Abstract
Outer membrane vesicles (OMVs) are continually released from a range of bacterial species. Numerous functions of OMVs, including the facilitation of horizontal gene transfer (HGT) processes, have been proposed. In this study, we investigated whether OMVs contribute to the transfer of plasmids between bacterial cells and species using Gram-negative Acinetobacter baylyi as a model system. OMVs were extracted from bacterial cultures and tested for the ability to vector gene transfer into populations of Escherichia coli and A. baylyi, including naturally transformation-deficient mutants of A. baylyi. Anti-double-stranded DNA (anti-dsDNA) antibodies were used to determine the movement of DNA into OMVs. We also determined how stress affected the level of vesiculation and the amount of DNA in vesicles. OMVs were further characterized by measuring particle size distribution (PSD) and zeta potential. Transmission electron microscopy (TEM) and immunogold labeling were performed using anti-fluorescein isothiocyanate (anti-FITC)-conjugated antibodies and anti-dsDNA antibodies to track the movement of FITC-labeled and DNA-containing OMVs. Exposure to OMVs isolated from plasmid-containing donor cells resulted in HGT to A. baylyi and E. coli at transfer frequencies ranging from 10−6 to 10−8, with transfer efficiencies of approximately 103 and 102 per μg of vesicular DNA, respectively. Antibiotic stress was shown to affect the DNA content of OMVs as well as their hydrodynamic diameter and zeta potential. Morphological observations suggest that OMVs from A. baylyi interact with recipient cells in different ways, depending on the recipient species. Interestingly, the PSD measurements suggest that distinct size ranges of OMVs are released from A. baylyi.
INTRODUCTION
Intercellular transfer of nucleic acids is an intrinsic feature of bacterial evolution (1). Such transfer is possible through the established mechanisms of natural transformation, conjugation, and transduction, as well as via the more recently identified functions of nanotubes and outer membrane vesicles (OMVs). For instance, recent studies have revealed that Bacillus subtilis forms tubular structures that can connect to neighboring cells and facilitate the exchange of cytoplasmic contents (2). It is still unclear whether nanotubes are similar to the nanopods that have been recently reported in Delftia sp., which are able to transfer membrane vesicles (MVs) to other recipient cells (3). Gene transfer via nanotubes and OMVs has gained particular interest because of their unique feature of intercellular transportation of cellular material. Long-distance transport of cytoplasmic contents is a distinctive feature of such mechanisms, for which the full set of biological functions remain to be revealed. One identified function of MVs is the dissemination of nucleic acids, possibly resulting in horizontal gene transfer (HGT) events occurring under conditions where other established mechanisms of gene exchange are not active.
OMVs have been reported to serve a number of biological functions, such as the delivery of proteins and toxins to target cells during infection, the transport of various effectors between bacterial cells in populations, including in biofilms, the protection of nucleic acids during intercellular transport, and bacterial defense (4–6). For instance, OMVs can adsorb antibacterial peptides and thereby possibly increase bacterial survival (5). MVs are commonly released from both Gram-positive and Gram-negative bacteria (6, 7). The production of MVs is a common phenomenon in growing bacterial populations and is not due to random cell death or lysis (8). OMVs of Gram-negative bacteria have been extensively studied due to their association with virulence factors (9). OMVs are produced by the bulging of the outer membrane, followed by constriction and subsequent release from the bacterial cell, a process referred to as vesiculation (10). OMVs contain outer membrane (OM) and periplasmic components, such as OM proteins, virulence proteins, phospholipids, and lipopolysaccharides (LPS). However, cytoplasmic content, such as genetic material, is also present in MVs (11, 12). The levels of MV formation differ depending on the strain and growth conditions, such as variations in temperature, exposure to antibiotics, the presence of oxygen, and nutrient availability (13–17).
OMVs are spherical and range in size from 50 to 250 nm in diameter (9). Once released from the parental bacterium, they can persist in an independent state until lysis. The bilayered structure of OMVs protects the lumen content from immediate degradation by extracellular enzymes, such as proteases and nucleases (18). OMVs can fuse with other cells, resulting in intercellular transfer of lumen contents, including nucleic acids (19, 20). The gene transfer potential of OMVs has been previously studied in various genera. For example, in Neisseria gonorrhoeae, a plasmid containing a penicillin resistance gene was transferred to penicillin-sensitive gonococci (7). The OMVs from an Escherichia coli O157:H7 strain harboring a gfp gene-containing plasmid were transferred to other E. coli members (21). In Ruminococcus spp., OMVs were able to transfer genes required for the ability to degrade crystalline cellulose (22). OMVs of Moraxella catarrhalis were capable of transferring β-lactamase proteins to Streptococcus pneumoniae and Haemophilus influenzae, thereby promoting their survival in the presence of amoxicillin (23). Recently, the horizontal transfer of a carbapenem resistance gene via OMVs was shown in Acinetobacter baumannii (24). The release of DNA-containing OMVs from pathogenic species of Acinetobacter has also been previously reported (25–27). Acinetobacter baylyi (previously also denoted Acinetobacter calcoaceticus) has been described to release OMVs when grown on hexadecane (28).
Several members of the Acinetobacter genus are now recognized as emerging threats to public health because of the frequent occurrence of multidrug-resistant strains in intensive care units worldwide (29–31). Approximately 80% of Acinetobacter isolates carry multiple plasmids of various sizes (32–34). Moreover, transposons and integrons carrying multiple antibiotic resistance genes are increasingly found in clinical isolates of Acinetobacter (35, 36) and can be transferred between species by natural transformation (37).
In this work, we characterized the production of OMVs by the model bacterium A. baylyi by vesicle extraction, transmission electron microscopy (TEM), particle size distribution (PSD) measurements, and zeta potential analysis. Moreover, we used immunogold labeling to follow the movement of double-stranded DNA (dsDNA) from the bacterial cytoplasm to the periplasm and subsequently into vesicles. We determined the potential of OMVs released by A. baylyi to contribute to HGT by transferring a plasmid-borne β-lactamase gene. Finally, we investigated whether stress induced by antibiotics or environmental parameters affects the characteristics of OMVs, including their DNA content.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and antimicrobial susceptibility testing.
In this study, we used A. baylyi JV26 (38, 39), which is a Trp+ derivative of the DSM588 strain obtained from the Deutsche Sammlung von Mikroorganismen and Zellkulturen (DSMZ, Braunschweig, Germany), and E. coli DH5α (40). The two strains were transformed by pMU125 as described in references 41 and 42, respectively. Bacteria were grown in Luria-Bertani (LB) broth (BD, Le Pont-de-Claix, France) at 37°C (E. coli) or 30°C (A. baylyi) with shaking. The plasmid was maintained by the addition of 100 μg ml−1 ampicillin in the broth or in solid LB medium. pMU125 is a broad-host-range shuttle vector containing genes for the green fluorescent protein (GFP) and for a β-lactamase conferring resistance to ampicillin (37). This plasmid is a high-copy-number replicon in E. coli, but it is a low-copy-number plasmid in A. baylyi (43). The MICs of gentamicin and chloramphenicol for the strain JV26(pMU125) were determined with Etest strips (AB Biodisk, Sweden). Experiments with environmental stressors, such as nutrient deprivation and UV light exposure, were carried out in M9 minimal medium (42). Experiments with temperature increases, desiccation, and subinhibitory concentrations (SICs) of antibiotics were performed with LB medium along with control experiments in which organisms were grown without stress using LB or M9 medium. SICs were determined from established bacterial growth curves recorded with increasing concentrations of the relevant antibiotic (44) using a microplate reader (VersaMax; Molecular Devices, USA). The inoculum used to establish the growth curves was 1 × 106 CFU ml−1, obtained by dilution (1:500) of an overnight culture with 2× LB medium.
Construction of transformation-deficient mutants and of the gentamicin-resistant strain of A. baylyi.
The transformation-deficient ΔcomA::(nptII sacB) (replacement of comA by a marker pair conferring kanamycin resistance and sucrose susceptibility) and ΔcomB-comF::dhfr (replacement of comB, ACIAD3317 [a putative pilX gene {38}], and comC, -E, and -F by a trimethoprim resistance gene) derivatives of A. baylyi JV26 were constructed as described previously (39, 45). The gentamicin-resistant JV26 derivative contained the ACIAD2756::aacC1 insertion (N. Hülter, unpublished).
Isolation and purification of OMVs.
OMVs were isolated from liquid cultures of JV26(pMU125) as previously described (46), with some modifications. Briefly, 8 ml or 1 ml of an overnight culture was used to inoculate 800 ml or 100 ml of LB broth containing 100 μg ml−1 ampicillin, respectively. To isolate vesicles from cultures grown under antibiotic stress, liquid cultures were grown with shaking with subinhibitory concentrations of chloramphenicol (0.1 μg ml−1 or 1 μg ml−1) or gentamicin (0.1 μg ml−1 or 0.3 μg ml−1) and, in all cases, also with ampicillin, for 15 h at 30°C. For temperature stress experiments, the temperature was raised to 37°C. Desiccation stress was induced by adding 0.5 M NaCl to the growth (LB) medium. Nutrient deprivation was achieved by growing cells in minimal medium with 2.2 mM succinate plus 5.2 mM glucose as the carbon source. For UV light stress, UV exposure was applied to log-phase cells growing in minimal medium using a germicidal lamp irradiating the top of the culture in a beaker while it was stirred (dose, 0.36 mW/cm2) for 5 or 20 min. The distance between the lamp and the liquid surface was 47.5 cm. After irradiation, the culture was further grown until 15 h was reached. Bacterial cells were removed by centrifugation at 12,000 × g at 4°C for 30 min. The supernatants were again centrifuged at 15,000 × g for 20 min at 4°C, followed by vacuum filtration using a Rapid-Flow polyethersulfone (PES) bottle top filter (0.2-μm size; Thermo Scientific, MA, USA). The filtrates were concentrated using Vivaspin centrifugal concentrators (Vivascience, Hannover, Germany) with a 10-kDa molecular mass cutoff. OMVs were recovered from the concentrates by ultracentrifugation at 130,000 × g for 3 h at 4°C using an Optima LE80k ultracentrifuge (SW 40 Ti rotor). The pellet was washed with HEPES buffer (50 mM, pH 6.8) and resuspended in the same buffer. The OMVs were isolated from 3 independent batches of cultures grown under stressed and nonstressed conditions. The solubilized OMVs were once again filtered through 0.22-μm-pore-size syringe filters (Pall Corporation, NY, USA) and treated with proteinase K (100 μg ml−1) to digest any phage coats, if present, and DNase I (100 ng ml−1). The suspensions were incubated at 37°C for 20 min, followed by deactivation of the DNase at 80°C for 10 min. One hundred microliters of the vesicle suspension was spread on LB agar and also inoculated in fresh LB medium and cultured for 24 h to confirm the absence of viable cells. The protein concentration of the vesicles was determined by the Bradford assay (Bio-Rad, USA) (24). The vesicles were stored at −20°C until further use. The bacterial cultures were also checked for the absence of bacteriophages according to previously described methods (47).
