Abstract
Vaccination has been one of the most important interventions in disease prevention and control. The impact of vaccination largely depends on the quality and suitability of the chosen vaccine. To determine the suitability of a vaccine strain, antigenic matching is usually studied by in vitro analysis. In this study, we performed three in vitro test methods to determine which one gives the lowest variability and the highest discriminatory capacity. Binary ethylenimine inactivated vaccines, prepared from 10 different foot-and-mouth disease (FMD) virus serotype A strains, were used to vaccinate cattle (5 animals for each strain). The antibody titers in blood serum samples 3 weeks postvaccination (w.p.v.) were determined by a virus neutralization test, neutralization index test, and liquid-phase blocking enzyme-linked immunosorbent assay (ELISA). The titers were then used to calculate relationship coefficient (r1) values. These r1 values were compared to the genetic lineage using receiver operating characteristic (ROC) analysis. In the two neutralization test methods, the median titers observed against the test strains differed considerably, and the sera of the vaccinated animals did not always show the highest titers against their respective homologous virus strains. When the titers were corrected for test strain effect (scaling), the variability (standard error of the mean per vaccinated group) increased because the results were on a different scale, but the discriminatory capacity improved. An ROC analysis of the r1 value calculated on both observed and scaled titers showed that only r1 values of the liquid-phase blocking ELISA gave a consistent statistically significant result. Under the conditions of the present study, the liquid-phase blocking ELISA showed less variation and still had a higher discriminatory capacity than the other tests.
INTRODUCTION
Foot-and-mouth disease (FMD) is highly contagious infectious disease that affects all cloven-hoofed animals and is one of the most economically important diseases of livestock. The virus belongs to the genus Aphthovirus, in the family Picornaviridae (1). It has seven different serotypes, namely, O, A, C, Asia 1, and Southern African Territories (SAT) 1, 2, and 3, with multiple subtypes within each serotype (2). FMD virus (FMDV) is a highly variable RNA virus (3–5), and in general, there is little or no cross-protection between serotypes and even between different strains of the same serotype (2, 6, 7). Serotype A is considered to be the most antigenically and genetically diverse of the FMDV serotypes, and new antigenic variants emerge frequently (8, 9).
An inactivated FMDV vaccine is being used to control FMD. This type of vaccine is commonly used in many parts of the world, namely, South America, Asia, and Africa, where FMD is endemic. Although the quality of the vaccine used is probably the most important factor for the success of a vaccination program, a reasonable antigenic match between the FMDV vaccine and the outbreak virus strains is also considered essential for the effectiveness of the vaccine (6). Initially, in vivo cross-protection studies were performed to test for antigenic differences. However, since serological tests became available, they are also being used for antigenic matching on the basis of the assumption/hypothesis that the level of protection is correlated with the antigenic match from the serological tests. In such serological tests, the antibody titers of serum samples collected from vaccinated animals against both the vaccine strain and field strain virus are determined (6, 10). The value that is used the most to express antigenic match is the relationship coefficient (r1 value), which is the ratio of the titers obtained in serological tests using the heterologous (field) strains and the vaccine strain (7, 10–12).
Serological test methods have been used to quantify antigenic differences between FMDV structural protein antibodies and thereby to estimate vaccine matching between a vaccine strain and a field isolate. Antigenic analysis of field isolates in relation to vaccine strains, based on virus neutralization tests (VNTs), plays a significant role in evaluating the suitability of existing vaccine strains (6, 10, 11), although significant variation has been reported with VNTs (13, 14). The genetic diversity in the P1-coding region within the SAT serotypes is reflected in the antigenic properties of these viruses; therefore, there are implications for the selection of vaccine strains that would provide the best vaccine match against emerging viruses (15). In the antigenically diverse FMDV serotype A strains, predicting antigenic differences using genetic sequences alone does not provide reliable information for vaccine matching (16). Therefore, others have added structural information on the location of the amino acid sequence in the virus to the sequence data and have shown that it was a powerful tool for predicting antigenic relationships and has the potential to be applied to a variety of different infectious agents (17).
In studies in which cross-protection was related to cross-reactions in serology (10, 18, 19), the relationship between cross-protection and cross-reactions varies. In some studies, protection was observed when high-potency (type A) vaccines were being used (10), even when the r1 values were low (range, 0.04 to 0.23). In other studies, no cross-protection was detected after vaccination with the A/Iran/05 FMDV vaccine and challenge with A/TUR/64/11 (18), while the r1 value of 0.1 fell within the range of the r1 values found in the previous studies (10). For the FMDV type O Manisa vaccine, a reasonable high r1 value of 0.6 was observed against O Campos, but a vaccine containing 15 μg of O Manisa antigen protected in 99% of the cases against homologous challenge but only in 54% of the cases against O Campos challenge (19). This shows that a highly potent O Manisa vaccine did not protect sufficiently against a heterologous O Campos challenge even though the r1 value indicated it should. This shows that r1 values are not always consistent with the challenge results. Since protection against a challenge with FMDV is well correlated with serology results (20), variation in r1 value determination might have caused this inconsistency. Cross-validation studies on r1 value determination between laboratories (18, 21) showed huge variation between the laboratories and techniques used. The exact cause of variation was not established, as there were too many variables. A recent study (22) analyzed the r1 value of the VNT using ROC analysis. They showed that the r1 value did not predict cross-protection. Other authors have linked genetic information to cross-reactions in serology (15–17) and have showed that the r1 value can be predicted from surface-exposed amino acid changes. These studies did not address the inherent variability of the r1 value, and it is therefore important to have a more comprehensive view on the usefulness of r1 values using results generated with various FMDV strains in one laboratory. One of the first questions is to identify which test should be used to test cross-reaction in serology.
In this study, we analyzed the cross-reactions in serology using serum samples from cattle vaccinated with 10 different FMDV serotype A strain vaccines using three serological test methods, with the objective of determining which serological test method provides the least variation within a group of vaccinated cattle and provides the best discrimination between vaccines.
MATERIALS AND METHODS
FMDV serotype A strains.
Different serotype A FMDV (n = 10) strains that were isolated in Africa, the Middle East, and Europe were selected from various genetic lineages (Table 1). The 10 strains were A/KEN/12/2005, A/ERI/2/98, A/SUD/2/84, A/ETH/13/2005, and A/MAU/1/2006, which were received from the OIE/FAO World Reference Laboratory for FMD (The Pirbright Institute, United Kingdom), and A10/Holland/42, A22/IRQ/24/64, A/TUR/20/2006, A/TUR/14/98, and A/IRN/2/97, obtained from the Central Veterinary Institute (CVI) (Wageningen UR, Lelystad, The Netherlands). For a control on the strain identity, we sequenced the VP1 gene of the 10 FMDV serotype A strains and compared them to data in the NCBI database.
TABLE 1.
