Abstract
Due to the limited sensitivities of stool-based microscopy and/or culture techniques for Strongyloides stercoralis, the detection of antibodies to this intestinal nematode is relied upon as a surrogate for determining exposure status or making a diagnosis of S. stercoralis infection. Here, we evaluated three immunoassays, including the recently released InBios Strongy Detect IgG enzyme-linked immunosorbent assay (ELISA) (InBios International, Inc., Seattle, WA), the SciMedx Strongyloides serology microwell ELISA (SciMedx Corporation, Denville, NJ), and the luciferase immunoprecipitation system (LIPS) assay performed at the National Institutes of Health (NIH), for their detection of IgG antibodies to S. stercoralis. A total of 101 retrospective serum samples, previously submitted for routine S. stercoralis antibody detection using the SciMedx assay, were also evaluated by the InBios and LIPS assays. The qualitative results from each assay were compared using a Venn diagram analysis, to the consensus result among the three assays, and each ELISA was also evaluated using the LIPS assay as the reference standard. By Venn diagram analysis, 65% (66/101) of the samples demonstrated perfect agreement by all three assays. Also, the numbers of samples considered positive or negative by a single method were similar. Compared to the consensus result, the overall percent agreement of the InBios, SciMedx, and LIPS assays were comparable at 87.1%, 84.2%, and 89.1%, respectively. Finally, the two ELISAs performed analogously but demonstrated only moderate agreement (kappa coefficient for the two assays, 0.53) with the LIPS assay. Collectively, while the two commercially available ELISAs perform equivalently, neither should be used independently of clinical evaluation to diagnose strongyloidiasis.
INTRODUCTION
Strongyloides stercoralis, a soil-transmitted intestinal nematode and the causative agent of strongyloidiasis, is widely endemic in both tropical and subtropical climates throughout Africa, Asia, and South America (1). Extension into temperate regions has also been reported, including pockets of endemicity in the southeastern United States (2, 3). Given its wide geographical distribution, S. stercoralis has been estimated to infect as many as 100 million people worldwide and is therefore an important infection to consider in patients who either reside in or have traveled to regions that are endemic for strongyloidiasis (4, 5).
S. stercoralis has a complicated life cycle, and infection can be asymptomatic, present with gastrointestinal symptoms, or in more severe cases, progress to disseminated disease. Briefly, the skin is penetrated by infective S. stercoralis L3 filariform larvae via direct exposure to contaminated soil. Initial infection can lead to pruritus and irritation at the site of entry (larva currens), typically along the lower extremities (6). Subsequent hematogenous dissemination to the lungs, migration up the bronchial tree, and passage into the gastrointestinal tract may present clinically as respiratory symptoms, diarrhea, and/or abdominal pain (6, 7). S. stercoralis is unique among the intestinal nematodes because of its ability to mature into the infective filariform stage without leaving the gastrointestinal tract. This creates the potential for continuous reinfection and “hyperinfection syndrome,” a potentially life-threatening condition, particularly among immunosuppressed individuals (7–10). As many cases of strongyloidiasis are subclinical and can persist for decades following exposure, a significant number of patients with undiagnosed strongyloidiasis are at risk for hyperinfection once initiated on immunosuppressive regimens (6, 11). Therefore, accurate diagnostic modalities are needed for both the diagnosis of symptomatic strongyloidiasis and the identification of asymptomatic infections in high-risk individuals prior to receiving immunosuppressants.
Diagnosing strongyloidiasis is particularly challenging. Unlike most other intestinal helminths, S. stercoralis does not produce characteristic ova within the intestinal tract, and therefore, direct observation of the larva is required. Classic techniques to identify Strongyloides include larval concentration from fecal specimens prior to microscopic examination, and agar plate culture of fresh stool specimens along with daily plate inspections for the presence of bacterial trails left by motile S. stercoralis larva. However, due to sporadic larval shedding and generally low larval concentrations, particularly among chronically infected individuals, the diagnostic sensitivities of these direct detection methods from single stool specimens are low (30 to 50%), and repeat sampling (up to seven specimens) may be necessary prior to ruling out infection (12–14). Additionally, these methods require prompt submission of fresh stool specimens to the laboratory, which may be impractical in some cases, and as S. stercoralis is infective on contact, the manipulation of this nematode poses a significant infection risk to laboratory personnel.
