Abstract
Outbreaks of low-pathogenicity avian influenza (LPAI) viruses of the H7N3 subtype were first detected in Italy in October 2002, and the virus continued to circulate between 2002 and 2004 in a densely populated poultry area in the northeast portion of that country. This virus circulated in unvaccinated and vaccinated poultry farms, and the infection was controlled in August 2003 by culling, control of movements, improved biosecurity, and heterologous vaccination. In 2004, H7N3 reoccurred in vaccinated poultry farms in which infection had been successfully controlled by the vaccination program. To shed light on this occurrence and the temporal pattern and genetic basis of antigenic drift for avian influenza viruses (AIVs) in the absence and presence of heterologous vaccination, a collection of H7N3 viruses isolated in 2002 and 2004 were characterized genetically and antigenically. Molecular analysis showed that viruses isolated in the 2004 outbreaks after the implementation of vaccination had acquired specific amino acid signatures, most of which were located at reported antibody binding sites of the hemagglutinin (HA) protein. Antigenic characterization of these 2004 isolates showed that they were antigenically different from those isolated prior to the implementation of vaccination. This is the first report on antigenic and genetic evolution of H7 LPAI viruses following the application of heterologous vaccination in poultry. These findings may have an impact on control strategies to combat AI infections in poultry based on vaccination.
INTRODUCTION
Avian influenza (AI) is a listed disease of the World Organisation for Animal Health (OIE) and has become one of the greatest concerns for animal and human health. Avian influenza viruses (AIVs) are distinguished as high-pathogenicity (HPAI) and low-pathogenicity (LPAI) viruses based on the intravenous pathogenicity index (IVPI) in specific-pathogen-free (SPF) chickens and the presence of multiple basic amino acids at the cleavage site of the hemagglutinin (HA) protein. To date, all influenza outbreaks of the highly pathogenic form have been caused by influenza A viruses of either subtype H5 or H7. Since 1959, only 24 primary outbreaks of subtypes H5 and H7 have been reported worldwide; a majority occurred in Europe and the Americas (1), and as a consequence, HPAI outbreaks in poultry were considered rare events.
The emergence and spread of A/goose/Guangdong/1/1996 (H5N1)-like HPAI virus in 1996 (1, 2) radically changed the perception of the disease in the veterinary and public health sectors. The outbreak of the H5N1 HPAI virus was responsible for the death and culling of millions of birds and infected a variety of nonavian hosts, including humans and pigs (3), spreading to 63 countries or regions (2, 4). In some countries, culling was successfully applied to control the disease, whereas in others, vaccination was added to control programs.
Due to the economic cost associated with the culling program, vaccination was first implemented for HPAI virus infections in Mexico against H5N2 (1995) (5, 6) and in Pakistan against H7N3 (1995) (7) viruses. Moreover, Italy used vaccination to control the H7N1 HPAI epidemic in 2000 (8). In 2002, vaccination was first implemented for H5N1 HPAI virus in Hong Kong, and in 2004, it was implemented in Indonesia and China (9). Additionally, vaccination was used to control LPAI virus infections in Mexico (10, 11), Italy, and the United States (12, 13).
It is well established that influenza virus naturally undergoes antigen drift and shift, but to date, few studies have documented the influence of a vaccination program on the antigenic diversity of the AIVs. It is important to consider that H5N1 HPAI viruses belonging to different genetic clades show considerable antigenic variation, suggesting a correlation between the genetic and antigenic characteristics (14). A recent study compared the genetic evolution of the HA genes of H5N1 HPAI viruses isolated in countries where vaccination was applied and not applied (15), highlighting that the HA genes of viruses isolated from countries applying vaccination evolved genetically more rapidly than those of viruses circulating in countries not applying vaccination. However, that study did not address whether such a genetic difference had any impact on the antigenic characteristics of the viruses under study. Additionally, several studies are available on the distinct antigenic characteristics of H5N1 HPAI viruses from Egypt, where vaccination was implemented; however, data on the antigenic characteristics of Egyptian H5N1 HPAI viruses isolated from unvaccinated and vaccinated farms are absent (15–18).
As vaccination of susceptible animals might be more advantageous than a culling program, the implication of the vaccination on driving antigenic drift is an important consideration when designing new vaccines and implication of control strategies. To date, there has been only one study investigating the antigenic difference of the H7 AIVs, with the aim to select challenge viruses for vaccine efficacy (19) and not examine selection pressure due to vaccination. Therefore, still-limited data are available on the antigenic characteristics of H7 AIVs.
Italy experienced an epidemic of LPAI virus subtype H7N3 in poultry from 2002 to 2004. At the end of 2002, a vaccination program was designed based on a differentiating infected from vaccinated animals (DIVA) strategy and was carried out using an inactivated heterologous H7 AIV vaccine (strain A/chicken/Italy/1067/1999 [H7N1]) from December 2002 to August 2003 (20). Nevertheless, from January 2003 to October 2003, H7N3 LPAI virus had spread in unvaccinated flocks but to a lesser extent in vaccinated flocks. Additional control measures, including culling and controlled marketing procedures, were enforced, and these efforts resulted in the absence of detectable virus circulation. One year after the depopulation of the LPAI virus-infected flock, the H7N3 subtype reemerged. Following reemergence, some vaccinated flocks contracted infection, as the population was only partially immune (20, 21). During the 2002-2004 Italian LPAI H7N3 epidemic, a longitudinal collection of H7N3 viruses was obtained and archived. These viruses represent a unique collection, as it allowed the generation of data on the antigenicity and genetic makeup of viruses obtained before, during, and after the implementation of the vaccination campaign. In this study, we have investigated whether vaccination practices can lead to selection and transmission of variants that exhibit different antigenic characteristics that lead to the reoccurrence of the virus after a control program. Our results clearly showed the occurrence of the antigenic and genetic variations for the H7N3 LPAI Italian strains, which circulated in a naive and vaccinated population between 2002 and 2004.
MATERIALS AND METHODS
Viruses.
AIVs of the H7N3 subtype were selected for this study according to the following criteria: (i) year of isolation, (ii) vaccination status of the flock from which they were isolated, and (iii) HA sequence data (Table 1). A flock was defined as vaccinated when a minimum of two vaccination interventions were carried out and at least 21 days had elapsed between the second vaccination intervention and the date of collection of positive samples in the field. A total of 44 isolates were selected. In particular, 16 H7N3 isolates collected during the 2002 epidemic (no vaccination campaign in place), 14 isolates collected in 2003 from unvaccinated (7 viruses) and vaccinated (7 viruses) flocks, and 11 H7N3 isolates sampled from vaccinated farms during the 2004 epidemic were analyzed (Table 1); the last were all available viruses isolated in 2004 and 3 genetically unrelated H7 viruses.
TABLE 1.
