Abstract
Traditional three-domain fungal and bacterial laccases have been extensively studied for their significance in various biotechnological applications. Growing molecular evidence points to a wide occurrence of more recently recognized two-domain laccase-like multicopper oxidase (LMCO) genes in Streptomyces spp. However, the current knowledge about their ecological role and distribution in natural or artificial ecosystems is insufficient. The aim of this study was to investigate the diversity and composition of Streptomyces two-domain LMCO genes in agricultural waste composting, which will contribute to the understanding of the ecological function of Streptomyces two-domain LMCOs with potential extracellular activity and ligninolytic capacity. A new specific PCR primer pair was designed to target the two conserved copper binding regions of Streptomyces two-domain LMCO genes. The obtained sequences mainly clustered with Streptomyces coelicolor, Streptomyces violaceusniger, and Streptomyces griseus. Gene libraries retrieved from six composting samples revealed high diversity and a rapid succession of Streptomyces two-domain LMCO genes during composting. The obtained sequence types cluster in 8 distinct clades, most of which are homologous with Streptomyces two-domain LMCO genes, but the sequences of clades III and VIII do not match with any reference sequence of known streptomycetes. Both lignocellulose degradation rates and phenol oxidase activity at pH 8.0 in the composting process were found to be positively associated with the abundance of Streptomyces two-domain LMCO genes. These observations provide important clues that Streptomyces two-domain LMCOs are potentially involved in bacterial extracellular phenol oxidase activities and lignocellulose breakdown during agricultural waste composting.
INTRODUCTION
Composting is a self-heating, aerobic biodecomposition process of organic waste that has advantages over other disposal strategies because it reduces waste volume by 40% to 50% and provides a final product that can be used as a soil conditioner or as good-quality fertilizer (1–3). As the important fraction of the agricultural wastes, lignin is highly resistant to microbial attack because of its complex cross-linked structure. The degradation of cellulose and hemicellulose is also strongly inhibited by the presence of lignin, which consequently makes lignin degradation the limiting step influencing the speed and quality of composting (4). Fungi such as Aspergillus spp., Penicillum spp., Trichoderma spp., and Phanerochaete chrysosporium can secrete industrial quantities of extracellular lignocellulolytic enzymes (5). However, bacterial enzymes have characteristics that are more advantageous than those of fungal enzymes. This is because bacteria grow more rapidly, produce multienzyme complexes with increased functionality and higher specificity, and can tolerate larger and more-diverse environmental stresses (6, 7).
The genus Streptomyces, composed of Gram-positive filamentous bacteria of the order Actinomycetales, with more than 600 described species (http://www.bacterio.cict.fr/), is an ecologically important group ubiquitous in natural or artificial ecosystems. Some Streptomyces isolates participate in the degradation of recalcitrant biopolymers such as lignin, hemicelluloses, or celluloses found in wood (8–10). Ulrich et al. (7) reported that Streptomyces spp. could dominate certain cellulolytic microbial communities after long-term manure application in a soil ecosystem. In the composting process, the pile temperature could rise quickly from room temperature to 70°C, leading to a transition of microbial communities from mesophilic to thermophilic ones, which provided superior conditions for the growth of streptomycetes (4). Thus, Streptomyces is among the most represented actinomycete genera for their pH and thermal stability during composting (11). Such observations suggest the potential importance of thermophilic streptomycetes with lignocellulolytic enzyme activities and lignocellulose breakdown abilities during agricultural waste composting.
As the vital functional enzymes for lignocellulose degradation, laccases (benzenediol/oxygen oxidoreductases; EC 1.10.3.2) or laccase-like multicopper oxidases (LMCOs) can catalyze the oxidation of various phenolic compounds and some nonphenolic substrates, concomitantly with the reduction of the molecular oxygen to water (12). Traditional three-domain fungal and bacterial laccases have been widely studied for their importance in various biotechnological applications (13–15). A novel lineage of relatively small LMCOs—the two-domain LMCOs—has now attracted the attention of researchers (16), and two-domain LMCOs were found to be typically present in Streptomyces spp., such as Streptomyces griseus, Streptomyces ipomoeae, and Streptomyces coelicolor (17–20). They lack the second of the three domains and yet exhibit significant activity in the restrictive environment (21). Such two-domain laccases also contain four copper ions/subunits, oxidize various phenolic and nonphenolic substrates, and have spectroscopic properties similar to those of common three-domain laccases (22). These new two-domain laccases may have advantages over traditional three-domain laccases: small laccase from Streptomyces coelicolor and EpoA from Streptomyces griseus could oxidize phenolic compounds under alkaline and high temperature conditions (21). Additionally, the copper binding sites are different from those of the common three-domain fungal and bacterial laccases, which allows the design of specific Streptomyces two-domain primers to investigate the diversity and composition of Streptomyces two-domain LMCO genes with potential polyphenol oxidative and lignocellulose decomposition capacities in agricultural waste composting systems.
In this present research, a PCR primer set was designed to target the two conserved copper binding regions (cbr II and IV) of Streptomyces two-domain LMCO genes. A molecular investigation of the diversity and distribution of Streptomyces two-domain LMCO genes was conducted among the representative samples in composting systems. Meanwhile, our study associated Streptomyces two-domain LMCO gene abundance with phenol oxidase activities and lignocellulose degradation to analyze the potential function of Streptomyces two-domain laccases during composting.
MATERIALS AND METHODS
Composting and sample collection.
The source materials for composting mainly consisted of rice straw, vegetable residues, soil, and wheat bran. Rice straw and vegetable residues were shredded to a size of 10 to 20 mm and dried in the open air. Soil from the surface humus soil on Yuelu Mountain (Changsha, China) was used as a source of indigenous microbial inoculants added into the compost pile. The physicochemical properties of composting materials used in this study are displayed in Table 1.
TABLE 1.
Summary of the materials used in the compostinga
| Material | pH | TOC (g kg−1) | TKN (g kg−1) | C/N | Moisture (%) |
|---|---|---|---|---|---|
| Rice straw | ND | 421.36 (14.90) | 9.28 (0.64) | 45.40 (2.16) | 10.89 (0.69) |
| Vegetable residue | ND | 84.25 (2.80) | 5.52 (0.15) | 15.26 (0.84) | 59.97 (2.47) |
| Bran | ND | 458.12 (14.10) | 46.14 (3.42) | 9.93 (0.35) | 14.36 (0.51) |
| Soil | 4.96 (0.86) | 62.13 (3.36) | 2.51 (0.24) | 24.75 (1.36) | 40.68 (0.41) |
Means are shown with standard deviations in parentheses (n = 3). TOC, total organic carbon; TKN, total Kjeldahl nitrogen; C/N, ratio of total organic carbon to total Kjeldahl nitrogen; ND, not determined.
Composting is a self-heating, aerobic solid-phase biodegradative natural process of organic materials. An experimental composting system with a dry weight of about 20 kg of materials was set up indoors in this study. The original materials were mixed thoroughly with organic matter content of about 61.8% (dry weight) and a ratio of total organic carbon to total Kjeldahl nitrogen (C/N ratio) of about 30:1. The composting process was conducted in a specially designed pilot-scale reactor. The reactor was a cylinder (50 cm in diameter, 45 cm in depth) equipped with a perforated plate at the bottom to distribute the air. Air from a compressor was supplied at a constant flow rate of 0.25 liters min−1 kg−1 (wet weight) to maintain aerobic conditions throughout the experimental run. Three replicates were sampled on days 0, 4, 8, 15, 30, and 50. Samples with a total weight of 10 g (wet weight) were periodically taken from the top, middle, and bottom depths in the different sections of the pile and were stored at −20°C for total DNA extraction. Moisture content was adjusted to about 50% to 60% during composting by adding sterile deionized water after sampling.
