Abstract
Burkholderia pseudomallei is a Gram-negative soil bacillus that is the etiological agent of melioidosis and a biothreat agent. Little is known about the biogeography of this bacterium in Australia, despite its hyperendemicity in the northern region of this continent. The population structure of 953 Australian B. pseudomallei strains representing 779 and 174 isolates of clinical and environmental origins, respectively, was analyzed using multilocus sequence typing (MLST). Bayesian population structure and network SplitsTree analyses were performed on concatenated MLST loci, and sequence type (ST) diversity and evenness were examined using Simpson's and Pielou's indices and a multivariate dissimilarity matrix. Bayesian analysis found two B. pseudomallei populations in Australia that were geographically distinct; isolates from the Northern Territory were grouped mainly into the first population, whereas the majority of isolates from Queensland were grouped in a second population. Differences in ST evenness were observed between sampling areas, confirming that B. pseudomallei is widespread and established across northern Australia, with a large number of fragmented habitats. ST analysis showed that B. pseudomallei populations diversified as the sampling area increased. This observation was in contrast to smaller sampling areas where a few STs predominated, suggesting that B. pseudomallei populations are ecologically established and not frequently dispersed. Interestingly, there was no identifiable ST bias between clinical and environmental isolates, suggesting the potential for all culturable B. pseudomallei isolates to cause disease. Our findings have important implications for understanding the ecology of B. pseudomallei in Australia and for potential source attribution of this bacterium in the event of unexpected cases of melioidosis.
INTRODUCTION
Melioidosis, a disease hyperendemic in northern Australia and Southeast Asia, is caused by the environmental bacterium Burkholderia pseudomallei (1). In the tropical Northern Territory, Australia, there have been >820 documented cases of melioidosis in the past 24 years, of which around 13% have been fatal (2, 3). Percutaneous inoculation is considered the most common route of infection; however, case reports associated with severe weather events and B. pseudomallei-contaminated water supplies highlight the potentially important roles of inhalation and ingestion (1, 2, 4). In October 2012, B. pseudomallei was upgraded to Tier 1 select agent categorization by the Centers for Disease Control and Prevention owing to fears of a deliberate release coupled with a high mortality rate, the lack of an available vaccine, and a nonspecific disease presentation.
B. pseudomallei can infect any organ in the body, leading to a plethora of clinical presentations ranging from localized skin infection without sepsis to rapidly progressive fatal septicemic shock. Pneumonia is universally the most common presentation, although prostatic abscesses and encephalomyelitis are more common in Australian melioidosis cases and parotitis and liver abscesses are more prevalent in Southeast Asia (1). To investigate these regional differences, previous studies have analyzed associations with B. pseudomallei genotypes based on housekeeping genes using multilocus sequence typing (MLST) (5–7). Ribotyping, BOX primer PCR, or pulsed-field gel electrophoresis (PFGE) have also been used to study the population structure of B. pseudomallei (8–10); however, these approaches either suffered from reduced resolution and low reproducibility or are labor-intensive. Multilocus variable-number tandem-repeat analysis (MLVA) has previously been used to study the fine-scale genetic diversity of epidemiologically linked B. pseudomallei isolates (11), but this method suffers from homoplasy issues across more distantly related isolates. Recently developed approaches, such as genomic island and 16S to 23S rRNA gene internal transcribed spacer analysis are useful for certain circumstances but, like MLVA, can be confounded by a high rate of lateral gene transfer (12, 13). Therefore, MLST was chosen for our study, as this method is currently the best tool for reconstructing phylogeographic relationships in B. pseudomallei (14, 15).
Despite considerable diversity among B. pseudomallei sequence types (STs) (0.57 and 0.46 unique ST per isolate from Australia and Thailand, respectively) (6, 7), which can confound population structure, broad geographic attribution patterns have nevertheless been identified; specifically, no environmental STs have been confirmed to be shared between these landmasses. Likewise, no shared environmental STs have been found between the adjacent Northern Territory and Queensland, Australia. The distinction between B. pseudomallei populations in Southeast Asia and Australia is evident with whole-genome sequencing data (14, 15) and may also be the case for the Northern Territory and Queensland. These intercontinental B. pseudomallei population differences suggest that there might also be an underlying population structure within Australia.