FMDV serotype A strains used in the study
| FMDV type A strain | Lineage/sublineagea | GenBank accession no. (VP1)b | Reference |
|---|---|---|---|
| A10/Holland/42 | EURO-SA | AY593751 | 41 |
| A22/IRQ/24/64 | ASIA | AY593764 | 41 |
| A/ERI/2/98 | G-IV | EU919238 | 42 |
| A/ETH/13/2005 | G-II | FJ798145 | 43 |
| A/IRN/2/97 | Iran-96 | KF152935 | 43 |
| A/KEN/12/2005 | G-I | KF112912 | 16 |
| A/MAU/1/2006 | G-VI | JF749842 | 44 |
| A/SUD/2/84 | G-IV | GU566067 | 45 |
| A/TUR/20/2006 | Iran-05 | FG755116 | 46 |
| A/TUR/14/98 | Iran-96 | DQ296537 | 47 |
Ten strains came from 8 different lineages (as defined by the World Reference Laboratory). Two lineages, Iran-96 and G-IV, contained two strains. Within lineage Iran-96, both strains were 97.7% similar in VP1 on the amino acid level, and within lineage G-IV, both strains were 93.1% similar in VP1 on the amino acid level.
Best matching VP1 sequence in NCBI.
Vaccine (antigen production).
The vaccines employed in this study were noncommercially produced at the CVI for this particular study. The vaccines were prepared from the aforementioned 10 different FMDV serotype A strains after growing them on monolayers of BHK-21 cells in 850-cm2 roller bottles (Corning tissue culture treated, nonpyrogenic; Corning Incorporated, NY). The viruses were subsequently inactivated with 10 mM binary ethylenimine (BEI) (23, 24). The BEI was neutralized by 0.79% (wt/vol) sodium thiosulfate, and the inactivated antigens were concentrated with one cycle of 8.0% (wt/vol) polyethylene glycol (PEG) precipitation. The antigens were checked by standard VP1 sequencing (25). The 146S antigen concentration was determined by quantitative sucrose density gradient analysis, as previously described (26, 27). A known 146S FMDV antigen reference standard was tested with each run. The aqueous vaccines were formulated using 2% aluminum hydroxide (Alhydrogel; Ph. Eur. Brenntag Biosector) and 10% (wt/vol) saponin (Quil-A solution in phosphate-buffered saline [PBS]) as the adjuvant. The vaccine payload was 10 μg of antigen per 2-ml dose and was expected to have ≥3× the 50% protective dose (PD50). The antigens were tested for the presence of live FMD virus both in vitro and in vivo.
Vaccination of cattle.
Vaccinations were performed in 2010. Healthy unvaccinated Eritrean local indigenous zebu breed (Bos indicus) cattle, 10 to 18 months of age, were used for this study. The cattle used originated from regions known to be free from FMD without vaccination. Before purchase and prior to vaccination, all cattle were ear tagged and bled twice at a 2-week interval and tested for presence of anti-FMDV nonstructural protein (NSP) antibodies using the FMD nonstructural enzyme-linked immunosorbent assay (NS-ELISA) (PrioCHECK) (28). Only cattle with a negative result with the FMD NS-ELISA were purchased. The cattle were housed in stables at the National Veterinary Laboratory, Asmara, Eritrea. The cattle were randomly allocated into 11 groups of 5 cattle. In total, we used 10 groups for the 10 different FMDV serotype A vaccines and 10 cattle as nonvaccinated controls to monitor incursions of FMD infection that would influence the experiment. The cattle in each vaccine group were vaccinated subcutaneously in the middle of the cervical area. Blood samples were collected at 0, 7, 14, 21, and 28 days postvaccination for serology testing.
Rabbit and guinea pig antisera for liquid-phase blocking ELISA.
In total, 20 rabbits and 20 guinea pigs (two for each FMDV serotype A strain antigens) were immunized as described by Hamblin et al. (29) using the same semipurified PEG precipitated antigens as those used to vaccinate the cattle in this study.
The vaccine was prepared by mixing an FMDV serotype A antigen with an equal volume of Montanide ISA 206 VG oil adjuvant to get a final concentration of 5 μg/ml. The rabbits were vaccinated subcutaneously and intramuscularly with 0.5 ml of vaccine at each site. The guinea pigs were vaccinated subcutaneously with 0.5 ml of vaccine. All animals were boosted 2 weeks postvaccination (w.p.v.) with the same protocol used for primary vaccination. The animals were bled 10 to 12 days after the booster vaccination by exsanguination under general anesthesia.
FMDV NS-ELISA.
The PrioCHECK FMDV NS-ELISA (Prionics) (28) is a blocking ELISA that detects antibodies directed against the nonstructural 3ABC protein of FMDV. This ELISA detects FMDV-infected animals independently of the serotype causing the infection. The serum samples from the animals were tested by the PrioCHECK FMDV NS-ELISA kit, and the test results were interpreted as indicated by the producer, i.e., <50 percent inhibition (PI) was considered negative to NS antibodies and ≥50 PI values was considered positive to NS antibodies.
Virus neutralization test.
A VNT was performed as described in the OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (7). It was performed using BHK-21 cells in flat-bottomed tissue culture-grade microtiter plates (Greiner Bio-One). Stock viruses for the VNT were grown in cell monolayers, titrated, and stored at −70°C until used. The experimental serum samples were inactivated at 56°C for 30 min before testing. Dulbecco's modified Eagle's medium (DMEM) (Gibco/Invitrogen) supplemented with 5% fetal calf serum (FCS) and 2% antibiotics was used to dilute the sera and virus, and also as a medium for growing the cells. All 3-w.p.v. sera were tested in duplicate by mixing 50 μl of 2-fold dilutions of the serum with a 50-μl virus suspension containing 100× the 50% tissue culture infective dose (TCID50) FMDV. This mixture was incubated for 1 h at 37°C and 5% CO2, in a humidified atmosphere. After 1 h of incubation, 150 μl BHK-21 cell suspension was added to each well, and the plates were incubated for 3 days at 37°C and 5% CO2, in a humidified atmosphere. After 3 days, the monolayers were stained with 50 μl amido black solution (1 g/liter in water containing 88 mM sodium acetate, 10% glycerol, and 5.4% acetic acid), and the cytopathic effect (CPE) was read macroscopically. The endpoint titers of the serum samples, tested against all 10 vaccine strains, were expressed as the logarithm (base 10) of the reciprocal of the last dilution of serum that neutralized 100× TCID50 of the virus in 50% of the wells (30).
Serum samples collected at 0, 1, 2, 3, and 4 w.p.v. were tested by VNT using the homologous test strain to see the response of neutralizing antibodies postvaccination. For a comparison of the tests, the 3-w.p.v. sera were tested against all 10 virus strains used for vaccination. In the calculation of the mean titers, VNT titers with a titer of <0.3 at the day of vaccination were considered to be 0. In the calculations of r1 values and scaled titers, the titers below the detection limit (0.3) were excluded, since no titer was measured.
Neutralization index test.
