Abstract
Monomethylamine (MMA, CH3NH2) can be used as a carbon and nitrogen source by many methylotrophic bacteria. Methylobacterium extorquens DM4 lacks the MMA dehydrogenase encoded by mau genes, which in M. extorquens AM1 is essential for growth on MMA. Identification and characterization of minitransposon mutants with an MMA-dependent phenotype showed that strain DM4 grows with MMA as the sole source of carbon, energy, and nitrogen by the N-methylglutamate (NMG) pathway. Independent mutations were found in a chromosomal region containing the genes gmaS, mgsABC, and mgdABCD for the three enzymes of the pathway, γ-glutamylmethylamide (GMA) synthetase, NMG synthase, and NMG dehydrogenase, respectively. Reverse transcription-PCR confirmed the operonic structure of the two divergent gene clusters mgsABC-gmaS and mgdABCD and their induction during growth with MMA. The genes mgdABCD and mgsABC were found to be essential for utilization of MMA as a carbon and nitrogen source. The gene gmaS was essential for MMA utilization as a carbon source, but residual growth of mutant DM4gmaS growing with succinate and MMA as a nitrogen source was observed. Plasmid copies of gmaS and the gmaS homolog METDI4690, which encodes a protein 39% identical to GMA synthetase, fully restored the ability of mutants DM4gmaS and DM4gmaSΔmetdi4690 to use MMA as a carbon and nitrogen source. Similarly, chemically synthesized GMA, the product of GMA synthetase, could be used as a nitrogen source for growth in the wild-type strain, as well as in DM4gmaS and DM4gmaSΔmetdi4690 mutants. The NADH:ubiquinone oxidoreductase respiratory complex component NuoG was also found to be essential for growth with MMA as a carbon source.
INTRODUCTION
Monomethylamine (MMA; methylamine) is a nitrogen-containing C1 compound released by natural sources such as the breakdown of proteins and amine osmolytes, as well as by human-made nitrogen-containing pesticides, pharmaceuticals, and herbicides (1). MMA is ubiquitous in the environment and can serve as the sole source of carbon and energy for methylotrophic bacteria, which grow on compounds with no C-C bonds (2) but also as a nitrogen source for a large variety of bacteria (3). MMA utilization by Gram-negative bacteria occurs either by oxidation of MMA into formaldehyde by MMA dehydrogenase (MADH) encoded by mau genes (4) found only in methylotrophic bacteria so far (5) or by the N-methylglutamate (NMG) pathway, the genes for which were first identified in the betaproteobacterium Methyloversatilis universalis FAM5 (6). This metabolism effects the condensation of MMA with glutamate to NMG, with γ-glutamylmethylamide (GMA) a possible intermediate (Fig. 1). The genes gmaS, mgsABC, and mgdABCD encode GMA synthetase (GMAS), NMG synthase, and NMG dehydrogenase, respectively (6). Of these, only gmaS was found not to be required for MMA metabolism in M. universalis FAM5 (6). In contrast, gmaS was also required for MMA utilization in the facultative methane utilizer Methylocella silvestris BL2 (1). In addition, the NMG pathway was recently demonstrated to be also involved in nitrogen assimilation by nonmethylotrophic bacteria (7).
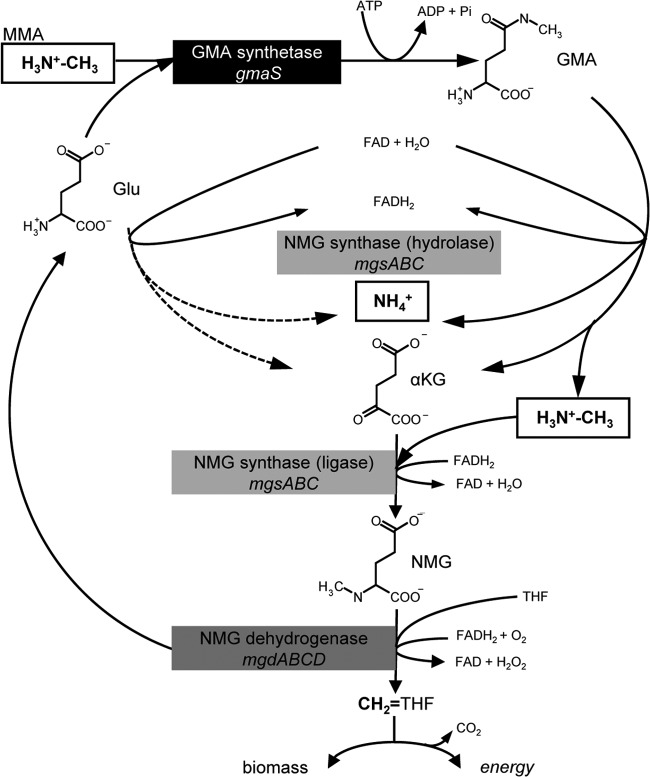
FIG 1.
Proposed pathway and genes for MMA utilization as the source of carbon, energy, and nitrogen on the basis of observed mutant phenotypes of M. extorquens DM4. MMA oxidation by the NMG pathway involves GMAS (EC 1.6.5.3, encoded by gmaS), NMG synthase (EC 2.4.2.2, encoded by mgsABC), and NMG dehydrogenase (EC 1.5.99.5, encoded by mgdABCD). α-Ketoglutarate and tetrahydrofolate are abbreviated as αKG and THF, respectively. In M. extorquens DM4, GMAS is required for growth with MMA as the carbon source. In the absence of gmaS, direct oxidative deamination of glutamate by NMG synthase (hydrolase) may yield sufficient ammonium for growth (broken lines). NMG dehydrogenase catalyzes the transformation of NMG to methylene tetrahydrofolate, which subsequently enters the serine cycle for carbon assimilation into biomass or is oxidized into CO2 for energy production via enzymes encoded by mdtA, fch, ftfL, and fdh (27). MMA utilization as a sole carbon and nitrogen source was shown to require the presence of both mgs and mgd (see the text).
Both strains DM4 and AM1 of the alphaproteobacterial species Methylobacterium extorquens are able to use MMA as the sole source of carbon and nitrogen. However, comparative genomic analysis demonstrated that the mau cluster essential for strain AM1 to grow on MMA as the sole source of carbon and energy (4) was absent from strain DM4 (8). A bank of several thousand mutants obtained by random mutagenesis was used to identify genes required for MMA utilization as a carbon and nitrogen source by M. extorquens DM4, and MMA-dependent gene expression studies and targeted-site mutagenesis were performed. The role of the NMG pathway for MMA oxidation by M. extorquens DM4 was assessed by comparisons with other methylotrophic strains with previously characterized genes for MMA oxidation by the NMG pathway (1, 6) and with the closely related strain M. extorquens AM1, which also contains canonical mau genes for MMA dehydrogenase in addition to genes for the NMG pathway (4).