Analysis of phenol oxidase activity and lignocellulose degradation.
Phenol oxidase activity was estimated using the substrate 2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulfononic acid) diammonium salt (ABTS) at pH 4.5 or l-3,4-dihydroxyphenylalanine (l-DOPA) at pH 6.0 and 8.0 (12, 23, 24). Compost samples (10 g) were shaken in flasks with 90 ml of sterile 0.9% NaCl solution at 25°C for 1 h. The resulting suspension was used for measuring the phenol oxidase activity. Each reaction mixture contained 150 μl of citrate phosphate buffer (pH 4.5, 6.0, and 8.0; 50 mM), 50 μl of compost suspension, and 50 μl of substrate (50 mM ABTS or 25 mM l-DOPA). The mixtures were incubated with shaking (100 rpm) at 25°C for 4 h, and the supernatant, after centrifugation at 3,500 rpm for 4 min, was transferred to a 96-well microplate. Negative-control samples were autoclaved beforehand and prepared in the same way. The absorbance was measured at 420 nm for ABTS and 475 nm for l-DOPA by a Multiscan Spectrum spectrophotometer (Thermo, Vantaa, Finland). The enzymatic activity was calculated with the extinction coefficient of ABTS (ε420 = 36,000 M−1 cm−1) (25) or l-DOPA (ε450 = 3,600 M−1 cm−1) (26). Enzyme activity was expressed as μmol h−1 g−1dry matter. Moisture content was determined by drying the sample in an oven at 105°C to a constant weight. Three replicates were used in all measurements.
Neutral detergent fiber (NDF), acid detergent fiber (ADF), and acid detergent lignin (ADL) were assayed according to the sequential extraction methods proposed by Van Soest et al. (27). Hemicellulose content was assessed by the difference between NDF and ADF, cellulose content by the difference between ADF and ADL, and lignin content by the difference between ADL and ash content. This detergent-fiber analytical method was used because of its convenience and consistency for agricultural wastes (28, 29), although the outcome might be slightly biased by proteins bound to lignin or by a contaminant, particularly cutin, present in the acid-insoluble residues.
Primer design.
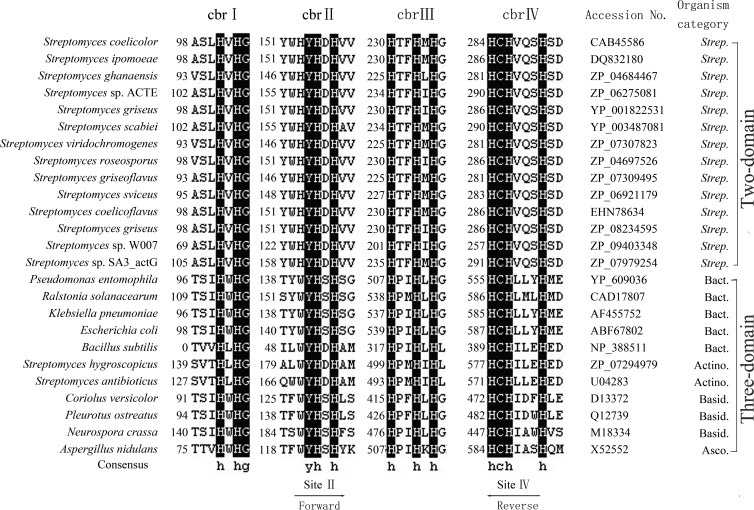
Prior to this study, no general primers were available and suitable for amplification of the Streptomyces two-domain LMCO genes in environmental samples. Twenty-five published laccase amino acid sequences from various microorganisms, including three-domain bacteria, actinomycetes, basidiomycetes, and ascomycetes, and 14 Streptomyces two-domain LMCO genes were aligned using DNAMAN 7.0. Primers were designed to be specific to Streptomyces two-domain LMCO genes and targeted the conserved sequences between copper binding regions II (YWHYHDH) and IV (HCHVQSH) of published Streptomyces two-domain LMCO sequences (Fig. 1). The forward (Cu2SF; 5′-TAC TGG MAC TAC CAC GAC CAY-3′) and reverse (Cu4SR; 5′-RTG VSW CTG SAC RTG RCA GTG-3′) specific degenerate primers amplified a product of about 426 bp. Ambiguous bases are defined as follows: M = C/A, Y = C/T, R = A/G, V = A/G/C, W = A/T, and S = G/C.
FIG 1.
Amino acid alignment of published Streptomyces (Strep.) two-domain LMCOs and some reference three-domain LMCOs between the conserved copper binding regions (including those of bacteria [Bact.], actinomycetes [Actino.], basidiomycetes [Basid.], and ascomycetes [Asco.]). Similarity shading was applied, with a 100% threshold indicated in black. Degenerate primers for this study were designed based on conserved copper binding regions two (II) and four (IV).
DNA extraction, positive controls, and PCR.
The compost samples on days 0, 4, 8, 15, 30, and 50 (defined as D0, D4, D8, D15, D30, and D50) were selected to study the diversity and succession of Streptomyces two-domain LMCO gene communities during agricultural waste composting. Genomic DNA from the six composting samples (fresh and wet) was extracted according to the method described by Yang et al. (30). The crude DNA was purified with a TIANquick Midi Purification kit (TianGen, Beijing, China) and then dissolved in 100 μl Tris-EDTA (TE) buffer and stored at −20°C before use.
Three Streptomyces strains (Streptomyces coelicolor ATCC 23899, Streptomyces griseus ATCC 13273, and Streptomyces ipomoeae ATCC 25462), with known two-domain LMCO genes, were bought from the China General Microbiological Culture Collection Center as positive controls in PCR amplifications. The PCR mixture contained about 150 ng of template DNA, 25 μl 2× Power Taq PCR MasterMix (TianGen, Beijing, China), and 1 μl (25 μM) of each forward and reverse primer and was adjusted to a final volume of 50 μl with sterilized Milli-Q water. PCR amplification was run on a MyCycler (Bio-Rad) using the following cycling conditions: 5 min at 94°C; 35 cycles consisting of 45 s at 94°C, 40 s at 60°C, and 40 s at 72°C; a final extension of 7 min at 72°C; and ending at 4°C.
Fragment purification and laccase clone library construction.
Amplifications of the expected size (about 426 bp between cbr II and cbr IV) were excised from the 2% agarose gel and purified with a TIANgel Midi Purification kit (TianGen, Beijing, China) according to the manufacturer's specifications. The fragments were then ligated into pGEM-T vector (Promega), and the plasmids were transformed into Escherichia coli competent cells (DH5α). A blue-white colony appearance and ampicillin resistance were both used to select the correct clones. The plasmid DNA was extracted from a transformant with a TIANpure Midi Plasmid kit (TianGen, Beijing, China) before sequencing. Finally, the nucleotide sequences obtained were deposited in the GenBank nucleotide sequence database of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/GenBank/).
qPCR analysis.