Given the limitations of these traditional techniques, serologic approaches to detect an immune response to S. stercoralis have emerged as valuable alternative diagnostic tools. Several different serologic based assays have been described, and while the majority detect anti-Strongyloides IgG, they differ in the antigen targets used for detection (crude lysate versus purified or recombinant proteins), in the applied methodology (enzyme-linked immunosorbent assays [ELISAs], dipstick methods, or luciferase immunoprecipitation systems [LIPS]), and whether the assay is commercially available or laboratory-developed test (4, 15–19). In cases of proven strongyloidiasis, the sensitivities of these serologic assays vary from 73% to 100%, with false negatives noted in immunosuppressed individuals. Specificity is likewise inconsistent (29% to 100%) between methods, with cross-reactions occurring primarily in patients with prior filarial infections (17, 18, 20, 21). Among these assays, the National Institutes of Health (NIH) LIPS method presents the highest combined sensitivity and specificity (97% and 100%, respectively) for the detection of anti-S. stercoralis antibodies (16, 17, 19).
The purpose of this study was to evaluate the performance of the recently released InBios Strongy Detect IgG ELISA (InBios International, Inc., Seattle, WA) compared to those of both the commercially available SciMedx Strongyloides serology microwell ELISA (SciMedx Corporation, Denville, NJ) and the LIPS assay performed at the NIH.
MATERIALS AND METHODS
Study overview.
One hundred one serum samples submitted to ARUP Laboratories (Salt Lake City, UT) for clinical testing to detect IgG-class antibodies to S. stercoralis by the SciMedx Strongyloides serology microwell ELISA (SciMedx Corporation, Denville, NJ) were retrospectively retrieved, tested by the InBios Strongy Detect IgG ELISA (InBios International, Inc., Seattle, WA) in our laboratory, and were also forwarded to the Parasitic Diseases Laboratory at the NIH for testing by the luciferase immunoprecipitation system (LIPS) assay. The selection of samples for this panel was based on (i) sufficient volume for analysis by all three methods and (ii) an approximately equal number of positive and negative specimens (determined as such by the SciMedx assay). The samples were deidentified, and clinical information (e.g., presentation, duration of symptoms, travel, and exposure history) was not available. An additional 20 serum samples, 10 from patients with confirmed strongyloidiasis by stool culture and 10 from healthy controls, were also tested by the InBios and LIPS assays. This study was approved by the institutional review board at the Mayo Clinic.
InBios Strongy Detect IgG ELISA.
The InBios Strongy ELISA is a one-step sandwich-format immunoassay for qualitative detection of IgG-class antibodies to the Strongyloides recombinant NIE antigen (SRA) in serum. Testing was performed on the Triturus automated enzyme immunoassay (EIA) analyzer (Grifols, Miami, FL) at the Mayo Clinic, according to the manufacturer's instructions. Briefly, the patient serum samples were diluted in a 1:200 ratio in dilution buffer, and 100 μl of the dilution was added into wells precoated with SRA. Following a 30-min incubation at 37°C, the wells were washed and incubated with a horseradish peroxidase-conjugated secondary antibody to detect bound anti-Strongyloides IgG-class antibodies. After repeat incubation and washing, 100 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) was added to the wells and incubated in the dark at room temperature (RT) for 10 min, followed by the addition of 50 μl of stop solution. The optical density (OD) was measured at a wavelength of 450/630 nm. The immune status ratio (ISR) for each sample was calculated as the ratio of the OD obtained with the test sample divided by the calculated cutoff value as determined by the cutoff control sample. ISR values of >1.1, 0.9 to 1.1, and <0.9 were interpreted as positive, equivocal, and negative, respectively, for the presence of IgG-class antibodies to S. stercoralis, as per the manufacturer's recommendations. This assay is currently labeled for research use only.
SciMedx Strongyloides serology microwell ELISA.
