, the purified protein was eluted with 5 bed volumes of elution buffer (50 mM potassium phosphate, 300 mM NaCl, 150 mM imidazole, pH 7.0). Amicon Ultra centrifugal filter units (10-kDa cutoff; Merck Millipore) were used to exchange the purified protein into storage buffer (50 mM potassium phosphate, 150 mM NaCl, 10% [vol/vol] glycerol, pH 8.0). Aggregates were removed by filtration through a sterile 0.22-μm filter (Millex-GV; Millipore). CaADH concentrations were quantified by measuring the A280 (ε = 33,920 M−1 cm−1, calculated as reported previously [30]). Aliquots of the purified protein were stored at −80°C. Activity assays verified that these storage conditions, combined with a freeze-thaw cycle, did not lead to any loss of activity.
Activity assays.
CaADH activity was measured by using a spectrophotometric assay based on a method described previously (31). Activity was quantified by monitoring the decrease in absorbance at 340 nm associated with the oxidation of NADPH (ε340 = 6,220 M−1 cm−1). Steady-state kinetic parameters were measured at 25°C by using a Cary 100 UV-visible (UV-Vis) spectrophotometer with a Peltier temperature controller. The standard assay mixture contained 50 mM Tris-HCl, 0.2 mM NADPH, and 1 mM DTT, pH 7.5. CaADH was present at 6 nM. Initial reaction rates were measured with at least six concentrations of each substrate, generally covering the range of 0 to 5 times the estimated Km value. The concentration ranges used were as follows: 0 to 50 mM for acetaldehyde, 0 to 240 mM for acetoin, 0 to 3 mM for acetone, and 0 to 6 mM for butanone. The ability of CaADH to oxidize each stereoisomer of 2,3-butanediol was tested under the same conditions, except that 0.2 mM NADP+ was used in place of NADPH and the enzyme concentration was increased to 50 nM. A concentration range of 0 to 150 mM was tested for each stereoisomer. All measurements were made in triplicate and corrected for background. Kinetic parameters (kcat and Km) were determined by fitting the data directly to the Michaelis-Menten equation, using nonlinear regression analysis in GraphPad Prism (GraphPad, La Jolla, CA). Values are reported as means ± standard errors.
Acetone-to-isopropanol reduction capability.
A number of strains were analyzed for the ability to reduce exogenous acetone to isopropanol. The strains were cultivated under strict anaerobic conditions (32) and at 37°C, except for C. ragsdalei, which was incubated at 30°C. C. beijerinckii and C. acetobutylicum were grown in RCM medium (33), and E. coli was grown in LB medium. All other organisms were grown autotrophically in modified PETC medium (ATCC medium 1754 with fructose omitted; for K. pneumoniae and C. aceticum, the pH was adjusted to 8.2 and 7.4, respectively). Steel mill waste gas (composition, 42% CO, 35% N2, 21% CO2, and 2% H2; collected from a New Zealand Steel site in Glenbrook, New Zealand) at a pressure of 200 kPa was the carbon and energy source for the acetogens.
Growth experiments were carried out in 50 ml of medium, using 125-ml serum bottles (Bellco Glass Inc.) with butyl rubber stoppers. All cultures were inoculated to an OD600 of 0.1 in 50 ml of the appropriate medium. The cultures were allowed to double (OD600 = 0.2) before acetone was added to approximately 170 mM (i.e., approximately 10 g liter−1). Immediately after acetone addition, a sample was taken and analyzed for acetone and isopropanol by GC. After 120 h of growth, samples were analyzed for acetone and isopropanol by GC and for acetate, ethanol, and 2,3-butanediol by HPLC. Cell density (OD600) was also measured. All growth experiments were performed at least in triplicate.
RESULTS
Pathway reconstruction and identification of a primary-secondary alcohol dehydrogenase.
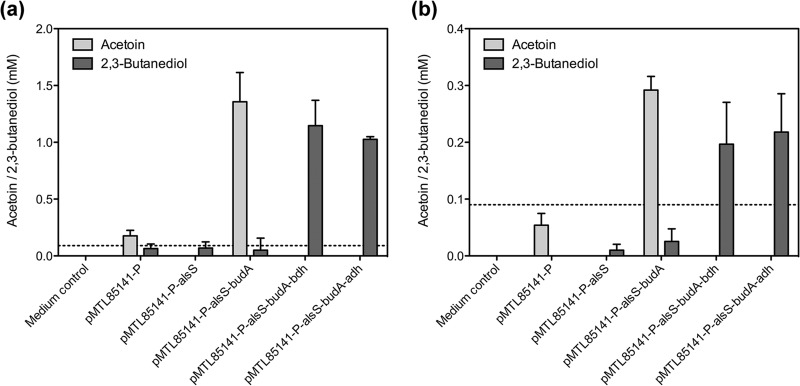
The first goal of this study was to verify the functions of the putative acetolactate synthase (AlsS), acetolactate decarboxylase (BudA), and 2,3-butanediol dehydrogenase (BDH) enzymes (Fig. 1) that we previously identified in silico (9). The candidate genes from C. autoethanogenum were cloned into the pMTL85141 vector and expressed in E. coli XL1-Blue. The genes were expressed under the control of the phosphotransacetylase-acetate kinase operon promoter (Ppta-ack) of C. autoethanogenum, which our preliminary studies had shown to be active in E. coli. The production of acetoin and 2,3-butanediol was monitored using HPLC and cultures grown anaerobically and aerobically (Fig. 2).
FIG 2.