Real-time qPCR was performed on an iCycler iQ5 thermocycler (Bio-Rad) to determine Streptomyces two-domain LMCO gene abundance in the compost. The corresponding primers were Cu2SF and Cu4SR. The 25-μl quantitative PCR (qPCR) mix contained 2 ng of compost microbial DNA, 4 pmol of each primer, 10 μl of 2.5× RealMasterMix (TianGen, China) and 1.5 μl of 20× SYBR solution (TianGen, China). The protocol was as follows: 2 min at 94°C; 35 cycles consisting of 10 s at 94°C, 30 s at 60°C, and 30 s at 72°C; a final extension of 7 min at 72°C; and ending at 4°C. The fluorescent signal was measured at the end of each extension step. Melting curve analysis was performed to verify amplification specificity. The plasmid previously obtained from each sample was extracted, and the concentration of the plasmid was then determined. Ten-fold serial dilutions of from 1.0 × 103 to 1.0 × 108 copies of plasmids were used as the templates in qPCR for generation of standard curves, as described by Blackwood et al. (14). qPCR was performed on all samples in triplicate runs, and each run included two sets of standards and no-template controls.
Sequence analysis and statistics.
For each gene library, operational taxonomic units (OTUs) were established at the nucleotide genetic distances of 0.01, 0.03, and 0.10. Then, the OTU-based analyses (rarefaction curves, Shannon-Wiener index [H], and Chao1 and ACE richness estimators) were performed using the mothur program (http://www.mothur.org/) (31). The detected nucleotide sequences were compared with those in the GenBank database in NCBI using the BLAST search algorithm. This comparison allowed confirmation of clones to be associated with Streptomyces two-domain LMCO sequences and selection of reference sequences for phylogenetic analysis. Mega 5 was used for sequence manipulation and phylogenetic tree construction by the neighbor-joining method (32). A bootstrap analysis with 1,000 replicates was applied to estimate the confidence values for the tree nodes. For the whole-gene sequences obtained, rarefaction curves were established at the nucleotide genetic distances of 0.01, 0.03, and 0.10.
Three replicates were performed for each sample day to assess the phenol oxidase activity and abundance of Streptomyces two-domain LMCO genes. The mean values determined for the three replicates were compared by one-way analysis of variance (ANOVA) using a post hoc Tukey's test to assess the significant differences (P < 0.05). All univariate data were analyzed using the software package SPSS 16.0 (SPSS, Germany).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained were deposited in the GenBank nucleotide sequence database of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/GenBank/) with accession numbers JQ954774 to JQ954825.
RESULTS
Composting and sampling.
The pile temperature was kept higher than 50°C for at least 8 days, which is the minimum requirement for a proper disinfection of waste materials from animal and plant pathogens. The pH value dropped slightly in the first several days due to the acid formation in the early stage of composting and then increased gradually in the following 2 weeks for the ammonification and mineralization of organic nitrogen and was finally maintained at 7.5 to 8.0 (data not shown). Six representative samples from the mesophilic stage (day 0), the thermophilic stage (day 4), the earlier cooling stage (days 8 and 15), and the maturation stage (days 30 and 50) were selected for the following experiments. The physical-chemical properties of composting samples used in this study are displayed in Table 2.
TABLE 2.
Physical-chemical parameters of the samples selecteda
| Sampling day | Temp | pH | C/N | Moisture (%) | WSC (mg liter−1) |
|---|---|---|---|---|---|
| 0 | 30.0 (0.5) | 6.98 (0.03) | 30.12 (0.32) | 60.83 (0.43) | 200.21 (3.11) |
| 4 | 62.9 (0.6) | 6.25 (0.02) | 26.82 (0.43) | 53.22 (0.36) | 215.46 (2.63) |
| 8 | 50.4 (0.2) | 7.12 (0.05) | 21.94 (0.22) | 54.65 (0.53) | 170.28 (2.24) |
| 15 | 26.8 (0.3) | 7.92 (0.04) | 19.56 (0.48) | 55.61 (0.38) | 105.24 (2.03) |
| 30 | 28.6 (0.4) | 7.88 (0.03) | 17.26 (0.35) | 51.92 (0.67) | 76.17 (1.72) |
| 50 | 27.5 (0.2) | 7.91 (0.04) | 16.81 (0.26) | 53.85 (0.81) | 70.23 (2.03) |
Means are shown with standard deviations in parentheses (n = 3). WSC, water-soluble carbon.
Degradation of cellulose, hemicellulose, and lignin.
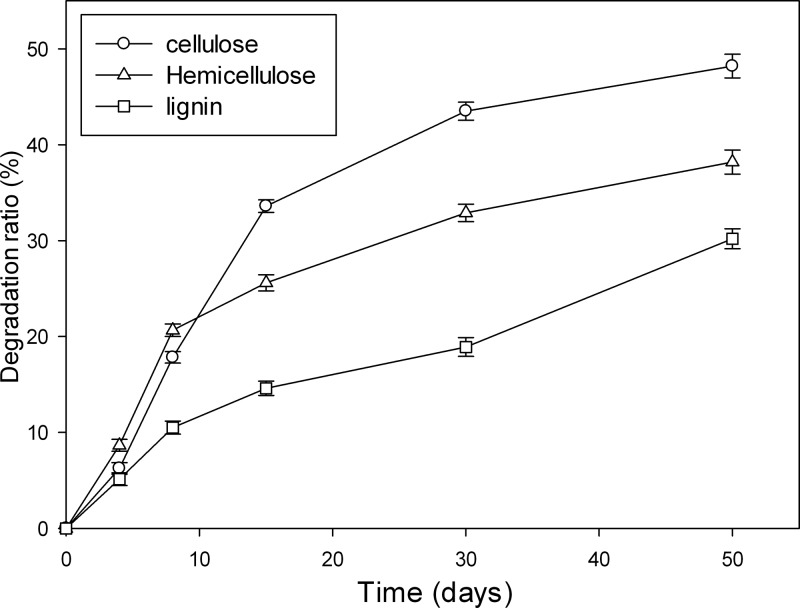
Lignocelluloses were gradually degraded during composting (Fig. 2). A high rate of degradation of cellulose was observed between days 0 and 15. The hemicellulose was mainly decomposed between days 0 and 30 except for the early cooling period. The degradation rate of recalcitrant lignin was lower than those of hemicelluloses and celluloses in the first 4 days, because lignin is more resistant to microbial attack.
FIG 2.
Lignocellulose degradation ratios during lignocellulosic waste composting.
Phenol oxidase activity and abundance of Streptomyces two-domain LMCO genes.
The phenol oxidase activity detected at various pH values and abundance of Streptomyces two-domain LMCO genes are displayed in Table 3. Generally, phenol oxidase activity was at a constantly high level throughout the whole composting process. The sample on the fourth day showed the maximal enzyme activity at pH 4.5, suggesting rapid growth in fungal microbes with phenol oxidase activity in the warming period. A sharp decline of phenol oxidase activity was observed from day 4 to day 8 (in the high-temperature period) and then a sudden increase in the cooling and maturation phase at pH 4.5. Phenol oxidase activities detected at pH 8.0 were generally higher than at pH 6.0. The maximal enzyme activity at pH 8.0 was observed on the eighth day, which indicated that the microbes producing alkaline phenol oxidase propagated quickly in the thermophilic phase. The copy numbers of Streptomyces two-domain LMCO genes increased quickly in the thermophilic phase, which might be ascribed to the rising temperature and high level of substrate availability in the pile.