The SciMedx Strongyloides ELISA is a one-step sandwich-format immunoassay for the qualitative detection of IgG-class antibodies to Strongyloides antigen. Testing was performed manually at ARUP. Briefly, serum was diluted at a 1:64 ratio in dilution buffer, and 100 μl was pipetted into microtiter wells coated with Strongyloides antigen and incubated at RT for 10 min. The wells were washed three times, and 2 drops of the supplied horseradish peroxidase-conjugated protein A was added and incubated for 5 min at RT. The wells were washed three times and 2 drops of TMB was added and incubated for 5 min at RT, followed by the addition of 2 drops of stop solution. The optical density was subsequently measured at a wavelength of 450/620 to 650 nm and divided by the OD of a calibrator to give an index value (IV). IV values of <1.5, 1.5 to 2.1, and >2.10 were considered negative, equivocal, and positive, respectively, for the presence of IgG-class antibodies to S. stercoralis. These cutoff values were established by ARUP using a panel of clinically defined patients with strongyloidiasis as well as healthy volunteers. This assay is currently labeled for research use only.
Luciferase immunoprecipitation system (LIPS) assay.
The LIPS assay was performed using shorter incubation times than those described previously (17). The assay detects antibodies against two antigens, the NIE, a 31-kDa recombinant antigen from the S. stercoralis L3 larva, and the S. stercoralis immunoreactive antigen (SsIR); each was fused to the Renilla luciferase (Ruc) and expressed and purified from COS cells. Briefly, patient serum was diluted (1:15) in buffer A and added to 50 μl of 1 × 107 luminometer units (LU) of each purified Ruc-antigen fusion construct. The reaction mixtures were incubated for 10 min on a rotary shaker. Subsequently, antigen-serum reaction mixtures and 7 μl of a 30% suspension of polyprotein A/G beads in phosphate-buffered saline (PBS) (Pierce Biotechnology, Rockford, IL) were added to a 96-well filter high-throughput-screening (HTS) plate and incubated for 10 min at room temperature on a rotary shaker. Following incubation, the protein A/G beads were isolated, washed, incubated with a coelenterazine substrate mix (Promega, Madison, WI), and the LU were determined using a Berthold Centro LB 960 microplate luminometer. Results of <711 LU, 711 to 2,000 LU, and >2,000 LU were considered negative, low positive, and positive, respectively. For the purposes of this study, any result of ≥711 LU was considered positive.
Statistical analysis.
Each of the three immunoassays was evaluated against the consensus result, which was defined as the result obtained by at least two of the three assays. Also, the performances of the two ELISA methods were compared, using the NIH LIPS assay as the reference standard. GraphPad software was used to calculate kappa values (κ), the positive, negative, and overall percent agreement values, and the associated 95% confidence intervals (GraphPad, La Jolla, CA). Overall agreement as determined by κ was interpreted as follows: near perfect, 0.81 to 1.0; substantial, 0.61 to 0.8; moderate, 0.41 to 0.6; fair, 0.21 to 0.4; slight, 0 to 0.2; and poor, <0 (22).
RESULTS
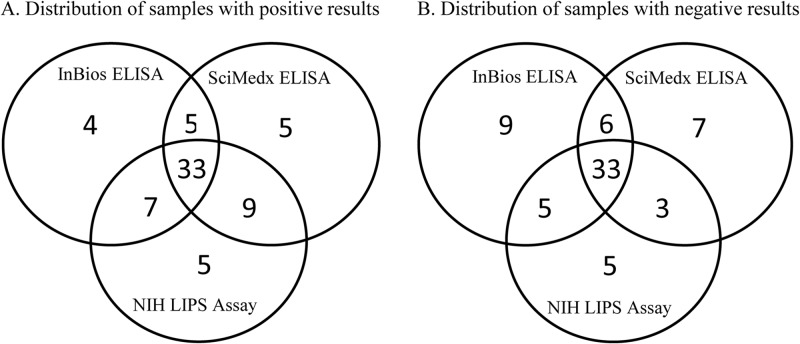
The distribution of positive and negative results by the InBios, SciMedx, and LIPS immunoassays for detection of IgG antibodies to S. stercoralis is shown in the Venn diagram in Fig. 1. Equivocal results by the InBios (n = 2) and SciMedx (n = 6) ELISAs were considered negative for this evaluation. All three assays were in perfect agreement for 65% of the samples (66/101; 33 positive and 33 negative). Also, a similar number of samples were either positive (4 by InBios, 5 by SciMedx, and 5 by LIPS) or negative (9 by InBios, 7 by SciMedx, and 5 by LIPS) by only one of the three assays.