MATERIALS AND METHODS
Strains and growth conditions.
M. extorquens DM4 was cultivated aerobically at 30°C in mineral medium M3 (9) unless specified otherwise. Escherichia coli strains DH5α and S17-1 (ATCC 47055) were cultivated aerobically at 37°C in Luria-Bertani medium (Difco Laboratories). M3N0 medium (N-depleted M3 medium) was used to assess the filter-sterilized nitrogen sources ammonium sulfate (1.5 mM), MMA (1.5 mM), and GMA (0.75 mM, i.e., 1.5 mM total nitrogen). Chemical synthesis of GMA was achieved by a protocol specifically developed for this study (see Fig. S1 in the supplemental material). Dichloromethane (Fluka) was added (300 μl) in a glass tube placed in a 3.3-liter glass jar, which was hermetically closed. Antibiotics were used at final concentrations of 10 μg · ml−1 (tetracycline) and 25 μg · ml−1 (kanamycin) as required.
Growth rates were determined as the average of at least two independent replicates in 20-ml liquid cultures in agitated (100 rpm) 100-ml Erlenmeyer flasks by measuring optical density at 600 nm (OD600) spectrophotometrically, with the exception of cultures with GMA as the growth substrate, which were performed in 5 ml medium in 16-ml Hungate tubes, and the OD was monitored directly in the tubes (Libra S6; Biochrom).
Screening and mapping of minitransposon insertion mutants.
The previously described mutant library (10) of random transcriptional gfp fusion minitransposon insertions in M. extorquens DM4 was conserved at −80°C in 96-well microtiter plates in M3 medium containing methanol, 25% glycerol, and kanamycin. Individual mutants were spotted onto solid M3 plates containing MMA at 20 mM, methanol at 20 mM, or both at 20 mM each, and their abilities to grow and emit fluorescence were tested after 5 days at 30°C. Control strains included the nonfluorescent wild-type strain, a mutant with constitutive expression of green fluorescent protein (GFP) carrying the minitransposon in coding sequence (CDS) METDI4743 of unknown function (10), and mutant DM4cycH, which is unable to grow with methanol and was selected and characterized during this work. Mutants with an MMA-dependent phenotype were then further assayed for growth and induction of GFP fluorescence in M3 medium containing MMA (20 and 80 mM), dichloromethane (1.4 mM), methanol or formate (20 mM), pyruvate or glycerol (7.5 mM), acetate or betaine (10 mM), succinate (5 mM), and combinations thereof. Growth and fluorescence were tested after 7 days of incubation at 30°C of 5-μl drops of serial dilutions of bacterial cultures by a two-step PCR method combining semidegenerate primers together with minitransposon-specific primers as previously described (10). The sequences obtained were compared with the DM4 genome sequence by using BLAST (11) to determine the minitransposon insertion site.
Site-direct mutagenesis.
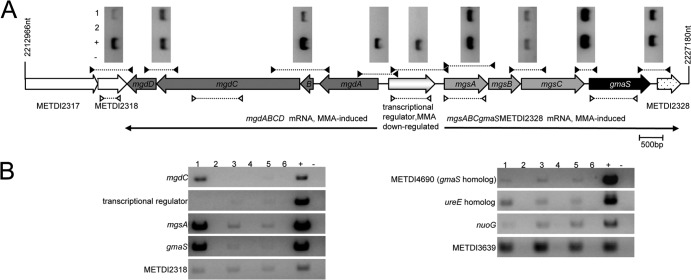
Wild-type and mutant alleles were reciprocally exchanged by using the sacB-based pCM433 vector for marker-free allelic exchange as previously described (12). Briefly, mutant alleles were constructed by a two-step PCR amplification process. The first step involved the use of two primer pairs to amplify approximately 0.5 kb upstream and downstream to the region targeted for whole CDS deletion (see right and left deletion primer pairs in Table S1 in the supplemental material). The two fragments, overlapping by 16-nucleotide (nt)-long complementary fragments, were PCR reamplified to yield a single PCR product in the second step. The resulting approximately 1-kb DNA fragment was digested with restriction enzymes to generate compatible cohesive ends and cloned into vector pCM433. The resulting plasmids, pME8282, pME8283, and pME8284 (for details, see Table S1), were electroporated into E. coli DH5α and then transferred to M. extorquens DM4 by triparental mating (12). Plasmid pME8282 was also introduced into the DM4gmaS mutant, in which gmaS was disrupted by minitransposon insertion to generate the double mutant DM4gmaSΔmetdi4690 strain. The mutants obtained (Fig. 2) were checked by sequencing the PCR-amplified genome deletion region.
FIG 2.
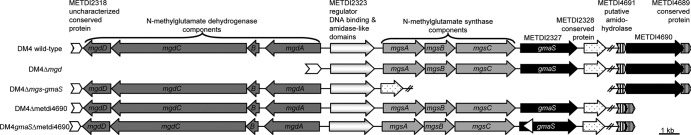
Gene organization of the NMG pathway-encoding genes in M. extorquens DM4 and constructed deletion mutants. The double slashes indicate distinct chromosome regions. The triangle shows the minitransposon insertion site and the orientation of the promoterless gfp gene relative to gmaS in mutant DM4gmaSΔmetdi4690.
Plasmid cloning of gmaS homologs.
Homologs of gmaS were PCR amplified from wild-type DNA and cloned into the XbaI site of the PmxaF promoter-based expression vector pCM80 (13), resulting in plasmids pME8280 and pME8285 harboring gmaS and the gene encoding METDI4690, respectively (for details, see Table S1). Insert sequences were verified to be of the wild type. Plasmids were transferred into Methylobacterium by conjugation as previously described (10).
Reverse transcription (RT)-PCR.
RNA was extracted from cell pellets frozen at −80°C that were obtained by the centrifugation (5,000 rpm, 10 min, 4°C) of 50-ml liquid cultures of strain DM4 harvested at mid-exponential phase (OD600 = 0.2) as previously described (14). cDNA synthesized by using SuperScript III reverse transcriptase (Invitrogen) was used as the template for PCR amplification with iProof Taq polymerase (Bio-Rad) with the primer pairs listed in Table S1 in the supplemental material. RNA and cDNA concentrations were determined with Qubit2.0 (Invitrogen) according to the manufacturer's instructions.