Particle size distribution and measurement of zeta potential.
Purified vesicles and bacterial cells were diluted with HEPES buffer (50 mM, pH 6.8; vesicles were filtered through filters with a 0.22-μm pore size) in a particle-free environment. The size distribution analysis was recorded by a Photocor (College Park, MD, USA) instrument at a 90° angle with a laser with a wavelength of 632 nm. The data were analyzed with the Dynals software to obtain the average hydrodynamic radius of a given particle. The measurements were conducted at 22°C with three runs of 30 min each for each sample, and the average intensity weighted diameter was calculated. The stability of the vesicles was determined by measuring the size distribution of the samples after they were stored for 1 week at room temperature. The zeta potential of both vesicles and bacterial cells was estimated using the Malvern Zetasizer Nano Z (Malvern Instruments, Malvern, United Kingdom). The average diameter and zeta potential were obtained for OMVs isolated from 3 independent batches.
Fatty acid analysis.
Fatty acids from 3 independent batches of overnight-grown stationary bacterial cell cultures and purified OMVs (proteins concentrations, 486, 562.4, and 442 μg ml−1) were extracted and transformed into fatty acid methyl esters (FAMEs) by the Sherlock microbial identification system (MIS) protocol (48) and identified by gas chromatography (GC). The sample processing included the following steps. (i) For saponification, 1 ml of 15% (wt/vol) NaOH in 50% methanol was added to the samples, followed by heating at 100°C for 30 min. (ii) For methylation, 2 ml of methanolic HCl was added to the above-described mixture after it was cooled to room temperature, and the mixture was vortexed and heated at 80°C for 10 min. (iii) For extraction, the FAMEs were extracted from the cooled mixture in 1.25 ml of 1:1 (vol/vol) ether and hexane. (iv) For the base wash, the organic extracts were washed with 3 ml of 1.2% (wt/vol) NaOH. The FAMEs were identified by GC (Agilent 6890N) with an autosampler, an Agilent 7683B injector, and a flame ionization detector (FID). Helium was used as the carrier gas in the column (a Varian wall-coated open tubular [WCOT] fused-silica column, 50 m by 0.25 mm [internal diameter]). The following temperature program was used in the column oven: 50°C at the starting point, which was held for 2 min and followed by an increase of 10°C/min to 150°C, 1°C/min to 205°C, and finally 15°C/min to 255°C (held for 10 min). The injection volume was 1 μl, and the inlet was held at a temperature of 240°C and the detector at 250°C. The FAMEs were identified and qualified by the Sherlock MIS software (v. 4.5).
β-NADH oxidase assay.
To detect the inner membrane β-NADH oxidase activity, purified OMVs from the stress and nonstress treatments were used in addition to bacterial cell lysate, which served as a positive control. The assay was carried out as described previously (49), with modifications. Briefly, the total reaction volume for the assay was set to 230 μl, and the reactions were carried out in a 96-well plate in triplicate with OMVs from 3 independent batches (protein concentration, 100 to 500 μg ml−1). Bacterial cell lysate was prepared by sonication and multiple freeze-thawing. The protein concentration range was kept the same as for the OMVs and served as a positive control. The 125-μl reaction mixture consisted of 0.4 mM dithiothreitol, 0.1 M Tris-HCl (pH 7.9), 75 μl of 0.1% NaHCO3, 20 μl of OMVs, and 20 μl of 0.2-mg-ml−1 β-NADH. The absorbance at 340 nm was measured every minute (total measurement time, 10 min) with a plate reader (VersaMax; Molecular Devices, USA).
Vesicular DNA (V-DNA) isolation and amplification of the β-lactamase gene.
DNA from purified vesicles was isolated as follows. Prior to DNA isolation, the vesicles were treated with DNase (Roche Diagnostics, Basel, Switzerland) (100 ng ml−1) at 37°C for 20 min, followed by inactivation at 80°C for 10 min to remove any extravesicular DNA. The DNA associated with or present within the lumen was isolated using the Zyppy plasmid miniprep kit (Zymo Research, USA). This was performed in triplicate from 3 independent batches of vesicle samples with a protein concentration equivalent to 100 μg ml−1 (for both stressed and nonstressed cultures) and also from gentamicin-treated batches with protein concentrations of 11 mg ml−1, 780 μg ml−1, 800 μg ml−1, and 708 μg ml−1. The extracted DNA was resuspended in 15 μl of Tris-EDTA (TE) buffer. Colony PCR was carried out with cells from bacterial colonies growing on ampicillin-containing medium obtained after exposure to vesicles containing pMU125. All PCRs were performed in a total volume of 20 μl containing 1 μM each primer, 10 μl of Dream Taq PCR master mix (2×) (Fermentas, Germany) (containing Taq buffer, 0.4 mM each deoxynucleoside triphosphate [dNTP], and 4 mM MgCl2). The primers (Sigma-Aldrich, USA) used to amplify the 650-bp product of the bla gene were bla-int-f (5′-GTAAGATCCTTGAGAGTTTTCG-3′) and bla-ORF-r (5′-TTACCAATGCTTAATCAGTGAGG-3′).
The DNA templates (2 μl) used for the PCR included purified vesicular DNA, the supernatant of the lysed cell suspension (after boiling at 80°C for 5 min) from pure colonies of vesiculants (transformants from vesicle-mediated gene transfer [VMGT]), donor bacteria containing a plasmid (positive control), or water (negative control). PCRs were carried out in a PTC-0200 thermal cycler (Bio-Rad, USA). The reaction conditions consisted of 1 cycle of 5 min at 94°C for the initial denaturation step, followed by 30 cycles of denaturation (92°C, 30 s), annealing (55°C, 30 s), and extension (72°C, 40 s), with a final extension step of 72°C for 5 min. The PCR products were analyzed by gel electrophoresis on 1% agarose and visualized with a Bio-Rad gel documentation system. For some control experiments, purified pMU125 plasmid DNA isolated from A. baylyi (JV26) using the Qiagen plasmid kit (Germany) was used.
Vesicular DNA quantification.
Purified vesicle DNA was quantified using the Quant-iT PicoGreen dsDNA assay (Molecular Probes, Invitrogen, USA). The measurement was determined in triplicate for each V-DNA sample isolated from 3 independent batches of OMVS from all the stress and nonstressed growth conditions. A PicoGreen assay was carried out according to the manufacturer's instructions by determining the fluorescence using a hybrid multimode microplate reader (BioTek Instruments, Inc. USA).
OMV-mediated gene transfer in liquid cultures.
Gene transfer experiments were based on a previously published report (21), with certain modifications. The recipient strains DH5α, JV26, JV26 ΔcomA, JV26 ΔcomB-comF, and JV26 ACIAD2756::aacC1 were grown in SOC broth (2% Bacto tryptone, 0.5% bacterial yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose) until log phase, harvested by centrifugation (5,000 × g for 10 min), washed, and diluted with saline to produce a bacterial suspension of 2 × 104 CFU ml−1. For the gene transfer incubation mix, 50 μl of diluted cells was added to 500 μl of SOC medium with 500 μl of purified OMVs with a protein concentration of 10.5 mg ml−1 (isolated and pooled from several batches), 620 μg ml−1, 780 μg ml−1, or 708 μg ml−1 (induced with 0.1 μg ml−1 gentamicin) and with approximately 54 ng μl−1, 2.3 ng μl−1, 6.6 ng μl−1, or 2.3 ng μl−1 of V-DNA, respectively, and 1 μl of 100 μg μl−1 DNase (final concentration, 100 ng ml−1). The mixtures were then incubated at 37°C for 1 h without shaking, followed by 2 h with shaking at 150 rpm. Next, 2 ml of SOC was added per assay, and the incubation was continued for an additional 21 h. Control experiments were carried out without vesicles or, for JV26, with purified pMU125 DNA at different concentrations (2.3, 6.6, 20, 54, and 150 ng μl−1). Correspondingly, E. coli DH5α cells were incubated with the pMU125 plasmid (1 μg μl−1). To confirm the activity of DNase over time in the presence of cells, assays were performed without vesicles but with DNase (final concentration, 100 ng ml−1), mixtures were incubated at 37°C, and purified pMU125 plasmid DNA (70 ng ml−1) was added after 17 h. Bacterial cells were pelleted by centrifugation after 24 h of incubation and resuspended in 1 ml SOC medium. The bacterial cells that had acquired ampicillin resistance were selected for on LB agar supplemented with ampicillin (100 μg ml−1), and the recipient titers were determined by plating cells on LB medium. The plates were incubated for 16 h at 37°C for E. coli or for 2 days at 30°C for A. baylyi and evaluated by colony counting. Per assay, up to 10 ampicillin-resistant isolates were screened for the presence of the bla gene by PCR. In rare cases, PCR-negative isolates were encountered, and the gene transfer frequencies were adjusted accordingly. Gene transfer frequencies were calculated from ≥3 independent experiments as the number of gene transfer events over the number of recipient cells.
TEM.
Ten microliters of purified OMVs from the JV26 strain was negatively stained with freshly prepared 3% uranyl acetate for 1 min on 300-mesh-size, carbon-coated Formvar copper grids (Electron Microscopy Sciences, USA). The excess stain was blotted, and the grids were washed once with distilled water and dried. The micrographs were obtained by screening approximately 50 fields of each grid (in triplicate) from 3 independent batches of OMVs with a JEOL JEM-2100 high-resolution transmission electron microscope (HRTEM) (Peabody, MA) at 100 kV.
Vesicle labeling and IEM.
Vesicles were labeled as previously described (50), with some modifications. The purified vesicles (with a protein concentration of 400 μg ml−1) were incubated for 1 h at 25°C, followed by overnight incubation at 4°C with 1:1 fluorescein isothiocyanate (FITC [Sigma-Aldrich, USA]; 1 mg ml−1 in 50 mM Na2CO3, 100 mM NaCl, pH 9.2). The vesicles were pelleted at 130,000 × g for 3 h, washed, and resuspended in HEPES buffer (50 mM, pH 6.8) to remove unbound FITC. Recipient cells in log phase were diluted (1:100) in HEPES buffer (50 mM, pH 6.8), and 100 μl of cells was coincubated with 50 μl of labeled vesicles in two batches. One batch was incubated for 1 h at 37°C and then fixed in 8% formaldehyde in HEPES buffer overnight. The other batch was fixed immediately after the recipient cells were mixed with vesicles. The method for immune electron microscopy (IEM) was adapted from a previously published report (51). The fixed samples were pelleted, treated with 0.12% glycine, and pelleted again, and then 12% liquid gelatin was added. The cell pellet was transferred to 2.3 M sucrose in phosphate-buffered saline (PBS). After 1 h on ice, the samples were mounted on specimen pins and frozen in liquid nitrogen. Ultrathin cryosections were cut and incubated with rabbit anti-FITC antibody 1:600 (Invitrogen, USA), followed by protein A conjugated to 5-nm gold particles.