FIG 1.
Venn diagram comparing results from the InBios ELISA, the SciMedx ELISA, and the LIPS immunoassay for detection of IgG-class antibodies to S. stercoralis.
Compared to the consensus result, defined as the result obtained from at least two of the three assays, all three methods showed comparable overall percent agreement, ranging from 84.2% (88/101) for the SciMedx assay to 89.1% (90/101) for the LIPS assay (Table 1). However, the SciMedx ELISA showed a notably lower negative agreement (82.6%) and kappa value (0.68) than those of the InBios ELISA (91.3% and 0.74, respectively) and LIPS assay (89.1% and 0.78, respectively). The performance characteristics of the InBios and SciMedx ELISAs were also evaluated using the NIH LIPS assay as the reference standard (Table 2). The two ELISAs demonstrated equivalent performance characteristics, including identical kappa values (0.53), collectively indicating only moderate correlation with the NIH LIPS method.
TABLE 1.
Comparison of the InBios, SciMedx, and LIPS assays to the consensus result for detection of IgG class antibodies to S. stercoralis
| Assay type | Assay result | Consensus result (no.)a |
% agreement (95% CIb) |
Kappa (95% CI) | |||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Overall | Positivec | Negatived | |||
| InBios Strongy Detect ELISA | Positive | 46 | 3 | 87.1(79.1–92.5) | 83.6 (71.5–91.4) | 91.3 (79.1–97.1) | 0.74 (0.61–0.87) |
| Negative | 8 | 42 | |||||
| Equivocal | 1 | 1 | |||||
| SciMedx Strongyloides ELISA | Positive | 47 | 5 | 84.2 (75.7–90.1) | 85.5 (73.6–92.7) | 82.6 (69.0–91.2) | 0.68 (0.54–0.82) |
| Negative | 5 | 38 | |||||
| Equivocal | 3 | 3 | |||||
| LIPS assay | Positive | 49 | 5 | 89.1 (81.4–94.0) | 89.1 (77.8–95.3) | 89.1 (76.5–95.7) | 0.78 (0.66–0.90) |
| Negative | 6 | 41 | |||||
The reference standard was defined as the result obtained from ≥2 of the 3 immunoassays.
CI, confidence interval.
Equivocal results were counted as negative.
Equivocal results were counted as positive.
TABLE 2.
Comparison of the InBios and SciMedx ELISAs to the NIH LIPS assay for detection of IgG-class antibodies to S. stercoralis
| ELISA used | Assay result | NIH LIPS assay result (no.) |
% agreement (95% CI) |
Kappa (95% CI) | |||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Overall | Positive | Negative | |||
| InBios Strongy Detect | Positive | 40 | 9 | 76.2 (67.0–83.5) | 74.1 (61.0–84.0) | 78.7 (64.9–88.2) | 0.53 (0.36–0.69) |
| Negative | 13 | 37 | |||||
| Equivocal | 1 | 1 | |||||
| SciMedx Strongyloides | Positive | 42 | 10 | 77.2 (68.1–84.4) | 77.8 (64.9–87.0) | 76.6 (62.6–86.7) | 0.53 (0.36–0.69) |
| Negative | 7 | 36 | |||||
| Equivocal | 5 | 1 | |||||
Finally, the InBios ELISA was also evaluated using serum samples from 20 patients that were either positive (n = 10) or negative (n = 10) by stool culture for S. stercoralis (Table 3). Compared to culture, the InBios assay showed a sensitivity of 80% (8/10) and a specificity of 90% (9/10). Among the three discordant samples, one was culture positive/InBios ELISA negative, and the remaining two samples were equivocal by the InBios ELISA. The LIPS assay showed 100% sensitivity and specificity using these specimens (data not shown), and the volumes were not sufficient for testing by the SciMedx ELISA.
TABLE 3.
Comparison of the InBios Strongy Detect ELISA and Strongyloides stool culturea
| InBios Strongy Detect ELISA result |
Strongyloides stool culture (NIH panel) result (no.) |
|
|---|---|---|
| Positive | Negative | |
| Positive | 8 | 0 |
| Negative | 1 | 9 |
| Equivocal | 1 | 1 |
Compared to culture, the sensitivity of the InBios Strongy Detect ELISA was 80% (8/10) (95% CI, 47.9 to 95.4%), the specificity was 90% (9/10) (95% CI, 57.4 to 100%), and the agreement was 85% (17/20) (95% CI, 63.1 to 95.6%).