Acetoin and 2,3-butanediol production in modified strains of E. coli XL1-Blue, normalized to biomass. The C. autoethanogenum genes for acetolactate synthase (alsS), acetolactate decarboxylase (budA), 2,3-butanediol dehydrogenase (bdh), and primary-secondary alcohol dehydrogenase (adh) were expressed from the pMTL85141 plasmid under the control of the C. autoethanogenum Ppta-ack promoter. Endpoint measurements were made after 47 h of growth. Data are means and standard deviations for three independent experiments. The reliable detection limit for each metabolite (90 μM) is indicated with a dashed line. (a) Anaerobic growth. (b) Aerobic growth.
Acetoin and 2,3-butanediol were occasionally detected in cultures harboring pMTL85141-P (i.e., the promoter-only control) and when AlsS alone was expressed. However, the levels of these metabolites were at or below the reliable detection limits of our HPLC protocol (Fig. 2), suggesting that they were trace contaminants. In contrast, when the AlsS and BudA enzymes were coexpressed, significant acetoin production was observed within 47 h of growth. Anaerobic cultures accumulated acetoin to 1.4 ± 0.3 mM per OD600 unit (Fig. 2a), whereas acetoin production was reduced nearly 5-fold (to 0.29 ± 0.02 mM per OD600 unit) under aerobic conditions (Fig. 2b). Addition of BDH led to complete reduction of all acetoin to 2,3-butanediol (Fig. 2).
Our previous gene expression studies (9) and the identification of NADPH-dependent acetoin reductase activity in C. autoethanogenum cell extracts (23) led us to hypothesize that a second dehydrogenase was also important for 2,3-butanediol production during gas fermentation. The sugar-fermenting, isopropanol-producing C. beijerinckii strain NRRL-B593 is known to contain an NADPH-dependent primary-secondary alcohol dehydrogenase (31, 34). Inspection of the C. autoethanogenum genome revealed a close homologue (locus tag CAETHG_0553). The C. autoethanogenum enzyme (CaADH) and the C. beijerinckii enzyme (CbADH) each comprise 351 amino acids, 302 of which (i.e., 86%) are identical. Conserved amino acids include those that determine specificity for the NADPH cofactor (Gly198, Ser199, Arg200, and Tyr218) and those that coordinate a catalytic zinc ion in the active site (Cys37, His59, and Asp150) (35). CaADH was tested for the ability to reduce acetoin by coexpression with AlsS and BudA in E. coli XL1-Blue. As observed for the 2,3-butanediol dehydrogenase, this enzyme was also able to completely reduce acetoin to 2,3-butanediol (Fig. 2).
C. autoethanogenum has been shown to produce 94% 2R,3R-butanediol and 6% meso-2,3-butanediol during gas fermentation (9). The absence of 2S,3S-butanediol implies that only R-acetoin, not S-acetoin, is likely to be produced by the C. autoethanogenum BudA enzyme (as shown in Fig. 1). This is consistent with the known mechanism of all other acetolactate decarboxylases, which also produce only the R enantiomer of acetoin (36). It also suggests that the C. autoethanogenum ADH and BDH enzymes are highly stereoselective and install an R stereocenter when they reduce acetoin to 2,3-butanediol. On the other hand, the K. pneumoniae budC gene encodes a 2,3-butanediol dehydrogenase that installs an S stereocenter (21, 27). To investigate the stereospecificity of CaADH, we compared it to the K. pneumoniae 2,3-butanediol dehydrogenase by coexpressing each enzyme with AlsS and BudA in E. coli XL1-Blue. Expression of the K. pneumoniae dehydrogenase led to production of optically inactive meso-2,3-butanediol, almost exclusively (Fig. 3b), as expected when a new S stereocenter is installed during the reduction of R-acetoin. A low level of optically active 2,3-butanediol was also detected. This was presumed to arise through the spontaneous racemization of R-acetoin (37), yielding a low level of S-acetoin that could be reduced to 2S,3S-butanediol. In contrast, E. coli cells expressing CaADH produced only the optically active form of 2,3-butanediol (Fig. 3a). This provided strong circumstantial evidence that CaADH stereospecifically reduces R-acetoin to 2R,3R-butanediol.
FIG 3.
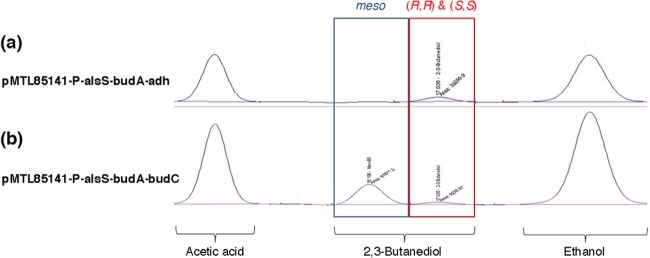
Stereospecific production of different 2,3-butanediol enantiomers by E. coli cells expressing either CaADH (a) or the K. pneumoniae 2,3-butanediol dehydrogenase (b).
Finally, expression of the putative C. autoethanogenum lactate dehydrogenase gene (ldhA) from plasmid pMTL85241-P-ldhA restored lactate production in a lactate dehydrogenase-negative E. coli strain, JW1375, when it was cultured anaerobically in M9 medium with glucose. After 24 h, lactate had accumulated to a normalized concentration of 2.9 ± 0.7 mM per OD600 unit (mean ± standard deviation; n = 2 independent experiments). After 47 h, the lactate concentration had increased to 4.4 ± 0.1 mM per OD600 unit. In contrast, no lactate was produced when E. coli JW1375 cells harboring the empty vector, pMTL85241-P, were cultured under identical conditions. Together, these results confirmed that the genome of C. autoethanogenum encodes active enzymes for the conversion of pyruvate to both lactate and 2,3-butanediol.