TABLE 3.
Phenol oxidase activity and gene copy numbers of Streptomyces two-domain LMCOs in composting samplesa
| Day | Phenol oxidase activity (μmol h−1 g−1) |
Streptomyces two-domain LMCO gene abundance (no. of copies g−1 × 106) | ||
|---|---|---|---|---|
| pH 4.5 | pH 6.0 | pH 8.0 | ||
| 0 | 4.22 (0.11) a | 4.21 (0.15) a | 5.84 (0.13) a | 1.35 (0.06) a |
| 4 | 6.96 (0.15) b | 6.35 (0.12) b | 10.06 (0.22) b | 2.68 (0.12) b |
| 8 | 2.95 (0.23) c | 8.91 (0.28) c | 13.49 (0.35) c | 3.56 (0.14) c |
| 15 | 6.78 (0.20) bd | 6.27 (0.21) b | 11.54 (0.19) d | 2.86 (0.09) b |
| 30 | 6.42 (0.18) de | 5.03 (0.19) d | 12.95 (0.24) e | 3.46 (0.10) c |
| 50 | 6.24 (0.13) e | 4.76 (0.23) d | 9.65 (0.18) b | 3.26 (0.15) c |
Means are shown with standard deviations in parentheses (n = 3). ABTS was used as the substrate at pH 4.5 and l-DOPA at pH 6.0 and 8.0. Means with different letters in the same column are significantly different (P < 0.05).
Diversity of Streptomyces two-domain LMCO genes during agricultural waste composting.
The newly designed primers (Cu2SF and Cu2SR) were used to amplify Streptomyces two-domain LMCO genes in composting samples. According to the available nucleotide sequences, the PCR amplification performed with this primer pair was expected to produce fragments of about 426 bp. The pure cultures of streptomycetes were analyzed, and all of them showed positive PCR products of about 426 bp. The amplified sequences have high similarities with those of known Streptomyces two-domain LMCO genes. Additionally, the whole deduced amino acids of the obtained sequences included copper binding regions two (cbr II), three (cbr III), and four (cbr IV).
Six gene libraries were constructed from composting samples D0, D4, D8, D15, D30, and D50, and 25 clones of each were analyzed. A total of 110 laccase gene sequences were retrieved from the six selected composting samples. They were distributed in the libraries as follows: D0 (19 sequences), D4 (23 sequences), D8 (20 sequences), D15 (18 sequences), D15 (16 sequences), and D50 (14 sequences). All the detected laccase genes having the same DNA sequence were set as identical laccase gene “types.” Thus, the 110 obtained sequences could be divided into 52 different gene types. The assumed amino acid sequences consist of about 142 amino acid residues. Despite of the repeated emergence of some Streptomyces two-domain LMCO gene types in different clone libraries, 29 gene types were solely found in one sample, indicating a diversity and dynamic succession of Streptomyces two-domain LMCO genes during agricultural waste composting.
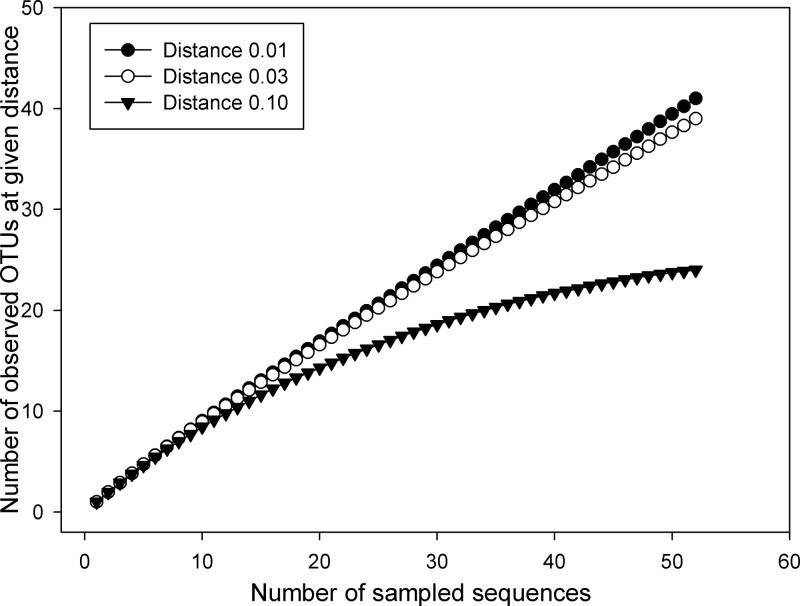
The diversity and richness of each clone library were assessed by calculating the Shannon-Wiener index (H) and the Chao1 and ACE richness estimators (Table 4). There was a steady increase in the number of observed OTUs in the warming period and then a rapid decrease in the cooling and maturation period. The Shannon diversity index, indicating the clone numbers of individual gene types found at each sampling date, varied consistently with the richness of LMCO gene population. The highest Shannon index for the Streptomyces two-domain LMCO genes was observed on day 8 (2.94 at the genetic distance of 0.01), whereas the lowest was 1.35 on day 50 at the genetic distance of 0.01. The occurrence of the highest Shannon index on day 8 also suggested that streptomycetes with two-domain LMCO genes propagated rapidly in the thermophilic period. The established rarefaction curves indicated relatively high coverage in the library from composting samples, and many more sequences would need to be sampled for the rarefaction curves to reach a plateau (Fig. 3).
TABLE 4.
OTU based analyses of Streptomyces two-domain LMCO gene sequences (Shannon diversity index, Chao1, and Ace richness) from each composting sample at the nucleotide genetic distance of 0.01 to 0.03a
| Day | No. of gene types | Distance | OTU | H | Chao1 richness | Ace richness |
|---|---|---|---|---|---|---|
| Day 0 | 19 | 0.01 | 17 | 2.83 (2.50–3.17) | 153 (71–360) | 262 (78–958) |
| 0.03 | 16 | 2.75 (2.41–3.09) | 69 (30–209) | 106 (36–722) | ||
| Day 4 | 23 | 0.01 | 18 | 2.89 (2.43–3.12) | 166 (75–327) | 232 (35–2,467) |
| 0.03 | 17 | 2.80 (2.03–2.94) | 72 (22–154) | 110 (25–966) | ||
| Day 8 | 20 | 0.01 | 19 | 2.94 (2.63–3.26) | 190 (90–430) | 221 (24–4,055) |
| 0.03 | 18 | 2.90 (2.57–3.22) | 126 (57–298) | 81 (23–643) | ||
| Day 15 | 18 | 0.01 | 6 | 1.54 (1.01–2.07) | 9 (6–28) | 15 (7–63) |
| 0.03 | 5 | 1.47 (0.97–1.92) | 7 (5–17) | 11 (6–40) | ||
| Day 30 | 16 | 0.01 | 6 | 1.48 (0.99–1.98) | 12 (7–43) | 14 (7–61) |
| 0.03 | 5 | 1.41 (0.95–1.80) | 8 (6–33) | 9 (6–35) | ||
| Day 50 | 14 | 0.01 | 5 | 1.35 (0.90–1.80) | 8 (5–29) | 9 (6–37) |
| 0.03 | 4 | 1.24 (0.83–1.72) | 7 (4–22) | 8 (5–33) | ||
| Total | 52 | 0.01 | 41 | 3.48 (3.20–3.76) | 782 (440–1,419) | 703 (313–2,591) |
| 0.03 | 39 | 3.43 (3.15–3.70) | 237 (107–615) | 378 (153–1,044) |
The numbers in parentheses represent 95% confidence intervals of the indices. H, Shannon diversity index.