List of H7N3 viruses isolated in Italy between 2002 and 2004 subjected to antigenic analysisa
| Virus | Date (day-mo-yr) of suspected outbreak | Date (day-mo-yr) of sample submission | Province of isolation | Species | No. of animals on farm | Antigenic group |
|---|---|---|---|---|---|---|
| A/turkey/Italy/7159/2002 (H7N3) | 12-Oct-02 | 16-Oct-02 | Fattening turkey | 33,019 | Unv | |
| A/turkey/Italy/7653/2002 (H7N3) | 28-Oct-02 | 29-Oct-02 | Verona | Fattening turkey | 8,896 | Unv |
| A/turkey/Italy/8000/2002 (H7N3) | 04-Nov-02 | 07-Nov-02 | Brescia | Fattening turkey | 7,430 | Unv |
| A/turkey/Italy/8912/2002 (H7N3) | 25-Nov-02 | 27-Nov-02 | Vicenza | Fattening turkey | 7,000 | Unv |
| A/turkey/Italy/8535/2002 (H7N3) | 08-Nov-02 | 15-Nov-02 | Brescia | Fattening turkey | 22,080 | Unv |
| A/turkey/Italy/8307/2002 (H7N3) | 15-Nov-02 | 13-Nov-02 | Verona | Fattening turkey | 14,000 | Unv |
| A/chicken/Italy/8093/2002 (H7N3) | 08-Nov-02 | 12-Nov-02 | Verona | Broiler breeder | 25,197 | Unv |
| A/turkey/Italy/8458/2002 (H7N3) | 13-Nov-02 | 15-Nov-02 | Verona | Fattening turkey | 18,500 | Unv |
| A/turkey/Italy/8651/2002 (H7N3) | 21-Nov-02 | 21-Nov-02 | Verona | Fattening turkey | 14,400 | Unv |
| A/turkey/Italy/8834/2002 (H7N3) | 23-Nov-02 | 25-Nov-02 | Verona | Fattening turkey | 7,000 | Unv |
| A/turkey/Italy/9369/2002 (H7N3) | 10-Dec-02 | 11-Dec-02 | Verona | Fattening turkey | 14,000 | Unv |
| A/turkey/Italy/9314/2002 (H7N3) | 06-Dec-02 | 09-dic-02 | Verona | Fattening turkey | 40,600 | Unv |
| A/turkey/Italy/9289/2002 (H7N3) | 05-Dec-02 | 06-Dec-02 | Verona | Fattening turkey | 5,090 | Unv |
| A/turkey/Italy/9737/2002 (H7N3) | 31-Dec-02 | 31-Dec-02 | Brescia | Fattening turkey | 28,716 | Unv |
| A/turkey/Italy/9374/2002 (H7N3) | 11-Dec-02 | 12-Dec-02 | Verona | Fattening turkey | 12,500 | Unv |
| A/turkey/Italy/9102/2002 (H7N3) | 29-Nov-02 | 03-Dec-02 | Verona | Fattening turkey | 12,860 | Unv |
| A/turkey/Italy/251/2003 (H7N3) | 11-Jan-03 | 15-Jan-03 | Brescia | Fattening turkey | 10,800 | Unv |
| A/chicken/Italy/682/2003 (H7N3) | 28-Jan-03 | 30-Jan-03 | Padova | Layers | 32,000 | Unv |
| A/turkey/Italy/1010/2003 (H7N3) | 04-Feb-03 | 12-Feb-03 | Brescia | Fattening turkey | 40,000 | Unv |
| A/guineafowl/Italy/1613/2003 (H7N3) | 13-Mar-03 | 12-Mar-03 | Padova | Guinea Fowl | 54,000 | Unv |
| A/turkey/Italy/2043/2003 (H7N3) | 21-Mar-03 | 03-Apr-03 | Brescia | Fattening turkey | 100,000 | Unv |
| A/chicken/Italy/2240/2003 (H7N3) | 16-Apr-03 | 17-Apr-03 | Verona | Fattening turkey | 13,000 | Unv |
| A/turkey/Italy/2684/2003 (H7N3) | 01-Apr-03 | 19-May-03 | Brescia | Fattening turkey | 12,000 | Unv |
| A/turkey/Italy/2856/2003 (H7N3) | 23-May-03 | 27-May-03 | Brescia | Fattening turkey | 79,800 | Vac |
| A/turkey/Italy/2987/2003 (H7N3) | 03-Jun-03 | 05-Jun-03 | Verona | Fattening turkey | 12,800 | Vac |
| A/turkey/Italy/2962/2003 (H7N3) | 03-Jun-03 | 04-Jun-03 | Verona | Fattening turkey | 13,700 | Vac |
| A/turkey/Italy/2963/2003 (H7N3) | 26-May-03 | 04-Jun-03 | Verona | Fattening turkey | 48,000 | Vac |
| A/turkey/Italy/4036/2003 (H7N3) | 15-Jul-03 | 16-Jul-03 | Verona | Fattening turkey | 16,000 | Vac |
| A/turkey/Italy/3620/2003 (H7N3) | 01-Jul-03 | 01-Jul-03 | Verona | Fattening turkey | 25,500 | Vac |
| A/turkey/Italy/4608/2003 (H7N3) | 08-Aug-03 | 12-Aug-03 | Verona | Fattening turkey | 5,600 | Vac |
| A/turkey/Italy/3337/2004 (H7N3) | 17-Sept-04 | 21-Sept-04 | Verona | Fattening turkey | 14,500 | Vac |
| A/quail/Italy/3347/2004 (H7N3) | 21-Sept-04 | 22-Sept-04 | Verona | Fattening turkey | 360,000 | Vac |
| A/turkey/Italy/3439/2004 (H7N3) | 24-Sept-04 | 27-Sept-04 | Verona | Fattening turkey | 23,000 | Vac |
| A/turkey/Italy/3477/2004 (H7N3) | 27-Sept-04 | 29-Sept-04 | Verona | Fattening turkey | 21,600 | Vac |
| A/turkey/Italy/3399/2004 (H7N3) | 20-Sept-04 | 23-Sept-04 | Verona | Fattening turkey | 7,000 | Vac |
| A/turkey/Italy/3807/2004 (H7N3) | 14-Oct-04 | 14-Oct-04 | Verona | Fattening turkey | 11,300 | Vac |
| A/turkey/Italy/3829/2004 (H7N3) | 15-Oct-04 | 14-Oct-04 | Verona | Fattening turkey | 22,200 | Vac |
| A/turkey/Italy/4042/2004 (H7N3) | 21-Oct-04 | 28-Oct-04 | Verona | Fattening turkey | 36,400 | Vac |
| A/turkey/Italy/4130/2004 (H7N3) | 26-Oct-04 | 04-Nov-04 | Verona | Fattening turkey | 6,500 | Vac |
| A/turkey/Italy/4479/2004 (H7N3) | 16-Nov-04 | 15-Nov-04 | Verona | Fattening turkey | 20,000 | Vac |
| A/turkey/Italy/4199/2004 (H7N3) | 04-Nov-04 | 09-Nov-04 | Verona | Fattening turkey | 7,200 | Vac |
A/chicken/Italy/1067/1999 (H7N1) (not shown) was the vaccine strain, and A/shoveler/Italy/377/2006 (H7N7) and A/turkey/ENGLAND/647/1977 (H7N7) (not shown) were genetically unrelated viruses used to produce turkey sera. Viruses from vaccinated farms are in bold. Vac, viruses from vaccinated farms; Unv, viruses from unvaccinated farms.
In summary, 21 H7N3 isolates from unvaccinated farms and 20 H7N3 isolates from vaccinated farms isolated during the 2002-2004 epidemics were studied. Three additional isolates were also included in this study: A/chicken/Italy/1067/1999 (H7N1), which was the vaccine strain used during the vaccination campaign, A/turkey/England/647/1977 (H7N7), and A/shoveler/Italy/377/2006 (H7N7) (Table 1), included as unrelated viruses. Viruses were propagated in 9- to 11-day-old SPF embryonated chicken eggs (Charles River Laboratories Inc., US) and inactivated with 0.05% (vol/vol) β-propiolactone (Sigma-Aldrich, US) for 3 h at 37°C to carry out cross-hemagglutination inhibition (HI) tests.
Production of turkey sera.