We have collected >3,000 unique B. pseudomallei isolates of clinical and environmental origins over the past 24 years at the Menzies School of Health Research (Menzies), Darwin, Australia, and many of these have been genotyped by MLST. Using this large collection of isolates, we analyzed the population structure and diversity of Australian B. pseudomallei STs, focusing on regions of endemicity in the Northern Territory and Queensland. We determined the molecular divergence of B. pseudomallei based on geographic location within Australia and on source type (clinical versus environmental). A high recombination rate has previously hindered the analysis of B. pseudomallei population structure using traditional algorithms such as eBURST (7, 16) or cladogram-based phylogenies (e.g., maximum likelihood or neighbor-joining trees) (5, 7). To accommodate the confounding effects of recombination, Bayesian analysis-based approaches have been successfully used to identify phylogeographic structure in the global B. pseudomallei population (14, 15). Based on these studies, three different approaches were chosen to analyze the Australian MLST population structure: first, a Bayesian population structure analysis; second, a phylogenetic Splits network analysis; and third, disregarding any phylogenetic relationships between STs, Simpson's and Pielou's indices and a multivariate dissimilarity matrix-based approach that focuses on ST diversity and abundance.
MATERIALS AND METHODS
Ethics approval.
This study was approved by the Human Research Ethics Committee of the Northern Territory Department of Health and Menzies School of Health Research (HREC approval number 02/38).
B. pseudomallei isolates.
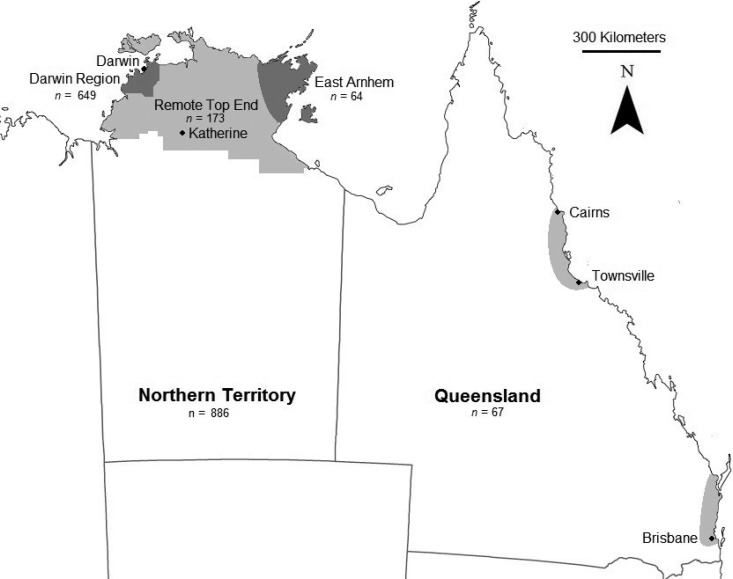
Nine-hundred fifty-three B. pseudomallei isolates from Australia (779 clinical isolates, including 736 human plus 43 animal isolates, and 174 environmental strains), collected between 1989 and 2012, were analyzed in this study (data set 1). This data set constitutes almost all isolates from Australia that have been submitted to the B. pseudomallei MLST database (http://bpseudomallei.mlst.net/), with the exception of isolates from Western Australia, where strain numbers were too low to provide statistical power. Isolates from the Top End of the Northern Territory (see Fig. 1) represented 886 of the 953 isolates and were cultured and curated at the Menzies School of Health Research. Northern Territory clinical isolates were obtained from the ongoing Darwin Prospective Melioidosis Study, which commenced in October 1989 and has included all known cases of melioidosis in the tropical Top End of the Northern Territory over the past 24 years, with B. pseudomallei isolates stored in over 95% of cases. Most cases originate from the urban Darwin region and surrounding rural Darwin locations, but cases also occur in remote, small, indigenous communities across the vast area of the tropical north of the Northern Territory and in the smaller regional towns of Katherine and Gove (2). In addition, our group and others have been actively collecting B. pseudomallei isolates from environmental and animal sources across Australia for various projects over the past few decades. The remaining 67 isolates (7%) were obtained from James Cook University or from collaborators in Southeast Queensland and were collected between 1961 and 2003 from two distinct Queensland regions (see Fig. 1). Nonparametric permutational analysis of similarity (ANOSIM) found no significant difference between the two sampling regions of Queensland (global R = 0.04, P = 0.7); therefore, Queensland isolates were regarded as a single group to bolster sample size. B. pseudomallei isolates were cultured using a modified Ashdown's broth (17). DNA extraction (9) and MLST (18) were performed as previously described. Existing and novel sequence types (STs) used in this study are included (see data set S1 in the supplemental material); these data can also be found at the B. pseudomallei MLST database (http://bpseudomallei.mlst.net/).