FIG 3.
Rarefaction curve for Streptomyces two-domain LMCO gene fragments. Operational taxonomic units (OTUs) were formed at genetic distances of 1%, 3%, and 10%.
Phylogenetic analysis of Streptomyces two-domain LMCO gene sequences.
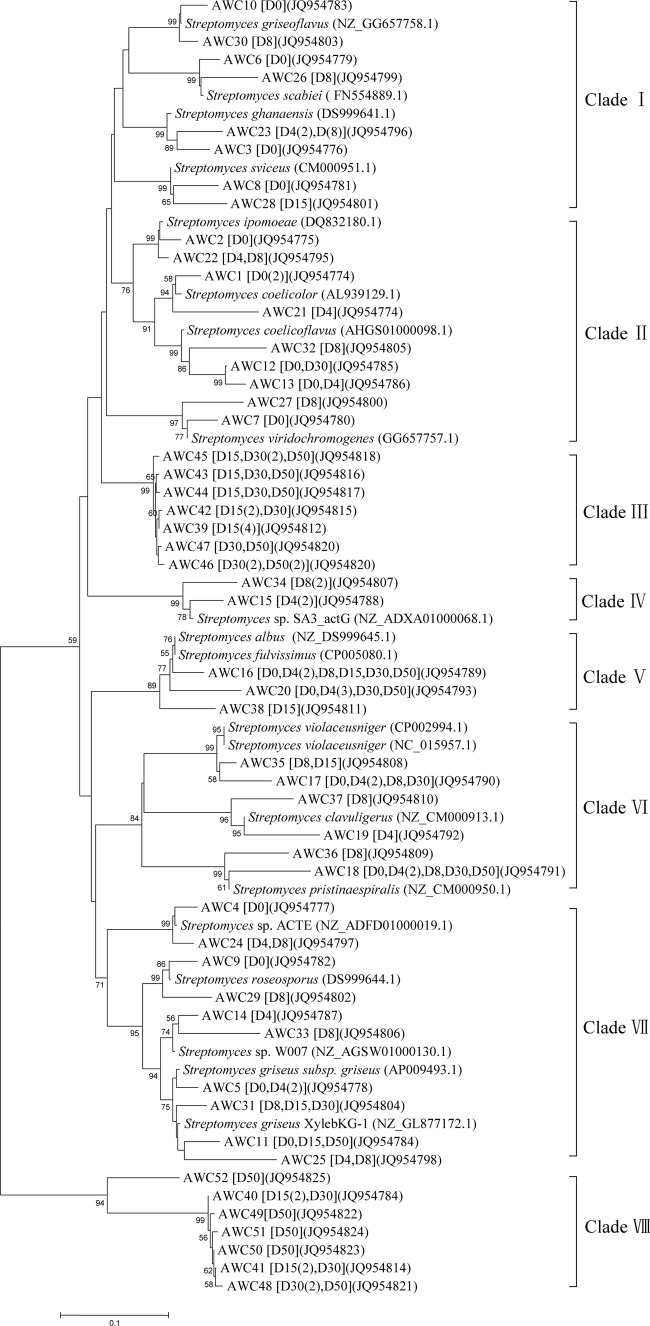
The sequences determined in this survey and the ones retrieved from GenBank resulted in an alignment of 72 LMCO gene sequences (Fig. 4). High diversity and a rapid succession of Streptomyces two-domain LMCO genes were found in the six representative samples with 8 distinct clades of sequence types, among which most sequences clustered with the reference two-domain LMCO genes of Streptomyces coelicolor, Streptomyces violaceusniger, Streptomyces griseus, Streptomyces ipomoeae, and so on. However, the sequences of clades III and VIII do not match with any reference sequence of known streptomycetes.
FIG 4.
Neighbor-joining (NJ) tree of Streptomyces two-domain LMCO gene fragments obtained from composting samples together with the reference sequences retrieved from GenBank. Values above branches indicate bootstrap support derived from 1,000 replicates. The occurrences of obtained laccase gene types are given in square brackets with their corresponding GenBank accession numbers. Day 0 (D0), D4, D8, D15, D30, and D50 indicate the sampling times; the number of occurrences at a given sampling time is given if >1. D0(3), for example, indicates that the sequence appeared three times in the sample from day 0.
Sequence groups of clades I and II were mainly found on days 0, 4, and 8, and the obtained sequences were clustered with the reference laccases of Streptomyces ipomoeae and Streptomyces coelicolor. AWC2 (day 0), AWC22 (days 4 and 8), and Streptomyces ipomoeae clustered to the same branch, which implied that Streptomyces ipomoeae probably existed in the thermophilic phase of composting. AWC1, AWC21 (appeared on days 0 and 4), and small laccase from Streptomyces coelicolor were classified to the same branch with a bootstrap value of 64. Moreover, the sequences in clades V, VI, and VII were detected on all sample days, and these sequences mainly belonged to Streptomyces fulvissimus, Streptomyces violaceusniger, and Streptomyces griseus. These observations suggested the potentially important role of the three groups in organic compound decomposition because they existed during the whole process of composting and could stand the extreme high temperature in the thermophilic phase.
The unknown clade III (including AWC39 and AWC42 to AWC47)- and clade VIII (including AWC40 to AWC41 and AWC48 to AWC52)-associated sequences did not cluster with reference two-domain multicopper oxidases from Streptomyces spp. probably due to the limited two-domain LMCO data available. The BLASTn search of sequence AWC43 (JQ954816) in clade III showed that it matches best with a multicopper oxidase gene from Streptomyces viridochromogenes (91% identity; E value = 9e−128), and the BLASTp search of translated amino acid showed it had the highest identity with a multicopper oxidase from Streptomyces coelicoflavus (89% identity; E value = 6e−88). The BLASTn search of AWC52 in clade VIII showed it had the highest identity with a multicopper oxidase gene from Streptomyces viridochromogenes (78% identity; E value = 3e−29), and the sequence of AWC48 is similar to that of a multicopper oxidase gene from Streptomyces rapamycinicus (76% identity; E value = 2e−49). The sequences clustered in these two groups mainly distributed on days 15, 30, and 50 (cooling and maturation stages), suggesting that the relevant microorganisms dominated in the maturation phase. In summary, this newly designed primer could amplify the potential unknown putative Streptomyces two-domain LMCO genes in environmental samples.
Phenol oxidase activity, lignocellulose degradation, and abundance of Streptomyces two-domain LMCO genes.