DISCUSSION
The diagnosis of strongyloidiasis is challenging due to the insensitivity of standard parasitologic methods, including agar plate stool culture and microscopic detection of larva following concentration. While serial collection and evaluation of stool samples significantly improve the sensitivity of these methods, few local clinical laboratories continue to offer these classic assays, and the requirement of unpreserved stool specimens precludes utilization by reference laboratories that still perform Strongyloides culture (4, 14). Additionally, these methods require hours to days to complete and pose a risk for laboratory-acquired strongyloidiasis. Serologic assays detecting anti-S. stercoralis antibodies have therefore largely supplanted these traditional techniques. They offer the convenience of being commercially produced, utilize methods familiar to most laboratories, are rapid, and require a readily accessible specimen source (serum). Here, we evaluate the recently released InBios Strongy Detect IgG ELISA in comparison to the commercially available SciMedx Strongyloides serology microwell ELISA and the LIPS assay that is currently performed at the NIH. The InBios and LIPS assays utilize Strongyloides-specific recombinant antigen(s) that provide a small (but important) increase in specificity, as crude antigen-based ELISAs suffer from a degree of cross-reactivity in filarial-infected patients (the antigen identity for the SciMedx assay has not been released by the manufacturer).
By Venn diagram analysis, all three assays were in perfect agreement for only 65% (66/101) of the samples tested (Fig. 1). The results among the remaining 35 samples demonstrated significant variability between the assays: 14 samples were positive and 21 samples were negative by only one of the three methods. Interestingly, the distribution of samples with individual discordant results was similar among the three assays (e.g., four samples were positive only by the InBios ELISA, five samples were positive only by the SciMedx ELISA, and five samples were positive only by the LIPS assay), indicating that no single assay was responsible for a majority of the discrepant results. Additionally, a similar number of samples were either positive or negative by each possible combination of two assays (e.g., five samples were positive by both ELISAs, seven samples were positive by the InBios ELISA and the LIPS assay, and nine samples were positive by the SciMedx ELISA and the LIPS assay). These observations suggest that of the assays evaluated here, no two can be used interchangeably, as significant variability exists between all three methods. Of interest for future consideration, InBios recently produced a second generation of this assay with a modified antigen preparation. Additional studies comparing the updated iteration of this assay should be conducted to determine whether the lack of consensus is still seen among the three methods.
In the absence of a gold standard reference method, we chose to initially evaluate these immunoassays against the consensus result from all three methods. We found a similar overall performance for each assay using this reference standard, and the substantial agreement was further supported by similar kappa coefficients (range, 0.68 to 0.78) (Table 1). While the SciMedx assay showed a notably lower kappa value (0.68) and percent negative agreement (82.6%) than both the InBios ELISA (0.74 and 91.3%, respectively) and the LIPS assay (0.78 and 89.1%, respectively), the 95% confidence intervals for these two parameters overlapped significantly for all three assays, and a larger-scale study with prospectively collected specimens is needed to determine if these differences are truly significant. It is not surprising that that there was more agreement between the InBios and NIH LIPS assays, as both utilize the same recombinant NIE antigen.
While not yet commercially available, the NIH LIPS assay is emerging as a premier serologic test for differentiating patients with stool culture-confirmed strongyloidiasis from healthy controls, and it has documented sensitivity and specificity values approaching 100% (4, 16, 17, 19). Notably, the InBios and SciMedx ELISAs showed only moderate agreement with the LIPS assay based on the associated kappa coefficients (0.53 for both ELISAs), indicating that while these two commercial systems perform similarly overall, their accuracy is low compared to that of the NIH method (Table 2). However, an additional evaluation of the new InBios ELISA using serum collected from patients with positive Strongyloides stool cultures showed improved sensitivity and specificity (80% and 90%, respectively), suggesting that this assay is often positive in patients with active strongyloidiasis, though infection cannot be ruled out based on a single negative result (Table 3).