FIG 1.
Origin of Burkholderia pseudomallei isolates used in this study. (Adapted from a map from the University of Melbourne Library Map Collection [http://www.lib.unimelb.edu.au/collections/maps/digital/outline-maps/aust-l.gif].)
Statistical analysis.
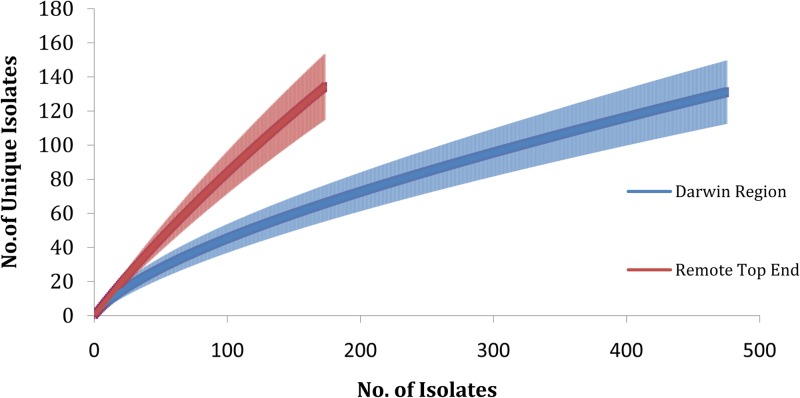
B. pseudomallei diversity and variance measures were calculated in the Primer-E v6 software package (Primer-E Ltd., United Kingdom) based on a Manhattan resemblance matrix of presence/absence data of STs. Diversity measures included Simpson's diversity index (D), Pielou's evenness, and the average contribution of an ST to the Sorensen group similarity, implemented in SIMPER, a module of Primer-E. The average contribution is based on ST group similarity averaged across all isolate pairs in a group over its standard deviation. The group similarity is calculated through the Sorensen index, which in this case equals 100 if both isolates share the same ST or 0 if they are different. D values approaching 1 indicate that the diversity of the ST population under examination is high; in other words, there is a greater chance of identifying a novel ST in regions with higher D values. Evenness values approaching 1 indicate that the ST population under examination is evenly distributed; i.e., there is no ST bias. The ANOSIM module was used to test the null hypothesis of no difference in ST composition with the generated dissimilarity metrics between geographic groups. A rarefaction index was calculated using EstimateS (v9.1.0; USA) to determine the relative diversity of STs captured in the Darwin region and Remote Top End. These two regions vary substantially in both geographic size and human population density, which has resulted in a greater number of isolates being obtained from the Darwin region (n = 473 versus n = 173 for the Remote Top End). Despite this difference, the rarefaction curve is not subject to bias based on the use of different isolate numbers from each cohort. EstimateS automatically performs 999 permutations (rearrangements) of each data set, which are then used to calculate the consensus curve and the corresponding 95% confidence intervals (CI). Therefore, random down-sampling of the Darwin isolates to match the Remote Top End (i.e., n = 173) would not alter the curve shown in Fig. 3.
FIG 3.
Rarefaction curves reflecting the sampling effort of Burkholderia pseudomallei isolates compared to the sequence type diversity for the Darwin region and Remote Top End. Vertical bars extending from each curve represent 95% confidence intervals.
For the Splits network and Bayesian cluster analysis, the seven-locus allelic data for each ST was concatenated and used as input. The Neighbor-Net function (SplitsTree v4.13.1; Germany) (19) was used to create a dendrogram representing the Splits network output. Bootstrap analysis was conducted with 1,000 resamplings of the data. Bayesian population structure analysis (BAPS) was used to assign isolates to populations without prior knowledge of the number of expected groups (BAPS v6; Finland). The set parameters were consistent with those of previous studies working with B. pseudomallei MLST data in BAPS (15).
