Abstract
Microbial communities in extreme environments often have low diversity and specialized physiologies suggesting a limited resistance to change. The McMurdo Dry Valleys (MDV) are a microbially dominated, extreme ecosystem currently undergoing climate change-induced disturbances, including the melting of massive buried ice, cutting through of permafrost by streams, and warming events. These processes are increasing moisture across the landscape, altering conditions for soil communities by mobilizing nutrients and salts and stimulating autotrophic carbon inputs to soils. The goal of this study was to determine the effects of resource addition (water/organic matter) on the composition and function of microbial communities in the MDV along a natural salinity gradient representing an additional gradient of stress in an already extreme environment. Soil respiration and the activity of carbon-acquiring extracellular enzymes increased significantly (P < 0.05) with the addition of resources at the low- and moderate-salinity sites but not the high-salinity site. The bacterial community composition was altered, with an increase in Proteobacteria and Firmicutes with water and organic matter additions at the low- and moderate-salinity sites and a near dominance of Firmicutes at the high-salinity site. Principal coordinate analyses of all samples using a phylogenetically informed distance matrix (UniFrac) demonstrated discrete clustering among sites (analysis of similarity [ANOSIM], P < 0.05 and R > 0.40) and among most treatments within sites. The results from this experimental work suggest that microbial communities in this environment will undergo rapid change in response to the altered resources resulting from climate change impacts occurring in this region.
INTRODUCTION
Biodiversity surveys have demonstrated that microorganisms are ubiquitous, inhabiting even the most extreme environments (1–3). Microbial communities in these habitats tend to have low diversity (2, 4, 5) and specialized physiologies (6), suggesting a potentially limited resistance and resilience to change (7). However, few experiments have been conducted in extreme environments to assess the susceptibility of these communities to disturbance. Since extreme environments harbor novel microorganisms with unique adaptations and metabolisms, these communities represent a pool of rare forms of biodiversity likely containing important genomic resources. Understanding how they respond to environmental change is an important first step in predicting the vulnerability of this resource to disturbance and developing conservation strategies.
The McMurdo Dry Valleys (MDV) region comprises the largest ice-free zone in continental Antarctica, represents one of the harshest environments on Earth, and is an ideal simplified environment in which to study the effects of disturbance on soil communities, as microbes are the dominant form of life. Higher plants and animals are absent, and a limited diversity of protozoans (8, 9) and invertebrates (10–12) is endemic to the region. Three important factors act to constrain biological activity in MDV dry mineral soils, in addition to temperature extremes: liquid water, organic carbon availability, and high salinity. Recent observations suggest that these factors are subject to climate-induced changes that are likely to intensify in the near future (13). In spite of a period of cooling of average air temperatures of 0.7°C per decade in this region of Antarctica starting in the 1980s (14), during the past 2 decades, solar radiation has steadily increased, with significant effects for the MDV landscape (13). Massive buried ice is melting, causing surface deflation and wetting of previously dried soils. Streams are laterally and vertically cutting through permafrost, liberating sequestered nutrients and salts and expanding stream margins (13). Episodic warming events and associated permafrost thaw and high flows have caused inundation of previously dry soils (15). This increase in moisture across the landscape is likely altering conditions for soil microbial communities by mobilizing nutrients and salts and stimulating primary productivity and carbon inputs to soils.
Recent lines of evidence suggest that the MDV soils harbor relatively diverse microbial communities (16, 17) that are active and responsive to change, with evidence of biologically produced fluxes of CO2, CH4, and N2O (18, 19), extracellular enzyme activities indicative of decomposition (20), and ATP associated with viable cells (21, 22) being detected. Three studies have reported that altered soil conditions influence microbial communities, concluding that water additions had little discernible impact on soil respiration (23) but that nutritional resource addition in various forms increased soil respiration (24, 25) and altered community composition (25). While these studies suggest that soil microbial communities in the MDV are responsive to change, a multisite study that simultaneously investigates the structural and functional responses of bacterial communities to disturbance is necessary to understand how both ecosystem functioning and community composition may respond to the environmental changes occurring and predicted to occur in the near future.