RESULTS
Comparative analysis of the fully assembled genome sequences of the closely related M. extorquens strains DM4 and AM1 (15) surprisingly showed that strain DM4 lacked the well-studied mau genes for MMA utilization, although strain DM4 had been observed early on to grow with MMA (16). Reassessment of the ability of strains AM1 and DM4 to grow with MMA as the sole source of carbon and nitrogen then revealed clear differences in the growth rates of the two strains, with generation times of approximately 6 and 18 h, respectively (under the conditions used in this study). We therefore embarked on a search to identify and characterize the genes that allowed the growth of strain DM4 with MMA as the sole source of carbon and energy. This involved (i) the isolation and characterization of random minitransposon insertion mutants with a specific MMA-dependent phenotype when MMA was provided as the sole C source, (ii) site-directed mutagenesis and complementation assays to study the roles of specific genes or gene clusters in the utilization of MMA as a carbon or nitrogen source, and (iii) MMA-dependent gene expression by RT-PCR.
Isolation and characterization of MMA minitransposon insertion mutants.
We started by screening a previously constructed library (10) of 6,054 mutants of strain DM4 capable of growing with 10 mM methanol and carrying random insertions of a minitransposon containing a promoterless gfp marker and a kanamycin resistance gene. Twenty-one mutants emitted MMA-dependent fluorescence or displayed impaired growth when tested on MMA, methanol, and a combination of the two compounds. Additional tests on M3 plates supplemented with a larger panel of one-carbon (dichloromethane, formate) and multicarbon (acetate, glycerol, pyruvate, betaine, and succinate) compounds were performed to identify mutants with specific MMA-dependent phenotypes. Seven mutants harboring a single minitransposon and displaying a specific MMA-dependent phenotype were obtained and classified into the following three groups: class 1, MMA-impaired growth with no detectable GFP fluorescence (three mutants); class 2, MMA-induced GFP fluorescence (two mutants); class 3, MMA-repressed GFP fluorescence (two mutants) (Table 1).
TABLE 1.
Characteristics of minitransposon mutants and reference strains with respect to growth on MMA
| Interrupted locusa | Strain name | Insertion positionb | Product of interrupted gene (gene) | GFP expression on MMAc | Growth on MMAd |
|---|---|---|---|---|---|
| Control strains | |||||
| None | DM4 | Absent | Wild type | ||
| METDI2066 | DM4cycH | 1953319 | Cytochrome c-type biogenesis protein (cycH) | Basal | Wild type |
| METDI4743 | 26F12 | 4639214 | Conserved protein of unknown function | Basal | Wild type |
| Class 1, MMA-impaired growth | |||||
| METDI2324 | DM4mgsA | 2222651 | NMG synthase component A (mgsA) | Absent | Altered |
| METDI2326 | DM4mgsC | 2224246 | NMG synthase component C (mgsC) | Absent | Altered |
| METDI2327 | DM4gmaS | 2225098 | GMA synthetase (gmaS) | Absent | Altered |
| Class 2, MMA-induced fluorescence | |||||
| METDI2328 | 07C5 | 2226639 | Hypothetical protein | Induced | Wild type |
| METDI1773 | DM4ureE | 1646785 | Putative urease accessory protein | Inducede | Wild type |
| Class 3, MMA-repressed fluorescence | |||||
| METDI1560 | DM4nuoG | 1424682 | NADH:quinone oxidoreductase chain G (nuoG) | Downregulated | Altered |
| IG METDI3639-METDI3640 | 04B1 | 3582478 | Downregulated | Wild type | |
Mutants DM4mgsA, DM4mgsC, and DM4gmaS harbor the minitransposon gfp expression marker in reverse orientation with respect to the interrupted gene. In mutant 04B1, the minitransposon is located in an intergenic (IG) region with the gfp marker gene in the same orientation as METDI3639 downstream.
Nucleotide position upstream of minitransposon I end.
Fluorescence on solid medium with MMA. Induced, fluorescence observed only in the presence of MMA; enhanced, basal fluorescence enhanced in the presence of MMA; downregulated, fluorescence decreased in the presence of MMA alone or in combination with methanol; absent, no fluorescence.
Wild-type or altered growth on solid M3 medium with MMA as the sole source of carbon and energy. All mutants displayed wild-type growth on methanol (20 mM), except mutant DM4cycH, which featured altered, weak growth on methanol.
Urea did not induce GFP fluorescence.
In four mutants, insertions colocalized within a 4-kb genomic region that encodes subunits of the NMG synthase (METDI2324, mgsA; METDI2325, mgsB; METDI2326, mgsC) and GMAS (METDI2327, gmaS) (Table 1; Fig. 2). The corresponding proteins displayed at least 66% amino acid sequence identity with characterized homologs in M. silvestris BL2 (1). Directly downstream of gmaS (Fig. 2), MMA-induced METDI2328 corresponds to a putative 169-residue-long protein with no homology to characterized proteins (mutant 07C5, Table 1). No GFP fluorescence emission was detected for mutants DM4gmaS, DM4mgsA, and DM4mgsC (Table 1, class 1), as expected, given that the minitransposon promoterless gfp gene was in reverse orientation with respect to the interrupted genes.
The other three mutants identified in this study showed MMA-dependent gene expression of the gfp minitransposon (Table 1), indicating differential expression of the genes at the minitransposon insertion site, i.e., genes ureE, nuoG, and those encoding the protein of unknown function METDI3639.
gmaS is essential for MMA utilization as the source of carbon but auxiliary for its utilization as the source of nitrogen.
Mutant DM4gmaS was unable to use MMA as the sole source of carbon and energy (Table 2). Minitransposon insertion into gmaS did not cause detrimental polar effects on the expression of adjacent genes, since wild-type gmaS provided in trans by plasmid pME8280 restored wild-type growth to mutant DM4gmaS (Table 3). We conclude that gmaS is required for MMA utilization as a carbon source by M. extorquens DM4.
TABLE 2.
Use of MMA or GMA as the sole carbon and/or nitrogen source for growth
| Strain(s) | Avg generation time (h)a ± SD with: |
|||
|---|---|---|---|---|
| MMA as C + N source | MMA as C source | MMA as N source | GMA as N source | |
| DM4 | 18.3 ± 0.3 | 17.6 ± 0.6 | 3.5 ± 0.5 | +c |
| 07C5b | 17.4 ± 0.7 | 17.7 ± 0.3 | 3.3 ± 0.1 | NDd |
| DM4Δmetdi4690 | 18.4 ± 0.8 | 17.8 ± 0.5 | 3.3 ± 0.6 | ND |
| DM4gmaS | No growth | No growth | 32.7 ± 1.8 | +c |
| DM4gmaSΔmetdi4690 | No growth | No growth | 38.5 ± 1.5 | +c |
| DM4Δmgs-gmaS, DM4mgsC | No growth | No growth | No growth | ND |
| DM4mgsA, DM4Δmgd | No growth | No growth | No growth | No growth |
| DM4nuoG | No growth | No growth | 3.3 ± 0.4 | ND |
| DM4ureE | 17.5 ± 0.4 | 17.4 ± 0.6 | 3.5 ± 0.2 | ND |
Growth in M3N0 medium was tested (see Materials and Methods). When tested as a C source, MMA was provided at 20 mM. None of the strains grew with GMA as the sole C source (20 mM). When tested as N sources, MMA and GMA were provided at 1.5 and 0.75 mM, respectively. In controls, the N source was (NH4)2SO4 at 1.5 mM and the C source was succinate at 5 mM. Growth with succinate and (NH4)2SO4 resulted in a generation time of 2.5 ± 0.8 h for the wild-type strain and all of the other strains tested.