Eight-week-old turkeys were used to produce 13 polyclonal antisera against H7N3 viruses. Prior to the immunization, all turkeys were tested virologically and serologically by real-time RT-PCR (RRT-PCR) for type A influenza virus and by commercial competitive type A enzyme-linked immunosorbent assay (ELISA) (ID Screen influenza A antibody competition; IDVet, France), respectively. Turkeys were immunized intravenously with 0.2 ml of inactivated H7 viruses and boosted 4 weeks after by the same route. All antigens used to produce sera had HA titers ranging between 27 and 28 log2, and their purity was confirmed by OIE standard techniques (22).
All turkeys received two immunizations to obtain HI titers above 26. Serum samples were collected 3 weeks after the second immunization. Turkey sera were generated using two turkeys per serum batch, except for the batch generated against A/chicken/Italy/1067/1999 (H7N1). Blood was collected, pooled per individual bird, and allowed to clot in plastic vessels at 37°C for 2 h. Vessels were centrifuged at 1,000 × g for 10 min, and 1-ml aliquots were placed in vials and stored at −20°C until use.
Prior to testing, sera were treated with receptor-destroying enzyme (RDE; Sigma-Aldrich, US) to remove any nonspecific reactions as described by the OIE manual (22). Viruses to immunize birds were selected according the year of isolation, vaccination status of the farm from which they were isolated, and phylogenetic analysis.
Turkey sera were produced against the H7N1 virus (vaccine strain), A/turkey/England/647/1977 (H7N7), two H7N3 strains isolated during the 2002 epidemic (A/turkey/Italy/8000/2002 and A/turkey/Italy/9369/2002), and two H7N3 strains isolated during the 2004 epidemic (A/turkey/Italy/4130/2004 and A/turkey/Italy/3399/2004) (Table 1). A total of 13 turkey serum samples were collected and used to carry out the cross-HI tests (Table 1). All animals were handled in strict accordance with the relevant national and local animal welfare bodies (24).
Cross-HI tests.
The HI test was carried out according to OIE guidelines (22). Briefly, 4 hemagglutinin units and 1% (vol/vol) chicken red blood cell suspension were used to carry out HI tests. Serum samples were tested singularly, resulting in 13 different titers for each virus isolate. The data set consisted of a table of 13 turkey sera and 44 viruses resulting in 572 individual HI measurements (Table 2). Each HI experiment was repeated twice, and the average value was calculated.
TABLE 2.
Cross-HI data obtained testing H7N3 LPAI viruses isolated between 2002 and 2004 from unvaccinated and vaccinated farms against representative H7N3 turkey seraa
| Antigen | HI titer obtained with serum |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/turkey/England/647/1977 | A/turkey/England/647/1977 | A/turkey/Italy/1067/1999 | A/turkey/Italy/8000/2002 | A/turkey/Italy/8000/2002 | A/turkey/Italy/3399/2004 | A/turkey/Italy/3399/2004 | A/turkey/Italy/9369/2002 | A/turkey/Italy/9369/2002 | A/turkey/Italy/8651/2002 | A/turkey/Italy/8651/2002 | A/turkey/Italy/4130/2004 | A/turkey/Italy/4130/2004 | |
| A/turkey/Italy/8000/2002 (H7N3) | 320 | 80 | 320 | 160 | 40 | 80 | 160 | 160 | 160 | 320 | 320 | 40 | 320 |
| A/shoveler/Italy/377/2006 (H7N7) | 40 | 20 | 40 | 20 | 0 | 20 | 80 | 80 | 40 | 160 | 80 | 40 | 160 |
| A/turkey/Italy/9289/2002 (H7N3) | 80 | 40 | 160 | 80 | 20 | 20 | 80 | 160 | 160 | 160 | 80 | 40 | 80 |
| A/turkey/Italy/8912/2002 (H7N3) | 160 | 80 | 320 | 160 | 160 | 80 | 160 | 320 | 160 | 320 | 160 | 40 | 160 |
| A/turkey/Italy/8535/2002 (H7N3) | 160 | 80 | 320 | 160 | 40 | 160 | 320 | 320 | 640 | 320 | 160 | 80 | 160 |
| A/turkey/Italy/9737/2002 (H7N3) | 80 | 40 | 160 | 40 | 0 | 20 | 80 | 160 | 160 | 320 | 80 | 40 | 320 |
| A/turkey/Italy/9374/2002 (H7N3) | 40 | 20 | 80 | 80 | 0 | 20 | 80 | 40 | 80 | 160 | 80 | 40 | 160 |
| A/turkey/Italy/7159/2002 (H7N3) | 80 | 20 | 20 | 80 | 0 | 20 | 40 | 160 | 80 | 160 | 80 | 80 | 320 |
| A/turkey/Italy/7653/2002 (H7N3) | 40 | 20 | 40 | 40 | 0 | 20 | 40 | 80 | 80 | 160 | 80 | 40 | 160 |
| A/turkey/Italy/8307/2002 (H7N3) | 80 | 40 | 80 | 320 | 0 | 40 | 80 | 80 | 80 | 160 | 80 | 80 | 160 |
| A/chicken/Italy/8093/2002 (H7N3) | 160 | 160 | 160 | 160 | 40 | 80 | 160 | 160 | 80 | 160 | 80 | 80 | 160 |
| A/turkey/Italy/8458/2002 (H7N3) | 320 | 40 | 160 | 160 | 0 | 20 | 80 | 320 | 80 | 320 | 160 | 40 | 160 |
| A/turkey/Italy/8651/2002 (H7N3) | 80 | 40 | 80 | 40 | 0 | 20 | 80 | 160 | 80 | 160 | 80 | 20 | 80 |
| A/turkey/Italy/8834/2002 (H7N3) | 40 | 20 | 40 | 40 | 0 | 0 | 40 | 80 | 80 | 160 | 80 | 40 | 160 |
| A/turkey/Italy/9369/2002 (H7N3) | 40 | 20 | 80 | 40 | 0 | 20 | 40 | 80 | 40 | 80 | 40 | 40 | 160 |
| A/turkey/Italy/9314/2002 (H7N3) | 80 | 40 | 80 | 80 | 0 | 40 | 80 | 160 | 80 | 160 | 80 | 160 | 640 |
| A/turkey/Italy/9102/2002 (H7N3) | 80 | 40 | 160 | 80 | 40 | 40 | 160 | 320 | 160 | 320 | 320 | 40 | 80 |
| A/turkey/ENGLAND/647/1977 (H7N7) | 80 | 160 | 40 | 20 | 20 | 20 | 20 | 80 | 80 | 320 | 80 | 40 | 80 |
| A/turkey/Italy/1067/1999 | 20 | 0 | 80 | 40 | 20 | 20 | 0 | 40 | 0 | 160 | 40 | 0 | 160 |
| A/turkey/Italy/251/2003 (H7N3) | 20 | 0 | 20 | 20 | 0 | 0 | 0 | 0 | 20 | 80 | 40 | 40 | 40 |
| A/chicken/Italy/682/2003 (H7N3) | 640 | 160 | 640 | 160 | 160 | 160 | 640 | 640 | 320 | 640 | 320 | 20 | 160 |
| A/turkey/Italy/1010/2003 (H7N3) | 320 | 80 | 320 | 320 | 0 | 160 | 320 | 320 | 320 | 1280 | 640 | 20 | 320 |
| A/guineafowl/Italy/1613/2003 (H7N3) | 160 | 40 | 80 | 80 | 20 | 40 | 80 | 160 | 80 | 320 | 80 | 80 | 640 |
| A/turkey/Italy/2043/2003 (H7N3) | 20 | 0 | 80 | 40 | 0 | 0 | 0 | 20 | 40 | 80 | 40 | 0 | 80 |
| A/chicken/Italy/2240/2003 (H7N3) | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 160 |
| A/turkey/Italy/2684/2003 (H7N3) | 80 | 40 | 80 | 80 | 0 | 20 | 80 | 80 | 40 | 320 | 80 | 40 | 160 |