Nonsectioned OMVs were also exposed to rabbit anti-OmpA (1:150) antibody followed by protein A conjugated to 5-nm gold particles. To examine the presence of DNA in OMVs, ultrathin sections of stationary-phase cells of JV26(pMU125) and OMV preparations, including nonsectioned OMVs, were exposed to monoclonal anti-dsDNA antibodies (1633p77) diluted 1:30 with protein A conjugated to gold particles (5 nm). Micrographs were taken from 3 independent sample grids (each in triplicate) at a 100-kV total magnification using a JEM-1010 transmission electron microscope (Tokyo, Japan).
Confocal microscopy.
Green fluorescent protein (GFP) fluorescence in bacterial cells was examined by confocal microscopy. The samples used were OMVs containing pMU125 coincubated and grown with DH5α and JV26 recipient cells for 24 h at 37°C and 30°C, respectively. The JV26(pMU125) strain grown to log phase at 30°C in LB broth supplemented with ampicillin (100 μg ml−1) served as a positive control. Cells were harvested by centrifugation (3,500 × g at 4°C for 10 min). The cell pellet was resuspended in 500 μl of HEPES buffer (50 mM, pH 6.8). One hundred microliters of cells was added to sterile 8-well chambers (Lab-Tek, USA) and observed with a Leica TCS SP5 microscope. The observation was repeated three times from three independent VMGT assays.
Statistical analysis.
Each data point was averaged from results of three independent experiments, each with three replicates. Measurements of fatty acids, protein concentrations, diameters, and zeta potential and quantitation of V-DNA of OMVs were obtained for each sample. Additionally, transformation frequencies were entered into Excel spreadsheets (Microsoft, USA). Frequency distribution, namely, the mean and standard deviation, was determined. Statistical analysis was performed by Student's t tests for paired samples for the mean. In the figures, one asterisk (*) represents a P value between 0.01 and 0.05, and three asterisks (***) represent a P value of <0.001. These P values were considered statistically significant.
RESULTS
Analysis of the membrane composition and size distribution of OMVs from A. baylyi.
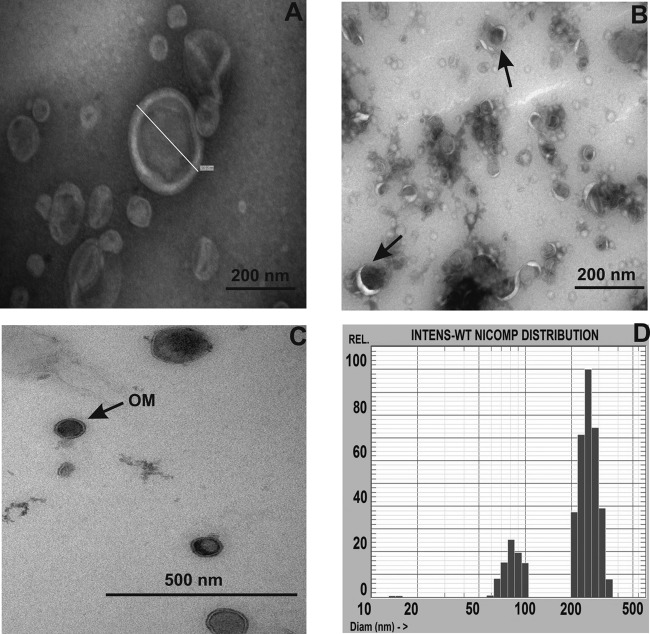
The presence and size of OMVs in late-stationary-phase cultures observed by TEM suggest that A. baylyi cells actively release OMVs during growth (Fig. 1A and B). The OMV micrographs revealed the bilayer, a spherical structure, and the presence of electron-dense material inside the vesicle lumen (Fig. 1C). The measurements of the sizes of the vesicles by dynamic light scattering show that the vesicles could be grouped into 3 size populations, with a mean diameter ranging from 13 to 304 nm (Fig. 1D; Table 1). The absence of viable cells in OMV preparations was confirmed by the absence of growth after inoculation of vesicles into LB broth or streaking of the samples on LB plates. Moreover, bacteriophages were not observed in a phage detection assay or by TEM.
FIG 1.
TEM images of OMVs isolated from A. baylyi JV26(pMU125) stained with uranyl acetate and the size distribution of OMVs. (A) Bilayer and spherical structure of a vesicle with a diameter of 237 nm. Bar, 200 nm. (B) Electron-dense area of the vesicle lumen in small-sized OMVs (arrows). Bar, 200 nm. (C) Ultrathin section of OMVs. The arrow indicates the outer membrane of a bilayer vesicle. Bar, 500 nm. (D) Mean size distribution of vesicles released from A. baylyi JV26(pMU125) cells without stress treatment, determined by the submicron particle sizing system. The x axis shows the particle diameter in nanometers against the relative percentages of intensity (INTENS).
TABLE 1.
Physical characteristics of OMVs isolated from A. baylyi strain JV26(pMU125)
| Treatment or stress | Antibiotic concn (μg ml−1) | Size distribution (avg diam [nm] ± SD), % intensity ± SD for peak or intensity: |
Zeta potential (mV)i | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| No treatment 1a | 304 ± 25, 83 ± 0.9 | 64 ± 26, 16 ± 10 | 13 ± 1.6, 0.7 ± 0.2 | −16.7 ± 1.1 | |
| Temp increase | 294 ± 46, 95 ± 2 | 45 ± 12, 3.6 ± 1.5 | −16.5 ± 1.0 | ||
| Desiccationb | 279 ± 42, 87 ± 3.5 | 46 ± 22, 9.3 ± 5.0 | 15 ± 3.4, 4.0 ± 1.8 | −18.3 ± 0.3 | |
| Gentamicin exposurec | 0.1 | 349 ± 11, 87 ± 5.7 | 81 ± 18, 4.2 ± 2.4 | 14 ± 1.5, 2.8 ± 1.0 | −27.6 ± 0.7 |
| 0.3 | 389 ± 16, 91 ± 4.6 | 128 ± 19, 7.8 ± 4.4 | 14 ± 0.8, 0.7 ± 0.1 | N.D | |
| Chloramphenicol exposured | 0.1 | 299 ± 12, 95 ± 1.6 | 54 ± 18, 3.0 ± 0.5 | −11.2 ± 1.1 | |
| 1.0 | 385 ± 42, 95 ± 3.5 | 83 ± 3.0, 4.7 ± 0.4 | ND | ||
| No treatment 2e | 237 ± 16, 92 ± 5 | 46 ± 19, 3.5 ± 0.4 | −19.4 ± 0.4 | ||
| Nutrient deprivationf | 270 ± 14, 88 ± 7 | 46 ± 8.5, 8.0 ± 2.7 | −19.2 ± 0.6 | ||
| UV exposure 1g | 229 ± 20, 95 ± 7.4 | 39 ± 7.7, 1.8 ± 0.7 | −19.4 ± 0.6 | ||
| UV exposure 2h | 239 ± 8.3, 93 ± 3.2 | 50 ± 17, 5.6 ± 2.0 | −21 ± 0.05 | ||
All A. baylyi cells were grown in the presence of ampicillin (100 μg ml−1), with resistance conferred by pMU125.
Cells were grown in medium containing 0.5 M NaCl.
The MIC of gentamicin is 0.5 μg ml−1.
The MIC of chloramphenicol is 4 μg ml−1.
Cells were grown in minimal medium containing succinate.
Cells were grown in minimal medium containing 2.2 mM succinate and 5.2 mM glucose.
UV light exposure was for 5 min.
UV light exposure was for 20 min.
The zeta potential for the A. baylyi JV26 recipient cells was −30 ± 0.7 mV, and that for the E. coli DH5α cells was −49.4 ± 0.5 mV. ND, not determined.
The stability of A. baylyi OMVs stored over a week at room temperature was monitored by PSD measurements and revealed an increase in the diameters of stored vesicles in comparison with those of freshly isolated OMVs (data not shown). The increase in the diameters might be due to the aggregation of some of the smaller OMVs. The OMVs did not sediment into closed packed beds, suggesting that A. baylyi OMVs retain their colloidal nature even after storage at room temperature.
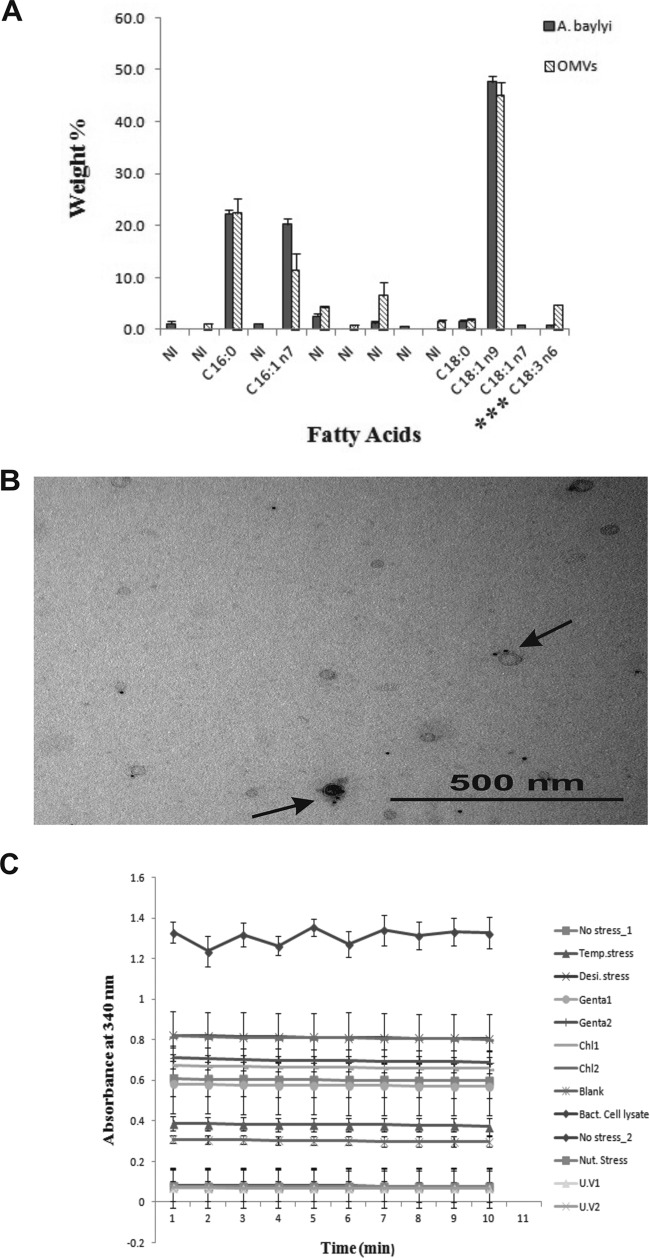
The average percentages of fatty acid profiles were identified from A. baylyi (JV26) cells and OMVs by gas chromatography and are presented in Fig. 2A. The fatty acids were found to be almost identical in A. baylyi cells and their released vesicles, with C18:1n9 (oleic acid) being the most dominant (47.7% ± 1.0% in cells and 44.8% ± 2.6% in released vesicles). The other major cellular fatty acids (CFAs) were C16:0 (palmitic acid), with 22.3% ± 0.6% in A. baylyi cells and 22.3% ± 2.8% in OMVs, and C16:1n7 (palmitoleic acid), with 20.3% ± 1.0% in cells and 11.4% ± 3.1% in OMVs. In addition, C18:0 (stearic acid) was identified at 1.6% ± 0.3% in cells and 1.8% ± 0.3% in OMVs, and C18:3n6 (α-linolenic acid) was identified at 0.7% ± 0.1% in cells and 4.7% ± 0.02% in OMVs. The quantities of α-linolenic acid were significantly different between cells and OMVs (P < 0.001). C18:1n7 (11-octadecenoic acid) was found in A. baylyi cells at 0.9% ± 0.1% but was not identified in OMVs. In addition to these compounds, two unidentified fatty acids were identified only in cells and three unidentified fatty acids were identified only in OMVs. Vesicles released from the A. baylyi cells with and without stress were checked for the inner membrane β-NADH oxidase activity (Fig. 2C). β-NADH oxidase activity was not observed in the OMV preparations, whereas activity was found in bacterial cell lysates due to the presence of inner membrane material, as expected. The Acinetobacter genus-specific outer membrane protein A (OmpA) was detected in the vesicles by IEM using rabbit anti-OmpA antibodies attached to gold particles (Fig. 2B). The bilayer structure of vesicles is similar to the OM of bacterial cells; the absence of inner membrane protein-specific activity and positive gold labeling of an OmpA marker suggest that the principal composition of the OMV coat is outer membrane material.