This study has several limitations that require discussion. First, the specimens used for this evaluation were retrospectively collected, and consequently, the associated performance characteristics may not be representative of their performance in routine practice. Second, these specimens were received through a reference laboratory, and neither patient clinical information nor additional laboratory results were available for correlations to be made with the results acquired by either of the evaluated immunoassays. Third, assay accuracy was determined in comparison to either the consensus result among the three evaluated tests or to the NIH LIPS assay alone. Both of these reference standards have drawbacks. The consensus result, despite having agreement of at least two of the assays, may still be incorrect, leading to result misclassifications, and the LIPS assay, for which exceptional accuracy has been documented among populations endemic for strongyloidiasis, may not perform similarly in regions of low prevalence. Finally, detailed analytical specificity studies were not undertaken in this evaluation, though serologic cross-reactivity among the anti-Strongyloides IgG assays has been noted in individuals with prior filarial infections (18). Interestingly, preliminary specificity analysis using sera positive for anti-Trichinella spiralis antibodies showed that the InBios ELISA was positive in all five tested specimens, whereas both the SciMedx and LIPS assays were negative (data not shown). Future studies will be pursued to better elucidate this and other cross-reactivities.
In summary, the InBios and SciMedx ELISAs showed similar overall performance but were only in moderate agreement with the NIH LIPS assay, a recently developed method with high accuracy for the detection of strongyloidiasis among populations endemic for the disease. Therefore, as these two commercially available ELISAs show significant variability with both the LIPS assay and each other (Fig. 1), their results must be interpreted alongside a clinical evaluation (e.g., pretest probability, exposure history, and symptoms) and other laboratory evidence (e.g., culture) prior to a confirmation or exclusion of strongyloidiasis diagnosis. Neither of these ELISAs should be relied upon solely to establish a diagnosis of S. stercoralis infection but rather should be viewed as additional tools available to support clinical practice. In future studies, it will be worthwhile to evaluate the commercially available ELISAs using prospectively collected samples from clinically defined cases for which additional clinical and laboratory evidences are available. This would provide valuable information on the role of these assays as screening tools, particularly among immunocompromised individuals at risk for Strongyloides hyperinfection.
ACKNOWLEDGMENTS
The NIE antigen used in the InBios Strongy Detect ELISA is licensed through Thomas Nutman.
We thank the technologists and assistants in the Infectious Disease Serology Laboratory at the Mayo Clinic who provided support throughout this study.
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1. Schär F, Trostdorf U, Giardina F, Khieu V, Muth S, Marti H, Vounatsou P, Odermatt P. 2013. Strongyloides stercoralis: global distribution and risk factors. PLoS Negl. Trop. Dis. 7:e2288. 10.1371/journal.pntd.0002288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Montes M, Sawhney C, Barros N. 2010. Strongyloides stercoralis: there but not seen. Curr. Opin. Infect. Dis. 23:500–504. 10.1097/QCO.0b013e32833df718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Starr MC, Montgomery SP. 2011. Soil-transmitted helminthiasis in the United States: a systematic review–1940–2010. Am. J. Trop. Med. Hyg. 85:680–684. 10.4269/ajtmh.2011.11-0214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Requena-Méndez A, Chiodini P, Bisoffi Z, Buonfrate D, Gotuzzo E, Muñoz J. 2013. The laboratory diagnosis and follow up of strongyloidiasis: a systematic review. PLoS Negl. Trop. Dis. 7:e2002. 10.1371/journal.pntd.0002002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston FH, Morris PS, Speare R, McCarthy J, Currie B, Ewald D, Page W, Dempsey K. 2005. Strongyloidiasis: a review of the evidence for Australian practitioners. Aust. J. Rural Health 13:247–254. 10.1111/j.1440-1584.2005.00710.x [DOI] [PubMed] [Google Scholar]