| A/turkey/Italy/2856/2003 (H7N3) | 40 | 0 | 80 | 80 | 0 | 0 | 20 | 40 | 40 | 160 | 80 | 80 | 160 |
| A/turkey/Italy/2987/2003 (H7N3) | 40 | 40 | 80 | 40 | 0 | 80 | 80 | 20 | 20 | 20 | 20 | 20 | 40 |
| A/turkey/Italy/4036/2003 (H7N3) | 0 | 0 | 20 | 20 | 0 | 20 | 40 | 40 | 0 | 80 | 20 | 20 | 40 |
| A/turkey/Italy/3620/2003 (H7N3) | 20 | 20 | 20 | 20 | 0 | 20 | 20 | 20 | 20 | 20 | 20 | 0 | 0 |
| A/turkey/Italy/4608/2003 (H7N3) | 0 | 0 | 20 | 20 | 0 | 20 | 40 | 40 | 0 | 40 | 20 | 0 | 80 |
| A/turkey/Italy/2962/2003 (H7N3) | 20 | 20 | 20 | 40 | 0 | 40 | 40 | 80 | 20 | 80 | 20 | 40 | 80 |
| A/turkey/Italy/2963/2003 (H7N3) | 40 | 80 | 80 | 80 | 0 | 80 | 160 | 80 | 40 | 80 | 80 | 0 | 80 |
| A/turkey/Italy/3337/2004 (H7N3) | 20 | 20 | 40 | 40 | 0 | 160 | 320 | 0 | 0 | 80 | 20 | 80 | 80 |
| A/quail/Italy/3347/2004 (H7N3) | 20 | 20 | 40 | 80 | 0 | 160 | 320 | 0 | 20 | 160 | 20 | 20 | 80 |
| A/turkey/Italy/3439/2004 (H7N3) | 0 | 20 | 20 | 40 | 0 | 80 | 160 | 0 | 0 | 80 | 0 | 40 | 80 |
| A/turkey/Italy/3477/2004 (H7N3) | 20 | 40 | 80 | 80 | 20 | 320 | 640 | 0 | 20 | 160 | 20 | 40 | 160 |
| A/turkey/Italy/3807/2004 (H7N3) | 0 | 20 | 40 | 40 | 0 | 80 | 320 | 0 | 20 | 80 | 20 | 20 | 40 |
| A/turkey/Italy/3829/2004 (H7N3) | 20 | 40 | 40 | 80 | 20 | 160 | 320 | 0 | 20 | 80 | 20 | 80 | 80 |
| A/turkey/Italy/4042/2004 (H7N3) | 20 | 20 | 40 | 80 | 0 | 80 | 320 | 0 | 0 | 80 | 20 | 80 | 160 |
| A/turkey/Italy/4130/2004 (H7N3) | 0 | 20 | 20 | 80 | 0 | 80 | 160 | 0 | 40 | 80 | 20 | 80 | 80 |
| A/turkey/Italy/4479/2004 (H7N3) | 0 | 20 | 20 | 80 | 20 | 80 | 160 | 0 | 40 | 40 | 20 | 40 | 160 |
| A/turkey/Italy/3399/2004 (H7N3) | 40 | 40 | 80 | 80 | 20 | 320 | 640 | 0 | 20 | 160 | 20 | 20 | 160 |
| A/turkey/Italy/4199/2004 (H7N3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 160 | 160 |
A/turkey/1067/1999 was the vaccine strain. Viruses from vaccinated farms are in bold.
Genomic sequencing.
Viral RNA was extracted from the infective allantoic fluid of SPF embryonated chicken eggs using the Nucleospin RNA II kit (Macherey-Nagel, Duren, Germany) and was reverse transcribed with the SuperScript III reverse transcriptase kit (Invitrogen, Carlsbad, CA). PCR amplification was performed by using specific primers (primer sequences available upon request). The complete coding sequences were generated using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). The products of the sequencing reactions were cleaned up using the Performa DTR Ultra 96-well kit (Edge BioSystems, Gaithersburg, MD) and sequenced in a 16-capillary ABI PRISM 3130xl genetic analyzer (Applied Biosystems). Sequence data were assembled and edited with SeqScape software v2.5 (Applied Biosystems).
Molecular characterization and evolutionary analyses.
Phylogenetic analyses were performed using maximum likelihood by GARLI (25) and bootstrap resampling analyses using PAUP* 4.0 Beta (26) to apply a neighborhood joining method, as described earlier (27). Positive-selection analyses were performed using PAML (28) as described elsewhere (27).
A total of 1,410 HA sequences of the H7 subtypes of influenza A viruses were downloaded from the Influenza Research Database (IRD; http://www.fludb.org), and the phylogenetic analysis was used to identify those HA genes belonging to the Eurasian lineage for further analysis. Subsequently, phylogenetic analyses were performed on a total of 77 HA full-length sequences of 30 H7N3 LPAI viruses isolated from the 2002-2004 Italian epidemic and 47 other viruses isolated from avian species in Europe and Asia from 1934 to 2011.
The protein sequences of these isolates were aligned with the H3 protein sequences as shown previously (29). The antibody binding sites were annotated based on those in H3N2 influenza A virus (30).
To investigate the impact of vaccinations on the evolutionary rate of the HA gene, evolutionary rates using Bayesian statistics were analyzed. To make the data analysis more accurate, only isolates with specific isolation dates were included. The evolutionary rates of HA genes were estimated using Bayesian Evolutionary Analysis Sampling Trees (BEAST) version 1.7.1 (31). The Bayesian skyline coalescent tree prior (10 piece-wise constant groups) is used in the analysis. In addition, we used the HKY substitution model with Markov Chain Monte Carlo runs of 50,000,000 steps and analyzed with Tracer with a discarded burn-in of 10%. The strict clock model was used in this analysis. Three independent tests were performed.
Construction of the antigenic map.
The antigenic map was constructed using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap) (32, 33), as described previously (16), with an HI titer of 1:10 as the threshold for a low reactor. Each entry in the HI table was normalized by the maximum value from each serum sample. The antigenic map utilized low-rank matrix completion to minimize the noises in the HI data and multiple dimensional scaling to generate the map reflecting the antigenic distances embedded in HI data. Each unit in the antigenic map corresponded to a log2 unit in the HI test.
Nucleotide sequence accession numbers.
The HA sequences obtained in this study are available in GenBank under the following accession numbers: CY024738, CY095562, CY029905, CY020605, GQ247853, CY095546, CY034750, CY021365, CY022613, CY020589, CY095554, JX515663, CY021493, CY021357, CY021485, CY020613, CY028684, CY020597, CY028676, CY029913, CY020581, and CY021501 and in the Global Initiative on Sharing All Influenza Data (GISAID; www.gisaid.org) database under the following accession numbers: EPI154974, EPI243276, EPI243277, EPI154967, EPI154966, EPI154969, EPI154970, EPI154973, EPI154972, EPI154971, EPI154959, EPI154960, EPI154958, EPI154963, EPI243279, EPI154962, EPI154964, EPI154961, and EPI243280.
RESULTS
Evolutionary dynamics of H7N3 viruses before, during, and after H7N3 outbreaks.