FIG 2.
Analysis of the membrane compositions of OMVs from A. baylyi JV26. (A) Histogram based on the mean values of cellular and vesicular fatty acids of A. baylyi. The fatty acid with three asterisks indicates that the values for this fatty acid were significantly different between cells and vesicles. Error bars indicate standard deviations. NI, not identified. (B) IEM image of unsectioned OMVs stained with uranyl acetate. Arrows indicate gold particles (black dots, 5 nm) attached to rabbit anti-OmpA antibodies. Bar, 500 nm. (C) Quantification of inner membrane-associated β-NADH oxidase activity. Bacterial cell lysate is the positive control. No activity was detected for OMVs released from A. baylyi (JV26) with no stress (No stress_1), temperature stress, desiccation stress, 0.1 μg ml−1 of gentamicin (Genta1), 0.3 μg ml−1 of gentamicin (Genta2), 0.1 μg ml−1 chloramphenicol (Chl1), 1 μg ml−1 chloramphenicol (Chl2), bacterial cell lysate grown with no stress in M9 minimal medium (No stress_2), nutrient stress, UV light exposure for 5 min (U.V1), UV light exposure for 20 min (U.V2), and a negative control (Blank). Error bars indicate standard deviations.
DNA translocates from A. baylyi cells to OMVs.
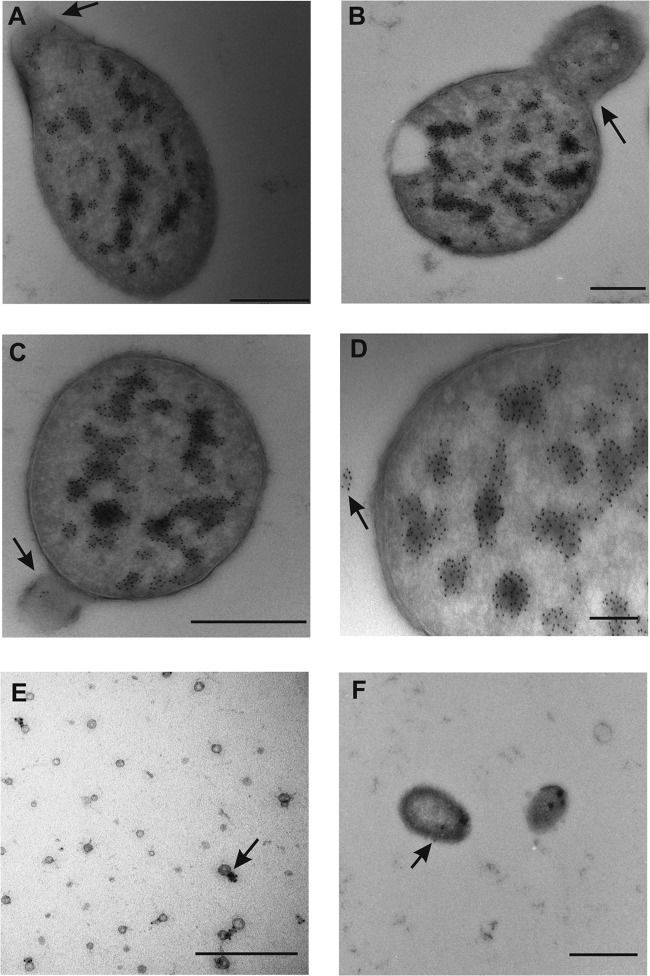
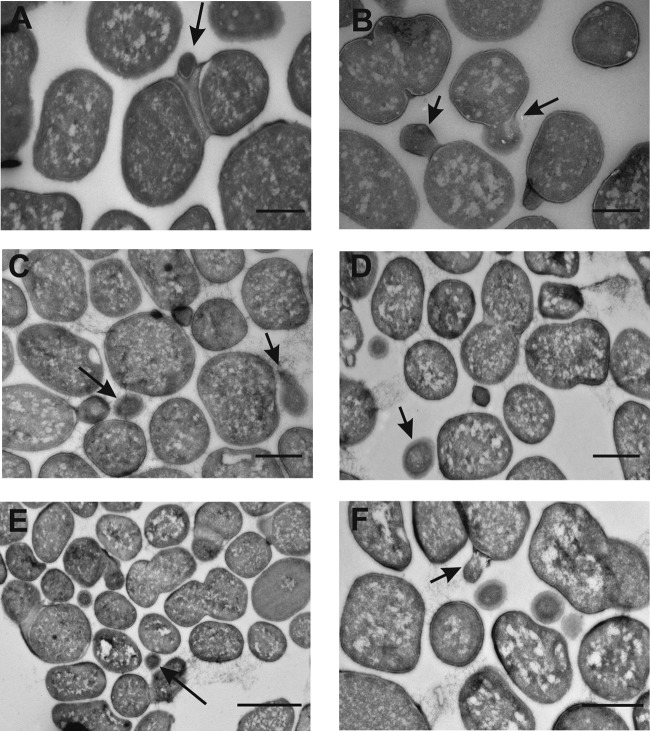
To investigate the location of and hypothesized transfer of DNA from the bacterial cytoplasm to OMVs, TEM with an immunogold labeling technique was carried out using dsDNA binding antibodies attached to gold particles. As observed in the Fig. 3 micrographs, movement of dsDNA from the cytoplasm to the periplasm occurred during OMV formation prior to the release from the bacterial surface. The dsDNA antibody-attached gold particles were found inside the lumen, as well as on the outer surface of the released OMVs (Fig. 3E and F). The antibodies are specific for dsDNA and bind to both plasmids and genomic DNA fragments. Some of the released OMVs (relatively few in number) did not bind the antibodies, indicating the absence of dsDNA in these vesicles, although the possibility of incomplete antibody binding during the sample preparation cannot be excluded. A total of 584 bacterial cells in log phase (grown without stress treatment) were observed by IEM, and out of these cells, 54 cells were found to be bulging with gold particles bound inside. In addition, a total of 58 released OMVs contained gold particles inside their lumens.
FIG 3.
TEM micrographs showing DNA immunolabeling of A. baylyi JV26(pMU125) cells and OMVs. (A) Micrograph of an A. baylyi cell with a bulging outer membrane. The arrow indicates the location of anti-dsDNA antibodies with gold particles (black dots, 5 nm). Bar, 500 nm. (B) Micrograph of an A. baylyi cell with the arrow pointing to a budding vesicle with the gold particles located inside the vesicle lumen. Bar, 1 μm. (C) Micrograph of an A. baylyi cell releasing a vesicle. The arrow points to the presence of 5-nm gold particles inside the vesicle. Bar, 500 nm. (D) Micrograph of an A. baylyi cell; the arrow points to the released OMV coupled to gold particles. Bar, 200 nm. (E) Micrograph of ultrathin sections of large-sized vesicles with gold particles inside the lumen. Bar, 1 μm. (F) Micrograph of small unsectioned vesicles. The arrow indicates gold particles (5 nm) attached to the outer surface of an OMV. Bar, 500 nm.
The presence of DNA in the OMVs of A. baylyi, so-called vesicular DNA (V-DNA), was determined after isolation from vesicles pretreated with DNase. The purified V-DNA was quantified using the PicoGreen assay (Table 2), and the presence of plasmid DNA was confirmed by PCR amplification of the β-lactamase gene (650 bp) (see Fig. S1A in the supplemental material).
TABLE 2.
Protein and vesicular DNA concentrations from OMVs isolated from A. baylyi strain JV26(pMU125)
| Treatment | Protein concn (μg ml−1)a | V-DNA concn (ng ml−1)b |
|---|---|---|
| No treatment (LB medium) | 403 ± 23 | 4.6 ± 2.2 |
| Temp increase | 354 ± 3.0 | 0.25 ± 0.1 |
| Desiccation | 382 ± 4.6 | 0.11 ± 0.1 |
| Gentamicin concn of: | ||
| 0.1 μg ml−1 | 437 ± 6.0 | 13 ± 9.0 |
| 0.3 μg ml−1 | 450 ± 22 | 51 ± 0.1 |
| Chloramphenicol concn of: | ||
| 0.1 μg ml−1 | 404 ± 28 | 9.4 ± 3.2 |
| 1.0 μg ml−1 | 495 ± 35 | 4.0 ± 3.7 |
| No treatment (minimal medium) | 126 ± 6.7 | 35 ± 7.0 |
| Nutrient deprivation | 151 ± 14 | 18 ± 11 |
| UV irradiation at: | ||
| 108 mJ cm−2 | 125 ± 5.0 | 8.0 ± 1.0 |
| 432 mJ cm−2 | 151 ± 3.6 | 12.6 ± 1.0 |
OMVs were isolated from three independent batches of cultures (100 ml) and quantified by the Bradford assay. General growth conditions were as described in Table 1.
The purified OMVs were adjusted to 100 μg ml−1 of the total protein concentration prior to vesicular DNA isolation. V-DNA was quantified with the PicoGreen assay.
OMV-mediated transfer of pMU125.