RESULTS AND DISCUSSION
Recent phylogenetic studies of B. pseudomallei populations have had varied success in evaluating the biogeography of this bacterium. To date, the only clear separation of B. pseudomallei genotypes into distinct geographic populations has been between Australian and Southeast Asian strains (6, 7, 15). Identifying B. pseudomallei population structure on a smaller geographic scale, such as within Australia, is important not only for understanding the ecology and evolution of this bacterium but also for identifying the putative origin of B. pseudomallei in the unlikely event that an Australian isolate is bioweaponized. Compounding this issue is the high rate of recombination of B. pseudomallei relative to its rate of mutation (14), which can affect patterns of relatedness when molecular typing methods that are based on a limited snapshot of the genome, such as MLST, are used. High rates of lateral gene transfer have been shown to disrupt phylogenetic signals, leading to homoplasy, and have led to misinterpretations in strain relatedness and a lack of robustness when clustering algorithms such as eBURST or traditional phylogenetic tools (e.g., maximum likelihood and neighbor-joining cladograms) are used (7, 16).
In the present study, we have utilized a well-curated, large collection of Australian B. pseudomallei isolates and associated MLST data to examine the diversity and structure of the Australian B. pseudomallei population. Given the high number of B. pseudomallei isolates collected from both clinical cases and the environment in the Darwin region (n = 649) (Fig. 1), the diversity and relative abundances between strains of clinical and environmental origins were determined with a view to potentially identify a subset of STs overrepresented in clinical disease. No significant differences in evenness or diversity (D) were found between clinical and environmental STs (Table 1), with the same STs equally abundant in clinical and environmental groups. Therefore, the relative abundances and composition of environmental STs in Darwin are directly correlated with the ST population associated with clinical disease. Intriguingly, this finding implies that all culturable environmental B. pseudomallei STs in Darwin appear potentially capable of causing disease. It remains to be determined whether this phenomenon is observed in other regions where melioidosis is endemic.
TABLE 1.
Diversity measures of B. pseudomallei populations in different geographic regions of northern Australia, based on ST abundance data
| Comparison of geographic locations or types of samplesa | No. of isolates (no. of unique STs) | Db | Evennessc | ANOSIMd global R value (P value) |
|---|---|---|---|---|
| Northern Territory | 712 (279) | 0.968 | 0.808 | 0.032 (<0.001) |
| Queensland | 67 (43) | 0.968 | 0.924 | |
| Darwin Region | 475 (131) | 0.947 | 0.776 | 0.035 (<0.001) |
| Remote Top End | 173 (134) | 0.995 | 0.968 | |
| Remote Top End | 173 (134) | 0.995 | 0.968 | 0.004 (0.200) |
| East Arnhem | 64 (36) | 0.987 | 0.967 | |
| Clinical (Darwin region) | 475 (131) | 0.947 | 0.776 | 0.002 (0.297) |
| Environmental (Darwin Region) | 174 (62) | 0.936 | 0.827 |
Isolates from the compared geographic locations were of clinical origin only.
Simpson's diversity index (D) was calculated with the 1 − lambda method.
By Pielou's evenness (J′) calculation, J′ = H/log(S), with H being the Shannon-Wiener diversity index and S being the ST richness, i.e., the total number of STs.
By ANOSIM, R = (r̄B − r̄W)/[n(n − 1)]/2, where r̄W is the average of rank similarities within groups, r̄B is the average of rank similarities between groups, and n is the number of isolates.