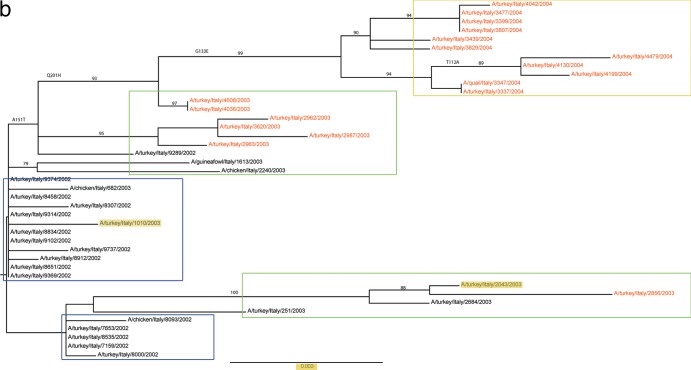
To investigate the natural history of Italian H7 viruses, a phylogenetic tree of the HA gene was constructed by including 1,235 H7 genes from a public database, analyzed after removing incomplete sequences and redundant sequences. These viruses included 171 H7 Italian AIVs, all of which belonged to the Eurasian H7 lineage (data not shown). The HA genes of H7N1 viruses from 1999 to 2001 genetically formed a sublineage; the vaccine strain A/chicken/Italy/1067/1999 (H7N1) used for the 2003-2004 campaign in Italy belonged to this branch (Fig. 1). The HA genes of the viruses isolated from the 2002-2004 outbreak form a distinct lineage and are associated with three H7N3 LPAI viruses from mallards, including A/mallard/Italy/33/2001 (H7N3), A/mallard/Italy/43/2001 (H7N3), and A/mallard/Italy/199/2001 (H7N3) (Fig. 1).
FIG 1.
(a) Phylogenetic analysis of HA gene segments of H7N3 influenza A viruses from 2002-2004 outbreak in Italy and of H7 viruses isolated elsewhere. The isolates from unvaccinated farms are marked in red and those from vaccinated farms in blue. The phylogenetic analyses were performed using maximum likelihood by GARLI (25) and bootstrap resampling analyses using PAUP* 4.0 Beta (26) to apply a neighborhood joining method, as described earlier (28). (b) Phylogenetic analysis of HA gene segments of H7N3 influenza A viruses from 2002-2004 outbreak in Italy. The isolates from unvaccinated farms are marked in black and those from vaccinated farms in red. Colored boxes are used to highlight the year of isolation: blue for viruses from 2002, green for viruses from 2003, and yellow for viruses from 2004.
The phylogenetic analysis conducted on the HA genes of the Italian H7N3 LPAI viruses showed that viruses isolated in 2004 from vaccinated farms and viruses isolated in 2002 and 2003 from unvaccinated farms form 2 distinct sublineages (Fig. 1b). There is an exception to that: A/turkey/Italy/2856/2003 was isolated from an unvaccinated farm.
Comparison of nucleotide sequences of 2002-2004 H7N3 isolates revealed the larger extent of genetic diversity that existed pre- and postvaccination. Viruses isolated from October 2002 to early 2003 from unvaccinated farms showed a relatively lower amino acid sequence divergence (identity ranged from 99.1% to 100%). Postvaccination viruses isolated during the second half of 2003 and in 2004 had higher sequence divergence (identity ranged from 98% to 100%). Only the virus A/turkey/Italy/2856/2003 isolated from vaccinated animals clustered separately: it grouped with the 2003 isolates from unvaccinated farms. A/turkey/Italy/2856/2003) was isolated on 27 May from the Brescia province, and most of the other viruses were identified after July of 2003.
The evolutionary rate analysis conducted on the HA genes of H7N3 LPAI isolates showed that strains isolated before the implementation of the vaccine campaign had an evolutionary rate of 12.20 × 10−3 substitutions/nucleotide per year (standard error of mean, 5.62 × 10−5; lower bound of the highest posterior density [HPD] interval [95% HPD lower], 7.56 × 10−3; upper bound of the highest posterior density interval [95% HPD upper], 16.50 × 10−3) with an effective sample size of 1,617. The evolutionary rate for the strains after vaccine campaigns was 5.37 × 10−3 substitutions/nucleotide per year (standard error of mean, 3.22 × 10−5; 95% HPD lower, 2.70 × 10−3; 95% HPD upper, 7.98 × 10−3) with an effective sample size of 1,801. Finally, the mean evolutionary rate was 8.04 × 10−3 substitutions/nucleotide per year (standard error of mean, 3.11 × 10−5; 95% HPD lower, 5.84 × 10−3, 95% HPD upper, 10.30 × 10−3) with an effective sample size of 1,384.
Molecular characterization of H7N3 viruses.
To investigate the amino acid mutations that might have occurred under vaccine pressure, the HA1 sequence of the first strain isolated at the beginning of the epidemic in 2002 (A/turkey/Italy/7159/2002) was compared to the HA1 sequences of all viruses used in this study. A total of five mutations were observed between isolates from unvaccinated farms and those from vaccinated farms, four of which were located in antibody binding sites: G133E (144, H3 numbering; antibody binding site A), A151T (160, H3 numbering; antibody binding site B), G177V (186, H3 numbering; antibody binding site B), Q201H (210, H3 numbering), and T112A (122, H3 numbering; antibody binding site A) (Fig. 1b).
Viruses isolated after the beginning of vaccination (May to August 2003) possessed an A151T change from the original isolate. Sequence from viruses isolated toward the end of the vaccination campaign, September and October 2004, in addition to A151T, had three distinct amino acid substitutions: G133E, G177V, and Q201H. In addition, 3 viruses (A/turkey/Italy/4130/2004, A/turkey/Italy/4479/2004, and A/turkey/Italy/4199/2004) from November 2004 showed one more additional mutation, T112A (122, H3 numbering; antibody binding site A). Moreover, our results demonstrated that there was positive selection on A151T after the start of the vaccination program. Interestingly, the mutations G133E, A151T, G177V, Q201H, and T112A found in the postvaccination isolate were not observed in the isolate A/turkey/Italy/2856/2003.
We analyzed the profiles of these amino acids among other H7 influenza A viruses, including the H7 viruses circulating in Italy before the 2002 H7N3 LPAI outbreaks, the H7N7 viruses circulating in Netherlands from 2003 to 2006, the H7N3 viruses circulating in Mexico during 2012 outbreaks and in Pakistan from 1995 to 2004, and the recently emerging H7N9 viruses in China (Table 3). Interestingly, these viruses predominantly showed T112, G133, A151, G177, and Q201, except for the H7N9 viruses isolated in China, which possessed A112 and V177.
TABLE 3.
Frequencies of amino acids at the positions in HA protein where the mutations were identified in the H7N3 influenza A viruses before and after heterologous vaccination was employed
| H7 position | H3 positiona | Relevant amino acidsb | No. of corresponding amino acids for indicated viruses (yr, no. of isolates)c |
|||||
|---|---|---|---|---|---|---|---|---|
| Italian H7N3 viruses (2002–2004, 41) | Italian H7 viruses (1999–2002, 44) | Netherland H7N7 viruses (2003–2006, 7) | Mexican H7N3 viruses (2012, 3) | Pakistan H7N3 viruses (1995–2004, 16) | China H7N9 viruses (2013, 33) | |||
| 112 | 122 (A) | T/A/S/E | 37/3/0/0 | 33/11/0/0 | 7/0/0/0 | 0/0/3/0 | 16/0/0/0 | 0/32/0/1 |
| 133 | 144 (A) | G/E | 30/11 | 42/2 | 7/0 | 3/0 | 16/0 | 33/0 |
| 151 | 160 (B) | A/T | 23/18 | 28/16 | 7/0 | 3/0 | 16/0 | 33/0 |
| 177 | 186 (B) | G/V | 28/13 | 31/13 | 6/1 | 3/0 | 15/1 | 0/33 |
| 201 | 210 | Q/H/R/L | 28/13/0/0 | 41/0/2/1 | 7/0/0/0 | 3/0/0/0 | 16/0/0/0 | 33/0/0/0 |
The alignments between H7 and H3 were performed based on the work of Yang et al. (29), and the antibody binding sites (in parentheses) were annotated by Wilson and Cox (30).