Bacterial cells were mixed with OMVs, and the transfer of the plasmid pMU125 from OMVs to cells was quantified after 24 h of incubation. The gene transfer frequencies are shown in Table 3. OMVs adjusted to a protein concentration of 10.5 mg ml−1 (containing 54 ng ml−1 DNA) were able to transform DH5α cells with a frequency of 3 × 10−8, corresponding to 80 vesiculants (transformed cells obtained through OMV exposure) per μg of V-DNA. For strain JV26, an OMV transfer frequency of 1 × 10−6 (equivalent to 9 × 103 vesiculants per μg of V-DNA) was found (Table 3). Further experiments were carried out exposing strain JV26 to different OMV batches; OMVs with 620 μg ml−1 protein and 2.3 ± 1.6 ng ml−1 DNA resulted in a transfer frequency of (2.1 ± 2.0) × 10−8 ml−1, and OMVs with 780 μg ml−1 protein and 6.6 ± 0.7 ng ml−1 DNA resulted in a transfer frequency of (1.4 ± 1.6) × 10−7 ml−1. OMVs obtained from cells exposed to sub-MIC levels of gentamicin during growth (protein concentration, 708 μg ml−1) and 2.3 ± 0.1 ng ml−1 DNA resulted in a transfer frequency of (1.3 ± 0.1) × 10−7 ml−1. The gene transfer frequency obtained with OMVs from gentamicin-treated cultures was significantly higher than that obtained with OMVs from untreated cultures (both OMVs contained the same amount of V-DNA [2.3 ng ml−1]; P < 0.05). Natural transformation experiments with purified pMU125 plasmid DNA in the absence of DNase were conducted for comparison. Exposure of strain JV26 to 2.3, 6.6, or 55 ng ml−1 pMU125 DNA yielded transformation frequencies of (1.4 ± 2.2) × 10−9, (1.5 ± 0.7) × 10−8, and (5.5 ± 4.0) × 10−7 ml−1, respectively. Ampicillin-resistant colonies were regularly screened for the presence of the β-lactamase (bla) gene by PCR (Fig. S1B in the supplemental material). Vesiculants displayed the same level of resistance to ampicillin (>256 μg ml−1) as the donor bacteria. These results indicate that OMV-mediated gene transfer occurs in a DNA concentration-dependent manner. Remarkably, in experiments using the transformation-deficient A. baylyi ΔcomA or A. baylyi ΔcomB-comF strains as recipients, no vesiculants were obtained (Table 2).
TABLE 3.
Gene transfer frequencies and antibiotic susceptibility profiles of vesiculants of A. baylyi and E. colid
| Recipient cells | Presence or absence of: |
Gene transfer frequencyc | MIC for Ampr vesiculants | PCR (bla) | ||
|---|---|---|---|---|---|---|
| OMVsa | DNase I | DNAb | ||||
| A. baylyi | ||||||
| JV26 | + | + | − | 1 × 10−6 | >256 | + |
| JV26 ΔcomB-comF | + | + | − | <3 × 10−9 | NA | NA |
| JV26 ΔcomA | + | + | − | <2 × 10−9 | NA | NA |
| JV26 | − | + | + | <4 × 10−10 | NA | NA |
| JV26 | − | − | − | <1.3 × 10−9 | NA | NA |
| JV26 (ACIAD2756::aacC1) | + | + | − | 1 × 10−7 | >256 | + |
| JV26 | − | − | + | 5.5 × 10−7 | >256 | + |
| E. coli | ||||||
| DH5α | + | + | − | 3 × 10−8 | >256 | + |
| DH5α | − | − | + | <1.8 × 10−9 | NA | NA |
The OMVs were isolated from A. baylyi donor strain JV26(pMU125) and had a protein concentration of 10.5 mg ml−1 and a DNA content of 54 ng ml−1. When A. baylyi (ACIAD2756::aacC1) was used as a recipient, OMVs isolated from gentamicin (0.1 μg ml−1)-stressed cells were used (protein concentration, 708 μg ml−1; V-DNA concentration, 2.3 ng ml−1).
Purified pMU125 plasmid DNA (55 ng ml−1) was used for natural transformation of A. baylyi cells.
The gene transfer experiments were carried out in three time-independent assays. The gene transfer frequencies are calculated as numbers of vesiculants or transformants over the number of recipient cells.
NA, not applicable.
In control experiments, no colonies were found in the absence of vesicle exposure or when the E. coli DH5α recipient was exposed to purified pMU125 plasmid DNA, confirming the supposition that the tested E. coli cultures are not competent for natural transformation by free DNA. Furthermore, exposure of A. baylyi JV26 recipient cells to purified pMU125 DNA in the presence of DNase did not yield transformants. DNase was found to be efficient in destroying free DNA added even after 17 h in medium without cells or in the presence of cells, and no transformant was obtained by natural transformation in these experiments. These results suggest that the addition of DNase prohibits transfer of free DNA but does not affect vesicle-mediated transfer.
pMU125 carries, in addition to the bla gene, a gfp (green fluorescent protein) gene. The qualitative observation of GFP activity in vesiculants by confocal microscopy is shown in Fig. S2 in the supplemental material. The ampicillin-resistant vesiculants exhibited green fluorescence, suggesting that OMV mediated the cotransfer of the bla and gfp genes and the presence of an intact plasmid.
Integration of OMVs with recipient cells.
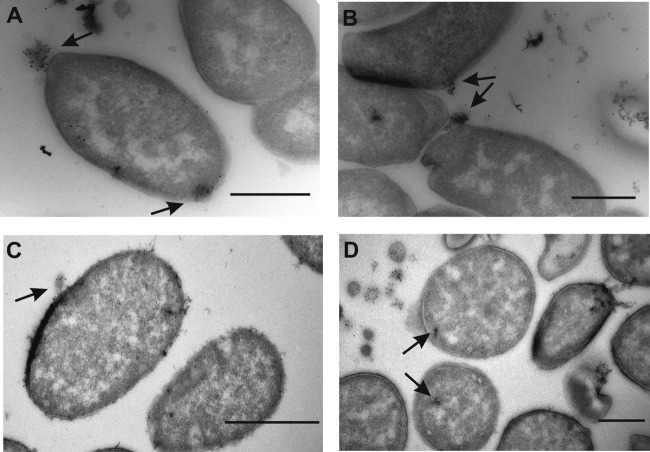
Immunogold labeling of ultrathin sections with an FITC-specific antibody enabled the detection of the interaction of OMVs with exposed bacterial cells. The TEM images visualized the association, fusion, and internalization of OMVs from JV26 (wild-type) with recipient (strains DH5α and JV26) cells. The localization of gold particles attached to FITC-labeled OMVs showed that the adhesion of vesicles to E. coli and A. baylyi JV26 cells occurred immediately. Interestingly, vesicles were attached to, as well as internalized by, E. coli cells immediately (at time zero [t0]) (Fig. 4A) and also after 1 h (t1) (Fig. 4B). In total, 250 E. coli cells were inspected in sample grids at t0, and OMVs were attached to 35 cells, while 15 OMVs were internalized. In addition, 250 E. coli cells were inspected at t1, at which time OMVs were attached to 30 cells and internalized in 27 cells. When A. baylyi JV26 cells were used as recipient cells, somewhat fewer internalization events were detected, but attachment of OMVs to the outer membrane was observed at both t0 and t1 (Fig. 4C). Gold particles were also detected inside the cytoplasm (Fig. 4D) of both E. coli and A. baylyi recipient cells, suggesting the transfer of vesicle content into the bacterial cytoplasm. All together, 233 JV26 cells were inspected on sample grids at t0. Gold particles were found inside the cytoplasm of 10 of these cells, and OMVs were found adhered to 49 cells. For t1 sample grids, 359 cells were inspected and OMVs were found attached to 45 cells, while gold particles were observed inside 17 cells.
FIG 4.
TEM micrographs of FITC-labeled OMVs. (A, B) IEM micrographs of E. coli DH5α cells incubated with FITC-labeled OMVs derived from A. baylyi. The arrows point to the attached or internalized OMVs coupled to gold particles (5 nm) at exposure time t0 in panel A and at exposure time t1 in panel B. (C, D) Same as described for panels A and B but with A. baylyi JV26 cells. Bars, 500 nm.
Effect of environmental and antibiotic stresses on bacterial growth, vesiculation, and DNA content in OMVs.
Temperature increase, desiccation, nutrient deprivation, UV light exposure, and low-level exposure to the antibiotics gentamicin and chloramphenicol resulted in changes in bacterial surfaces and increased release of vesicles (Fig. 5 and see Fig. S3 in supplemental material). TEM micrographs revealed an intact outer membrane with few surface bulging sites in the absence of stress (Fig. 5A), whereas a moderate difference was observed in temperature- and desiccation-stressed cells (Fig. S3). Cells stressed with antibiotic exposure were found to have substantially more sites releasing vesicles and the vesicles were larger (Fig. 5B and C) than in untreated cells. From ultrathin sections of differently treated JV26 cultures, the fraction of vesiculating cells was determined by TEM. From 433 bacterial cells grown without stress, 54 cells were bulging. Similarly, 64 cells were found to be in a bulging stage out of 446 cells when the temperature was increased. When cells were grown under desiccation stress, 53 vesiculating cells were counted in 426 cells. When cultures were grown in the presence of subinhibitory antibiotic concentrations, the results were as follows: 68 bulging cells were identified out of 434 cells when they were grown with gentamicin (0.1 μg ml−1), and 73 out of 453 cells were bulging when they were incubated with chloramphenicol (0.1 μg ml−1). There was no observable difference in the cell surfaces when the cells were grown in minimal medium without stress (Fig. 5D) or after UV irradiation (Fig. 5F), whereas a moderate OMV size alteration was observed in nutrient-deprived cells (Fig. 5E). Out of 420 cells, 65 cells exhibited bulging when grown in minimal medium without stress. When nutrient stress was applied, 70 out of 415 cells were vesiculating. Cells irradiated with UV light (20-min exposure) were grown for 1 h and then inspected for vesiculation, and 78 out of 410 cells produced OMVs.
FIG 5.
TEM micrographs showing the effect of stress on the morphology of A. baylyi cells. (A) Cells grown under normal conditions in rich medium. Bar, 1 μm. (B) Cells grown with gentamicin stress (0.1 μg ml−1). Bar, 1 μm. (C) Cells grown with chloramphenicol stress (0.1 μg ml−1). Bar, 1 μm. (D) Cells grown under normal conditions in minimal medium. Bar, 1 μm. (E) Cells grown in minimal medium with nutrient stress. Bar, 2 μm. (F) Cells grown in minimal medium and exposed to UV light for 20 min. Bar, 1 μm. The arrows indicate budding vesicles in panels A, B, and F and released OMVs in panels C, D, and E.