Expression and purification of CaADH.
In order to confirm its cofactor and substrate specificities, the CaADH gene was reamplified and subcloned into pBAD. This facilitated arabinose-inducible expression of the His6-tagged enzyme in E. coli strain LMG194. The recombinant protein was purified from soluble cell lysates by using immobilized-metal affinity chromatography. Yields of purified CaADH were typically ∼10 mg per liter of culture medium. The enzyme was >95% pure and migrated as a single band corresponding to the expected subunit mass (40.3 kDa) on SDS-PAGE gels (Fig. 4). As with CbADH (31), the purified enzyme was stable in air indefinitely when stored frozen at −80°C and for at least 1 day when thawed. Therefore, we measured its activity under aerobic conditions.
FIG 4.
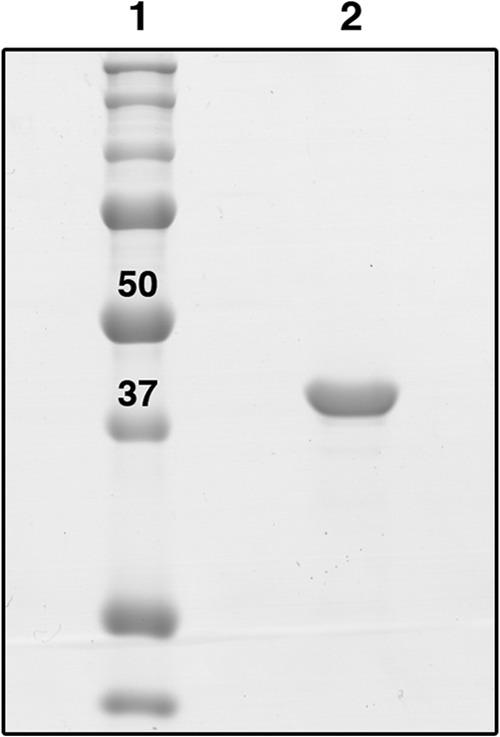
His6-tagged CaADH was purified using immobilized-metal affinity chromatography and visualized with Coomassie blue after separation in a 12% SDS-PAGE gel. Lane 1, molecular size marker (Bio-Rad Precision Plus; 50- and 37-kDa markers are indicated); lane 2, purified CaADH.
Kinetics with physiological substrates.
As predicted from the sequence, CaADH exhibited no activity with NADH in spectrophotometric assays. In contrast, the enzyme was able to catalyze the NADPH-linked reduction of a range of two-, three-, and four-carbon substrates (Table 2). C. autoethanogenum produced high levels of ethanol (>20 mM) during fermentative growth on steel mill waste gas (9). We hypothesized that CaADH might play a key role in this process, by converting acetaldehyde to ethanol. As expected, the enzyme was highly active with acetaldehyde as the substrate, and the specificity constant of CaADH for this reaction (kcat/Km = 1.7 × 104 M−1 s−1) is similar to that reported previously for CbADH (kcat/Km = 3.2 × 104 M−1 s−1) (31). Unlike C. beijerinckii, and as discussed above, C. autoethanogenum also produces 2R,3R-butanediol by reduction of R-acetoin. CaADH catalyzes this conversion with an efficiency (kcat/Km = 2.0 × 103 M−1 s−1) that is ∼9-fold lower than its activity toward acetaldehyde. Despite the size difference between two-carbon acetaldehyde and four-carbon acetoin, the turnover numbers (kcat) for the two reactions are comparable (Table 2). The increased specificity constant for acetaldehyde reflects a significantly decreased Michaelis constant (Km) for the smaller substrate.
TABLE 2.
Steady-state kinetic parameters of CaADHa
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Acetaldehyde | 93 ± 6 | 5.5 ± 1.4 | 1.7 × 104 |
| Acetoin | 145 ± 4 | 71 ± 5 | 2.0 × 103 |
| 2R,3R-Butanediol | 2.1 ± 0.2 | 65 ± 12 | 3.3 × 101 |
| meso-2,3-Butanediol | <1 | ||
| 2S,3S-Butanediol | ND | ND | ND |
| Acetone | 51.4 ± 0.8 | 0.60 ± 0.02 | 8.6 × 104 |
| Butanone | 44 ± 2 | 1.2 ± 0.1 | 3.8 × 104 |
ND, not detected.
To confirm the stereoselectivity of CaADH, we also measured its NADP+-dependent ability to oxidize each of the stereoisomers of 2,3-butanediol. There was no detectable activity with 2S,3S-butanediol as the substrate. In contrast, reproducible but weak activity was detected with 2R,3R-butanediol (Table 2). While the Km value for 2R,3R-butanediol was the same as that for acetoin, the kcat value for its oxidation was substantially lower. Overall, the catalytic efficiency of CaADH for 2R,3R-butanediol oxidation (kcat/Km = 33 M−1 s−1) was therefore 60-fold lower than its efficiency for acetoin reduction. The third stereoisomer, meso-2,3-butanediol, was an even worse substrate for CaADH. Activity was barely detectable above background noise, and we estimated it to be at least 50-fold weaker than the activity observed for 2R,3R-butanediol oxidation (i.e., kcat/Km of <1 M−1 s−1).
Kinetics with nonphysiological substrates.