Minitransposon insertion in METDI2328, the last CDS of the mgsABC-gmaS-METDI2328 operon.
+, growth with flocculation prevented evaluation of generation times.
ND, not determined.
TABLE 3.
Characterization of growth of M. extorquens DM4 and mutants with MMA when provided with plasmid copies of GMAS homologs in trans
| Interruptedb CDS(s) and plasmidc | Avg generation time (h) ± SD with MMAa provided as: |
||
|---|---|---|---|
| C + N source | C source | N source | |
| None | |||
| pCM80 | 18.9 ± 0.9 | 18.0 ± 0.4 | 4.4 ± 0.2 |
| pME8280 | 16.2 ± 0.6 | 15.3 ± 0.3 | 3.3 ± 0.2 |
| pME8285 | 17.5 ± 0.9 | 18.8 ± 1.6 | 3.5 ± 0.2 |
| METDI2327 | |||
| pCM80 | No growth | No growth | 42.6 ± 1.2 |
| pME8280 | 16.9 ± 0.7 | 15.9 ± 0.3 | 4.7 ± 0.3 |
| pME8285 | 20.2 ± 0.8 | 16.5 ± 1.3 | 4.0 ± 0.3 |
| METDI2327, METDI4690 | |||
| pCM80 | No growth | No growth | 41.7 ± 1.2 |
| pME8285 | 21.5 ± 0.5 | 19.2 ± 1.5 | 4.0 ± 0.1 |
MMA was added as the C or N source at 20 or 1.5 mM, respectively. In controls, succinate at 5 mM and (NH4)2SO4 at 1.5 mM were provided as C and N sources, respectively. Similar growth of all strains on succinate (5 mM) and (NH4)2SO4 (1.5 mM) was observed (generation time, 3.4 ± 0.3 h).
Interruption of METDI4690 alone conferred no phenotype with MMA as the C or N source (Table 2, mutant DM4Δmetdi4690).
pCM80, empty expression vector; pME8280, pCM80 for gmaS expression of METDI2327; pCM8285, pCM80 for expression of METDI4690.
Regarding the assimilation of nitrogen, in contrast, when MMA was the only source of nitrogen in the presence of another carbon source, DM4gmaS grew about 10 times more slowly than the wild-type strain (generation times of 32.7 and 3.5 h, respectively; Table 2). However, no difference in growth ability between mutant DM4gmaS and the wild-type strain was observed when MMA was replaced with GMA, the product of MMA transformation by GMAS (Fig. 1). This suggested that GMA, which cannot be used by wild-type strain DM4 as the sole carbon source (Table 2, footnote a), can nevertheless enter the cell and serve as an alternative source of nitrogen, thereby alleviating the severe growth defect observed in the gmaS mutant.
The residual ability of the mutant DM4gmaS to grow with MMA as the sole nitrogen source suggested that other uncharacterized enzymes with GMAS activity may exist in M. extorquens DM4. We found another GMAS-like protein, METDI4690, which displayed 39% identity to GMAS (METDI2327) at the protein sequence level. To test whether METDI4690 could play a role in MMA utilization, additional mutants were generated by precisely deleting METDI4690 in both the wild-type and mutant DM4gmaS backgrounds, yielding the mutants DM4Δmetdi4690 and DM4gmaSΔmetdi4690, respectively. Mutant DM4Δmetdi4690 itself grew identically to the wild type under all of the conditions tested (Table 2). Only in mutant DM4gmaSΔmetdi4690 lacking both gmaS homologs, when MMA was provided as the sole nitrogen source, did the lack of METDI4690 have a detectable effect on growth (generation time of 38.5 h for DM4gmaSΔmetdi4690 versus 32.7 h for DM4gmaS, Table 2). Thus, MMA utilization as the sole nitrogen source involves both gmaS homologs, although METDI2327 (GMAS) plays a major role compared to that of METDI4690 in the wild-type context (Table 3).
mgsABC and mgdABCD are essential genes for the growth of M. extorquens DM4 with MMA as the source of carbon and nitrogen.
NMG synthase is a poorly characterized enzyme that is homologous to glutamate synthase (17), which, by analogy, may have the capacity to transform both MMA and GMA obtained from MMA by GMAS with the concomitant release of ammonia (Fig. 1). Genes homologous to mgsABC encoding NMG synthase were found to be essential for growth with MMA in strain DM4, as transposon insertion mutants DM4mgsA and DM4mgsC were unable to grow with MMA as either a carbon or a nitrogen source (Table 2). Similarly, mutant DM4mgsA was also unable to grow with GMA as a nitrogen source in the presence of another carbon source.
The following reaction in the NMG pathway is catalyzed by NMG dehydrogenase encoded by the genes mgdABCD and funnels the methyl group carbon of NMG into methylenetetrahydrofolate (Fig. 1). On the basis of sequence similarity, as well as experimental data, the mgdABCD genes constitute the last specific step of the NMG pathway (Fig. 1) (6). Very similar genes are known to encode sarcosine (N-methylglycine) oxidase and are found in many bacteria, including Methylobacterium strains, which contain at least two sets of homologs. Homologs of the mgdABCD genes, sharing 73, 27, 55, and 21% amino acid sequence identity with characterized components A, B, C, and D of the NMG dehydrogenase of M. silvestris BL2 (1), were found upstream of the mgsABC-gmaS region in strain DM4 (Fig. 2). Somewhat surprisingly, no mutants with minitransposon insertions in the mgdABCD genes were detected. Therefore, we constructed a mutant with all of the mgdABCD genes deleted in order to assess if the mgd cluster plays a role in MMA utilization by M. extorquens DM4. The resulting DM4Δmgd mutant was unable to grow in the presence of MMA as the sole source of carbon and energy and also as the sole source of nitrogen in the presence of another carbon source for growth (Table 2). In conclusion, both the mgs and mgd gene clusters are essential for MMA utilization as a carbon, energy, and nitrogen source by M. extorquens DM4.
The nuoG gene is essential for strain DM4 growth with MMA as the carbon and energy source.