In the neutralization index test (NIT), 50 μl of 10-fold dilutions of the virus (6 wells per dilution) was incubated with 50 μl of a fixed dilution of a 1:25 dilution of test serum for 1 h at 37°C and 5% CO2, in a humidified atmosphere. In each test, a titration mixed with 50 μl of dilution medium (DMEM with 2% fetal bovine serum and antibiotics) instead of the test serum was included. After 1 h, 150 μl BHK-21 cell suspension was added to each well, and the plates were incubated for 3 days at 37°C and 5% CO2, in a humidified atmosphere. After 3 days, the plates were washed and the monolayers were stained with 50 μl of amido black solution (see above), and the CPE was read macroscopically. The log10 titers of the virus with and without serum were calculated (30). The neutralization index log10 titer was calculated by subtracting the virus log10 titer of the strain with test serum from the log10 titer of the strain without serum. All 3-w.p.v. sera were tested against all 10 FMDV serotype A strains used for vaccination.
LPB-ELISA.
The liquid-phase blocking ELISA (LPB-ELISA) was performed as described by Hamblin et al. (29). Specific rabbit sera for coating and the guinea pig sera for detection were produced for each of the 10 strains. The rabbit and guinea pig antisera were partially purified by precipitation using saturated aluminum sulfate. The dilutions of coating antibody and detecting antibodies were optimized in the laboratory, as well as the rabbit anti-guinea pig conjugate (P0141; Dako).
Antigens were prepared from the 10 FMDV type A strains by growing them on monolayers of BHK-21 cells. After freeze-thawing the cell culture and centrifuging for 10 min at 250 × g, the unpurified supernatants were titrated, and the final dilution that gave an optical density (OD) at 450 nm of approximately 1.2 to 1.5 in the ELISA was used. ELISA plates (Corning Costar) were coated with a predetermined dilution of 100 μl/well rabbit anti-FMDV antibodies diluted in bicarbonate buffer (pH 9.6) and left overnight at room temperature. On the same day, in U-bottomed microtiter plates (dummy plate), each test serum sample was diluted in a 2-fold dilution series in duplicate wells, starting with a 1:5 dilution (60 μl/well) using phosphate-buffered saline containing 0.05% Tween 80, 0.5 M NaCl, and 5% FCS (ELISA buffer). Sixty microliters of a predetermined concentration of antigen, diluted in ELISA buffer, was added, mixed, and incubated overnight at 4°C. The next day, the ELISA plates were washed 6 times with tap water containing 0.05% Tween 80. Next, 100 μl of test serum-antigen mixture was transferred from dummy plates into the corresponding wells of the rabbit serum-coated ELISA plates and incubated at 37°C for 1 h. After washing, 100 μl of the guinea pig antiserum (in a predetermined dilution) homologous to the viral antigen used was added to each well and incubated at 37°C for 1 h. The plates were washed, and 100 μl of a predetermined dilution of rabbit anti-guinea pig immunoglobulins conjugated with horseradish peroxidase was added to all wells and incubated for 1 h at 37°C. The guinea pig antiserum and the rabbit anti-guinea pig conjugate were diluted in ELISA buffer. After washing, 100 μl of a substrate-chromogen solution (BioFX TMBS-1000-01) was added, and the plates were left to develop color for 10 min at room temperature. The reaction was stopped by adding 100 μl of 0.5-M sulfuric acid. The plates were read at 450 nm, and the log10 antibody titers were expressed as the log10 of the reciprocal of the final dilution of serum giving 50% of the mean OD value recorded in the maximum signal control wells (two wells with ELISA buffer instead of serum).
Data analysis. (i) Mean titers.
The mean titers were computed by adding the log10 titers and dividing by the number of observations.
(ii) Scaled titer (correction for strain effect).
Scaled titers were used to minimize the variations observed between neutralization tests with different viruses. For each specified serotype A strain, the titer was standardized using the formula (observed titer − mean titer for all sera tested against a specified test strain)/(standard deviation of all sera tested against a specified test strain). Therefore, for each specified strain, the mean scaled titer is 0, with a standard deviation of 1.0 (31).
(iii) Determination of r1 values.
A one-way antigenic relationship (r1 value) for each of the 3-w.p.v. serum samples was determined using VNT, NIT, and LPB-ELISA titers, as described in the OIE manual (7) using the equation r1 = (serum titer against heterologous virus)/(serum titer against homologous virus), where the heterologous virus is a field strain and the homologous virus is a vaccine strain.
To test whether the original and scaled titers of the sera from vaccinated cattle were able to discriminate between the various vaccine strains, we used an analysis of variance (ANOVA). In the ANOVA, we tested whether the titers induced by each vaccine were similar. If the ANOVA showed that one or more of the vaccine groups was significantly different, all possible pairwise comparisons were tested with a t test using Holm's adjustment for multiple testing. A P value of <0.05 was considered statistically significant. Statistical analyses were carried out using R version 2.14 (32).
(iv) Receiver operating characteristic analysis.
An ROC analysis was carried out to determine which serological technique and the r1 value derived from that technique had the best sensitivity and specificity in determining differences between the test strains. In the first analysis, we assumed that the two strains from the lineage Iran-96 have the same antigenic make-up (gold standard for sensitivity and specificity). In this case, the amino acid homology in VP1 was 97.5%. In the second, we assumed that strains within the Iran-96 and the G-IV lineages were antigenically similar. In that case, the amino acid homology in VP1 was 93.1%. The ROC analysis was performed using the pROC library in R (33).
RESULTS
FMDV vaccine strains.
The antigens used for vaccination came from 8 different lineages defined by the World Reference Laboratory in Pirbright (Table 1). In two lineages, we had two vaccine candidates. In the lineage Iran-96, we selected A/IRN/2/79 and A/TUR/14/98, which are 97.7% similar in VP1 at the amino acid level. In lineage G-IV, we selected A/ERI/2/98 and A/SUD/2/84, which are 93.1% similar in VP1 at the amino acid level. The identity at the amino acid level ranged from 84.3 to 91.4% for strains that did not belong to the same lineage. The VP1 sequences were compared to known sequences in GenBank. For most strains, a match (99 to 100% nucleotide identity) was found, and these are shown in Table 1. For our A/KEN/12/2005 VP1 sequence only, the best match (93% nucleotide identity) was with A/KEN/22/2009 (GenBank accession no. KF112912). Both the in vitro and in vivo innocuity tests did not detect live FMDV in the BEI-inactivated antigens produced from these 10 strains.
FMDV NS-ELISA.
All sera collected at the start of the experiment were negative in the Prionics NS-ELISA (28). In total, 7 cattle were positive in the NS-ELISA postvaccination, and in 4 of them only at one time point postvaccination (1, 3, or 4 w.p.v.), in 2 cattle at two time points, and in one cow at 2, 3, and 4 w.p.v. The maximum percent inhibition observed was 58%.
Postvaccination antibody response.
Serum samples collected from all cattle at 1, 2, 3, and 4 w.p.v. were tested using VNT against the homologous vaccine test strain. In some cattle, a low reaction was detected in the VNT before vaccination (Fig. 1); this was also seen in the sera of the control animals, especially with test strain A10/Holland/42. All vaccinated cattle responded to vaccination, with an increase in the log10 antibody titer in the first week ranging from 0.3 (2-fold) to 3.0 (1,000-fold), with a median of 1.1. At 2 to 4 w.p.v., the increase was ≥0.6 (4-fold) compared to the log10 titer before vaccination, with a median increase of 2.1 log10. The mean 3-w.p.v. homologous log10 titer in the VNT ranged from 1.5 to 3.5 (Fig. 1). The mean VNT titer 3 w.p.v. against the homologous and 9 heterologous virus test strains used in the VNT in the different vaccine groups ranged from 1.6 for A/MAU/1/2006 to 2.0 for A/TUR/20/2006, with an overall mean titer of 1.8.