In C. beijerinckii NRRL-B593, CbADH is responsible for converting acetone to isopropanol (31). C. autoethanogenum lacks the enzymes for acetone production. Nevertheless, the highest specificity constant that we measured for CaADH was in the conversion of acetone to isopropanol (Table 2). This was due to the decreased Michaelis constant (Km = 0.6 mM) for acetone compared with those for the physiological substrates. These data indicate that CaADH has a higher affinity for the nonphysiological, three-carbon substrate acetone than it does for physiological substrates that are either larger (acetoin) or smaller (acetaldehyde). Similarly, a low Km for butanone means that CaADH is 19-fold more efficient at catalyzing the reduction of this substrate to 2-butanol than it is at reducing acetoin (Table 2).
Whole-cell biocatalytic conversion of acetone to isopropanol.
Our in vitro data showed that CaADH was highly active toward acetone. This led us to predict that C. autoethanogenum cells would be able to convert exogenous acetone to isopropanol. Recently, reduction of acetone to isopropanol was also demonstrated in cultures of a closely related acetogen, C. ragsdalei (38), although no attempt was made to identify the enzyme that was responsible. We added acetone (∼170 mM) to early-log-phase cultures of C. autoethanogenum, C. ragsdalei, and other acetogens, as well as Clostridium species that are known to produce isopropanol or acetone. Reduction of acetone to isopropanol was monitored by GC.
Among the acetogens, only C. autoethanogenum, C. ragsdalei, and C. ljungdahlii were able to convert acetone to isopropanol (Table 3). These species have pairwise 16S rRNA gene sequence identities of >99% and thus form a tight subcluster within the Clostridium rRNA gene homology cluster I (39). Perhaps unsurprisingly, the sequenced genome of C. ljungdahlii (40) contains a homologue that is 100% identical to the C. autoethanogenum adh gene (locus tag CLJU_24860), and sequencing revealed that the exact same gene is also present in the C. ragsdalei genome. Consistent with the kinetic data for CaADH (Table 2), C. autoethanogenum was a highly effective whole-cell biocatalyst and converted ∼90% of the available acetone to isopropanol. Comparisons with cultures lacking exogenous acetone in the medium revealed that none of the three acetogens coupled acetone reduction to increased growth (Table 4). Instead, there were effects on the production of other metabolites, associated with the off-load of reducing equivalents onto acetone as an electron acceptor. In particular, production of the reduced alcohols ethanol and 2,3-butanediol was diminished, while the more oxidized compound, acetate, accumulated to marginally higher levels when acetone was added (Table 4).
TABLE 3.
Capability of various strains to reduce exogenous acetone to isopropanol
| Organism | Growth medium | Concn (mM) upon acetone addition |
Concn (mM) 120 h after acetone addition |
OD600 120 h after acetone addition | ||
|---|---|---|---|---|---|---|
| Acetone | Isopropanol | Acetone | Isopropanol | |||
| Acetogens | ||||||
| A. woodii DSM 1030 | PETC, pH 8.2 | 186 ± 15 | 0 | 186 ± 17 | 0 | 0.33 ± 0.05 |
| C. aceticum DSM 1496 | PETC, pH 7.4 | 173 ± 5 | 0 | 174 ± 10 | 0 | 0.31 ± 0.04 |
| C. autoethanogenum DSM 10061 | PETC, pH 5.9 | 159 ± 7 | 0 | 19 ± 17 | 138 ± 15 | 0.49 ± 0.03 |
| C. carboxidivorans DSM 15243 | PETC, pH 5.9 | 180 ± 12 | 0 | 178 ± 12 | 0 | 0.54 ± 0.07 |
| C. ljungdahlii DSM 13528 | PETC, pH 5.9 | 176 ± 10 | 0 | 64 ± 19 | 112 ± 17 | 0.47 ± 0.02 |
| C. ragsdalei ATCC BAA-622 | PETC, pH 5.9 | 194 ± 17 | 0 | 171 ± 28 | 22 ± 21 | 0.41 ± 0.05 |
| Isopropanol producer | ||||||
| C. beijerinckii NRRL-B593 | RCM | 171 ± 3 | 0 | 132 ± 29 | 43 ± 17 | 1.32 ± 0.06 |
| Acetone producers | ||||||
| C. beijerinckii NCIMB 8052 | RCM | 181 ± 10 | 0 | 182 ± 7 | 0 | 1.44 ± 0.08 |
| C. acetobutylicum ATCC 824 | RCM | 186 ± 10 | 0 | 188 ± 10 | 0 | 1.61 ± 0.12 |
| Controls | ||||||
| E. coli DH5α | LB | 201 ± 15 | 0 | 202 ± 17 | 0 | 1.82 ± 0.11 |
| Blank medium | PETC | 181 ± 7 | 0 | 182 ± 12 | 0 | |
| RCM | 179 ± 12 | 0 | 183 ± 12 | 0 | ||
| LB | 192 ± 16 | 0 | 194 ± 9 | 0 | ||
TABLE 4.
Impact of exogenous acetone on growth and metabolite synthesis
| Organism | Condition | OD600a | Concn (mM) of metabolitea |
||
|---|---|---|---|---|---|
| Ethanol | 2,3-Butanediol | Acetate | |||
| C. autoethanogenum DSM 10061 | No acetone | 0.47 ± 0.06 | 19 ± 2 | 0.6 ± 0.4 | 28 ± 2 |
| With acetone | 0.49 ± 0.03 | 4 ± 3 | 0.3 ± 0.2 | 34 ± 2 | |
| C. ljungdahlii DSM 13528 | No acetone | 0.48 ± 0.04 | 18 ± 1 | 0.4 ± 0.1 | 30 ± 3 |
| With acetone | 0.47 ± 0.02 | 6 ± 2 | 0.3 ± 0.1 | 36 ± 2 | |
| C. ragsdalei ATCC BAA-622 | No acetone | 0.44 ± 0.04 | 14 ± 1 | 0.4 ± 0.1 | 26 ± 3 |
| With acetone | 0.41 ± 0.05 | 7 ± 2 | 0.1 ± 0.1 | 31 ± 4 | |
Measured 120 h after acetone addition.