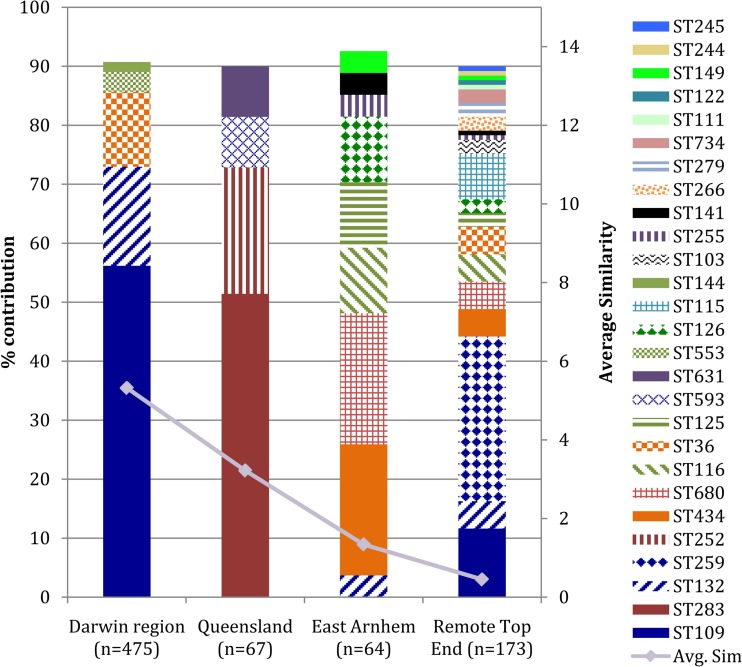
Given the lack of a significant difference between clinical and environmental STs in Darwin, the region-specific clinical and environmental data sets were combined to increase sample size in phylogenetic tests (BAPS and SplitsTree). Based on clinical notes and patient interviews, detailed epidemiological information on the origin of infection in the selected clinical cases allowed robust assignments of clinical isolates to their probable geographical origin (2). Using this approach, we examined diversity metrics for B. pseudomallei isolates of clinical origin from the Northern Territory and Queensland and within the Northern Territory, specifically, the Darwin region, East Arnhem, and other remote areas of the Top End (Fig. 1). The D value, which is dependent on the abundance of dominant STs (20) and indicates the probability that two isolates selected at random will have different STs, was found to be high for all regions. D ranged from 0.947 for the Darwin region to 0.995 for the Remote Top End (Table 1), indicating respective 94.7% and 99.5% probabilities that any two randomly selected isolates would yield different STs. In contrast, the Pielou evenness measure is sensitive to the addition of rare species, particularly in a sample set with a low overall number of STs (20). The Darwin region showed the lowest evenness measure of 0.776, indicating an overrepresentation of a few STs compared with those in the other regions, which had evenness values of >0.920 (P < 0.001) (Table 1). However, as the evenness measure is sensitive to a potential underestimate of the ST richness (21), this measure might not accurately reflect the ST diversity in areas with relatively small sample numbers, such as Queensland and East Arnhem. Therefore, the relative contribution of each ST for each region to the average group similarity was investigated, as shown in Fig. 2. In the Darwin region, five STs comprised 90% of the overall abundance. Similarly, the Queensland and East Arnhem regions showed four and nine STs, respectively, comprising 90% of the overall abundance. In contrast, 21 STs in the Remote Top End made up 90% of the overall abundance (Fig. 2). This analysis revealed a clear trend of diversity with regions from smallest to largest (Darwin, Queensland, East Arnhem, and Remote Top End), showing an increasing number of STs contributing to within-group similarity (Fig. 2). Nonparametric ANOSIM also confirmed that the ST compositions between the Darwin region and Remote Top End were significantly different (Table 1).
FIG 2.
Relative contribution of each Burkholderia pseudomallei sequence type (ST) to the average group similarity in various geographic regions across the Northern Territory and Queensland, Australia. The relative contribution of an ST is averaged across all pairs of isolates within a group over its standard deviation. The Bray-Curtis (or Sorensen) similarity of the ith ST between the jth and kth isolates [Sjk(i)] equals 100[2min(yij,yik)/Σi=1p(yij, yik)] or, simply, it is 100 if both isolates share the same ST or 0 if they do not. The more abundant an ST is in a group, the higher its contribution to the within-group similarity. The combined STs shown make up approximately 90% of the overall ST contribution for each sampled geographic region. As shown by the average group similarity (gray line), the larger the region sampled, the smaller the group similarity and greater the ST diversity.
These data indicate that geographic sampling size might be a critical metric when assessing population diversity in regions where B. pseudomallei is endemic. In other words, as the sampling area expands, an increasing number of STs are identified. Collectively, these findings suggest that B. pseudomallei populations in the environment in northern Australia are generally localized and are not frequently disseminated over a large area, such as through anthropogenic influences or from large-scale environmental disturbances, such as tropical cyclones.