FIG 1.
VNT homologous mean titers, including standard error of the mean values of FMDV serotype A strains postvaccination responses in cattle at 0, 1, 2, 3, and 4 w.p.v.
Analyzing differences between test strains using different serological test methods.
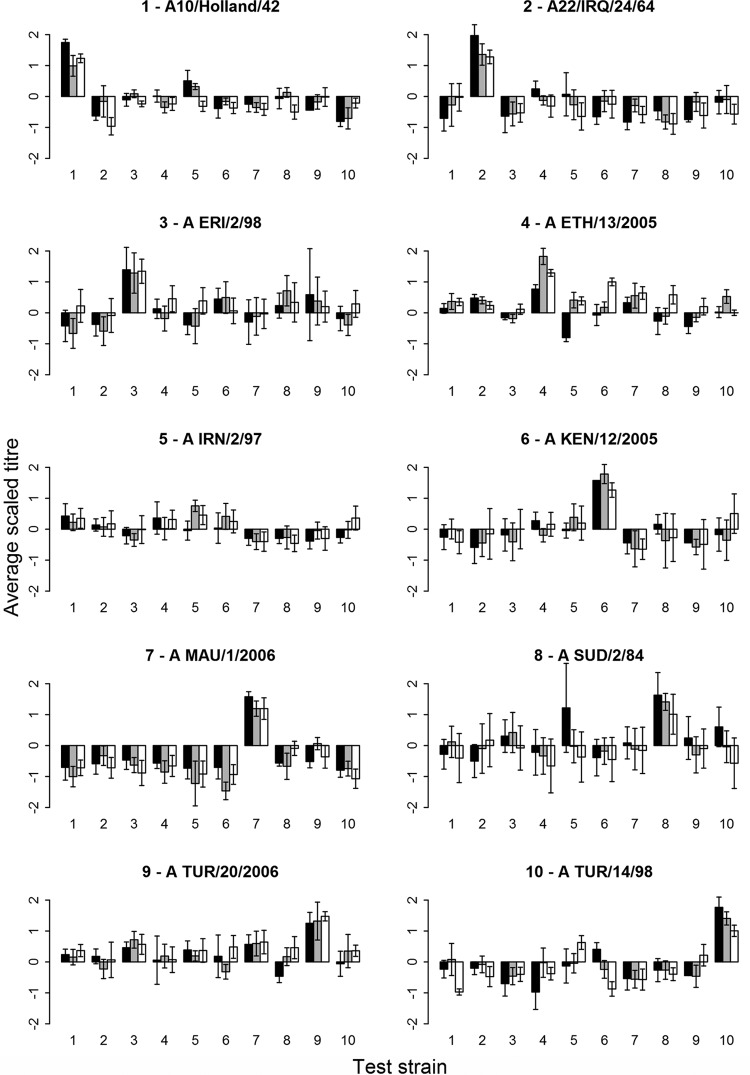
All 3-w.p.v. sera were tested and analyzed by the three serological assays (VNT, NIT, and LPB-ELISA) against the homologous and heterologous virus strains. The mean titers and standard error of the mean (SEM) were calculated for each vaccine (5 sera per vaccine) (Fig. 2). The correlation between VNT and NIT was higher (0.72) than the correlation between NIT and LPB-ELISA (0.55) or VNT and LPB-ELISA (0.49). Some virus test strains were more easily neutralized than others. Consequently, the mean titer against the homologous strain is not always the highest one. With the test strains A/IRN/2/97, A/SUD/2/84, and A/TUR/20/2006, low responses were observed with all serum samples from vaccinated animals for both VNT and NIT, similar to the homologous strain shown in Fig. 1. Because of these inherent variations between the test strains, many sera were found to have homologous titers that were lower than some of the heterologous ones. Figure 2 shows that the mean VNT (in 4 of 10 strains), the mean NIT (in 3 out of 10 strains), and the mean LPB-ELISA (in 2 out of 10 strains) homologous titers were not the highest.
FIG 2.
Mean titers and the standard error of the mean (SEM) values of the three serological methods, i.e., VNT (black), NIT (gray), and LPB-ELISA (white) for each vaccine and tested against the corresponding test strain (same number means the same strain). The individual graphs show the log10 mean titers and the standard error of the mean of the sera of each group of vaccinated cattle (5 cattle) tested against the 10 different FMDV type A test strains (x axis). The homologous responses are those for which the number of the test strain (x axis) is equal to the number in the title of a particular panel.
Scaled titers (correction for strain effect).
Because some virus strains were more easily neutralized than others, as mentioned above, we used scaled titers to correct and standardize for the test strain effect. Using scaled titers, the mean homologous response was in general the highest one (Fig. 3). Table 2 shows the range of the standard error of the mean (SEM) determined for each group of 5 vaccinated cattle tested against each of the test strains for both the observed and scaled titers. Before scaling, the LPB-ELISA had the lowest maximum SEM. After scaling, both the NIT and LPB-ELISA had the lowest maximum SEM values.
FIG 3.
Mean scaled titers, including the standard error of the mean values for the VNT (black), NIT (gray), and LPBE (white). The homologous responses are those for which the number of the test strain (x axis) is equal to the number in the title of a particular panel.
TABLE 2.
Comparison of the 3 serological test methods using titers and scaled titers, and a comparison of the SEM range within groups of 5 cattle
| Test method | SEM range of observed titers | SEM range of scaled titers |
|---|---|---|
| VNT | 0–0.98 | 0–1.49 |
| NIT | 0.06–0.78 | 0.10–0.88 |
| LPB-ELISA | 0.04–0.40 | 0.09–0.87 |
r1 value.
One would expect the r1 value to range between 0 and 1, but that was only observed in 78%, 92%, and 96% of VNT, NIT, and LPB-ELISA analyses, respectively (Table 3). The maximum r1 values were 178, 1,148, and 2.8 for VNT, NIT, and LPB-ELISA, respectively. After scaling, a higher proportion of r1 values were found that were between 0 and 1 (Table 3).
TABLE 3.
Percentage of r1 values within the range 0 to 1 before and after scaling the titers
| Test | Before scaling titer (%) | After scaling titer (%) |
|---|---|---|
| VNT | 78 | 93 |
| NIT | 83 | 94 |
| LPBE | 96 | 96 |
Discriminating between vaccines.
As stated above, the mean titers induced by the different vaccines were similar; therefore, we used an ANOVA to determine whether there were significant differences between vaccines. Using titers and scaled titers obtained by VNT, no significant differences were found between the vaccines. Using titers and scaled titers obtained by NIT, a significant difference between the test strains was observed in the ANOVA and in 1 (based on titer) or 3 (based on scaled titers) out of the 45 (Table 4) possible pairwise comparisons. Using titers and scaled titers obtained by the LPB-ELISA, a significant difference between the test strains was observed in the ANOVA and in 7 (based on titers) or 12 (based on scaled titers) out of the 45 possible pairwise comparisons (Table 4).