Analysis of mutant DM4nuoG showed that disruption of nuoG prevented the growth of strain DM4 with MMA as the sole carbon source but not as the sole nitrogen source when growing with succinate as the carbon source (Table 2). The gene nuoG is 1 of the 14 genes of the nuo cluster that encodes NADH:ubiquinone oxidoreductase, a respiratory complex 1 enzyme that catalyzes the transfer of electrons from NADH to the quinone pool, coupled with translocation of protons across the membrane. Disruption of individual nuo genes impairs respiratory complex 1 in E. coli (18). The inability of mutant DM4nuoG to grow with MMA as the carbon and energy source suggests that NuoG may be involved in accepting electrons from the oxidation of MMA by the NMG pathway in strain DM4. In contrast, mutant DM4cycH, in which the disrupted gene is involved in cytochrome c biogenesis (19), was able to grow with MMA by the NMG pathway but not with methanol. Thus, M. extorquens DM4 appears to require the cytochrome c electron transfer system for oxidation of the C1 alcohol methanol but not of the corresponding C1 amine MMA.
MMA-dependent gene expression.
By using RT-PCR amplifications targeting intergenic regions in the vicinity of the genes mgd, mgs, and gmaS, the expected operonic structure of the divergent mgdABCD and mgsABC-gmaS-METDI2328 gene clusters was confirmed (Fig. 3A). MMA-dependent regulation was assessed with cultures of strain DM4 grown with MMA, methanol, or succinate as the sole carbon source. The genes mgdC, mgsA, and gmaS and the MMA-associated ureE homolog displayed MMA-dependent upregulation, as shown by RT-PCR with intragenic gene primer pairs (Fig. 3B). Conversely, nuoG and the putative transcriptional regulator METDI2323 located between the two operons were downregulated by MMA (Fig. 3B). Taken together, the data obtained showed that the genome region encoding the NMG pathway (Fig. 2) harbors two MMA-induced, divergently transcribed operons (mgdABCD and mgsABC-gmaS-METDI2328) separated by a putative regulator gene whose expression was downregulated by MMA or by a metabolite generated in the course of MMA utilization. In contrast, for genes encoding the GMAS homolog METDI4690 and two proteins of unknown functions, METDI2318 and METDI3639 (located downstream of the insertion site in minitransposon mutant 04B1; Table 1), constitutive low-level expression irrespectively of the presence of MMA in the medium was found (Fig. 3B, right panel).
FIG 3.
Transcription studies of MMA-dependent gene expression in M. extorquens DM4 by PCR on reverse transcriptase-produced total cDNA. RNA was isolated from cultures grown with MMA (lines 1 and 2) or with (NH4)2SO4 as a nitrogen source with either methanol (lines 3 and 4) or succinate (lines 5 and 6) as a carbon source and reverse transcribed, and the cDNA was quantified before PCR amplification. Controls were performed for each primer pair with M. extorquens DM4 genomic DNA (1.5 ng) (lane +), water (lane −), and samples from which RT was omitted prior to PCR amplification (lanes 2, 4, and 6). (A) PCR amplification of cDNA (4 ng) was performed with primer pairs spanning intergenic regions (black triangles). Thick arrows denote experimentally confirmed operonic structures. (B) PCR amplification from cDNA of individual genes (primer pairs are indicated by white triangles) of the NMG pathway (left) and of other genes investigated in this study (right), starting from 8 ng total cDNA, except for the transcriptional regulator (16 ng cDNA).
Effect of plasmid-driven expression of gmaS homologs.
The constitutive low-level expression of METDI4690 suggested by RT-PCR experiments (Fig. 3B) may explain why deletion of the gene that encodes it had little effect on the ability of strain DM4 to utilize MMA (Table 2). To test this hypothesis, plasmid pME8285 harboring cloned METDI4690 under the control of promoters Ptac and PmxaF of vector pCM80, which favor strong constitutive expression of downstream genes in pCM plasmids (9, 15, 20), was introduced into wild-type and gmaS mutant strain DM4. No significant difference in growth with MMA as the sole source of carbon and nitrogen was observed in the wild-type strain harboring plasmid pME8285 or the empty expression vector pCM80 (Table 3). Similarly, providing GMAS (METDI2327) in trans from plasmid pME8280 allowed only slightly faster growth of the wild-type strain with MMA as the source of carbon or nitrogen (Table 3) and fully complemented the growth defect of mutant DM4gmaS (Table 2). However, providing METDI4690 in trans on pME8285 restored the ability of mutants lacking one or both gmaS homologs to use MMA as a source of carbon and nitrogen for growth (Table 3). Thus, METDI4690 represents a bona fide functional GMAS that may, however, be too weakly expressed from its native chromosomal promoter to sustain the utilization of MMA as the sole carbon source by M. extorquens DM4. We concluded that both the gene encoding METDI2327 and METDI4690 gmaS homologs express functional GMAS enzymes but that their expression is the limiting factor for optimal MMA utilization under the conditions tested.
Phylogenetic analysis of gmaS homologs and comparison of gene clusters.
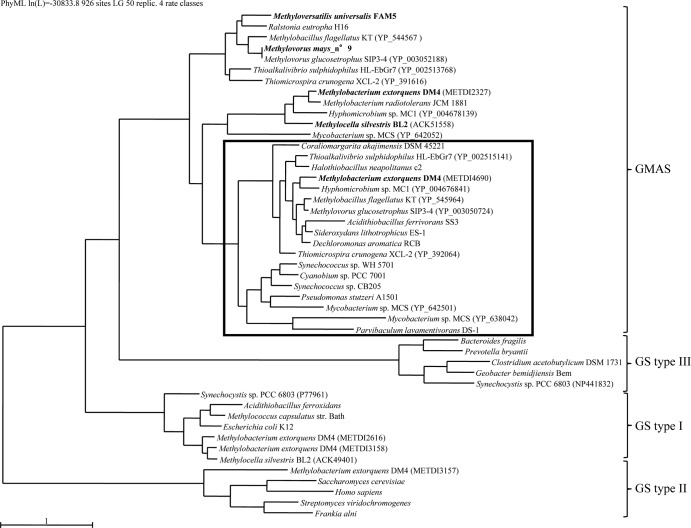
Proteins encoded by gmaS homologs belong to the larger protein family of glutamine synthetases, not all of which are able to transform MMA (1, 21). The sequences of M. extorquens DM4 gmaS homologs METDI2327 and METDI4690 were compared to those of strain DM4 homologs of the three known families of glutamine synthetases (METDI2616, METDI3157, and METDI3158) to experimentally characterized GMAS enzymes and to representative homologs of other prokaryotes and eukaryotes, as well as to sequences closely related to METDI4690 from representative sequenced genomes (at least 44% identity at the protein level, lengths of 444 to 472 amino acids; see Table S2 in the supplemental material). The resulting phylogenetic tree (Fig. 4) confirms the previously documented four types of glutamine synthetase homologs (1, 22), i.e., the three glutamine synthetase types and the group containing all of the GMAS enzymes experimentally characterized so far. DM4 homologs METDI2616, METDI3157, and METDI3158 can be assigned to glutamine synthetase types I and II, but the MMA-associated gmaS homologs METDI2327 and METDI4690 investigated here clearly cluster within the GMAS group. Notably, METDI4690-like sequences (framed in Fig. 4) are found mainly in members of the class Proteobacteria (see Table S2) but are only loosely associated with methylotrophic metabolism, in contrast to the closely related homologs of the experimentally characterized GMAS and METDI2327 of strain DM4.