Given the large number of samples collected in the Darwin and Remote Top End regions, we were interested in determining the effectiveness of our sampling efforts in capturing actual population diversity. A rarefaction curve of ST diversity as a function of sampling effort indicated that the ST diversity in the Darwin region is approaching saturation. In comparison, the rarefaction curve for the Remote Top End is not yet approaching saturation, indicating that there is a much larger diversity of STs within this region yet to be identified (Fig. 3). The 95% confidence intervals of the ST diversity for the Darwin region and Remote Top End do not overlap when calculated with the same number of samples (n = 173) (Fig. 3), indicating that the levels of diversity of B. pseudomallei STs between these regions are different (Darwin region 95% CI, 55 to 76, versus Remote Top End 95% CI, 115 to 153). Therefore, unlike in the Darwin region, the diversity of B. pseudomallei in the Remote Top End has not yet been fully explored, an observation that is consistent with the much larger geographic size of the Remote Top End. This finding has implications from a biodefense perspective, as currently it would be difficult to trace the origin of a novel B. pseudomallei strain originating from this region. More-intensive sampling efforts are required in the Remote Top End region to address this knowledge gap.
The final goal of the study was to examine different methods for identifying population structure in our Australian B. pseudomallei ST data set. First, a network algorithm method was tested due to its ability to account for the effects of recombination, which can otherwise hinder interpretation of phylogenetic results (22). Network analysis of B. pseudomallei STs using the SplitsTree network analysis found no evidence for clustering of populations within geographic regions (see Fig. S1 in the supplemental material). In addition, all nodes received low bootstrap support. Our findings show that network analysis of B. pseudomallei STs, similarly to neighbor-joining analysis (7), is not a useful method for identifying population structure of this bacterium in Australia.
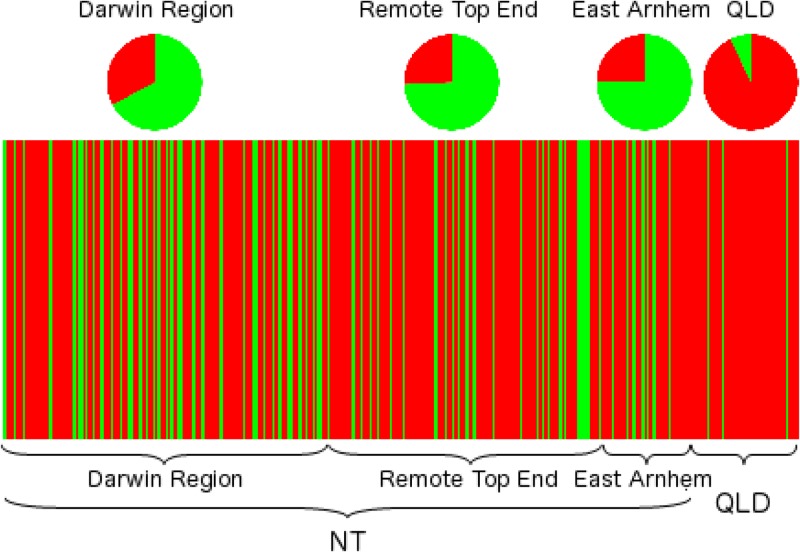
Next, we tested a Bayesian population structure analysis approach, as this method has been used successfully to identify population structure among global B. pseudomallei isolates (14, 15). Using BAPS, the ideal number of populations (K) was determined in the data set. A population number of 2 was found to consistently result in the highest log marginal likelihood for 10 out of 10 best-visited partitions, consistent across 20 repeated analyses. Isolates with >90% probability of belonging to population 1 consisted of 60%, 68%, 69%, and 7% of the isolates from the Darwin, Remote Top End, East Arnhem, and Queensland regions, respectively (Fig. 4). The majority (93%) of the Queensland isolates were placed into population 2 (red), whereas the majority of the Darwin region, Remote Top End, and East Arnhem isolates were placed into population 1 (green). A divergence in the ST compositions between the Top End and Queensland was also confirmed by a significant ANOSIM outcome (P = 0.001) refuting the null hypothesis that there are no differences in ST composition between these two regions. Overall, the BAPS and ANOSIM results confirm previous findings (7) that the Northern Territory and Queensland have unique B. pseudomallei populations.
FIG 4.
Burkholderia pseudomallei population structure based on Bayesian analysis of MLST-concatenated sequences. Each colored vertical line represents a B. pseudomallei isolate. Green, population 1; red, population 2. Isolates are in order of location, as indicated by the labeled braces. NT, Northern Territory; QLD, Queensland.