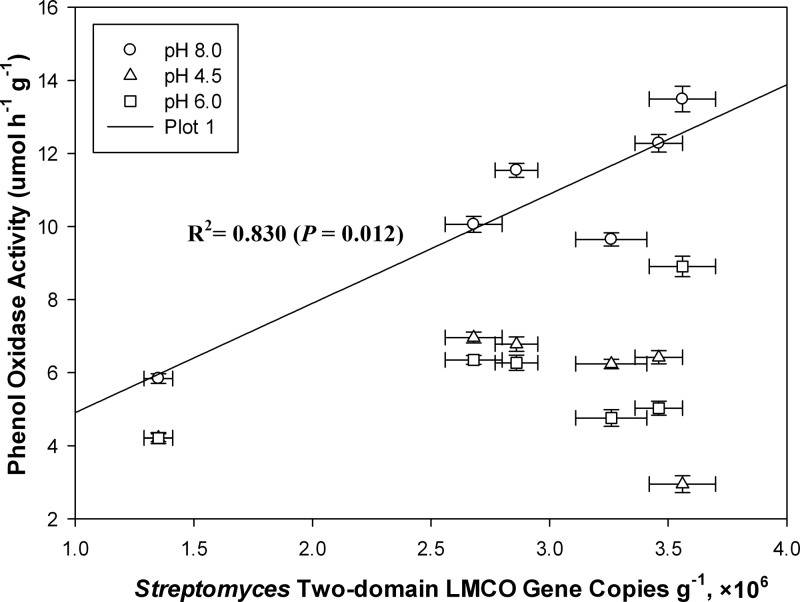
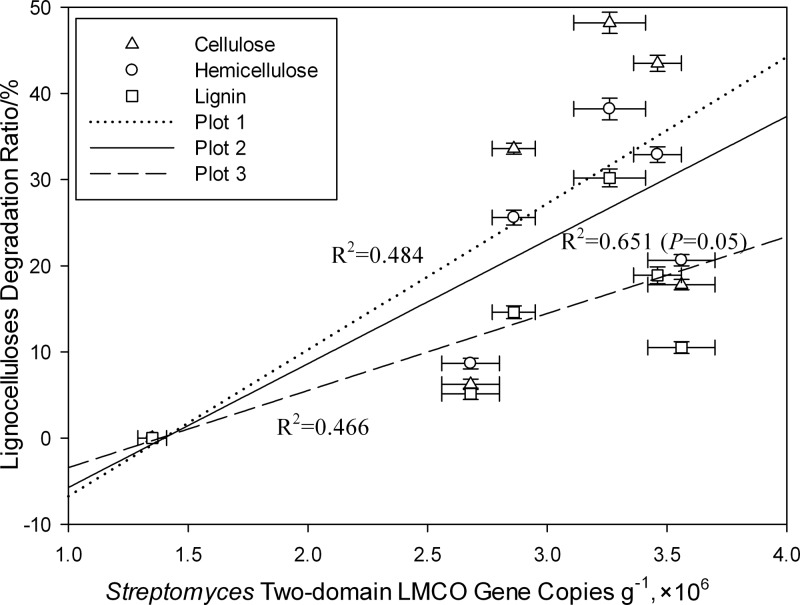
The relationships between phenol oxidase activity at different pHs, lignocellulose degradation rates, and LMCO gene abundance were estimated using a linear model analysis of covariance. Phenol oxidase activity at pH 8.0 was linearly positively associated with Streptomyces two-domain LMCO gene abundance (R2 = 0.830; P = 0.012) (Fig. 5). However, no such simple association was observed between LMCO gene abundance and phenol oxidase activity at pH 4.5 and 6.0. The variations in Streptomyces two-domain LMCO gene abundance seemed also positively associated with lignocellulose degradation ratios during composting, which indicated that the Streptomyces spp. probably participated in the lignocellulose degradation during composting (Fig. 6).
FIG 5.
Relationship between Streptomyces two-domain LMCO gene abundance and phenol oxidase activity. The regression line shown is for phenol oxidase activity at a pH value of 8.0.
FIG 6.
Relationship between Streptomyces two-domain LMCO gene abundance and lignocellulose degradation ratios. Cellulose, dotted line; hemicellulose, solid line; lignin, dashed line.
DISCUSSION
Traditional three-domain fungal and bacterial LMCO genes potentially participating in soil organic matter cycling and litter degradation were extensively investigated in previous studies (33, 34). The present survey relied on a similar molecular approach for analyzing Streptomyces two-domain LMCO gene diversity in a composting system. Based on the phylogenetic analysis and the deduced amino acids, the amplified genes were unambiguously identified as Streptomyces two-domain LMCO genes. The method used in a previous study found only very limited numbers of Streptomyces two-domain laccases (20); however, the metagenetic method in this study identified a wide species range in Streptomyces spp., including Streptomyces violaceusniger, Streptomyces griseus, and Streptomyces ipomoeae. The analysis revealed relatively high diversity and marked fluctuation in Streptomyces two-domain LMCO gene populations at different stages of agricultural waste composting. In summary, the present study demonstrated that direct amplification of Streptomyces two-domain LMCO gene sequences from compost DNA extracts allows tracing the distributions and changes of streptomycetes with a potential role in lignocellulose degradation.
The apparent increase of Streptomyces two-domain LMCO gene abundance in the initial phase of composting may be ascribed to the proliferation of streptomycetes stimulated by the high content of degradable organic compounds in the initial compost mixture. The highest Streptomyces two-domain LMCO gene abundance was observed on day 8, indicating that the corresponding organisms showed resistance to the changing temperature. Temperature-resistant Streptomyces spores are usually easy to propagate, giving rise to branching hyphal filaments in composting environments. Moreover, this growth adaptation assists in adherence to and penetration into the insoluble organic remains of other organisms, which are then depolymerized by multiple exoenzymes to provide nutrients (35). Xiao et al. (11) also suggested in their study that temperatures higher than 50°C in composting benefited the growth of actinomycetes (11). Especially thermostable streptomycetes are considered the predominant actinomycetes existing in the whole composting process (36). They can tolerate the higher temperature and alkaline environment in composting. Additionally, antibiotic production confers crucial advantages to streptomycetes in ecological niche occupancy by exerting a high selective pressure on a wide range of other organisms (36, 37).
There was a significant linear relationship between Streptomyces two-domain LMCO gene abundance and phenol oxidase activity at pH 8.0. This obvious relationship, however, was not present under acidic conditions. This may have been due to pH and temperature changes in the composting process that regulate enzyme activity and, in addition, to the relevant functional groups in Streptomyces spp. being important drivers of enzyme activity during composting. The significant relationship between phenol oxidase activity and Streptomyces two-domain LMCO gene abundance suggests that streptomycetes are involved in the extracellular phenol oxidase activities. Streptomyces two-domain LMCOs offer advantages over counterparts of conventional three-domain laccases, such as resistance to higher temperatures and alkaline pH values, which make them active under the extreme conditions; thus, we expected that Streptomyces spp. could dominate phenol oxidase activity under alkaline conditions during composting. However, the phenol oxidase activity at pH 8.0 was not completely attributed to the Streptomyces two-domain LMCO, for compost is a complicated ecosystem with high environmental and microbial variability and some other thermostable bacteria were also verified to be important good producers of alkaline laccases (4, 38, 39).
A positive relationship was observed between Streptomyces two-domain LMCO gene abundance and lignocellulose degradation rates. High lignin degradation at the thermophilic stage was probably caused by the greatly induced expression of Streptomyces two-domain LMCO genes at elevated temperatures and their symbiotic involvement with many other microorganisms (35). Schlatter et al. (37) also verified that the addition of cellulose or lignin results in relatively high densities of streptomycetes, suggesting that the lignocellulose degradation could promote the reproduction of streptomycetes to an extent. The potential lignocellulose degradation by streptomycetes was not due only to laccase because there are data indicating that streptomycetes are also good producers of other lignocellulolytic enzymes (40), such as cellulases from Streptomyces sp. strain G12 in compost (41), CBM33 polysaccharides-monooxygenases from Streptomyces sp. strain SirexAA-E (9), endo-β-1,4-xylanase from Streptomyces sp. strain SWU10 (42), lignin peroxidase from Streptomyces viridosporus (43), and dioxygenase (44).