Amino acids present in the corresponding position in HA sequences of H7 influenza A viruses.
The virus sequences were downloaded from influenza virus resources at GenBank. The corresponding amino acids were counted, and the numbers of predominant residues are in bold.
Antigenic characterization of H7N3 LPAI viruses in Italy.
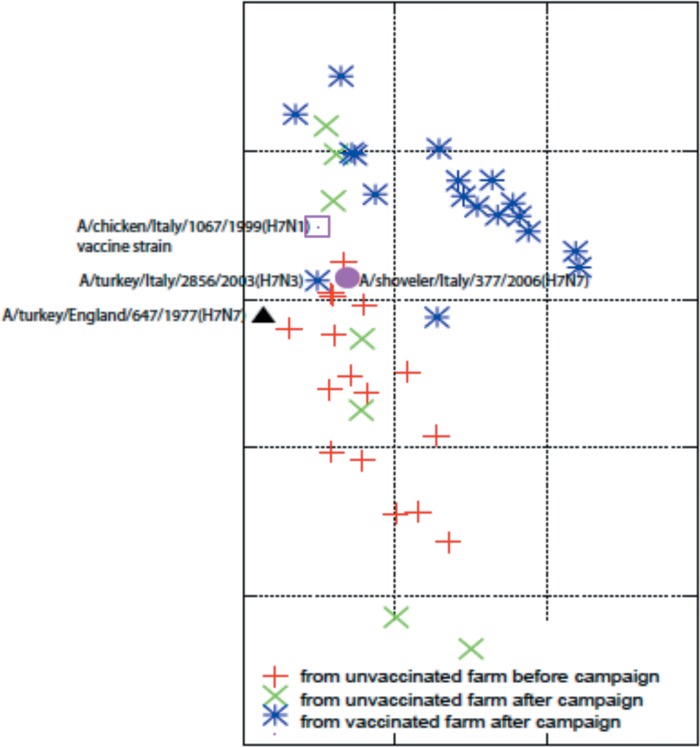
Antigenic cartography showed that Italian LPAI H7N3 viruses isolated during the 2002-2004 epidemic were separated in two antigenically diverse groups pre- and postvaccination. One group is formed by 18 viruses from vaccinated farms (group Vac) (Table 1 and Fig. 2, blue stars) while the second group (group Unv) is formed by 16 viruses isolated (Table 1, Fig. 2, red crosses) from unvaccinated farms. The average antigenic distance among the isolates of group Unv was estimated to be 1.16 units (standard deviation, 0.62 unit), and that among the viruses from group Vac was 0.95 unit (standard deviation, 0.43 unit). However, the average antigenic distance between viruses of the two groups was 1.54 units (standard deviation, 0.54 unit). There were 7 exceptions to the clustering of viruses into pre- and postvaccination virus isolates: A/turkey/Italy/2864/2003 (H7N3), A/chicken/Italy/682/2003 (H7N3), A/turkey/Italy/1010/2003 (H7N3), A/guineafowl/Italy/1613/2003 (H7N3), A/turkey/Italy/2043/2003 (H7N3), A/chicken/Italy/2240/2003 (H7N3), and A/turkey/Italy/251/2003 (H7N3) (Table 1 and Fig. 2). All were isolated in 2003 from unvaccinated flocks following the implementation of the vaccination campaign within Italy (Fig. 2, green crosses). Four of these viruses isolated between January and March 2003 (A/turkey/Italy/2684/2003, A/chicken/Italy/682/2003, A/turkey/Italy/1010/2003, and A/guineafowl/Italy/1613/2003) were more antigenically similar to the Unv group, while in contrast, 3 viruses isolated in April and May 2003 (A/turkey/Italy/2043/2003, A/chicken/Italy/2240/2003, and A/turkey/Italy/251/2003) were antigenically closer to the Vac group. The Student t test demonstrated that a statistical difference existed between these two groups of viruses (P < 0.01) but not within each group (P > 0.05).
FIG 2.
Antigenic cartography for H7N3 LPAI virus isolates from the 2002-2004 Italian epidemic. The map was constructed using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap) based on HI data from Table 2.
DISCUSSION
In this study, H7N3 LPAI viruses isolated during the Italian epidemic between 2002 and 2004 were analyzed to investigate the influence vaccination programs have on virus antigenic drift. Phylogenetic and evolutionary analyses were conducted on the HA gene to highlight that there were genetic changes which occurred in a 2-year period. The influence of these changes on antigenic change was supported in part by the cross-HI data, in which the viruses clustered into unvaccinated (Unv) and vaccinated (Vac) groups. Interestingly, even though the isolates were in two distinct groups, there was greater antigenic heterogeneity in the Unv group than in the Vac group, which were more antigenically related to each other.
The genetic analysis undertaken in this study confirmed previous work which suggested that the H7 LPAI viruses causing the 1999-2001 and 2002-2004 Italian outbreaks in poultry were likely the result of the separate introduction of AIV into domestic poultry from wild birds (34, 35). Moreover, the characterization showed that higher genetic divergence of the HA gene existed for viruses from the Vac group than for viruses of the Unv group. These findings place further emphasis on the role of vaccination in driving the genetic differentiation and evolution of H7N3 LPAI viruses.
Evolutionary rates of H7 influenza A viruses were estimated to be 8.04 × 10−3 substitutions/nucleotide per year, which is similar to 4.77 × 10−3 substitutions/site/year for HA genes of H5N1 highly pathogenic avian influenza viruses (36).
Although the evolutionary rates of H7 influenza A viruses were not significantly different between the viruses before and those after vaccination implementation, there were a number of amino acid substitutions that were conserved with the Vac group HA genes, and positive selection was observed in the antibody binding sites of these H7 viruses.
For example, the mutation A151T was observed in the Vac group compared to the Unv group, and this amino acid was under positive selection pressure after the introduction of the vaccination program. Interestingly, amino acid mutations on the HA1 located at potential antibody binding sites were observed only among the H7N3 isolates from vaccinated farms in a progressive manner. Thus, it appeared that the postvaccination antigenic change was mirrored by the simultaneous appearance of a progressive genetic change in immunodominant sites, explaining the correlation of the antigenic and genetic clustering. Interestingly, these five substitutions were not observed in other H7 influenza A viruses found in public databases, except A112 and V177 in the emerging H7N9 viruses in China. The impacts of these mutations on the antigenicity of H7N9 viruses were not clear.
There was one isolate (A/turkey/Italy/2856/2003) from a vaccinated farm that clustered genetically with viruses from unvaccinated farms but was antigenically closer to viruses of the Vac group. Genetic analysis did not show any particular substitutions which could explain the antigenic difference observed. This virus could have spread from unvaccinated farms and been able to infect a vaccinated population.
To the best of our knowledge, this is the first study on the antigenic characterization of H7 LPAI viruses collected throughout an epidemic which was controlled also by using heterologous vaccination. Therefore, the collection of H7N3 viruses in a longitudinal way allowed the comparison of genetic and antigenic characteristics of AIVs that circulated in absence and presence of vaccination. The limitation of this study was the lower number of viruses isolated in 2004 from vaccinated farms than of viruses isolated in 2002 and 2003. The reason for that was the prompt application of control measures and surveillance efforts which were in place in areas at risk that limited the spread of the infection after its reoccurrence in 2004.