Despite these results, we recognize that using MLST for definitive B. pseudomallei source attribution should be met with caution. Recently, five clinical isolates from islands in the Torres Strait region of Queensland were shown to overlap STs from the Northern Territory (STs 109, 255, 468, 470, and 594) (23). Although these STs have not yet been isolated from the environment in the Torres Strait and the travel history of these patients to the Northern Territory was not ascertained, the highly recombinogenic nature of the B. pseudomallei genome means that overlapping STs between geographically distinct regions is an eventual inevitability. Such cases cannot be resolved using MLST alone, and higher-resolution methods, such as whole-genome sequencing, are needed to deduce the true evolutionary relatedness of these isolates. Nevertheless, our study demonstrates the utility of MLST data for narrowing down the probable geographic origins of isolates within mainland Australia.
Conclusions.
This study has taken advantage of existing MLST data available for a large number of Australian B. pseudomallei strains to address several knowledge gaps with regard to the biogeography of this bacterium in northern Australia. First, we show that there is no significant difference between the environmental and clinical B. pseudomallei ST populations in the Northern Territory, with the implication that any culturable strain has the potential to cause disease. Second, our data indicate that the ST diversity in the populated Darwin region is similar to those of less populated geographic regions of similar size. Third, the diversity of B. pseudomallei populations increased with the geographic size being sampled, implying that this organism is ecologically established as localized populations that are not subject to frequent, widespread dissemination. Fourth, we found that there remains untapped diversity in the Remote Top End region, with the observed diversity and evenness of STs across remote areas of the Northern Territory and Queensland supporting the ancient history of B. pseudomallei in Australia (14). Finally, we show that Bayesian analysis of the MLST data is a useful tool for assessing the biogeography of B. pseudomallei in northern Australia, with BAPS analysis suggesting that there are distinct B. pseudomallei populations between the Northern Territory and Queensland that are undetectable using neighbor-joining or SplitsTree analyses. Our study is an important addition to our understanding of B. pseudomallei ecology in the region of endemicity of northern Australia and, more broadly, provides a framework for source attribution analysis in the unlikely event of B. pseudomallei bioweaponization.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by funding from the Australian National Health and Medical Research Council (project grants 605820 and 1046812), the Menzies School of Health Research, and Charles Darwin University.
We thank our colleagues in the microbiology laboratory at Royal Darwin Hospital for their support and expertise in identifying B. pseudomallei. Statistical support was provided by Mark Chatfield, based at Menzies School of Health Research.
Footnotes
Published ahead of print 21 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00128-14.
REFERENCES
- 1.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N. Engl. J. Med. 367:1035–1044. 10.1056/NEJMra1204699 [DOI] [PubMed] [Google Scholar]
- 2.Currie BJ, Ward L, Cheng AC. 2010. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl. Trop. Dis. 4:e900. 10.1371/journal.pntd.0000900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parameswaran U, Baird RW, Ward LM, Currie BJ. 2012. Melioidosis at Royal Darwin Hospital in the big 2009–2010 wet season: comparison with the preceding 20 years. Med. J. Aust. 196:345–348. 10.5694/mja11.11170 [DOI] [PubMed] [Google Scholar]