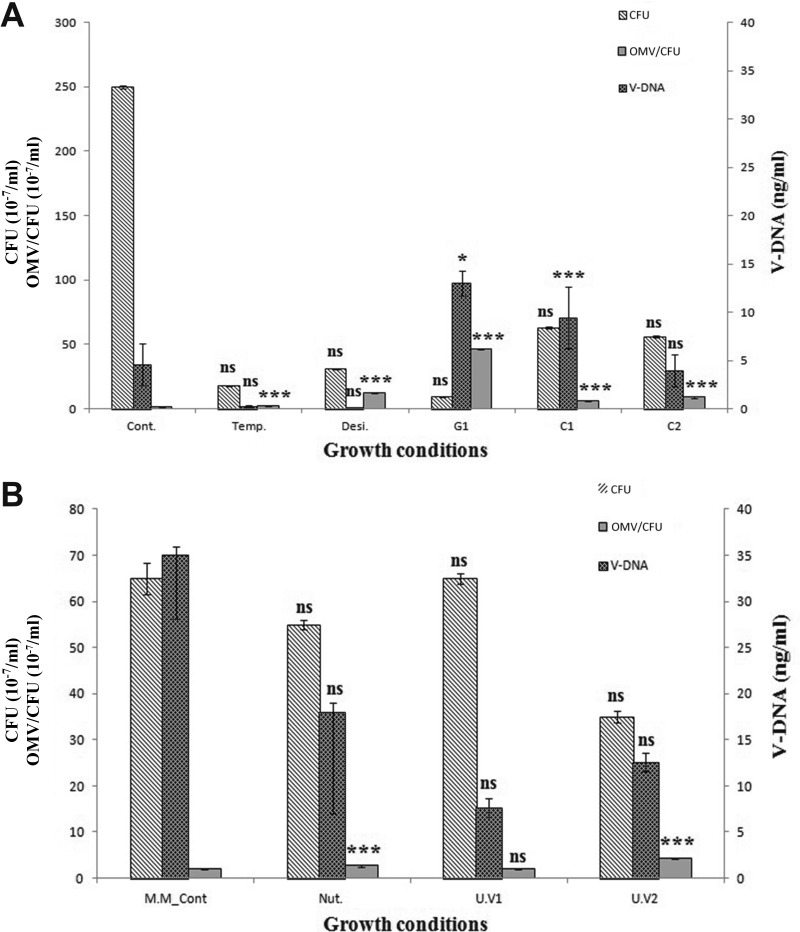
The effect of stress conditions affected the overall cell growth compared with that of untreated cultures (see Fig. S3A in supplemental material), determined through photometry. The culture grown without stress in the presence of ampicillin entered stationary phase after approximately 8 h and remained stable, giving an end titer of (2.5 ± 1.1) × 109 ml−1 after 15 h (t15). The isolated OMVs had a total protein content of 403 μg ml−1 (Table 2). Depending on the stress condition applied, one or more of the following occurred: a prolonged lag phase, an increase in generation time, a lower stationary-phase density, or an increase in the death rate in stationary phase (Fig. S3A). While the viable cell titers decreased 10- to 10,000-fold depending on the treatment (Table 1), the amount of vesicles produced (quantified by total protein content) was relatively constant in rich medium (Table 2). A significant increase (P < 0.05 and P < 0.01) was observed in vesiculation and in V-DNA concentration (3- to 11-fold) when cells were grown with 0.1 and 0.3 μg ml−1 gentamicin (Table 2; Fig. 6). In addition to this, OMVs derived from the cultures with 0.1 μg ml−1 chloramphenicol contained significantly larger amounts of V-DNA (P < 0.001). The total amount of vesicles produced in rich medium was approximately 2.5-fold higher (P < 0.001) than in minimal medium. In contrast, the V-DNA concentration was significantly increased in minimal medium (P < 0.01).
FIG 6.
Total quantities of OMVs (numbers of OMVs per CFU ml−1) released relative to the mean number of viable cells (CFU ml−1) and mean amount of V-DNA (ng ml−1) obtained from OMVs (protein concentration, 100 μg ml−1). Error bars indicate standard deviations. (A) Cont., cells grown under normal (control) conditions in LB medium; Temp., cells grown under temperature stress at 37°C; Desi, cells grown under desiccation stress with 0.5 M NaCl; G1, C1, and C2, cells grown with gentamicin at 0.1 μg ml−1, with chloramphenicol at 0.1 μg ml−1, and with chloramphenicol at 1 μg ml−1, respectively. (B) M.M. cont, cells grown under normal conditions in minimal medium; Nut., cells grown under nutrient stress (2.2 mM succinate and 5.2 mM glucose as a carbon source); U.V1 and U.V2, cells grown with UV light exposure for 5 min and 20 min, respectively. Significant differences of the various stress conditions from the normal growth condition are indicated as follows: *, P < 0.05; ***, P < 0.001; ns, no significant difference.
The total number of OMVs released was calculated with respect to the cell population (OMVs per viable recipient cell) under stress and nonstress conditions. There was a significant increase in the total number of OMVs released (P < 0.001) in all the stress treatments except for the short UV light exposure (108 mJ cm−2 for 5 min) (Fig. 6).
Effect of stress on particle size distribution and zeta potential.
OMVs released from the bacterial populations experiencing stress varied in size distribution and intensities. The particle size distribution measurements revealed a shift in the size distribution of OMVs released from antibiotic and nutrient stress treatments (Table 1) in comparison with that of the OMVs derived from cultures experiencing no stress. The presence of gentamicin (0.1 and 0.3 μg ml−1) and chloramphenicol (1 μg ml−1) led to a significant increase in the diameters of OMVs (P < 0.05, P < 0.001, and P < 0.001, respectively). OMVs from nutrient-deprived cells grown in minimal medium were also significantly larger (P < 0.001) than OMVs from LB medium.
The zeta potential of the A. baylyi cells was significantly more negative (P < 0.001) than that of the OMVs isolated from these cells (Table 1). The zeta potentials of OMVs produced under different stress conditions were not significantly different from those of their respective controls (Table 1), except with OMVs under gentamicin (0.1 μg ml−1)-induced stress. This treatment resulted in OMVs with the most noticeable morphological change and a significantly more negative surface charge (P < 0.001) than those of the other OMVs. In contrast, OMVs from chloramphenicol-treated cells had a significantly less negative zeta potential (P < 0.001) than those of OMVs from untreated cells.
DISCUSSION
OMVs have been reported to contain DNA within their lumens (52–55). However, not all OMV-producing cells have been found to enable vesicle-mediated HGT (56), and the mechanism of gene delivery at the host cell surface remains unclear. In this study, we identify and describe OMV production and OMV-mediated DNA transfer in the model bacterium A. baylyi. We show that A. baylyi exposure to OMVs results in interspecies gene transfer in the presence of DNase, possibly through a different DNA delivery mechanism that depends on the recipient species. We also describe how antibiotic and environmental stresses affect the production of and DNA content of OMVs in A. baylyi. We conclude that the antibiotic exposure and the other stress factors, except for the short-term UV exposure, had a significant effect on vesiculation and increased OMV production per viable recipient cells. Gentamicin-induced stress resulted in OMVs with significantly more DNA than in other OMVs (Fig. 6).
Particle size measurements showed that A. baylyi cells release OMVs of different sizes during growth (Fig. 1D). The biological significance of this novel observation remains to be determined. The distinct OMV size range may be growth phase dependent or indicative of different biological functions for the different sizes. Distinct sizes also suggest that vesicle release is a physiologically controlled and reproducible process. The differences in zeta potential between the cell membrane and OMVs also provide support for the hypothesis of distinct roles of various size groups of OMVs. The released OMVs were predominantly observed as spherical (Fig. 1A to C), but they were also found to be elongated at the time of budding (Fig. 3B and C) and elliptical (Fig. 3F) in the thin sections (55).
The analysis of fatty acid composition identified the major fatty acids present in both OMVs and bacterial cell membranes to be oleic acid, palmitic acid, and palmitoleic acid (Fig. 2A). Palmitoleic acid was less abundant in OMVs, whereas α-linolenic acid was more abundant in OMVs than in cell membranes. The presence of oleic and palmitic acids in the cell membranes of A. baylyi is consistent with the findings of previous studies of A. calcoaceticus (57). The OMVs were also further characterized for their membrane composition. Bacterial inner membrane-specific β-NADH oxidase activity (58) was used as an inner membrane marker. Vesicles isolated from both stressed and nonstressed bacterial cultures failed to show β-NADH oxidase activity, indicating the absence of inner membranes in vesicles released by A. baylyi (Fig. 2C). OmpA is an outer membrane protein that is commonly found in Acinetobacter species (59), and IEM with anti-OmpA antibodies attached to gold particles confirmed the presence of OmpA in the OMVs (Fig. 2B). Together with the lipid profiles, these results suggest that OMVs are formed largely from the outer membrane of A. baylyi.
The results from IEM studies using a dsDNA antibody revealed that dsDNA from the cytosol entered the periplasm and subsequently the OMVs during the process of vesiculation (Fig. 3). The transfer of DNA to the periplasm might also be causally linked to the liberation of DNA that occurs during competence expression in Acinetobacter (60). Thus, it remains unclear if DNA is specifically targeted to OMVs. This observation confirms earlier reports of association of DNA to the inner and outer surfaces of OMVs (55, 61, 62). It is tempting to speculate that the affinity between DNA and OMVs can contribute to biofilm structures as well (63, 64).
Members of the Acinetobacter genus are known to share genes horizontally via conjugation, transformation, and transduction (65–67). Our experimental data provide support for another route of HGT that occurs through OMVs. The recent report on OMVs from A. baumannii that are able to transfer the OXA-24 carbapenemase gene strengthens the notion that OMVs can be of importance in the dissemination of antimicrobial resistance in Acinetobacter (24). Although the frequency of vesicle-mediated gene transfer is low, and lower than, for instance, the transfer frequency obtained by virus-like particles (VLPs) in marine environments (68), it is not fundamentally different from HGT frequencies observed in this genus (69, 70). The gene transfer obtained with the OMVs yielded up to 103 vesiculants per μg of V-DNA, which is comparable to earlier observations in Pseudomonas spp. (56). Our experimental model focused on characterizing the transfer of a low-copy-number plasmid (pMU125) in Acinetobacter, also demonstrating that limited amounts of DNA in the bacterial cytoplasm were able to reach OMVs and reach OMV-exposed bacterial cells. DNase was also found to be effective even after 17 h of incubation at 37°C, rendering the possibility of natural transformation of small amounts of free DNA due to random OMV lysis very low. As the gene transfer mechanism via OMVs differs from natural transformation, we propose to refer to the transformed cells obtained by vesicle-mediated gene transfer (VMGT) as “vesiculants.”
To shed more light on the mechanism of vesicle-mediated DNA delivery in A. baylyi, we also exposed comA- and comB-comF-deficient mutants to OMVs. Interestingly, no vesiculant was recovered when com mutants were used as recipient cells, indicating that competence proteins play a role in the uptake of DNA delivered by OMVs. This observation suggests that vesicles are lysed upon contact with the outer membrane of A. baylyi cells followed by type IV pilus-mediated transport of DNA. ComA plays a crucial role in transporting DNA through the inner membrane in Acinetobacter (71), whereas ComB, ComE, and ComF are structural subunits of a DNA translocator and responsible for transport of DNA through the periplasm (72, 73). In particular, ComC functions as a DNA binding protein, is essential for pilus-mediated uptake of DNA (74), and is absent in the ΔcomB-comF strain.