TABLE 4.
P values of the pairwise t test results from the serological tests in which the ANOVA showed a significant difference between vaccine strains
| Vaccine strain |
P value for each vaccine strain |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| A10/Holland/42 | A22/IRQ/24/64 | A/ERI/2/98 | A/ETH/13/2005 | A/IRN/2/97 | A/KEN/12/2005 | A/MAU/1/2006 | A/SUD/2/84 | A/TUR/20/2006 | |
| Observed NIT titers | |||||||||
| A22/IRQ/24/64 | 1 | ||||||||
| A/ERI/2/98 | 1 | 1 | |||||||
| A/ETH/13/2005 | 1 | 0.672 | 1 | ||||||
| A/IRN/2/97 | 1 | 1 | 1 | 1 | |||||
| A/KEN/12/2005 | 1 | 1 | 1 | 1 | 1 | ||||
| A/MAU/1/2006 | 1 | 1 | 1 | 0.006a | 1 | 1 | |||
| A/SUD/2/84 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| A/TUR/20/2006 | 1 | 1 | 1 | 1 | 1 | 1 | 0.056 | 1 | |
| A/TUR/14/98 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Scaled NIT titers | |||||||||
| A22/IRQ/24/64 | 1 | ||||||||
| A/ERI/2/98 | 1 | 1 | |||||||
| A/ETH/13/2005 | 0.967 | 0.274 | 1 | ||||||
| A/IRN/2/97 | 1 | 1 | 1 | 1 | |||||
| A/KEN/12/2005 | 1 | 1 | 1 | 0.828 | 1 | ||||
| A/MAU/1/2006 | 0.261 | 0.938 | 0.064 | 0.000 | 0.074 | 0.680 | |||
| A/SUD/2/84 | 1 | 1 | 1 | 1 | 1 | 1 | 0.034 | ||
| A/TUR/20/2006 | 1 | 0.682 | 1 | 1 | 1 | 1 | 0.000 | 1 | |
| A/TUR/14/98 | 1 | 1 | 1 | 0.740 | 1 | 1 | 0.373 | 1 | 1 |
| Observed LPB-ELISA titers | |||||||||
| A22/IRQ/24/64 | 1 | ||||||||
| A/ERI/2/98 | 0.975 | 0.087 | |||||||
| A/ETH/13/2005 | 0.122 | 0.005 | 1 | ||||||
| A/IRN/2/97 | 1 | 1 | 1 | 1 | |||||
| A/KEN/12/2005 | 1 | 1 | 1 | 1 | 1 | ||||
| A/MAU/1/2006 | 1 | 1 | 0.008 | 0.000 | 0.333 | 0.690 | |||
| A/SUD/2/84 | 1 | 1 | 1 | 0.135 | 1 | 1 | 1 | ||
| A/TUR/20/2006 | 0.110 | 0.005 | 1 | 1 | 1 | 1 | 0.000 | 0.121 | |
| A/TUR/14/98 | 1 | 1 | 0.299 | 0.027 | 1 | 1 | 1 | 1 | 0.024 |
| Scaled LPB-ELISA titers | |||||||||
| A22/IRQ/24/64 | 1 | ||||||||
| A/ERI/2/98 | 0.365 | 0.032 | |||||||
| A/ETH/13/2005 | 0.033 | 0.001 | 1 | ||||||
| A/IRN/2/97 | 1 | 1 | 1 | 0.810 | |||||
| A/KEN/12/2005 | 1 | 1 | 1 | 0.718 | 1 | ||||
| A/MAU/1/2006 | 1 | 1 | 0.001 | 0.000 | 0.09 | 0.300 | - | - | - |
| A/SUD/2/84 | 1 | 1 | 0.349 | 0.030 | 1 | 1 | 1 | ||
| A/TUR/20/2006 | 0.039 | 0.002 | 1 | 1 | 0.814 | 0.717 | 0.000 | 0.035 | |
| A/TUR/14/98 | 1 | 1 | 0.152 | 0.011 | 1 | 1 | 1 | 1 | 0.014 |
Values in bold type are the P values that are <0.05.
ROC analysis.
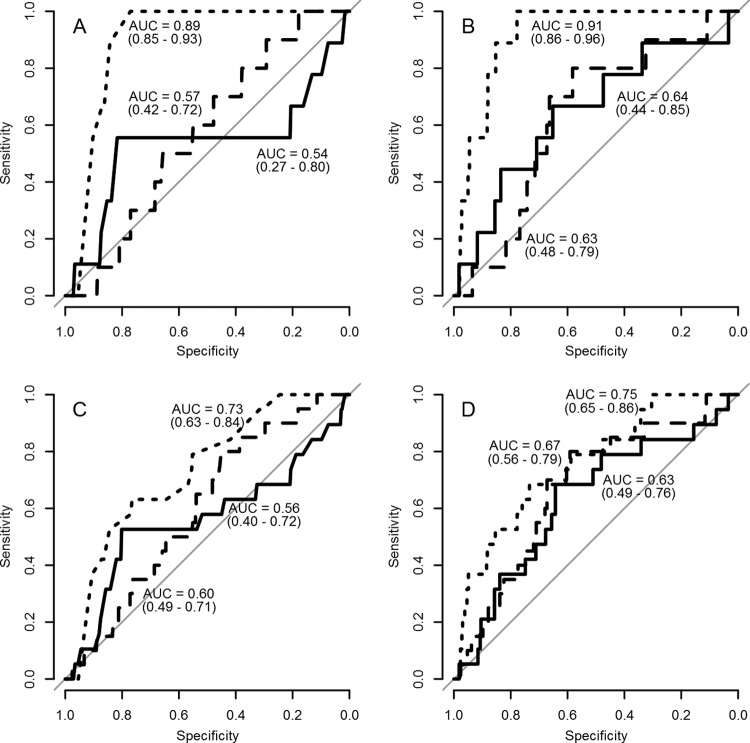
In the data set, we had 2 genetic lineages (Iran-96 and G-IV) that each contained 2 strains. In the ROC analysis, we compared the sensitivity and specificity of the r1 values of the different test methods before and after scaling. In the first analysis, we assumed that only strains A/IRN/2/97 and A/TUR/14/98 were antigenically similar (Fig. 4A and B for titers and scaled titers, respectively). In the second analysis, we assumed that strains A/ERI/2/98 and A/SUD/2/84 also were antigenically similar (Fig. 4C and D for titers and scaled titers, respectively). The 95% confidence interval of the area under the curve (AUC) of the ROC curve of the r1 value based on the LPB-ELISA titers and scaled titers did not include 0.5, hence showing a statistically significant result. However, for VNT and NIT and only when using the assumption that the strains in both lineages are antigenically similar, the AUC of the ROC curve of the r1 values based on scaled NIT titers (AUC, 0.67; 95% confidence interval [CI], 0.56 to 0.79; Fig. 4D) had a 95% confidence interval that did not include 0.5. Therefore, the dashed line in Fig. 4D represents the only statistically significant result for NIT in the ROC analysis.