However, the antigenic diversity of 2004 H7 viruses suggests that the antigenic diversification of viruses following the vaccination campaign was not a limiting factor for the control of the infection. This highlights that the successful management of AI outbreaks was not only a result of vaccination itself but also due to the proper application of other control measures. This is typified during the H5N1 HPAI outbreaks, which progressively resulted in the adoption of vaccination as a complementary measure to control the infection in several countries. Vaccination was implemented successfully in some countries, while in others, AI viruses continued to spread notwithstanding massive vaccination in place, like in Egypt (37, 38). Although an increasing amount of data is available on the management of AI outbreaks using, among other measures, vaccination, data on the impact of poultry vaccination on the evolutionary dynamics of AIVs in natural situations is limited.
Based on the influence on antigenic drift that the vaccination strategy adopted by Mexico using a homologous vaccine strain, i.e., a vaccine preparation containing an H5N2 field isolate to control the LPAI (H5N2), had, a DIVA heterologous vaccine strategy was developed and used in Italy. This involved using a vaccine strain with the same HA (H7) as was antigenically and genetically distinct to the field H7 with a different neuraminidase (NA) (N3 versus N1) (39, 40). This strategy allowed the differentiation of infected from vaccinated animals by the presence of antibodies to the different NA protein (39, 40). Thus, the samples in this study are unique, as the vaccine strain would elicit selective pressure only on the HA rather than the NA. Therefore, importantly, this study is complementary to that of Lee et al. (10) and adds new data on the impact of heterologous vaccination for the control of LPAI outbreaks in poultry.
The absence of antigenic studies on LPAI viruses that circulated in the presence and absence of vaccination makes comparison with other results difficult. Similarities can be identified between this study and research investigating H5N1 HPAI viruses isolated from countries where vaccination was adopted (Egypt and Indonesia) or not adopted (Nigeria, Turkey, and Thailand) (15). This study demonstrated that an LPAI virus rapidly evolved with increased selection in vaccinated populations (15). However, the direct correlation of antigenic and genetic evolution of AIVs and the use of vaccination remains difficult, as other factors may contribute to virus evolution in poultry populations, such as density, species diversity, and strain variability.
The 2002-2004 program for vaccination of poultry using a heterologous vaccine alongside more traditional controls such as culling and controlled animal movement enabled the eradication of the LPAI H7N3 virus from the population. The reemergence of the virus in 2003 was predominantly in unvaccinated farms, and vaccination of these flocks and revaccination of flocks of previously vaccinated birds with a heterologous vaccine assisted in control. Our genetic and antigenic characterization of a subset of isolates demonstrated that there was some positive pressure on the virus to change after vaccination. Interestingly, this pressure was selecting for virus identified in vaccinated populations to become more homogenous, with greater genetic difference observed in the unvaccinated population. This would suggest that the selection of vaccine strain is a critical factor in the selection pressure applied to the circulating strain. The use of a heterologous vaccine strain was successfully applied in Italy to control the infection (12, 41) and, if managed properly, might be able to deal with the antigenic changes of field isolates as suggested by the data presented.
Our study suggests that LPAI should be monitored with the longitudinal collection of viruses together with all relevant epidemiological information that would allow studies on the genome sequence and antigenic typing of LPAI viruses. This would enable a greater understanding of the dynamics of subtype evolution within different populations.
This is highlighted by the recent outbreaks of LPAI H7N9 that have been noted to occur in pigeons and chickens present in markets, all located in Shanghai, China, and neighboring provinces. Poultry that have tested positive for the presence of H7N9 and are also suspected of being the source of reported human cases did not show any visible signs of disease, making it very difficult to detect this virus in poultry. In addition to culling, the key options for controlling the virus at the animal source in the medium term, and preventing its regional and global spread, could be the implementation of a suitably adapted vaccination policy of limited duration, especially in areas where the culling policy is difficult to apply. The experience gained during the H5N1 HPAI virus crisis and other major epidemics caused by HPAI and LPAI viruses may provide the global community with a benefit in the field of vaccines and vaccination strategies. As a matter of fact, studies conducted on H7N3 LPAI viruses that caused several outbreaks in a 2-year period in Italy demonstrate that surveillance and retrospective investigation on genetic and antigenic characteristics of notifiable LPAI viruses are useful to aid decision makers in controlling avian influenza virus infections.
ACKNOWLEDGMENTS
This study was funded by the European project FLUAID SSPE-CT-2005-022417, from the European Union's Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 278433. Yifei Xu, Li-Ping Long, and Xiu-Feng Wan were supported partially by RC1AI086830 and P20RR032694 from the U.S. National Institutes of Health.
We thank Manuela Dalla Pozza and Marica Toson for assisting with the selection of vaccinated and unvaccinated turkey farms and Nadia Micoli and Meredith Stewart for proofreading the manuscript and providing valuable comments.
Footnotes
Published ahead of print 19 February 2014
REFERENCES
- 1. Werner O, Harder TC. 2006. Avian influenza, p 48–88 In Kamps BS, Hoffmann C, Preiser W. (ed), Influenza report 2006. Flying Publisher, Paris, France: . [Google Scholar]
- 2. Xu X, Subbarao Cox NJ, Guo Y. 1999. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261:15–19. 10.1006/viro.1999.9820 [DOI] [PubMed] [Google Scholar]
- 3. Perdue ML, Swayne DE. 2005. Public health risk from avian influenza viruses. Avian Dis. 49:317–327. 10.1637/7390-060305R.1 [DOI] [PubMed] [Google Scholar]
- 4.WHO/OIE/FAO H5N1 Evolution Working Group. 2012. Continued evolution of highly pathogenic avian influenza A (H5N1): updated nomenclature. Influenza Other Respir. Viruses 6:1–5. 10.1111/j.1750-2659.2011.00298.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Villarreal C. 2009. Avian influenza in Mexico. Rev. Sci. Tech. 28:261–265 [DOI] [PubMed] [Google Scholar]
- 6. Villarreal CL. 2006. Control and eradication strategies of avian influenza in Mexico. Dev. Biol. (Basel) 124:125–126 [PubMed] [Google Scholar]
- 7. Naeem K, Siddique N. 2006. Use of strategic vaccination for the control of avian influenza in Pakistan. Dev. Biol. (Basel) 124:145–150 [PubMed] [Google Scholar]
- 8. Capua I, Marangon S, dalla Pozza M, Terregino C, Cattoli G. 2003. Avian influenza in Italy 1997–2001. Avian Dis. 47:839–843. 10.1637/0005-2086-47.s3.839 [DOI] [PubMed] [Google Scholar]
- 9. Swayne DE, Kapczynski D. 2008. Strategies and challenges for eliciting immunity against avian influenza virus in birds. Immunol. Rev. 225:314–331. 10.1111/j.1600-065X.2008.00668.x [DOI] [PubMed] [Google Scholar]