The next most closely related acetogen to C. autoethanogenum, C. ragsdalei, and C. ljungdahlii was C. carboxidivorans (16S rRNA gene sequence identity of 93% with C. autoethanogenum). C. carboxidivorans was unable to convert acetone to isopropanol (Table 3). Its genome is known to contain two genes for primary alcohol dehydrogenases, which are important for producing ethanol and 1-butanol (41). However, a BLAST search showed that its genome (42) lacks a homologue of the C. autoethanogenum primary-secondary adh gene, explaining why C. carboxidivorans was unable to produce isopropanol in our experiment. The more distantly related acetogens (K. pneumoniae and C. aceticum) were also unable to produce isopropanol, indicating that they also lack dehydrogenases that are able to accept acetone as a substrate. No adh homologues were detected in the draft sequence of the K. pneumoniae genome (43).
As expected, the isopropanol-producing strain C. beijerinckii NRRL-B593 (44) had the ability to reduce externally added acetone to isopropanol by the action of CbADH. Neither a different strain of C. beijerinckii, NCIMB 8052, which lacks this enzyme, nor the acetone producer C. acetobutylicum ATCC 824 was able to convert acetone to isopropanol. The same was also true for the negative control, E. coli DH5α (Table 3). These results demonstrate the potential utility of adh-containing organisms as whole-cell biocatalysts for the conversion of acetone to isopropanol.
DISCUSSION
The native ability of C. autoethanogenum to ferment gaseous substrates and produce the pyruvate-derived metabolite 2,3-butanediol offers a promising route for the sustainable and deployable production of a range of valuable C4 chemicals. In order to develop a viable biotechnological process for the production of 2,3-butanediol from CO-containing industrial waste gases, we have initiated the characterization of the enzymes required to convert pyruvate into 2,3-butanediol.
To begin, we confirmed the activities of the C. autoethanogenum AlsS, BudA, and BDH enzymes (Fig. 1), which had been identified previously by sequence similarity searches (9). When the three genes were cloned and then expressed together in anaerobically cultured E. coli cells, we observed the production of 1.1 ± 0.2 mM 2,3-butanediol per OD600 unit over a growth period of 47 h (Fig. 2a). This corresponds to an average production rate of 23 μM h−1 OD unit−1. This is comparable to the level of 2,3-butanediol (2 mM, over 120 to 190 h) that was produced natively by C. autoethanogenum when it was grown on steel mill waste gas (9). The implication is that our E. coli system will be a useful model for testing engineered variants of the C. autoethanogenum pathway, albeit with the caveat that E. coli cannot be grown in PETC medium with steel mill waste gas (and, likewise, the growth of acetogens in M9 medium has never been demonstrated). Nevertheless, both media are minimal and defined, thus allowing the most robust analyses that are currently tractable.
In comparison to our study, Oliver et al. recently achieved 2,3-butanediol yields of ∼150 mM in 40 h, using an optimized, aerobically cultured strain of E. coli that expressed AlsS from Bacillus subtilis, BudA from either Enterobacter aerogenes or Aeromonas hydrophila, and CbADH (22). Our system relied on the activity of the C. autoethanogenum Ppta-ack promoter; in contrast, theirs utilized an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter that drove high-level overexpression of the pathway components. We also made no attempt to optimize the codons of the C. autoethanogenum genes for expression in E. coli. These observations suggest that there is considerable scope for increasing the yield of 2,3-butanediol from our E. coli system, perhaps by as much as 2 orders of magnitude. Moreover, Oliver et al. (22) observed the production of a mixture of stereoisomers (∼78% 2R,3R-butanediol and ∼22% meso-2,3-butanediol); in contrast, our E. coli strain produced 2R,3R-butanediol exclusively (Fig. 3a). While our primary goal is to understand and then manipulate chemical production in waste gas-fermenting strains of C. autoethanogenum, our experiments demonstrate the utility of a new set of enzymes, analogous to those described previously (21, 22), for the stereoselective synthesis of 2R,3R-butanediol in E. coli.
An important aspect of this study was the discovery that C. autoethanogenum possesses two dehydrogenases (BDH and CaADH) that are able to reduce acetoin to 2,3-butanediol. Each was highly active in our E. coli system, catalyzing the complete conversion of acetoin to 2,3-butanediol and leaving no detectable acetoin (Fig. 2). This contrasts with a previous study in which four dehydrogenases were tested (22). The authors found that the primary-secondary alcohol dehydrogenase from C. beijerinckii (CbADH) was the most effective by far, but it was still capable of converting only ∼94% of acetoin to 2,3-butanediol.
The acetoin reductase activity of C. autoethanogenum BDH was expected, based on its similarity (∼60% amino acid identity) to the 2,3-butanediol dehydrogenases from Bacillus strains that are known to produce 2,3-butanediol (45). Another BDH homologue, from C. beijerinckii NCIMB 8052 (78% amino acid identity), was also expressed heterologously in C. acetobutylicum, leading to the synthesis of 2,3-butanediol from natively produced acetoin (20). Furthermore, the acetoin reductase activity of the C. beijerinckii homologue was recently confirmed in vitro (46).