FIG 4.
ROC curves of the r1-value titers of the VNT (solid line), NIT (dashed line), and LPB-ELISA (dotted line). In the graphs, the area under the curve (AUC) is given, with the 95% confidence interval in parentheses. (A and B) Strains A/IRN/2/97 and A/TUR/14/98 are assumed to be antigenically the same, using the observed titers (A) and scaled titers (B). (C and D) Strains A/IRN/2/97 and A/TUR/14/98 as well as A/ERI/2/98 and A/SUD/2/84 are assumed to be antigenically the same, using the observed titers (C) and scaled titers (D).
DISCUSSION
The study was performed to determine which serological test method provides the least variation within a group of vaccinated cattle and provides the best discrimination between vaccines. We selected 10 FMDV serotype A strains from 8 different lineages (Table 1). Sequencing of the VP1 region of the genome confirmed the identity of the strains used. Although Eritrea is not free from FMD, the cattle used in this study were obtained from a region where no outbreaks of FMD had been recorded. All cattle were free of antibodies to nonstructural proteins of FMD virus at the start of the study, implying that they had not been exposed to FMDV. Before vaccination, the cattle were tested for neutralizing antibodies against 5 other serotypes (O, C, Asia 1, SAT-1, and SAT-2), with a negative result. Some cattle showed low antibody responses to NS protein postvaccination, but no FMDV clinical signs were observed. The observed 58% inhibition is just above the cutoff of 50% for the test. It is probable that the NS response was due to some remaining NS proteins present in the vaccine, as has been described before (34). The low titers found before vaccination in some of the cattle in both the VNT and LPB-ELISA is similar to the results reported for cattle in The Netherlands (35). Using the 10 strains as vaccine antigens, we observed a good homologous neutralizing antibody response, with at least a 4-fold increase in the titers at two w.p.v. The small differences, with mean titers ranging from 1.6 to 2.0, were observed in the mean response of each group of vaccinated cattle tested against all test strains; this shows that the quality of the vaccines was similar. Therefore, this experimental design, in which cattle are vaccinated with vaccines containing a similar amount of antigen, offers an excellent opportunity to compare the different serological test methods.
The major difficulty in interpreting the results with both the VNT and NIT was the difference in the levels of neutralization observed using different virus test strains. Some virus test strains, like A10/Holland/42, were neutralized easily, with high titers in the homologous and heterologous serum samples. For this reason, we also evaluated the scaled titers. Even though a difference in titer levels were observed, the mean response within the groups of 5 vaccinated cattle was consistent, resulting in a small range of observed SEMs in all the 3 test methods. The range of the observed SEM, however, was lowest in the LPB-ELISA, showing that the smallest amount of variation was observed with the LPB-ELISA results. Lower variation in the LPB-ELISA than in the VNT has also been reported by Van Maanen and Terpstra (36). After scaling, the range of the SEM observed in the groups of cattle increased, but the lowest maximum SEM was still found using the LPB-ELISA, but after scaling, the results obtained with the NIT were similar.
The titer differences between the test strains resulted in r1 values that were often higher than one. This was mainly observed with the seven test strains that had high homologous neutralizing-antibody titers. After scaling, lower r1 values were calculated; this is consistent with the fact that the strains were obtained from 8 distinct genetic lineages, from which we assume that they are also antigenically different. Using the LPB-ELISA, more significant differences between vaccines were observed by ANOVA, followed by pairwise t tests. The four strains from 2 genetic lineages were still similar in the pairwise t test (Table 4). Not only were the LPB-ELISA results more capable to distinguish between vaccines, they were also more reproducible, as they are not influenced by variations in tissue culture susceptibility. Although neutralizing antibodies are often considered in relation to protection, LPB-ELISA titers are also correlated with protection (36, 37). Therefore, the LPB-ELISA seems to perform better than the neutralization tests to discriminate between vaccines.
To further build on the assumption that genetically related strains should also be antigenically related, we did an ROC analysis using the r1 values derived from the different tests before and after scaling. An ROC curve with an AUC of 0.5 indicates that the test does not differentiate the differences defined by the gold standard (in our case, the sequence data). Before scaling, only the r1 values based on titers from the LPB-ELISA had an AUC of the ROC curve that was significantly different from 0.5. Therefore, without scaling the titers, the LPB-ELISA is the only test that can discriminate between strains that are genetically different and that are therefore also assumed to be antigenically different. After scaling, the AUC of the ROC curve of the NIT, when assuming the strains in both the Iran-96 and the G-IV lineage were similar, was significantly different from 0.5. This shows that scaling can help improve the NIT to discriminate strains antigenically, but the LPB-ELISA is the best test for this.
In our analysis, we used the r1 value as a measurement of the antigenic relationship, but the reason we do the analysis is to see whether a vaccine can protect against infection. Previous studies have shown that there is a strong relationship between antibody response and protection (36–38). The precision of this relationship can be improved slightly by including other immunological techniques (39, 40). However, the degree of the titer that relates with protection is not the same for different strains. Therefore, the use of an r1 value to estimate protection against challenge is not logical, because the difference in titer needed for protection is not taken into consideration. Hence, if the objective is to choose a vaccine that will best protect against infection with a field isolate, it is better to look for the vaccine that induces the highest titer against the field virus strain instead of looking at the r1 value. In a recent study that analyzed the serological results of cross-protection tests (22), it was shown that the r1 value of a serum sample from a vaccinated and heterologous-challenged cow did not predict protection, but the serum titer against the challenge strain in the VNT did. In fact, it was shown that an analysis of the humoral immune response using IgG1 and IgG2 ELISAs even improved this prediction (22). This also indicates that VNT or ELISA titers are probably more important in a prediction of protection than is the r1 value.
As stated before, the cattle serum samples used in this study are unique, as they were produced using the same amount of antigen in each vaccine batch. These cattle sera can be very valuable for further research on the antigenic differences of new serotype A FMDV isolates, as they cover a large part of the genetic differences observed in Europe, the Middle East, and Africa. We conclude that the LPB-ELISA, under the conditions in the present study, is the best test for detecting antigenic differences between FMDV strains. The smaller the variability within a vaccinated group of animals, the greater is its discriminatory capacity in the neutralization tests. However, one should realize that in our study, we used strain-specific guinea pig and rabbit antibodies. Nevertheless, neutralizing antibody tests also have their place, as VNT titers are closely related to protection (36, 38), but in that case, the use of r1 values is not feasible, and the suitability of a vaccine strain should be evaluated based on the titer against the outbreak strain. The higher the titer, the better suited the vaccine.
ACKNOWLEDGMENTS
This work was supported by International Atomic Energy Agency (IAEA) grant C6/ERI/07010, Vienna, Austria. Production of the guinea pig and rabbit antisera was supported by the EuFMD.