FIG 4.
Phylogenetic tree representation of protein sequences encoded by homologs of gmaS and representatives of the three known types of glutamine synthetase (GS). The tree was constructed by using PhyML. Organisms with experimentally characterized GMAS are in bold. Close homologs of METDI4690 are framed. For nucleotide sequence accession numbers, protein sequence lengths, and percentages of identity with M. extorquens GMAS and METDI4690, see Table S2 in the supplemental material. The accession number of each bacterium in which more than one gmaS homolog was found is shown in parentheses.
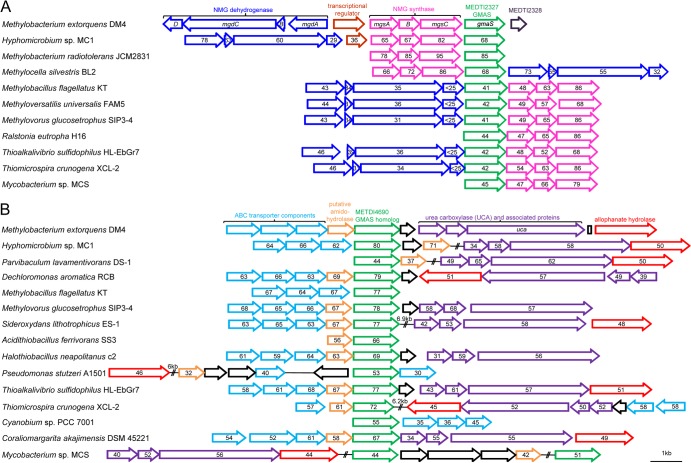
However, METDI237- and METDI4690-like gmaS homologs could be distinguished by taking their genetic organization context into account (Fig. 5). Previously experimentally characterized, bona fide gmaS genes colocate with genes encoding enzymes of the NMG pathway of MMA oxidation, especially with mgs genes (1, 6) (Fig. 5B). In contrast, homologs of METDI4690, shown here to restore the growth of strain DM4 with MMA as a carbon and nitrogen source, in mutants impaired in the NMG pathway (Table 3) were often associated with a conserved gene encoding an uncharacterized amidohydrolase (conserved domain COG1402) described as potentially acting on nonpeptidic C-N bonds (Fig. 5B). Genes encoding components of an ABC-type transporter and proteins involved in urea-related metabolism were also often found in close proximity (Fig. 5B). Thus, genes encoding GMAS homologs closely similar to METDI4690 appear to be associated with a conserved uncharacterized gene cluster involved in nitrogen metabolism.
FIG 5.
Gene organization and conservation of gmaS homolog-containing genomic regions for sequences closely related to METDI2327 (A) and METI4690 (B), respectively. Gene orientation and linkage are indicated with arrows, and double slashes indicate a distance of at least 10 kb between the genome regions considered. UCA-associated proteins 1 and 2 are encoded by a tandem of homologous, uncharacterized genes with conserved domains TIGR03424 and TIGR03425, respectively. Alphaproteobacteria class members (complete genome accession numbers are in parentheses): M. extorquens DM4 (NC_012988), Hyphomicrobium sp. strain MC1 (NC_015717), Methylobacterium radiotolerans JCM2831 (NC_010505), M. silvestris BL2 (NC_011666), and Parvibaculum lavamentivorans DS-1 (NC_009719). Betaproteobacteria class members: Dechloromonas aromatica RCB (NC_007298), Methylobacillus flagellatus KT (NC_007947), M. universalis FAM5 (NZ_AFHG00000000), Methylovorus glucosetrophus SIP3-4 (NC_012969), Sideroxydans lithotrophicus ES-1 (NC_013959), and Ralstonia eutropha H16 (NC_008314). Gammaproteobacteria class members: Acidithiobacillus ferrivorans SS3 (NC_015942), Halothiobacillus neapolitanus c2 (NC_013422), Pseudomonas stutzeri A1501 (NC_009434), Thioalkalivibrio sulfidophilus HL-EbGr7 (NC_011901), and Thiomicrospira crunogena XCL-2 (NC_007520). Cyanobacteria class member: Cyanobium sp. strain PCC 7001 (NZ_DS990556). Verrucomicrobia class member: Coraliomargarita akajimensis DSM 45221 (NC_014008). Actinobacteria class member: Mycobacterium sp. strain MCS (NC_008147).
DISCUSSION
We demonstrated in this work that the two closely adjacent, divergent operons, mgdABCD and mgsABC-gmaS-METDI2328, involved in the NMG pathway are induced by MMA and allow M. extorquens DM4 to use MMA as a sole carbon, nitrogen, and energy source. The two operons are separated by only a putative regulator gene (the gene for METDI2323) whose expression is downregulated by MMA (Fig. 3) and which belongs to the AraC family of transcriptional regulators with a C-terminal DNA-binding helix-turn-helix domain (PROSITE PS01124). In addition, METDI2323 features an N-terminal class I glutamine amidotransferase-like domain that may serve to sense a chemical effector such as MMA or another compound associated with MMA metabolism. The last gene of the cluster cotranscribed with gmaS encodes the 169-residue-long protein METDI2328 of unknown function with close homologs so far found only in Methylobacterium genomes. This gene is not essential for MMA metabolism, since its disruption had no effect on the ability of strain DM4 to grow with MMA under any of the conditions tested (Fig. 3; Table 1, mutant 07C5). Nevertheless, its expression is induced by MMA, as is that of a ureE homolog for a urease accessory protein (23). The transcription of urease genes can be regulated by nitrogen availability (24), and we speculate that this MMA-induced urease-like operon may contribute in some way to MMA-associated nitrogen assimilation in strain DM4. Nevertheless and as for METDI2328, disruption of ureE by minitransposon insertion in mutant DM4ureE is not associated with any detected growth defect (Table 1). The presence of two homologs of the seven-gene urease operon ureEFABCGD in strain DM4 may explain this observation.