It was more surprising to discover a second alcohol dehydrogenase that can play an important role in the production of 2,3-butanediol. This enzyme, CaADH, is strictly NADPH dependent and is a close homologue of the C. beijerinckii primary-secondary alcohol dehydrogenase, CbADH. However, C. autoethanogenum synthesizes acetoin (but not acetone) (9), whereas C. beijerinckii produces acetone (but not acetoin) (44). CaADH and CbADH therefore serve different physiological roles, participating in the production of 2,3-butanediol and isopropanol, respectively. These different roles appear to be reflected in the kinetic parameters of CaADH (Table 2) and CbADH (21, 31). In particular, CaADH and CbADH share similar overall catalytic efficiencies for the reduction of acetoin (kcat/Km values of 2.0 × 103 M−1 s−1 and 9.9 × 102 M−1 s−1, respectively); however, CaADH has significantly higher values for kcat (145 s−1, compared to 8.2 s−1 for CbADH) and Km (71 mM, compared to 8.3 mM for CbADH). This is consistent with an evolutionary model in which selection has acted on CaADH to maximize the rate of 2,3-butanediol production from acetoin in C. autoethanogenum. While the evolutionary optimization of enzyme-transition state complementarity can increase the kcat/Km value, for any given kcat/Km relationship, the reaction rate (i.e., molecules of product per molecule of enzyme per second) is maximal when kcat and Km are both high (47). In particular, rate maximization requires that the Michaelis constant be greater than the intracellular substrate concentration, and this is clearly the case for CaADH. On the other hand, CbADH has kcat (140 s−1) and Km (0.98 mM) values for acetone (31) that are both higher than the respective values of CaADH (Table 2). CbADH has been used as a component of new 2,3-butanediol pathways in a variety of engineered microorganisms (19–22). The high activity of CaADH toward acetoin suggests that it will prove a superior alternative to CbADH for these biotechnological applications.
During gas fermentation, C. autoethanogenum produces 2R,3R-butanediol almost exclusively, although a low level of meso-2,3-butanediol (∼6% of the total) can also be detected (9). Acetoin is extremely prone to spontaneous racemization via an enolate intermediate. Indeed, this propensity hampered our attempts to synthesize enantiomerically pure R-acetoin by several previously published routes (37, 48; data not shown). Therefore, it is likely that a small fraction of the R-acetoin produced by the C. autoethanogenum BudA enzyme spontaneously racemizes to S-acetoin in vivo (Fig. 1). Like CbADH (21), BDH and CaADH appear to be capable of installing R stereocenters exclusively during acetoin reduction, so both are candidates for reducing S-acetoin to meso-2,3-butanediol. However, CaADH-catalyzed oxidation of meso-2,3-butanediol was barely detectable in our assays (Table 2), suggesting that BDH is the most likely S-acetoin reductase. Consistent with this hypothesis, the C. beijerinckii BDH homologue is almost as active with meso-2,3-butanediol as it is with 2R,3R-butanediol as a substrate for oxidation (46).
While evolution has apparently acted to improve the R-acetoin reductase activity of CaADH, its highest activity in vitro was with acetone as the substrate (Table 2). Only a few microorganisms, such as C. beijerinckii NRRL-B593, are known to produce isopropanol, and acetone-to-isopropanol converting enzymes are rare (31, 49). A recent study showed that C. ragsdalei cells are capable of reducing acetone to isopropanol (38), even though this organism does not synthesize acetone or isopropanol naturally. Its ability to synthesize isopropanol was discovered in a fermentation with producer gas that was contaminated with acetone, which had been used as a scrubber after the gasification process. Here we have shown that C. ragsdalei, C. autoethanogenum, and their close relative, C. ljungdahlii, are all able to reduce exogenous acetone to isopropanol (Table 3). The data are consistent with a model in which the most recent common ancestor of C. autoethanogenum, C. ljungdahlii, and C. ragsdalei acquired the gene for a primary-secondary alcohol dehydrogenase from a sugar-fermenting isopropanol producer, such as C. beijerinckii NRRL-B593. The low, promiscuous activity of the acquired CbADH-like enzyme for reducing acetoin would have provided a selective advantage for its maintenance in the new host, and mutations that improved this activity would subsequently have been favored.
In addition to acetoin, CaADH was also efficient at catalyzing the reduction of the C4 compound butanone to 2-butanol (Table 2). In fact, the activity of the enzyme in vitro was greatest with the two nonphysiological ketone substrates, acetone and butanone. Our data suggest that CaADH will be a useful target for further engineering, with the goal of using variants of the enzyme in pathways for the stereo- and/or regioselective production of chemicals, including isopropanol, 2,3-butanediol, and butanol. Overall, this study represents an important step in the ongoing effort to realize the biotechnological potential of gas-fermenting bacteria. Not only have we functionally validated the pathway for 2,3-butanediol production by C. autoethanogenum, but also we have illustrated the potential of this acetogen as a platform for sustainable chemical manufacture.
ACKNOWLEDGMENTS
Work in the laboratory of M.L.G. and W.M.P. was funded by the University of Otago's Priming Partnerships Fund and a Smart Ideas grant from the New Zealand Ministry of Business, Innovation and Employment. W.M.P. is a Rutherford Discovery Fellow, and D.J.M. was supported by the Fanny Evans Postgraduate Scholarship for Women.