OMVs isolated from bacterial cells grown in the presence of subinhibitory gentamicin concentrations were able to transfer plasmid DNA into A. baylyi JV26 cells 10 times more efficiently than OMVs isolated from unexposed cells, despite containing the same amount of V-DNA. This may be due to a more efficient interaction between OMVs and the recipient cells due to of the altered surface potential of OMVs from gentamicin-treated cultures (75).
Taken together, our results suggest that OMV-mediated gene transfer occurs by two distinct and possibly species-specific pathways: (i) lysis of OMVs close to, or associated with, the outer membrane of target cells (20) as proposed for A. baylyi recipient cells or (ii) adhesion to the outer membrane followed by internalization into the cytoplasm, proximal lysis, or fusion (19) as proposed for the E. coli recipients. The latter finding is unexpected but is supported by the following TEM-based observations: when FITC-labeled OMVs were incubated with E. coli recipients, OMVs with the anti-FITC antibody-conjugated gold particles became associated with the outer membrane (Fig. 4A), some were internalized (Fig. 4B), and gold particles were also observed inside the cytoplasm.
Our IEM studies visualized vesicle-mediated DNA delivery events at the single-cell level (Fig. 4). To our knowledge, no experimental data exist where OMV internalization has been observed in bacteria, although such mechanisms have been observed in epithelial cells (50). The initial binding between OMVs and the outer membranes of recipient cells may be due to electrostatic interactions (76), salt bridging by Ca2+ or Mg2+ ions (19), or the presence of adhesins on the outer membranes facing toward OMVs. According to previous studies, Gram-negative bacteria possess autolysins in the periplasm (77) that are capable of cleaving covalent bonds in the peptidoglycan layer. In Gram-positive cells, lysis of the OMV membrane may occur after interaction with the positively charged peptidoglycan layer (19, 20).
Stress can lead to the accumulation of misfolded and toxic proteins in the periplasm of bacteria (78), and the release of OMVs has been proposed to be a way of responding to stress (78). We therefore compared the levels of OMV production in A. baylyi populations grown under different stressful conditions, such as desiccation, nutrient deprivation, UV irradiation, heat, and subinhibitory concentrations of the antibiotics chloramphenicol and gentamicin. The OMV concentration was approximated by measurements of the total vesicle protein concentrations relative to the bacterial end titer (assuming that only viable cells can release OMVs), as well as by quantifying released OMVs. A significant alteration in the release of OMVs per viable cell was observed for some stress conditions, with the protein synthesis inhibitor gentamicin resulting in the strongest effect (Fig. 6 A and B). In contrast, temperature, desiccation, short-time UV light exposure, and chloramphenicol (0.1 μg ml−1) had no significant effect on vesiculation when released OMVs were quantified directly (Table 2). During stress, the accumulation of damaged proteins can increase turgor pressure and therefore induce more disruption of lipoprotein bridges, leading to increased vesiculation (78). TEM images of ultrathin sections of bacterial cells grown under stress may display more surface bulging sites than those of cells grown without stress (Fig. 5; see also Fig. S3B and C in the supplemental material). However, this has not been statistically analyzed.
Both gentamicin and chloramphenicol are known to have bacteriostatic properties but are lethal at high concentrations (79, 80). A sub-MIC of gentamicin (0.3 μg ml−1) increased the overall amount of OMVs released per cell, despite the 4-order-of-magnitude reduction in the number of viable cells over that of the control. The OMVs obtained from gentamicin-stressed cultures also had a more negative zeta potential than those obtained from the control culture. We note that vesicles produced under gentamicin exposure have previously been suggested to occur by a mechanism different from that of naturally produced vesicles (19, 75). Surface-associated DNA causes an increase in the zeta potential (62); however, in our study, OMVs were DNase treated before the zeta potential was determined.
The presence of DNA in OMVs may be because A. calcoaceticus releases DNA in exponential phase during the induction of competence by cell lysis (60, 81) or due to active excretion of DNA (60). Prominent cell lysis during bacterial growth under stressful conditions may therefore also lead to increased amounts of DNA on the surfaces of OMVs. Alternatively, increased DNA excretion from cells via the periplasm during periods of competence development would facilitate DNA being present inside the lumens of OMVs (62). A. baylyi is salt stress tolerant, and other members of the Acinetobacter genus are known for their capacity to survive desiccation (82, 83), possibly due to the production of extracellular polysaccharides (84). In this study, desiccation stress had little effect on vesiculation (Table 2) but significantly reduced the amount of V-DNA, which may be a consequence of the shrinkage of cells due to water loss.
Taken together, our results provide evidence that A. baylyi releases OMVs of distinct-size populations that offer a nuclease-insensitive mode of gene transfer within and between species. Environmental stress factors can affect the level of vesicle release, V-DNA content, vesicle size, surface properties, and OMV-mediated HGT frequencies. Further studies should be conducted to determine the biological relevance of the various vesicle size categories, the surface and adherence properties, and the lumen content of OMVs.
Supplementary Material
ACKNOWLEDGMENTS
S.F. is grateful to the Research Council of Norway for awarding an Yggdrasil scholarship and the University of Pune (University with Potential for Excellence), India, for providing a fellowship for her doctoral research work.
We thank Randi Olsen (University of Tromsø) for her excellent guidance in TEM, Helga-Marie Bye and Tom-Ivar Eilertsen for their technical help, Tony Marion (University of Tennessee) and Heinz Schwarz (Max-Planck-Institut für Entwicklungsbiologie) for providing the monoclonal anti-dsDNA 1633p77 antibody, and Luis Actis (Ohio University) for the plasmid pMU125. We also thank the Protein Research Group, UiT, for the use of their bioimaging and confocal microscopy facility. We are also thankful to Ragnar Olsen and Guro Kristine Edvinsen (University of Tromsø) for the fatty acid analysis and to Nils Hülter for the A. baylyi ACIAD2756::aacC1 strain.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 21 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04248-13.
REFERENCES
- 1.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721. 10.1038/nrmicro1234 [DOI] [PubMed] [Google Scholar]
- 2.Dubey GP, Ben-Yehuda S. 2011. Intercellular nanotubes mediate bacterial communication. Cell 144:590–600. 10.1016/j.cell.2011.01.015 [DOI] [PubMed] [Google Scholar]
- 3.Shetty A, Chen S, Tocheva EI, Jensen GJ, Hickey WJ. 2011. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS One 6:e20725. 10.1371/journal.pone.0020725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, Clarke A, Beveridge TJ. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 180:5478–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defence. BMC Microbiol. 11:258. 10.1186/1471-2180-11-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mashburn-Warren L, McLean RJ, Whiteley M. 2008. Gram-negative outer membrane vesicles: beyond the cell surface. Geobiology 6:214–219. 10.1111/j.1472-4669.2008.00157.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorward DW, Garon CF, Judd RC. 1989. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J. Bacteriol. 171:2499–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655. 10.1101/gad.1299905 [DOI] [PubMed] [Google Scholar]
- 9.Beveridge TJ. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee SN, Das J. 1967. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J. Gen. Microbiol. 49:1–11. 10.1099/00221287-49-1-1 [DOI] [PubMed] [Google Scholar]
- 11.Horstman AL, Kuehn MJ. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489–12496. 10.1074/jbc.275.17.12489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25–35. 10.1016/S0092-8674(03)00754-2 [DOI] [PubMed] [Google Scholar]
- 13.Katsui N, Tsuchido T, Hiramatsu R, Fujikawa S, Takano M, Shibasaki I. 1982. Heat-induced blebbing and vesiculation of the outer membrane of Escherichia coli. J. Bacteriol. 151:1523–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadurugamuwa JL, Beveridge TJ. 1998. Delivery of the non-membrane-permeative antibiotic gentamicin into mammalian cells by using Shigella flexneri membrane vesicles. Antimicrob. Agents Chemother. 42:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabra W, Lunsdorf H, Zeng AP. 2003. Alterations in the formation of lipopolysaccharide and membrane vesicles on the surface of Pseudomonas aeruginosa PAO1 under oxygen stress conditions. Microbiology 149:2789–2795. 10.1099/mic.0.26443-0 [DOI] [PubMed] [Google Scholar]
- 16.Vasilyeva NV, Tsfasman IM, Suzina NE, Stepnaya OA, Kulaev IS. 2009. Outer membrane vesicles of Lysobacter sp. Dokl. Biochem. Biophys. 426:139–142. 10.1134/S1607672909030041 [DOI] [PubMed] [Google Scholar]
- 17.Thompson SS, Naidu YM, Pestka JJ. 1985. Ultrastructural localization of an extracellular protease in Pseudomonas fragi by using the peroxidase-antiperoxidase reaction. Appl. Environ. Microbiol. 50:1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolling GL, Matthews KR. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadurugamuwa JL, Beveridge TJ. 1996. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J. Bacteriol. 178:2767–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadurugamuwa JL, Beveridge TJ. 1999. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other gram-negative bacteria. Microbiology 145:2051–2060. 10.1099/13500872-145-8-2051 [DOI] [PubMed] [Google Scholar]
- 21.Yaron S, Kolling GL, Simon L, Matthews KR. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66:4414–4420. 10.1128/AEM.66.10.4414-4420.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klieve AV, Yokoyama MT, Forster RJ, Ouwerkerk D, Bain PA, Mawhinney EL. 2005. Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl. Environ. Microbiol. 71:4248–4253. 10.1128/AEM.71.8.4248-4253.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaar V, Nordstrom T, Morgelin M, Riesbeck K. 2011. Moraxella catarrhalis outer membrane vesicles carry beta-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob. Agents Chemother. 55:3845–3853. 10.1128/AAC.01772-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:3084–3090. 10.1128/AAC.00929-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon SO, Gho YS, Lee JC, Kim SI. 2009. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol. Lett. 297:150–156. 10.1111/j.1574-6968.2009.01669.x [DOI] [PubMed] [Google Scholar]
- 26.Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. 2011. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 6:e17027. 10.1371/journal.pone.0017027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahu PK, Iyer PS, Oak AM, Pardesi KR, Chopade BA. 2012. Characterization of eDNA from the clinical strain Acinetobacter baumannii AIIMS 7 and its role in biofilm formation. ScientificWorldJournal 2012:973436. 10.1100/2012/973436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borneleit P, Hermsdorf T, Claus R, Walther P, Kleber HP. 1988. Effect of hexadecane-induced vesiculation on the outer membrane of Acinetobacter calcoaceticus. J. Gen. Microbiol. 134:1983–1992 [DOI] [PubMed] [Google Scholar]