Unlike M. extorquens strains DM4, PA1, and BJ001 (8, 25), M. extorquens strains such as AM1 and CM4 contain mau genes and grow much faster with MMA as the sole carbon, nitrogen, and energy source (approximately 3-fold growth rate difference; C. Gruffaz and F. Bringel, unpublished data). Moreover, M. extorquens DM4 grew less efficiently with MMA than with methanol (generation times of 18.3 and 3.4 h, respectively; Table 2) (10). It is possible that energy production by the oxidation of one-carbon compounds involves different pathways of electron transfer for MMA oxidation (by the NMG pathway) and for methanol (by methanol dehydrogenase), as suggested by the different growth phenotypes of mutants DM4nuoG and DM4cycH (Tables 1 and 2). Still, the genetic organization of NMG pathway genes in M. extorquens DM4 is strongly conserved within the sequenced genomes of M. extorquens (8, 25), suggesting that possession of this gene set would allow all Methylobacterium strains to grow with MMA. That is not the case, since inactivation of mau genes for MADH in the closely related strain AM1 completely abolished its ability to grow on MMA as the sole C source (4). Nevertheless, NMG dehydrogenase activity was detected in M. extorquens AM1 (4, 19), suggesting that other factors beyond the mere possession of the required genes are required for the growth of Methylobacterium strains with MMA by the NMG pathway.
The observed growth phenotypes of NMG pathway mutants have shed some light on the still somewhat elusive roles of GMAS and NMG synthase in the growth of strain DM4 with MMA (Table 3). The fact that a mutant with impaired mgsA is unable to use MMA or GMA as either a carbon or a nitrogen source for growth represents the most clear-cut result obtained in this work (Table 2). It demonstrates the absolutely essential role of NMG synthase in the NMG pathway (Fig. 1), most likely involving the same glutamate–α-ketoglutarate redox couple as in the homologous enzyme glutamate synthase (17) for condensation of amines to α-ketoglutarate with concomitant release of ammonia, which can then be used as a nitrogen source for growth. We have also demonstrated that in the absence of a functional gmaS gene (mutant DM4gmaS), M. extorquens DM4 lacks the ability to utilize MMA as the sole carbon and energy source, suggesting that GMA is an obligate intermediate of MMA oxidation by the NMG pathway when MMA is the sole source of carbon and energy (Fig. 1). Nevertheless, GMA as the sole source of carbon and energy did not sustain the growth of wild-type strain DM4, unlike when it was provided as the sole nitrogen source in the presence of another carbon source. It remains to be tested whether GMA is transported efficiently enough within the cell to fulfill the higher carbon rather than nitrogen assimilation requirements for cell growth. When considering published studies, the question of whether GMA is an obligate intermediate of the NMG pathway (Fig. 1) is still debated, given the contradictory experimental evidence obtained so far for the role of GMAS in M. universalis FAM5 and M. sylvestris BL2. In M. silvestris BL2, gmaS is essential for MMA utilization as a C and energy source (1), whereas this gene is unexpectedly dispensable in M. universalis FAM5 (6). Intriguingly, M. universalis FAM5 and M. silvestris BL2 are bacteria that contain only one gmaS homolog (see Table S2 in the supplemental material), in contrast to all of the strains of M. extorquens sequenced so far (8, 25), which contain two gmaS homologs (Fig. 5). In M. extorquens DM4, GMAS (METDI2327) was found to be required for MMA utilization as a carbon source. This suggests that the direct reaction of MMA with glutamate catalyzed by NMG synthase is not sufficiently efficient to bypass mutation in gmaS with respect to carbon requirements for growth. In other words, the indirect pathway through gmaS and GMA as an intermediate may be a more efficient way to metabolize carbon from MMA. Deletion of the second gmaS homolog, METDI4690, further decreased the residual capacity of the DM4gmaS mutant to grow with MMA as a nitrogen source, and deletion of the entire gene cluster encoding NMG synthase and GMAS completely abolished the ability of strain DM4 to utilize MMA as a nitrogen source for growth (Table 3). This demonstrates that no enzymatic systems beyond the two GMAS homologs exist in strain DM4 to extract nitrogen from MMA in a growth-conducive fashion. Most notably, complementation with either GMAS METDI2327 or METDI4690 on a multicopy plasmid in both the DM4gmaS and DM4gmaSΔmetdi4690 backgrounds allowed the utilization of MMA as the sole source of carbon and nitrogen for growth (Table 3) and conclusively demonstrated that both GMAS homologs of strain DM4 were bona fide GMAS proteins. In addition, plasmid expression of either GMAS homolog was found to significantly improve the ability of M. extorquens DM4 to utilize MMA as the sole source of carbon, energy, and nitrogen for growth (Table 3). Thus, expression of gmaS homologs may represent a major bottleneck in the ability of methylotrophic bacteria to grow with methylated amines. The distinct expression levels of the two gmaS homologs in wild-type strain DM4 (Fig. 3) suggest that they are involved in different metabolic pathways associated with the transformation of compounds containing amino groups and that MMA-induced METDI2327 plays a predominant role in MMA oxidation. METDI4690, in contrast, is expressed at low constitutive levels and lies immediately adjacent to genes encoding components of the urea carboxylase (UCA)/allophanate hydrolase pathway, as in many bacteria known to use urea as the sole nitrogen source (26) (Fig. 5). The uca gene adjacent to METDI4690 may therefore be involved in the interconversion of methylated amides and amines for nitrogen metabolism. Indeed, in Oleomonas sagaranensis, a member of the class Alphaproteobacteria, ammonia is produced by UCA not only from urea degradation but also from acetamide and formamide (26). Thus, gmaS homologs, which are widely distributed in bacterial genomes, may contribute to the microbial metabolism of a large variety of amides and amines found in the environment.
A final point of the present study is that the NMG pathway downstream of NMG involves aspects that are not yet understood. Unexpectedly, mutant DM4Δmgd was unable to utilize MMA or GMA as a nitrogen source when succinate was supplied as a carbon source (Table 2), even though ammonium as a growth-supporting source of nitrogen is not generated directly through NMG dehydrogenase activity (Fig. 1). One of several possible explanations for the phenotype of mutant DM4Δmgd is that lack of NMG dehydrogenase somehow inactivates NMG synthase. This hypothesis remains to be tested, as well as the possible key role of glutamate as an intermediate, substrate, and/or product of all three steps of the NMG pathway.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Région Alsace for the Ph.D. grant of E.E.L.M.
Help in mutant library screening from S. El Hassoun and constructive discussions with S. Vuilleumier are acknowledged.
Footnotes
Published ahead of print 28 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04160-13.