However, only a portion of the laccase-encoding genes detected in the community could be expressed and translated into active enzymes, thereby affecting lignin decomposition (22). And it is worth noting that some laccase-encoding genes are not clearly assignable to extracellular enzymes involved in lignocellulose degradation but may relate to a variety of other functions, including morphogenesis, biochemical interaction with other organisms, and production of antibiotics (8). Thus, there must be further research to gain deeper insights into the ecological functions of streptomycetes with potential extracellular laccase activity and ligninolytic capacity in future studies. The challenge at this point is to strengthen investigations for detection of clearly verifiable extracellular laccases (e.g., by screening of ecologically relevant microbes for all potential laccases and linking their genetic potential to produce laccase exoenzymes under laboratory and natural conditions), with combined analysis of gene expression and protein synthesis in order to clarify which genes correspond to which functions (45).
ACKNOWLEDGMENTS
We thank the reviewers for very helpful comments and suggestions.
This study was financially supported by the National Natural Science Foundation of China (51378190, 50908079, and 51039001), the Hunan Provincial Natural Science Foundation of China (10JJ7005), the Scholarship Award for Excellent Doctoral Student granted by the Ministry of Education, the Hunan Provincial Innovation Foundation for Postgraduate (CX2012B137), and the Zhejiang Provincial Key Laboratory of Solid Waste Treatment and Recycling open fund (SWTR-2012-07).
Footnotes
Published ahead of print 21 March 2014
REFERENCES
- 1.Zeng G, Zhang J, Chen Y, Yu Z, Yu M, Li H, Liu Z, Chen M, Lu L, Hu C. 2011. Relative contributions of archaea and bacteria to microbial ammonia oxidation differ under different conditions during agricultural waste composting. Bioresour. Technol. 102:9026–9032. 10.1016/j.biortech.2011.07.076 [DOI] [PubMed] [Google Scholar]
- 2.He Y, Xie K, Xu P, Huang X, Gu W, Zhang F, Tang S. 2013. Evolution of microbial community diversity and enzymatic activity during composting. Res. Microbiol. 164:189–198. 10.1016/j.resmic.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 3.Xie K, Jia X, Xu P, Huang X, Gu W, Zhang F, Yang S, Tang S. 2012. Improved composting of poultry faeces via supplementation with ammonia oxidizing archaea. Bioresour. Technol. 120:70–77. 10.1016/j.biortech.2012.06.029 [DOI] [PubMed] [Google Scholar]
- 4.Tuomela M, Vikman M, Hatakka A, Itavaara M. 2000. Biodegradation of lignin in a compost environment: a review. Bioresour. Technol. 72:169–183. 10.1016/S0960-8524(99)00104-2 [DOI] [Google Scholar]
- 5.Adams AS, Jordan MS, Adams SM, Suen G, Goodwin LA, Davenport KW, Currie CR, Raffa KF. 2011. Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J. 5:1323–1331. 10.1038/ismej.2011.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maki M, Leung KT, Qin W. 2009. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 5:500. 10.7150/ijbs.5.500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulrich A, Klimke G, Wirth S. 2008. Diversity and activity of cellulose-decomposing bacteria, isolated from a sandy and a loamy soil after long-term manure application. Microb. Ecol. 55:512–522. 10.1007/s00248-007-9296-0 [DOI] [PubMed] [Google Scholar]
- 8.Bontemps C, Toussaint M, Revol PV, Hotel L, Jeanbille M, Uroz S, Turpault MP, Blaudez D, Leblond P. 2013. Taxonomic and functional diversity of Streptomyces in a forest soil. FEMS Microbiol. Lett. 342:157–167. 10.1111/1574-6968.12126 [DOI] [PubMed] [Google Scholar]
- 9.Takasuka TE, Book AJ, Lewin GR, Currie CR, Fox BG. 2013. Aerobic deconstruction of cellulosic biomass by an insect-associated Streptomyces. Sci. Rep. 3:1030. 10.1038/srep01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunne M, Urlacher VB. 2012. Characterization of the alkaline laccase Ssl1 from Streptomyces sviceus with unusual properties discovered by genome mining. PLoS One 7:e52360. 10.1371/journal.pone.0052360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao Y, Zeng GM, Yang ZH, Ma YH, Huang C, Xu ZY, Huang J, Fan CZ. 2011. Changes in the actinomycetal communities during continuous thermophilic composting as revealed by denaturing gradient gel electrophoresis and quantitative PCR. Bioresour. Technol. 102:1383–1388. 10.1016/j.biortech.2010.09.034 [DOI] [PubMed] [Google Scholar]
- 12.Ausec L, van Elsas JD, Mandic-Mulec I. 2011. Two- and three-domain bacterial laccase-like genes are present in drained peat soils. Soil Biol. Biochem. 43:975–983. 10.1016/j.soilbio.2011.01.013 [DOI] [Google Scholar]
- 13.Taprab Y, Johjima T, Maeda Y, Moriya S, Trakulnaleamsai S, Noparatnaraporn N, Ohkuma M, Kudo T. 2005. Symbiotic fungi produce laccases potentially involved in phenol degradation in fungus combs of fungus-growing termites in Thailand. Appl. Environ. Microbiol. 71:7696–7704. 10.1128/AEM.71.12.7696-7704.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackwood CB, Waldrop MP, Zak DR, Sinsabaugh RL. 2007. Molecular analysis of fungal communities and laccase genes in decomposing litter reveals differences among forest types but no impact of nitrogen deposition. Environ. Microbiol. 9:1306–1316. 10.1111/j.1462-2920.2007.01250.x [DOI] [PubMed] [Google Scholar]
- 15.Chen M, Zeng G, Tan Z, Jiang M, Li H, Liu L, Zhu Y, Yu Z, Wei Z, Liu Y. 2011. Understanding lignin-degrading reactions of ligninolytic enzymes: binding affinity and interactional profile. PLoS One 6:e25647. 10.1371/journal.pone.0025647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komori H, Miyazaki K, Higuchi Y. 2009. Crystallization and preliminary X-ray diffraction analysis of a putative two-domain-type laccase from a metagenome. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:264–266. 10.1107/S1744309109002462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endo K, Hayashi Y, Hibi T, Hosono K, Beppu T, Ueda K. 2003. Enzymological characterization of EpoA, a laccase-like phenol oxidase produced by Streptomyces griseus. J. Biochem. 133:671–677. 10.1093/jb/mvg086 [DOI] [PubMed] [Google Scholar]
- 18.Dubé E, Shareck F, Hurtubise Y, Daneault C, Beauregard M. 2008. Homologous cloning, expression, and characterisation of a laccase from Streptomyces coelicolor and enzymatic decolourisation of an indigo dye. Appl. Microbiol. Biotechnol. 79:597–603. 10.1007/s00253-008-1475-5 [DOI] [PubMed] [Google Scholar]