Footnotes
Published ahead of print 21 March 2014
REFERENCES
- 1.Monastersky R. 2013. Global carbon dioxide levels near worrisome milestone. Nature 497:13–14. 10.1038/497013a [DOI] [PubMed] [Google Scholar]
- 2.Horncastle A, Sastry A, Corrigan J, Gotpagar J. 2012. Future of chemicals: rebalancing global feedstock disruptions with “on-purpose” technologies. Booz & Company, New York, NY: http://www.booz.com/media/file/BoozCo_Future-of-Chemicals-Rebalancing-Global-Feedstock-Disruptions.pdf [Google Scholar]
- 3.Schiel-Bengelsdorf B, Dürre P. 2012. Pathway engineering and synthetic biology using acetogens. FEBS Lett. 586:2191–2198. 10.1016/j.febslet.2012.04.043 [DOI] [PubMed] [Google Scholar]
- 4.Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET. 2012. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr. Opin. Biotechnol. 23:364–381. 10.1016/j.copbio.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 5.Latif H, Zeidan AA, Nielsen A, Zengler K. 2014. Trash to treasure: production of biofuels and commodity chemicals via syngas fermenting microorganisms. Curr. Opin. Biotechnol. 27:79–87. 10.1016/j.copbio.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 6.Ragsdale SW, Pierce E. 2008. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784:1873–1898. 10.1016/j.bbapap.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood HG. 1991. Life with CO or CO2 and H2 as a source of carbon and energy. FASEB J. 5:156–163 [DOI] [PubMed] [Google Scholar]
- 8.Köpke M, Mihalcea C, Bromley JC, Simpson SD. 2011. Fermentative production of ethanol from carbon monoxide. Curr. Opin. Biotechnol. 22:320–325. 10.1016/j.copbio.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 9.Köpke M, Mihalcea C, Liew F, Tizard JH, Ali MS, Conolly JJ, Al-Sinawi B, Simpson SD. 2011. 2,3-Butanediol production by acetogenic bacteria, an alternative route to chemical synthesis, using industrial waste gas. Appl. Environ. Microbiol. 77:5467–5475. 10.1128/AEM.00355-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celińska E, Grajek W. 2009. Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnol. Adv. 27:715–725. 10.1016/j.biotechadv.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 11.Ji XJ, Huang H, Ouyang PK. 2011. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol. Adv. 29:351–364. 10.1016/j.biotechadv.2011.01.007 [DOI] [PubMed] [Google Scholar]
- 12.Xiu ZL, Zeng AP. 2008. Present state and perspective of downstream processing of biologically produced 1,3-propanediol and 2,3-butanediol. Appl. Microbiol. Biotechnol. 78:917–926. 10.1007/s00253-008-1387-4 [DOI] [PubMed] [Google Scholar]
- 13.Biebl H, Zeng AP, Menzel K, Deckwer WD. 1998. Fermentation of glycerol to 1,3-propanediol and 2,3-butanediol by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 50:24–29. 10.1007/s002530051251 [DOI] [PubMed] [Google Scholar]
- 14.De Mas C, Jansen NB, Tsao GT. 1988. Production of optically active 2,3-butanediol by Bacillus polymyxa. Biotechnol. Bioeng. 31:366–377. 10.1002/bit.260310413 [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Yang Y, Sun J, Shen Y, Wei D, Zhu J, Chu J. 2010. Microbial production of 2,3-butanediol by a mutagenized strain of Serratia marcescens H30. Bioresour. Technol. 101:1961–1967. 10.1016/j.biortech.2009.10.052 [DOI] [PubMed] [Google Scholar]
- 16.Syu MJ. 2001. Biological production of 2,3-butanediol. Appl. Microbiol. Biotechnol. 55:10–18. 10.1007/s002530000486 [DOI] [PubMed] [Google Scholar]
- 17.Hugenholtz J, Starrenburg MJC. 1992. Diacetyl production by different strains of Lactococcus lactis subsp. lactis var. diacetylactis and Leuconostoc spp. Appl. Microbiol. Biotechnol. 38:17–22 [Google Scholar]
- 18.Byun TG, Zeng AP, Deckwer WD. 1994. Reactor comparison and scale-up for the microaerobic production of 2,3-butanediol by Enterobacter aerogenes at constant oxygen transfer rate. Bioprocess Eng. 11:167–175. 10.1007/BF00518739 [DOI] [Google Scholar]
- 19.Collas F, Kuit W, Clément B, Marchal R, López-Contreras AM, Monot F. 2012. Simultaneous production of isopropanol, butanol, ethanol and 2,3-butanediol by Clostridium acetobutylicum ATCC 824 engineered strains. AMB Express. 2:45. 10.1186/2191-0855-2-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siemerink MA, Kuit W, López Contreras AM, Eggink G, van der Oost J, Kengen SW. 2011. d-2,3-Butanediol production due to heterologous expression of an acetoin reductase in Clostridium acetobutylicum. Appl. Environ. Microbiol. 77:2582–2588. 10.1128/AEM.01616-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan Y, Lee CC, Liao JC. 2009. Enantioselective synthesis of pure (R,R)-2,3-butanediol in Escherichia coli with stereospecific secondary alcohol dehydrogenases. Org. Biomol. Chem. 7:3914–3917. 10.1039/b913501d [DOI] [PubMed] [Google Scholar]
- 22.Oliver JW, Machado IM, Yoneda H, Atsumi S. 2013. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc. Natl. Acad. Sci. U. S. A. 110:1249–1254. 10.1073/pnas.1213024110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Huang H, Kahnt J, Mueller AP, Köpke M, Thauer RK. 2013. NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J. Bacteriol. 195:4373–4386. 10.1128/JB.00678-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertram J, Dürre P. 1989. Conjugal transfer and expression of streptococcal transposons in Clostridium acetobutylicum. Arch. Microbiol. 151:551–557. 10.1007/BF00454874 [DOI] [Google Scholar]