- 6. Greaves D, Coggle S, Pollard C, Aliyu SH, Moore EM. 2013. Strongyloides stercoralis infection. BMJ 347:f4610. 10.1136/bmj.f4610 [DOI] [PubMed] [Google Scholar]
- 7. Keiser PB, Nutman TB. 2004. Strongyloides stercoralis in the immunocompromised population. Clin. Microbiol. Rev. 17:208–217. 10.1128/CMR.17.1.208-217.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roxby AC, Gottlieb GS, Limaye AP. 2009. Strongyloidiasis in transplant patients. Clin. Infect. Dis. 49:1411–1423. 10.1086/630201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muennig P, Pallin D, Sell RL, Chan MS. 1999. The cost effectiveness of strategies for the treatment of intestinal parasites in immigrants. N. Engl. J. Med. 340:773–779. 10.1056/NEJM199903113401006 [DOI] [PubMed] [Google Scholar]
- 10. Marcos LA, Terashima A, Dupont HL, Gotuzzo E. 2008. Strongyloides hyperinfection syndrome: an emerging global infectious disease. Trans. R. Soc. Trop. Med. Hyg. 102:314–318. 10.1016/j.trstmh.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 11. Sheorey H, Biggs BA, Traynor P. 2011. Nematodes, p 2206–2209 In Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW. (ed), Manual of clinical microbiology, 10th ed. ASM Press, Washington, DC [Google Scholar]
- 12. Khieu V, Schär F, Marti H, Sayasone S, Duong S, Muth S, Odermatt P. 2013. Diagnosis, treatment and risk factors of Strongyloides stercoralis in schoolchildren in Cambodia. PLoS Negl. Trop. Dis. 7:e2035. 10.1371/journal.pntd.0002035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dreyer G, Fernandes-Silva E, Alves S, Rocha A, Albuquerque R, Addiss D. 1996. Patterns of detection of Strongyloides stercoralis in stool specimens: implications for diagnosis and clinical trials. J. Clin. Microbiol. 34:2569–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nielsen PB, Mojon M. 1987. Improved diagnosis of Strongyloides stercoralis by seven consecutive stool specimens. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 263:616–618 [DOI] [PubMed] [Google Scholar]
- 15. Loutfy MR, Wilson M, Keystone JS, Kain KC. 2002. Serology and eosinophil count in the diagnosis and management of strongyloidiasis in a non-endemic area. Am. J. Trop. Med. Hyg. 66:749–752 [DOI] [PubMed] [Google Scholar]
- 16. Krolewiecki AJ, Ramanathan R, Fink V, McAuliffe I, Cajal SP, Won K, Juarez M, Di Paolo A, Tapia L, Acosta N, Lee R, Lammie P, Abraham D, Nutman TB. 2010. Improved diagnosis of Strongyloides stercoralis using recombinant antigen-based serologies in a community-wide study in northern Argentina. Clin. Vaccine Immunol. 17:1624–1630. 10.1128/CVI.00259-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramanathan R, Burbelo PD, Groot S, Iadarola MJ, Neva FA, Nutman TB. 2008. A luciferase immunoprecipitation systems assay enhances the sensitivity and specificity of diagnosis of Strongyloides stercoralis infection. J. Infect. Dis. 198:444–451. 10.1086/589718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Doorn HR, Koelewijn R, Hofwegen H, Gilis H, Wetsteyn JCFM, Wismans PJ, Sarfati C, Vervoort T, van Gool T. 2007. Use of enzyme-linked immunosorbent assay and dipstick assay for detection of Strongyloides stercoralis infection in humans. J. Clin. Microbiol. 45:438–442. 10.1128/JCM.01735-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bisoffi Z, Buonfrate D, Sequi M, Mejia R, Cimino RO, Krolewiecki AJ, Albonico M, Gobbo M, Bonafini S, Angheben A, Requena-Mendez A, Muñoz J, Nutman TB. 2014. Diagnostic accuracy of five serologic tests for Strongyloides stercoralis infection. PLoS Negl. Trop. Dis. 8:e2640. 10.1371/journal.pntd.0002640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abdalla J, Saad M, Myers JW, Moorman JP. 2005. An elderly man with immunosuppression, shortness of breath, and eosinophilia. Clin. Infect. Dis. 40:1464,1535–1466. 10.1086/429724 [DOI] [PubMed] [Google Scholar]
- 21. Bon B, Houze S, Talabani H, Magne D, Belkadi G, Develoux M, Senghor Y, Chandenier J, Ancelle T, Hennequin C. 2010. Evaluation of a rapid enzyme-linked immunosorbent assay for diagnosis of strongyloidiasis. J. Clin. Microbiol. 48:1716–1719. 10.1128/JCM.02364-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Landis JR, Koch GG. 1977. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics 33:363–374. 10.2307/2529786 [DOI] [PubMed] [Google Scholar]