We thank The Pirbright Institute, United Kingdom, for providing FMDV type A strains. We also thank Tsghe Kfleyesus, Ghebremichael G. Egziabhier, and the late Tesfu Yemane (animal health staff at MoA, Asmara, Eritrea) for assisting in sample collection during the animal experiments. We also thank Boonstra Christine for sequencing the FMDV serotype A strains, and the FAO Representative Office in Eritrea for providing internet access during the study period.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1. Pereira HG. 1981. Foot-and-mouth disease, p 333–363 In Gibbs EPJ. (ed), Virus disease of food animals. Academic Press, Inc., London, United Kingdom [Google Scholar]
- 2. Brooksby JB. 1982. Portraits of viruses: foot-and-mouth disease virus. Intervirology 18:1–23. 10.1159/000149299 [DOI] [PubMed] [Google Scholar]
- 3. Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. 1982. Rapid evolution of RNA genomes. Science 215:1577–1585. 10.1126/science.7041255 [DOI] [PubMed] [Google Scholar]
- 4. Holland JJ, De La Torre JC, Steinhauer DA. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1–20 [DOI] [PubMed] [Google Scholar]
- 5. longjam N, Tayo T. 2011. Antigenic variation of foot and mouth disease virus–an overview. Vet. World 4:475–479. 10.5455/vetworld.2011.475-479 [DOI] [Google Scholar]
- 6. Paton DJ, Valarcher JF, Bergmann I, Matlho OG, Zakharov VM, Palma EL, Thomson GR. 2005. Selection of foot and mouth disease vaccine strains–a review. Rev. Sci. Tech. 24:981–993 [PubMed] [Google Scholar]
- 7.OIE. 2012. Chapter 2.1.5. Foot and mouth disease. In Manual of diagnostic tests and vaccines for terrestrial animals. OIE, Paris, France: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.01.05_FMD.pdf [Google Scholar]
- 8. Knowles NJ, Samuel AR. 2003. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 91:65–80. 10.1016/S0168-1702(02)00260-5 [DOI] [PubMed] [Google Scholar]
- 9. Mittal M, Tosh C, Hemadri D, Sanyal A, Bandyopadhyay SK. 2005. Phylogeny, genome evolution, and antigenic variability among endemic foot-and-mouth disease virus type A isolates from India. Arch. Virol. 150:911–928. 10.1007/s00705-004-0469-6 [DOI] [PubMed] [Google Scholar]
- 10. Brehm KE, Kumar N, Thulke HH, Haas B. 2008. High potency vaccines induce protection against heterologous challenge with foot-and-mouth disease virus. Vaccine 26:1681–1687. 10.1016/j.vaccine.2008.01.038 [DOI] [PubMed] [Google Scholar]
- 11. Jangra RK, Tosh C, Sanyal A, Hemadri D, Bandyopadhyay SK. 2005. Antigenic and genetic analyses of foot-and-mouth disease virus type A isolates for selection of candidate vaccine strain reveals emergence of a variant virus that is responsible for most recent outbreaks in India. Virus Res. 112:52–59. 10.1016/j.virusres.2005.03.021 [DOI] [PubMed] [Google Scholar]
- 12. Mattion N, Goris N, Willems T, Robiolo B, Maradei E, Beascoechea CP, Perez L, Smitsaart E, Fondevila N, Palma E, De Clercq K, La Torre J. 2009. Some guidelines for determining foot-and-mouth disease vaccine strain matching by serology. Vaccine 27:741–747. 10.1016/j.vaccine.2008.11.026 [DOI] [PubMed] [Google Scholar]
- 13. Rweyemamu MM, Booth JC, Head M, Pay TW. 1978. Microneutralization tests for serological typing and subtyping of foot-and-mouth disease virus strains. Epidemiol. Infect. 81:107–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rweyemamu MM. 1984. Antigenic variation in foot-and-mouth disease: studies based on the virus neutralization reaction. J. Biol. Stand. 13:323–337. 10.1016/S0092-1157(84)80013-X [DOI] [PubMed] [Google Scholar]
- 15. Maree FF, Blignaut B, Esterhuysen JJ, de Beer TA, Theron J, O'Neill HG, Rieder E. 2011. Predicting antigenic sites on the foot-and-mouth disease virus capsid of the South African Territories types using virus neutralization data. J. Gen. Virol. 92:2297–2309. 10.1099/vir.0.032839-0 [DOI] [PubMed] [Google Scholar]
- 16. Ludi AB, Horton DL, Li Y, Mahapatra M, King DP, Knowles NJ, Russell CA, Paton DJ, Wood JL, Smith DJ, Hammond JN. 2013. Antigenic variation of foot-and-mouth disease virus serotype A. J. Gen. Virol. 95(Pt 2):384–392. 10.1099/vir.0.057521-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reeve R, Blignaut B, Esterhuysen JJ, Opperman P, Matthews L, Fry EE, de Beer TAP, Theron J, Rieder E, Vosloo W, O'Neill HG, Haydon DT, Maree FF. 2010. Sequence-based prediction for vaccine strain selection and identification of antigenic variability in foot-and-mouth disease virus. PLoS Comput. Biol. 6:e1001027. 10.1371/journal.pcbi.1001027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FMD-DISCONVAC. 2013. Assessment and improvement of heterologous protection by FMD vaccines. Work package 3. Veterinary and Agrochemical Research Centre, Brussels, Belgium [Google Scholar]
- 19. Nagendrakumar SB, Srinivasan VA, Madhanmohan M, Yuvaraj S, Parida S, Di Nardo A, Horsington J, Paton DJ. 2011. Evaluation of cross-protection between O1 Manisa and O1 Campos in cattle vaccinated with foot-and-mouth disease virus vaccine incorporating different payloads of inactivated O1 Manisa antigen. Vaccine 29:1906–1912. 10.1016/j.vaccine.2010.12.127 [DOI] [PubMed] [Google Scholar]
- 20. Pay TW, Hingley PJ. 1987. Correlation of 140S antigen dose with the serum neutralizing antibody response and the level of protection induced in cattle by foot-and-mouth disease vaccines. Vaccine 5:60–64. 10.1016/0264-410X(87)90011-9 [DOI] [PubMed] [Google Scholar]
- 21. Hammond J. 2010. OIE/FAO FMD Reference Laboratory Network: annual report 2012. The Pirbright Institute, Pirbright, United Kingdom: http://www.fao.org/fileadmin/user_upload/eufmd/docs/Pirbright_reports/OIE-FAO_FMD_Reference_Laboratory_Network_report_2012__.pdf [Google Scholar]
- 22. Brito BP, Perez AM, Capozzo AV. 2014. Accuracy of traditional and novel serology tests for predicting cross-protection in foot-and-mouth disease vaccinated cattle. Vaccine 32:433–436. 10.1016/j.vaccine.2013.12.007 [DOI] [PubMed] [Google Scholar]
- 23. Bahnemann HG. 1975. Binary ethylenimine as an inactivant for foot-and-mouth disease virus and its application for vaccine production. Arch. Virol. 47:47–56. 10.1007/BF01315592 [DOI] [PubMed] [Google Scholar]