REFERENCES
- 1.Chen Y, Scanlan J, Song L, Crombie A, Rahman MT, Schäfer H, Murrell JC. 2010. γ-Glutamylmethylamide is an essential intermediate in the metabolism of methylamine by Methylocella silvestris. Appl. Environ. Microbiol. 76:4530–4537. 10.1128/AEM.00739-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony C. 1982. The biochemistry of methylotrophs. Academic Press, Inc., New York, NY [Google Scholar]
- 3.Bicknell B, Owens JD. 1980. Utilization of methyl amines as nitrogen sources by non-methylotrophs. J. Gen. Microbiol. 117:89–96 [Google Scholar]
- 4.Chistoserdov AY, Chistoserdova LV, McIntire WS, Lidstrom ME. 1994. Genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J. Bacteriol. 176:4052–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistoserdova L. 2011. Modularity of methylotrophy, revisited. Environ. Microbiol. 13:2603–2622. 10.1111/j.1462-2920.2011.02464.x [DOI] [PubMed] [Google Scholar]
- 6.Latypova E, Yang S, Wang Y-S, Wang T, Chavkin TA, Hackett M, Schäfer H, Kalyuzhnaya MG. 2010. Genetics of the glutamate-mediated methylamine utilization pathway in the facultative methylotrophic beta-proteobacterium Methyloversatilis universalis FAM5. Mol. Microbiol. 75:426–439. 10.1111/j.1365-2958.2009.06989.x [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, McAleer KL, Murrell JC. 2010. Monomethylamine as a nitrogen source for a nonmethylotrophic bacterium, Agrobacterium tumefaciens. Appl. Environ. Microbiol. 76:4102–4104. 10.1128/AEM.00469-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vuilleumier S, Chistoserdova L, Lee M-C, Bringel F, Lajus A, Zhou Y, Gourion B, Barbe V, Chang J, Cruveiller S, Dossat C, Gillett W, Gruffaz C, Haugen E, Hourcade E, Levy R, Mangenot S, Muller E, Nadalig T, Pagni M, Penny C, Peyraud R, Robinson DG, Roche D, Rouy Z, Saenampechek C, Salvignol G, Vallenet D, Wu Z, Marx CJ, Vorholt JA, Olson MV, Kaul R, Weissenbach J, Médigue C, Lidstrom ME. 2009. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One 4:e5584. 10.1371/journal.pone.0005584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roselli S, Nadalig T, Vuilleumier S, Bringel F. 2013. The 380 kb pCMU01 plasmid encodes chloromethane utilization genes and redundant genes for vitamin B12- and tetrahydrofolate-dependent chloromethane metabolism in Methylobacterium extorquens CM4: a proteomic and bioinformatics study. PLoS One 8:e56598. 10.1371/journal.pone.0056598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller EEL, Hourcade E, Louhichi-Jelail Y, Hammann P, Vuilleumier S, Bringel F. 2011. Functional genomics of dichloromethane utilization in Methylobacterium extorquens DM4. Environ. Microbiol. 13:2518–2535. 10.1111/j.1462-2920.2011.02524.x [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7:203–214. 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- 12.Marx CJ. 2008. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res. Notes 1:1. 10.1186/1756-0500-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marx CJ, Lidstrom ME. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065–2075 [DOI] [PubMed] [Google Scholar]
- 14.Marchal M, Briandet R, Halter D, Koechler S, DuBow MS, Lett M-C, Bertin PN. 2011. Subinhibitory arsenite concentrations lead to population dispersal in Thiomonas sp. PLoS One 6:e23181. 10.1371/journal.pone.0023181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayser MF, Ucurum Z, Vuilleumier S. 2002. Dichloromethane metabolism and C1 utilization genes in Methylobacterium strains. Microbiology 148:1915–1922 [DOI] [PubMed] [Google Scholar]
- 16.Gälli R. 1986. Optimierung des mikrobiellen Abbaus von Dichlormethan in einem Wirbelschicht-Bioreaktor. Eidgenössische Technische Hochschule, Zürich, Switzerland [Google Scholar]
- 17.Vanoni MA, Curti B. 2008. Structure-function studies of glutamate synthases: a class of self-regulated iron-sulfur flavoenzymes essential for nitrogen assimilation. IUBMB Life 60:287–300. 10.1002/iub.52 [DOI] [PubMed] [Google Scholar]
- 18.Erhardt H, Steimle S, Muders V, Pohl T, Walter J, Friedrich T. 2012. Disruption of individual nuo-genes leads to the formation of partially assembled NADH:ubiquinone oxidoreductase (complex I) in Escherichia coli. Biochim. Biophys. Acta 1817:863–871. 10.1016/j.bbabio.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 19.Page MD, Ferguson SJ. 1995. Cloning and sequence analysis of cycH gene from Paracoccus denitrificans: the cycH gene product is required for assembly of all c-type cytochromes, including cytochrome c1. Mol. Microbiol. 15:307–318 [DOI] [PubMed] [Google Scholar]
- 20.Marx CJ, Lidstrom ME. 2004. Development of an insertional expression vector system for Methylobacterium extorquens AM1 and generation of null mutants lacking mtdA and/or fch. Microbiology 150:9–19. 10.1099/mic.0.26587-0 [DOI] [PubMed] [Google Scholar]
- 21.Brown JR, Masuchi Y, Robb FT, Doolittle WF. 1994. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J. Mol. Evol. 38:566–576 [DOI] [PubMed] [Google Scholar]
- 22.Merrick MJ, Edwards RA. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrugia MA, Macomber L, Hausinger RP. 2013. Biosynthesis of the urease metallocenter. J. Biol. Chem. 288:13178–13185. 10.1074/jbc.R112.446526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobley HL, Hausinger RP. 1989. Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53:85–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marx CJ, Bringel F, Chistoserdova L, Moulin L, Farhan Ul Haque M, Fleischman DE, Gruffaz C, Jourand P, Knief C, Lee M-C, Muller EEL, Nadalig T, Peyraud R, Roselli S, Russ L, Goodwin LA, Ivanova N, Kyrpides N, Lajus A, Land ML, Médigue C, Mikhailova N, Nolan M, Woyke T, Stolyar S, Vorholt JA, Vuilleumier S. 2012. Complete genome sequences of six strains of the genus Methylobacterium. J. Bacteriol. 194:4746–4748. 10.1128/JB.01009-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanamori T, Kanou N, Atomi H, Imanaka T. 2004. Enzymatic characterization of a prokaryotic urea carboxylase. J. Bacteriol. 186:2532–2539. 10.1128/JB.186.9.2532-2539.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Gomez NC, Nguyen S, Lidstrom ME. 2013. Elucidation of the role of the methylene-tetrahydromethanopterin dehydrogenase MtdA in the tetrahydromethanopterin-dependent oxidation pathway in Methylobacterium extorquens AM1. J. Bacteriol. 195:2359–2367. 10.1128/JB.00029-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.