- 4.Limmathurotsakul D, Kanoksil M, Wuthiekanun V, Kitphati R, deStavola B, Day NP, Peacock SJ. 2013. Activities of daily living associated with acquisition of melioidosis in northeast Thailand: a matched case-control study. PLoS Negl. Trop. Dis. 7:e2072. 10.1371/journal.pntd.0002072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng AC, Godoy D, Mayo M, Gal D, Spratt BG, Currie BJ. 2004. Isolates of Burkholderia pseudomallei from northern Australia are distinct by multilocus sequence typing, but strain types do not correlate with clinical presentation. J. Clin. Microbiol. 42:5477–5483. 10.1128/JCM.42.12.5477-5483.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vesaratchavest M, Tumapa S, Day NP, Wuthiekanun V, Chierakul W, Holden MT, White NJ, Currie BJ, Spratt BG, Feil EJ, Peacock SJ. 2006. Nonrandom distribution of Burkholderia pseudomallei clones in relation to geographical location and virulence. J. Clin. Microbiol. 44:2553–2557. 10.1128/JCM.00629-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng AC, Ward L, Godoy D, Norton R, Mayo M, Gal D, Spratt BG, Currie BJ. 2008. Genetic diversity of Burkholderia pseudomallei isolates in Australia. J. Clin. Microbiol. 46:249–254. 10.1128/JCM.01725-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trakulsomboon S, Dance DA, Smith MD, White NJ, Pitt TL. 1997. Ribotype differences between clinical and environmental isolates of Burkholderia pseudomallei. J. Med. Microbiol. 46:565–570. 10.1099/00222615-46-7-565 [DOI] [PubMed] [Google Scholar]
- 9.Currie B, Gal D, Mayo M, Ward L, Godoy D, Spratt B, LiPuma J. 2007. Using BOX-PCR to exclude a clonal outbreak of melioidosis. BMC Infect. Dis. 7:68. 10.1186/1471-2334-7-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng AC, Day NP, Mayo MJ, Gal D, Currie BJ. 2005. Burkholderia pseudomallei strain type, based on pulsed-field gel electrophoresis, does not determine disease presentation in melioidosis. Microbes Infect. 7:104–109. 10.1016/j.micinf.2004.08.020 [DOI] [PubMed] [Google Scholar]
- 11.Currie BJ, Haslem A, Pearson T, Hornstra H, Leadem B, Mayo M, Gal D, Ward L, Godoy D, Spratt BG, Keim P. 2009. Identification of melioidosis outbreak by multilocus variable number tandem repeat analysis. Emerg. Infect. Dis. 15:169–174. 10.3201/eid1502.081036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liguori AP, Warrington SD, Ginther JL, Pearson T, Bowers J, Glass MB, Mayo M, Wuthiekanun V, Engelthaler D, Peacock SJ, Currie BJ, Wagner DM, Keim P, Tuanyok A. 2011. Diversity of 16S-23S rDNA internal transcribed spacer (ITS) reveals phylogenetic relationships in Burkholderia pseudomallei and its near-neighbors. PLoS One 6:e29323. 10.1371/journal.pone.0029323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, Currie BJ, Wagner DM, Keim P. 2008. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics 9:566. 10.1186/1471-2164-9-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, Price EP, Glass MB, Leadem B, Beckstrom-Sternberg JS, Allan GJ, Foster JT, Wagner DM, Okinaka RT, Sim SH, Pearson O, Wu Z, Chang J, Kaul R, Hoffmaster AR, Brettin TS, Robison RA, Mayo M, Gee JE, Tan P, Currie BJ, Keim P. 2009. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 7:78. 10.1186/1741-7007-7-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dale J, Price EP, Hornstra H, Busch JD, Mayo M, Godoy D, Wuthiekanun V, Baker A, Foster JT, Wagner DM, Tuanyok A, Warner J, Spratt BG, Peacock SJ, Currie BJ, Keim P, Pearson T. 2011. Epidemiological tracking and population assignment of the non-clonal bacterium, Burkholderia pseudomallei. PLoS Negl. Trop. Dis. 5:e1381. 10.1371/journal.pntd.0001381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30. 10.1186/1471-2180-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashdown LR, Clarke SG. 1992. Evaluation of culture techniques for isolation of Pseudomonas pseudomallei from soil. Appl. Environ. Microbiol. 58:4011–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068–2079. 10.1128/JCM.41.5.2068-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryant D, Moulton V. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255–265. 10.1093/molbev/msh018 [DOI] [PubMed] [Google Scholar]
- 20.Hubalek Z. 2000. Measures of species diversity in ecology: an evaluation. Folia Zool. 49:241–260 [Google Scholar]
- 21.Alatalo RV. 1981. Problems in the measurement of evenness in ecology. Oikos 37:199–204 [Google Scholar]
- 22.Hanage WP, Fraser C, Spratt BG. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3:6. 10.1186/1741-7007-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker A, Mayo M, Owens L, Burgess G, Norton R, McBride WJ, Currie BJ, Warner J. 2013. Biogeography of Burkholderia pseudomallei in the Torres Strait Islands of northern Australia. J. Clin. Microbiol. 51:2520–2525. 10.1128/JCM.00418-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.