- 19.Molina-Guijarro JM, Perez J, Munoz-Dorado J, Guillen F, Moya R, Hernandez M, Arias ME. 2009. Detoxification of azo dyes by a novel pH-versatile, salt-resistant laccase from Streptomyces ipomoea. Int. Microbiol. 12:13–21. 10.2436/20.1501.01.77 [DOI] [PubMed] [Google Scholar]
- 20.Lu L, Zeng G, Fan C, Ren X, Wang C, Zhao Q, Zhang J, Chen M, Chen A, Jiang M. 2013. Characterization of a laccase-like multicopper oxidase from newly isolated Streptomyces sp. C1 in agricultural waste compost and enzymatic decolorization of azo dyes. Biochem. Eng. J. 72:70–76. 10.1016/j.bej.2013.01.004 [DOI] [Google Scholar]
- 21.Nakamura K, Kawabata T, Yura K, Go N. 2003. Novel types of two-domain multi-copper oxidases: possible missing links in the evolution. FEBS Lett. 553:239–244. 10.1016/S0014-5793(03)01000-7 [DOI] [PubMed] [Google Scholar]
- 22.Ausec L, Zakrzewski M, Goesmann A, Schluter A, Mandic-Mulec I. 2011. Bioinformatic analysis reveals high diversity of bacterial genes for laccase-like enzymes. PLoS One 6:e25724. 10.1371/journal.pone.0025724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach CE, Warnock DD, Van Horn DJ, Weintraub MN, Sinsabaugh RL, Allison SD, German DP. 2013. Measuring phenol oxidase and peroxidase activities with pyrogallol, L-DOPA, and ABTS: effect of assay conditions and soil type. Soil Biol. Biochem. 67:183–191. 10.1016/j.soilbio.2013.08.022 [DOI] [Google Scholar]
- 24.Floch C, Alarcon-Gutiérrez E, Criquet S. 2007. ABTS assay of phenol oxidase activity in soil. J. Microbiol. Methods 71:319–324. 10.1016/j.mimet.2007.09.020 [DOI] [PubMed] [Google Scholar]
- 25.Luis P, Kellner H, Zimdars B, Langer U, Martin F, Buscot F. 2005. Patchiness and spatial distribution of laccase genes of ectomycorrhizal, saprotrophic, and unknown basidiomycetes in the upper horizons of a mixed forest cambisol. Microb. Ecol. 50:570–579. 10.1007/s00248-005-5047-2 [DOI] [PubMed] [Google Scholar]
- 26.Gandía-Herrero F, Jiménez-Atiénzar M, Cabanes J, García-Carmona F, Escribano J. 2005. Differential activation of a latent polyphenol oxidase mediated by sodium dodecyl sulfate. J. Agric. Food Chem. 53:6825–6830. 10.1021/jf050505e [DOI] [PubMed] [Google Scholar]
- 27.Van Soest PJ, Robertson J, Lewis B. 1991. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74:3583–3597. 10.3168/jds.S0022-0302(91)78551-2 [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Zeng G, Chen Y, Yu M, Huang H, Fan C, Zhu Y, Li H, Liu Z, Chen M. 2013. Impact of Phanerochaete chrysosporium inoculation on indigenous bacterial communities during agricultural waste composting. Appl. Microbiol. Biotechnol. 97:3159–3169. 10.1007/s00253-012-4124-y [DOI] [PubMed] [Google Scholar]
- 29.Yu H, Zeng G, Huang H, Xi X, Wang R, Huang D, Huang G, Li J. 2007. Microbial community succession and lignocellulose degradation during agricultural waste composting. Biodegradation 18:793–802. 10.1007/s10532-007-9108-8 [DOI] [PubMed] [Google Scholar]
- 30.Yang ZH, Xiao Y, Zeng G, Xu ZY, Liu YS. 2007. Comparison of methods for total community DNA extraction and purification from compost. Appl. Microbiol. Biotechnol. 74:918–925. 10.1007/s00253-006-0704-z [DOI] [PubMed] [Google Scholar]
- 31.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luis P, Kellner H, Martin F, Buscot F. 2005. A molecular method to evaluate basidiomycete laccase gene expression in forest soils. Geoderma 128:18–27. 10.1016/j.geoderma.2004.12.023 [DOI] [Google Scholar]
- 34.Kellner H, Luis P, Schlitt B, Buscot F. 2009. Temporal changes in diversity and expression patterns of fungal laccase genes within the organic horizon of a brown forest soil. Soil Biol. Biochem. 41:1380–1389. 10.1016/j.soilbio.2009.03.012 [DOI] [Google Scholar]
- 35.Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H. 2010. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34:171–198. 10.1111/j.1574-6976.2009.00206.x [DOI] [PubMed] [Google Scholar]
- 36.Kirby R. 2006. Actinomycetes and lignin degradation. Adv. Appl. Microbiol. 58:125–168. 10.1016/S0065-2164(05)58004-3 [DOI] [PubMed] [Google Scholar]
- 37.Schlatter D, Fubuh A, Xiao K, Hernandez D, Hobbie S, Kinkel L. 2009. Resource amendments influence density and competitive phenotypes of Streptomyces in soil. Microb. Ecol. 57:413–420. 10.1007/s00248-008-9433-4 [DOI] [PubMed] [Google Scholar]
- 38.Kellner H, Luis P, Zimdars B, Kiesel B, Buscot F. 2008. Diversity of bacterial laccase-like multicopper oxidase genes in forest and grassland Cambisol soil samples. Soil Biol. Biochem. 40:638–648. 10.1016/j.soilbio.2007.09.013 [DOI] [Google Scholar]
- 39.Sánchez C. 2009. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol. Adv. 27:185–194. 10.1016/j.biotechadv.2008.11.001 [DOI] [PubMed] [Google Scholar]
- 40.Schrempf H. 2007. Biology of streptomycetes, p 21–28 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed. Springer-Verlag, New York, NY [Google Scholar]
- 41.Amore A, Pepe O, Ventorino V, Birolo L, Giangrande C, Faraco V. 2012. Cloning and recombinant expression of a cellulase from the cellulolytic strain Streptomyces sp. G12 isolated from compost. Microb. Cell Fact. 11:164. 10.1186/1475-2859-11-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deesukon W, Nishimura Y, Sakamoto T, Sukhumsirichart W. 2013. Purification, characterization of GH11 endo-β-1,4-xylanase from thermotolerant Streptomyces sp. SWU10 and overexpression in Pichia pastoris KM71H. Mol. Biotechnol. 54:37–46. 10.1007/s12033-012-9541-8 [DOI] [PubMed] [Google Scholar]
- 43.Ramachandra M, Crawford DL, Hertel G. 1988. Characterization of an extracellular lignin peroxidase of the lignocellulolytic actinomycete Streptomyces viridosporus. Appl. Environ. Microbiol. 54:3057–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bianchetti CM, Harmann CH, Takasuka TE, Hura GL, Dyer K, Fox BG. 2013. Fusion of dioxygenase and lignin-binding domains in a novel secreted enzyme from cellulolytic Streptomyces sp. SirexAA-E. J. Biol. Chem. 288:18574–18587. 10.1074/jbc.M113.475848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theuerl S, Buscot F. 2010. Laccases: toward disentangling their diversity and functions in relation to soil organic matter cycling. Biol. Fert. Soils 46:215–225. 10.1007/s00374-010-0440-5 [DOI] [Google Scholar]