- 24. Bahnemann HG. 1990. Inactivation of viral antigens for vaccine preparation with particular reference to the application of binary ethylenimine. Vaccine 8:299–303. 10.1016/0264-410X(90)90083-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knowles NJ, Samuel AR. 1998. RT-PCR and sequencing protocols for the molecular epidemiology of exotic virus diseases of animals. OIE/FAO World Reference Laboratory for Foot-and-Mouth Disease, Institute for Animal Health, Pirbright, Surrey, United Kingdom [Google Scholar]
- 26. Doel TR, Baccarini PJ. 1981. Thermal stability of foot-and-mouth disease virus. Arch. Virol. 70:21–32. 10.1007/BF01320790 [DOI] [PubMed] [Google Scholar]
- 27. Wagner GG, Card JL, Cowan KM. 1970. Immunochemical studies of foot-and-mouth disease. VII. Characterization of foot-and-mouth disease virus concentrated by polyethylene glycol precipitation. Arch. Gesamte Virusforsch. 30:343–352 [DOI] [PubMed] [Google Scholar]
- 28. Sørensen KJ, Madsen KG, Madsen ES, Salt JS, Nqindi J, Mackay DKJ. 1998. Differentiation of infection from vaccination in foot-and-mouth disease by the detection of antibodies to the non-structural proteins 3D, 3AB and 3ABC in ELISA using antigens expressed in baculovirus. Arch. Virol. 143:1461–1476. 10.1007/s007050050390 [DOI] [PubMed] [Google Scholar]
- 29. Hamblin C, Barnett IT, Hedger RS. 1986. A new enzyme-linked immunosorbent assay (ELISA) for the detection of antibodies against foot-and-mouth disease virus I. Development and method of ELISA. J. Immunol. Methods 93:115–121. 10.1016/0022-1759(86)90441-2 [DOI] [PubMed] [Google Scholar]
- 30. Karber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 162:480–483 (Article in German.) [Google Scholar]
- 31. Becker RA, Chambers JM, Wilks AR. 1988. The new S language: a programming environment for data analysis and graphics. Wadsworth & Brooks/Cole, Computer Science Series, Pacific Grove, CA [Google Scholar]
- 32.The R Core Team. 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://web.mit.edu/r_v3.0.1/fullrefman.pdf [Google Scholar]
- 33. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Doel TR. 2003. FMD vaccines. Virus Res. 91:81–99. 10.1016/S0168-1702(02)00261-7 [DOI] [PubMed] [Google Scholar]
- 35. Moonen P, Miedema GKW, van Hemert-Kluitenberg F, Chenard G, Dekker A. 2000. Comparison of FMDV neutralisation tests using three different cell lines: validation of the new FAO reference sera, p 241–246 Session of the research group of the European Commission for the Control of Foot-and-Mouth Disease, Borovets, Bulgaria: http://www.fao.org/ag/againfo/commissions/docs/research_group/borovet/app30.pdf [Google Scholar]
- 36. Van Maanen C, Terpstra C. 1989. Comparison of a liquid-phase blocking sandwich ELISA and a serum neutralization test to evaluate immunity in potency tests of foot-and-mouth disease vaccines. J. Immunol. Methods 124:111–119. 10.1016/0022-1759(89)90192-0 [DOI] [PubMed] [Google Scholar]
- 37. Maradei E, La Torre J, Robiolo B, Esteves J, Seki C, Pedemonte A, Iglesias M, D'Aloia R, Mattion N. 2008. Updating of the correlation between lpELISA titers and protection from virus challenge for the assessment of the potency of polyvalent aphtovirus vaccines in Argentina. Vaccine 26:6577–6586. 10.1016/j.vaccine.2008.09.033 [DOI] [PubMed] [Google Scholar]
- 38. Hingley PJ, Pay TW. 1987. Sources of variability in foot and mouth disease vaccine potency estimates based on serum neutralizing antibody assay. J. Biol. Stand. 15:127–142. 10.1016/0092-1157(87)90036-9 [DOI] [PubMed] [Google Scholar]
- 39. McCullough KC, De Simone F, Brocchi E, Capucci L, Crowther JR, Kihm U. 1992. Protective immune response against foot-and-mouth disease. J. Virol. 6:1835–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lavoria MÁ, Di-Giacomo S, Bucafusco D, Franco-Mahecha OL, Pérez-Filgueira DM, Capozzo AV. 2012. Avidity and subtyping of specific antibodies applied to the indirect assessment of heterologous protection against foot-and-mouth disease virus in cattle. Vaccine 30:6845–6850. 10.1016/j.vaccine.2012.09.011 [DOI] [PubMed] [Google Scholar]
- 41. Carrillo C, Tulman ER, Delhon G, Lu Z, Carreno A, Vagnozzi A, Kutish GF, Rock DL. 2005. Comparative genomics of foot-and-mouth disease virus. J. Virol. 79:6487–6504. 10.1128/JVI.79.10.6487-6504.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chitray M. 2008. Investigation potential factors affecting foot-and-mouth disease virus internalisation. M.S. thesis University of Pretoria, Pretoria, South Africa [Google Scholar]
- 43. Ayelet G, Mahapatra M, Gelaye E, Egziabher BG, Rufeal T, Sahle M, Ferris NP, Wadsworth J, Hutchings GH, Knowles NJ. 2009. Genetic characterization of foot-and-mouth disease viruses, Ethiopia, 1981-2007. Emerg. Infect. Dis. 15:1409–1417. 10.3201/eid1509.090091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu L, Hurtle W, Rowland JM, Casteran KA, Bucko SM, Grau FR, Valdazo-González B, Knowles NJ, King DP, Beckham TR, McIntosh MT. 2013. Development of a universal RT-PCR for amplifying and sequencing the leader and capsid-coding region of foot-and-mouth disease virus. J. Virol. Methods 189:70–76. 10.1016/j.jviromet.2013.01.009 [DOI] [PubMed] [Google Scholar]
- 45. Habiela M, Ferris NP, Hutchings GH, Wadsworth J, Reid SM, Madi M, Ebert K, Sumption KJ, Knowles NJ, King DP, Paton DJ. 2010. Molecular characterization of foot-and-mouth disease viruses collected from Sudan. Transbound. Emerg. Dis. 57:305–314. 10.1111/j.1865-1682.2010.01151.x [DOI] [PubMed] [Google Scholar]
- 46. Knowles NJ, Nazem Shirazi MH, Wadsworth J, Swabey KG, Stirling JM, Statham RJ, Li Y, Hutchings GH, Ferris NP, Parlak U, Ozyörük F, Sumption KJ, King DP, Paton DJ. 2009. Recent spread of a new strain (A-Iran-05) of foot-and-mouth disease virus type A in the Middle East. Transbound. Emerg. Dis. 56:157–169. 10.1111/j.1865-1682.2009.01074.x [DOI] [PubMed] [Google Scholar]
- 47. Klein J, Parlak U, Ozyörük F, Christensen LS. 2006. The molecular epidemiology of foot-and-mouth disease virus serotypes A and O from 1998 to 2004 in Turkey. BMC Vet. Res. 2:35. 10.1186/1746-6148-2-35 [DOI] [PMC free article] [PubMed] [Google Scholar]