- 25.Brown SD, Nagaraju S, Utturkar S, DeTissera S, Segovia S, Mitchell W, Land M, Dassanayake A, Köpke M. 2014. Comparison of single-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant clostridia. Biotechnol. Biofuels 7:40. 10.1186/1754-6834-7-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79–85. 10.1016/j.mimet.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 27.Zhang GL, Wang CW, Li C. 2012. Cloning, expression and characterization of meso-2,3-butanediol dehydrogenase from Klebsiella pneumoniae. Biotechnol. Lett. 34:1519–1523. 10.1007/s10529-012-0933-4 [DOI] [PubMed] [Google Scholar]
- 28.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411–2423. 10.1002/pro.5560041120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ismaiel AA, Zhu CX, Colby GD, Chen JS. 1993. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J. Bacteriol. 175:5097–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hungate RE. 1969. A roll-tube method for cultivation of strict anaerobes, p 117–132 In Norris JR, Ribbons DW. (ed), Methods in microbiology, vol 3B Academic Press, New York, NY [Google Scholar]
- 33.Hirsch A, Grinsted E. 1954. Methods for the growth and enumeration of anaerobic spore-formers from cheese, with observations on the effect of nisin. J. Dairy Res. 21:101–110. 10.1017/S0022029900007196 [DOI] [Google Scholar]
- 34.Peretz M, Bogin O, Tel-Or S, Cohen A, Li G, Chen JS, Burstein Y. 1997. Molecular cloning, nucleotide sequencing, and expression of genes encoding alcohol dehydrogenases from the thermophile Thermoanaerobacter brockii and the mesophile Clostridium beijerinckii. Anaerobe 3:259–270. 10.1006/anae.1997.0083 [DOI] [PubMed] [Google Scholar]
- 35.Korkhin Y, Kalb AJ, Peretz M, Bogin O, Burstein Y, Frolow F. 1998. NADP-dependent bacterial alcohol dehydrogenases: crystal structure, cofactor-binding and cofactor specificity of the ADHs of Clostridium beijerinckii and Thermoanaerobacter brockii. J. Mol. Biol. 278:967–981. 10.1006/jmbi.1998.1750 [DOI] [PubMed] [Google Scholar]
- 36.Marlow VA, Rea D, Najmudin S, Wills M, Fülöp V. 2013. Structure and mechanism of acetolactate decarboxylase. ACS Chem. Biol. 8:2339–2344. 10.1021/cb400429h [DOI] [PubMed] [Google Scholar]
- 37.Tolasch T, Sölter SR, Tóth M, Ruther J, Francke W. 2003. (R)-acetoin-female sex pheromone of the summer chafer Amphimallon solstitiale (L.). J. Chem. Ecol. 29:1045–1050. 10.1023/A:1022992516854 [DOI] [PubMed] [Google Scholar]
- 38.Ramachandriya KD, Wilkins MR, Delorme MJ, Zhu X, Kundiyana DK, Atiyeh HK, Huhnke RL. 2011. Reduction of acetone to isopropanol using producer gas fermenting microbes. Biotechnol. Bioeng. 108:2330–2338. 10.1002/bit.23203 [DOI] [PubMed] [Google Scholar]
- 39.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812–826. 10.1099/00207713-44-4-812 [DOI] [PubMed] [Google Scholar]
- 40.Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, Wollherr A, Ehrenreich A, Liebl W, Gottschalk G, Dürre P. 2010. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U. S. A. 107:13087–13092. 10.1073/pnas.1004716107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ukpong MN, Atiyeh HK, De Lorme MJ, Liu K, Zhu X, Tanner RS, Wilkins MR, Stevenson BS. 2012. Physiological response of Clostridium carboxidivorans during conversion of synthesis gas to solvents in a gas-fed bioreactor. Biotechnol. Bioeng. 109:2720–2728. 10.1002/bit.24549 [DOI] [PubMed] [Google Scholar]
- 42.Bruant G, Lévesque MJ, Peter C, Guiot SR, Masson L. 2010. Genomic analysis of carbon monoxide utilization and butanol production by Clostridium carboxidivorans strain P7. PLoS One 5:e13033. 10.1371/journal.pone.0013033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poehlein A, Schmidt S, Kaster A-K, Goenrich M, Vollmers J, Thürmer A, Bertsch J, Schuchmann K, Voigt B, Hecker M, Daniel R, Thauer RK, Gottschalk G, Müller V. 2012. An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS One 7:e33439. 10.1371/journal.pone.0033439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.George HA, Johnson JL, Moore WE, Holdeman LV, Chen JS. 1983. Acetone, isopropanol, and butanol production by Clostridium beijerinckii (syn. Clostridium butylicum) and Clostridium aurantibutyricum. Appl. Environ. Microbiol. 45:1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicholson WL. 2008. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl. Environ. Microbiol. 74:6832–6838. 10.1128/AEM.00881-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raedts J, Siemerink MA, Levisson M, van der Oost J, Kengen SW. 2014. Molecular characterization of an NADPH-dependent acetoin reductase/2,3-butanediol dehydrogenase from Clostridium beijerinckii NCIMB 8052. Appl. Environ. Microbiol. 80:2011–2020. 10.1128/AEM.04007-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fersht A. 1999. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W. H. Freeman and Company, New York, NY [Google Scholar]
- 48.Crout DHG, Morrey SM. 1983. Synthesis of (R)- and (S)-acetoin (3-hydroxybutan-2-one). J. Chem. Soc. Perkin Trans. 1:2435–2440 [Google Scholar]
- 49.Peretz M, Burstein Y. 1989. Amino acid sequence of alcohol dehydrogenase from the thermophilic bacterium Thermoanaerobium brockii. Biochemistry 28:6549–6555. 10.1021/bi00442a004 [DOI] [PubMed] [Google Scholar]