- 29.Patwardhan RB, Dhakephalkar PK, Niphadkar KB, Chopade BA. 2008. A study on nosocomial pathogens in ICU with special reference to multiresistant Acinetobacter baumannii harbouring multiple plasmids. Indian J. Med. Res. 128:178–187 [PubMed] [Google Scholar]
- 30.Rello J, Diaz E. 2003. Acinetobacter baumannii: a threat for the ICU? Intensive Care Med. 29:350–351 [DOI] [PubMed] [Google Scholar]
- 31.Roca I, Espinal P, Vila-Farres X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. 10.3389/fmicb.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen TL, Lee YT, Kuo SC, Hsueh PR, Chang FY, Siu LK, Ko WC, Fung CP. 2010. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents Chemother. 54:4575–4581. 10.1128/AAC.00764-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardesi KR, Yavankar SP, Chopade BA. 2007. Plasmid distribution and antimicrobial susceptibility patterns of Acinetobacter genospecies isolated from healthy human skin of tribal population. Indian J. Med. Res. 125:77–86 [PubMed] [Google Scholar]
- 34.Povilonis J, Seputiene V, Krasauskas R, Juskaite R, Miskinyte M, Suziedelis K, Suziedeliene E. 2013. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 68:1000–1006. 10.1093/jac/dks499 [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez G, Sossa K, Bello H, Dominguez M, Mella S, Zemelman R. 1998. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol. Lett. 161:125–128. 10.1111/j.1574-6968.1998.tb12937.x [DOI] [PubMed] [Google Scholar]
- 36.Livermore DM, Woodford N. 2006. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 14:413–420. 10.1016/j.tim.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 37.Domingues S, Harms K, Fricke WF, Johnsen PJ, Silva GD, Nielsen KM. 2012. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 8:e1002837. 10.1371/journal.ppat.1002837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L, Cruveiller S, Robert C, Duprat S, Wincker P, Ornston LN, Weissenbach J, Marliere P, Cohen GN, Medigue C. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766–5779. 10.1093/nar/gkh910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overballe-Petersen S, Harms K, Orlando LA, Mayar JV, Rasmussen S, Dahl TW, Rosing MT, Poole AM, Sicheritz-Ponten T, Brunak S, Inselmann S, de Vries J, Wackernagel W, Pybus OG, Nielsen R, Johnsen PJ, Nielsen KM, Willerslev E. 2013. Bacterial natural transformation by highly fragmented and damaged DNA. Proc. Natl. Acad. Sci. U. S. A. 110:19860–19865. 10.1073/pnas.1315278110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 41.Harms K, Schön V, Kickstein E, Wackernagel W. 2007. The RecJ DNase strongly suppresses genomic integration of short but not long foreign DNA fragments by homology-facilitated illegitimate recombination during transformation of Acinetobacter baylyi. Mol. Microbiol. 64:691–702. 10.1111/j.1365-2958.2007.05692.x [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch ED, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed, p 49–55 Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 43.Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51. 10.1016/0378-1119(90)90494-C [DOI] [PubMed] [Google Scholar]
- 44.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl 1):5–16. 10.1093/jac/48.suppl_1.5 [DOI] [PubMed] [Google Scholar]
- 45.Bacher JM, Metzgar D, Crecy-Lagard VD. 2006. Rapid evolution of diminished transformability in Acinetobacter baylyi. J. Bacteriol. 188:8534–8542. 10.1128/JB.00846-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosen G, Naor R, Rahamim E, Yishai R, Sela MN. 1995. Proteases of Treponema denticola outer sheath and extracellular vesicles. Infect. Immun. 63:3973–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kropinski AM, Clokie MR. 2009. Methods for the isolation of viruses from environmental samples. Methods Mol. Biol. 502:13–22. 10.1007/978-1-60327-164-6_1 [DOI] [PubMed] [Google Scholar]
- 48.Sasser M. 2009. Microbial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME). MIDI technical note 101. MIDI Inc., Newark, DE: http://www.midi-inc.com/pdf/MIS_Technote_101.pdf [Google Scholar]
- 49.Shang ES, Skare JT, Exner MM, Blanco DR, Kagan B, Miller JN, Lovett MA. 1998. Isolation and characterization of the outer membrane of Borrelia hermsii. Infect. Immun. 66:1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538–4549. 10.1038/sj.emboj.7600471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster P, Webster A. 2007. Cryosectioning fixed and cryoprotected biological material for immunocytochemistry. Methods Mol. Biol. 369:257–289. 10.1007/978-1-59745-294-6_13 [DOI] [PubMed] [Google Scholar]
- 52.Deich RA, Hoyer LC. 1982. Generation and release of DNA-binding vesicles by Haemophilus influenzae during induction and loss of competence. J. Bacteriol. 152:855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dorward DW, Garon CF. 1990. DNA is packaged within membrane derived vesicles of Gram-negative but not Gram-positive bacteria. Appl. Environ. Microbiol. 56:1960–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagemann S, Stöger L, Kappelmann M, Hassl I, Ellinger A, Velimirov B. 2013. DNA-bearing membrane vesicles produced by Ahrensia kielensis and Pseudoalteromonas marina. J. Basic Microbiol 53:1–11. 10.1002/jobm.201300376 [DOI] [PubMed] [Google Scholar]
- 55.Pérez-Cruz C, Carrión O, Delgado L, Martinez G, López-Iglesias C, Mercadea E. 2013. New type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl. Environ. Microbiol. 79:1874–1881. 10.1128/AEM.03657-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renelli M, Matias V, Lo RY, Beveridge TJ. 2004. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150:2161–2169. 10.1099/mic.0.26841-0 [DOI] [PubMed] [Google Scholar]
- 57.Yang C, Guo ZB, Du ZM, Yang HY, Bi YJ, Wang GQ, Tan YF. 2012. Cellular fatty acids as chemical markers for differentiation of Acinetobacter baumannii and Acinetobacter calcoaceticus. Biomed. Environ. Sci. 25:711–717. 10.3967/0895-3988.2012.06.014 [DOI] [PubMed] [Google Scholar]
- 58.Scott CCL, Makula RA, Finnerty WR. 1976. Isolation and characterization of membranes from a hydrocarbon-oxidizing Acinetobacter sp. J. Bacteriol. 127:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moon DC, Choi CH, Lee JH, Choi CW, Kim HY, Park JS, Kim SI, Lee JC. 2012. Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J. Microbiol. 50:155–160. 10.1007/s12275-012-1589-4 [DOI] [PubMed] [Google Scholar]
- 60.Palmen R, Hellingwerf KJ. 1995. Acinetobacter calcoaceticus liberates chromosomal DNA during induction of competence by cell lysis. Curr. Microbiol. 30:7–10. 10.1007/BF00294516 [DOI] [PubMed] [Google Scholar]
- 61.Kahn ME, Maul G, Goodgal SH. 1982. Possible mechanism for donor DNA binding and transport in Haemophilus. Proc. Natl. Acad. Sci. U. S. A. 79:6370–6374. 10.1073/pnas.79.20.6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schooling SR, Hubley A, Beveridge TJ. 2009. Interactions of DNA with biofilm-derived membrane vesicles. J. Bacteriol. 191:4097–4102. 10.1128/JB.00717-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schooling SR, Beveridge TJ. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945–5957. 10.1128/JB.00257-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, Hanawa T, Kamiya S. 2009. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 9:197. 10.1186/1471-2180-9-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chopade BA, Wise PJ, Towner KJ. 1985. Plasmid transfer and behaviour in Acinetobacter calcoaceticus EBF65/65. J. Gen. Microbiol. 131:2805–2811 [DOI] [PubMed] [Google Scholar]
- 66.Herman NJ, Juni E. 1974. Isolation and characterization of a generalized transducing bacteriophage for Acinetobacter. J. Virol. 13:46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juni E. 1972. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J. Bacteriol. 112:917–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiura HX, Kogure K, Hagemann S, Ellinger A, Velimirov B. 2011. Evidence for particle-induced horizontal gene transfer and serial transduction between bacteria. FEMS Microbiol. Ecol. 76:576–591. 10.1111/j.1574-6941.2011.01077.x [DOI] [PubMed] [Google Scholar]
- 69.Nielsen KM, Kornelia S, Elsas JV. 2000. Natural transformation of Acinetobacter sp. strain BD413 with cell lysates of Acinetobacter sp., Pseudomonas fluorescens, and Burkholderia cepacia in soil microcosms. Appl. Environ. Microbiol. 66:206–212. 10.1128/AEM.66.1.206-212.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ray JL, Harms K, Wikmark OG, Starikova I, Johnsen PJ, Nielsen KM. 2009. Sexual isolation in Acinetobacter baylyi is locus-specific and varies 10,000-fold over the genome. Genetics 182:1165–1181. 10.1534/genetics.109.103127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Averhoff B, Graf I. 2008. The natural transformation system of Acinetobacter baylyi ADP1: a unique DNA transport machinery, p 119–139 In Gerischer U. (ed), Acinetobacter, molecular biology. Caister Academic, Norfolk, United Kingdom [Google Scholar]
- 72.Busch S, Rosenplanter C, Averhoff B. 1999. Identification and characterization of ComE and ComF, two novel pilin-like competence factors involved in natural transformation of Acinetobacter sp. strain BD413. Appl. Environ. Microbiol. 65:4568–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Herzberg C, Friedrich A, Averhoff B. 2000. ComB, a novel competence gene required for natural transformation of Acinetobacter sp. BD413: identification, characterization, and analysis of growth-phase-dependent regulation. Arch. Microbiol. 173:220–228. 10.1007/s002039900134 [DOI] [PubMed] [Google Scholar]
- 74.Link C, Eickernjager S, Porstendorfer D, Averhoff B. 1998. Identification and characterization of a novel competence gene, comC, required for DNA binding and uptake in Acinetobacter sp. strain BD413. J. Bacteriol. 180:1592–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tashiro Y, Ichikawa S, Shimizu M, Toyofuku M, Takaya N, Nakajima-Kambe T, Uchiyama H, Nomura N. 2010. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 76:3732–3739. 10.1128/AEM.02794-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams D, Vicogne J, Zaitseva I, McLaughlin S, Pessin JE. 2009. Evidence that electrostatic interactions between vesicle-associated membrane protein 2 and acidic phospholipids may modulate the fusion of transport vesicles with the plasma membrane. Mol. Biol. Cell 20:4910–4919. 10.1091/mbc.E09-04-0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vollmer W, Bertsche U. 2008. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1778:1714–1734. 10.1016/j.bbamem.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 78.McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63:545–558. 10.1111/j.1365-2958.2006.05522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kadurugamuwa JL, Clarke AJ, Beveridge TJ. 1993. Surface action of gentamicin on Pseudomonas aeruginosa. J. Bacteriol. 175:5798–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rahal JJJ, Simberkoff MS. 1979. Bactericidal and bacteriostatic action of chloramphenicol against meningeal pathogens. Antimicrob. Agents Chemother. 16:13–18. 10.1128/AAC.16.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lorenz MG, Gerjets D, Wackernagel W. 1991. Release of transforming plasmid and chromosomal DNA from two cultured soil bacteria. Arch. Microbiol. 156:319–326. 10.1007/BF00263005 [DOI] [PubMed] [Google Scholar]
- 82.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36:1938–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jawad A, Snelling AM, Heritage J, Hawkey PM. 1998. Exceptional desiccation tolerance of Acinetobacter radioresistens. J. Hosp. Infect. 39:235–240. 10.1016/S0195-6701(98)90263-8 [DOI] [PubMed] [Google Scholar]
- 84.Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.