- 10. Lee CW, Senne DA, Suarez DL. 2004. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J. Virol. 78:8372–8381. 10.1128/JVI.78.15.8372-8381.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Escorcia M, Carrillo-Sanchez K, March-Mifsut S, Chapa J, Lucio E, Nava GM. 2010. Impact of antigenic and genetic drift on the serologic surveillance of H5N2 avian influenza viruses. BMC Vet. Res. 6:57. 10.1186/1746-6148-6-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capua I, Marangon S. 2007. The use of vaccination to combat multiple introductions of notifiable avian influenza viruses of the H5 and H7 subtypes between 2000 and 2006 in Italy. Vaccine 25:4987–4995. 10.1016/j.vaccine.2007.01.113 [DOI] [PubMed] [Google Scholar]
- 13. Swayne DE. 2009. Avian influenza vaccines and therapies for poultry. Comp. Immunol. Microbiol. Infect. Dis. 32:351–363. 10.1016/j.cimid.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 14. Ducatez MF, Cai Z, Peiris M, Guan Y, Ye Z, Wan XF, Webby RJ. 2011. Extent of antigenic cross-reactivity among highly pathogenic H5N1 influenza viruses. J. Clin. Microbiol. 49:3531–3536. 10.1128/JCM.01279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cattoli G, Fusaro A, Monne I, Coven F, Joannis T, El-Hamid HS, Hussein AA, Cornelius C, Amarin NM, Mancin M, Holmes EC, Capua I. 2011. Evidence for differing evolutionary dynamics of A/H5N1 viruses among countries applying or not applying avian influenza vaccination in poultry. Vaccine 29:9368–9375. 10.1016/j.vaccine.2011.09.127 [DOI] [PubMed] [Google Scholar]
- 16. Beato MS, Mancin M, Yang J, Buratin A, Ruffa M, Maniero S, Fusaro A, Terregino C, Wan XF, Capua I. 2013. Antigenic characterization of recent H5N1 highly pathogenic avian influenza viruses circulating in Egyptian poultry. Virology 435:350–356. 10.1016/j.virol.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El-Shesheny R, Kayali G, Kandeil A, Cai Z, Barakat AB, Ghanim H, Ali MA. 2012. Antigenic diversity and cross-reactivity of avian influenza H5N1 viruses in Egypt between 2006 and 2011. J. Gen. Virol. 93:2564–2574. 10.1099/vir.0.043299-0 [DOI] [PubMed] [Google Scholar]
- 18. Balish AL, Davis CT, Saad MD, El-Sayed N, Esmat H, Tjaden JA, Earhart KC, Ahmed LE, Abd El-Halem M, Ali AH, Nassif SA, El-Ebiary EA, Taha M, Aly MM, Arafa A, O'Neill E, Xiyan X, Cox NJ, Donis RO, Klimov AI. 2010. Antigenic and genetic diversity of highly pathogenic avian influenza A (H5N1) viruses isolated in Egypt. Avian Dis. 54:329–334. 10.1637/8903-042909-Reg.1 [DOI] [PubMed] [Google Scholar]
- 19. Abbas MA, Spackman E, Fouchier R, Smith D, Ahmed Z, Siddique N, Sarmento L, Naeem K, McKinley ET, Hameed A, Rehmani S, Swayne DE. 2011. H7 avian influenza virus vaccines protect chickens against challenge with antigenically diverse isolates. Vaccine 29:7424–7429. 10.1016/j.vaccine.2011.07.064 [DOI] [PubMed] [Google Scholar]
- 20. Capua I, Marangon S. 2007. The challenge of controlling notifiable avian influenza by means of vaccination. Avian Dis. 51:317–322. 10.1637/7560-033106R.1 [DOI] [PubMed] [Google Scholar]
- 21. Cattoli G, Terregino C, Bertoli E, Ortali G, Marangon S, Capua I. 2003. The H7N3 avian influenza outbreak in Northern Italy in 2002, p 23–25 In Proceedings of the Fifty-Second Western Poultry Disease Conference, Sacramento, CA [Google Scholar]
- 22.World Organisation for Animal Health. 2010. Terrestrial animal health code. World Organisation for Animal Health, Paris, France [Google Scholar]
- 23. Reference deleted.
- 24.Ministry of Health. 1992. Decreto legislative 27 gennaio 1992, no. 116. Attuazione della Direttiva 86/609/EEC in materia di protezione degli animali utilizzati a fini sperimentali o ad altri fini scientifici. Gazzetta Ufficiale no. 40 (Suppl. ord.). Ministry of Health, Rome, Italy [Google Scholar]
- 25. Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. thesis. The University of Texas at Austin, Austin, TX [Google Scholar]
- 26. Wilgenbusch JC, Swofford D. 2003. Inferring evolutionary trees with PAUP*. Curr. Protoc. Bioinformatics 00:6.4.1–6.4.28. 10.1002/0471250953.bi0604s00 [DOI] [PubMed] [Google Scholar]
- 27. Wan XF, Nguyen T, Davis CT, Smith CB, Zhao ZM, Carrel M, Inui K, Do HT, Mai DT, Jadhao S, Balish A, Shu B, Luo F, Emch M, Matsuoka Y, Lindstrom SE, Cox NJ, Nguyen CV, Klimov A, Donis RO. 2008. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One 3:e3462. 10.1371/journal.pone.0003462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 29. Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. 2010. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 6:e1001081. 10.1371/journal.ppat.1001081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson IA, Cox NJ. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8:737–771. 10.1146/annurev.iy.08.040190.003513 [DOI] [PubMed] [Google Scholar]
- 31. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barnett JL, Yang J, Cai Z, Wan XF. 2012. AntigenMap 3D: an online antigenic cartography resource. Bioinformatics 28:1292–1293. 10.1093/bioinformatics/bts105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cai Z, Zhang T, Wan XF. 2010. A computational framework for influenza antigenic cartography. PLoS Comput. Biol. 6:e1000949. 10.1371/journal.pcbi.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, Facchini M, Chiapponi C, Foni E, Cordioli P, Webby R, Barigazzi G, Webster RG, Donatelli I. 2004. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology 323:24–36. 10.1016/j.virol.2004.02.015 [DOI] [PubMed] [Google Scholar]
- 35. Cilloni F, Toffan A, Giannecchini S, Clausi V, Azzi A, Capua I, Terregino C. 2010. Increased pathogenicity and shedding in chickens of a wild bird-origin low pathogenicity avian influenza virus of the H7N3 subtype following multiple in vivo passages in quail and turkey. Avian Dis. 54:555–557. 10.1637/8919-050809-Reg.1 [DOI] [PubMed] [Google Scholar]
- 36. Vijaykrishna D, Bahl J, Riley S, Duan L, Zhang JX, Chen H, Peiris JS, Smith GJ, Guan Y. 2008. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 4:e1000161. 10.1371/journal.ppat.1000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peyre M, Samaha H, Makonnen YJ, Saad A, Abd-Elnabi A, Galal S, Ettel T, Dauphin G, Lubroth J, Roger F, Domenech J. 2009. Avian influenza vaccination in Egypt: limitations of the current strategy. J. Mol. Genet. Med. 3:198–204. 10.4172/1747-0862.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abdelwhab EM, Hafez HM. 2011. An overview of the epidemic of highly pathogenic H5N1 avian influenza virus in Egypt: epidemiology and control challenges. Epidemiol. Infect. 139:647–657. 10.1017/S0950268810003122 [DOI] [PubMed] [Google Scholar]
- 39. Cattoli G, Terregino C, Brasola V, Rodriguez JF, Capua I. 2003. Development and preliminary validation of an ad hoc N1-N3 discriminatory test for the control of avian influenza in Italy. Avian Dis. 47:1060–1062. 10.1637/0005-2086-47.s3.1060 [DOI] [PubMed] [Google Scholar]
- 40. Capua I, Terregino C, Cattoli G, Mutinelli F, Rodriguez JF. 2003. Development of a DIVA (Differentiating Infected from Vaccinated Animals) strategy using a vaccine containing a heterologous neuraminidase for the control of avian influenza. Avian Pathol. 32:47–55. 10.1080/0307945021000070714 [DOI] [PubMed] [Google Scholar]
- 41. Capua I, Marangon S. 2007. Control and prevention of avian influenza in an evolving scenario. Vaccine 25:5645–5652. 10.1016/j.vaccine.2006.10.053 [DOI] [PubMed] [Google Scholar]