While low temperature limits MDV soil microbial biomass and productivity, gradients in soil chemistry, moisture content, and organic matter pose additional stresses on microbial function and diversity. Soil salinity represents one such gradient, with high-salinity soils containing reduced metazoan community abundance and diversity (26). Soil salinity has also been shown to act as a microbial stressor in other environments. Salinity exerts a primary limitation on water availability, as total water potential is the sum of matrix potential, which is determined by soil composition and texture, and osmotic potential, which is controlled by total ion concentrations. Soil salinity can also lead to high internal levels of ions that are toxic to metabolic activities (27) and can denature the extracellular enzymes necessary for carbon and nutrient acquisition. Thus, increased salinity represents a biological stress that requires evolutionary or energetically expensive solutions, has been shown to be an important determinant of microbial community composition in other systems (28, 29), and creates additional stress for biological life in MDV soils.
The goal of this study was to determine the responses of soil microbes in the MDV to altered resources that approximate changes likely to occur as climate change impacts this region. The specific objectives of this study were to (i) investigate the activity and community composition of bacterial communities along a natural salinity-induced severity gradient, (ii) determine if the alleviation of primary limitations (water and organic matter) would alter the composition and function of microbial communities in dry mineral soils, and (iii) determine how additional stress in the form of soil salinity alters the responses of bacterial communities to increased soil moisture and organic matter.
MATERIALS AND METHODS
Site description.
The study took place at three plots near the southern shore of Lake Fryxell (77°37′S, 163°12′ to 13′E) in Taylor Valley of MDV, Antarctica. Annual soil surface temperatures around the lake average −18.4°C, with only 25.5 days above freezing occurring each year (14). The amount of precipitation at Lake Fryxell is 20 to 37 mm annually (30); however, sublimation rates throughout the MDV are significantly greater than precipitation inputs (31). Liquid water inputs occur during a brief glacial melt period during the austral summer due to increased temperatures and solar radiation (32).
The soils of the Lake Fryxell basin are Typic Haploturbel glacial tills and Ross Sea drift deposits (33, 34). Historically, the basin was inundated by Glacial Lake Washburn in the late Wisconsin period (35), leaving behind legacy organic matter (36). Much of the contemporary inputs are from the aerial deposition of carbon produced in nearby lakes and streams during brief summer melt events (32). Thus, concentrations of soil organic matter (SOM) are low, averaging 0.03% organic carbon (37). Conductivity (as a proxy for soil salinity) in the Lake Fryxell basin averages 549 ± 77 μS cm−1 (38), with maximum reported values exceeding 5,000 μS cm−1 in lake margin sediments and fine sediment deposits (26, 39).
The MDV contain complex microbial communities, with bacterial diversity levels being approximately half as great as those found in other soils (17, 40). A variety of edaphic characteristics, including soil pH, salinity, and soil organic matter content, are correlated with bacterial diversity and community composition (16, 17); however, these relationships appear to be complex, with differences being found across sampling locations (17). The MDV soils also contain fungal communities containing limited diversity, but their abundance compared to that of bacteria is unknown (41, 42).
Soil sampling and treatments.
Field experiments were conducted during the austral spring and summer of 2010 and 2011. Three locations were selected to represent low-salinity (LS), moderate-salinity (MS), and high-salinity (HS) sites. The conductivity of the plots spanned approximately 1.5 orders of magnitude, from 105 ± 4 μS in the LS plot, 532 ± 63 μS in the MS plot, and 4,800 ± 470 μS in the HS plot. The soil pH decreased with increasing salinity (9.0 ± 0.1, 8.8 ± 0.1, and 7.9 ± 0.05 in the LS, MS, and HS plots, respectively), while percent nitrogen (0.006 ± 0.002, 0.007 ± 0.002, and 0.014 ± 0.002 in the LS, MS, and HS plots, respectively) and percent organic carbon (0.027 ± 0.006, 0.058 ± 0.013, and 0.08 ± 0.017 in the LS, MS, and HS plots, respectively) increased with increasing salinity. Twelve cylindrical mesocosms (10 cm in diameter by 17 cm deep) were installed in a 3-by-4 grid spaced 50 cm apart at each of the three plots on 23 and 24 November 2010, for a total of 36 samples. At each plot, soil was removed from each mesocosm location with a sterile scoop to a depth of ∼12 cm, homogenized, and used to fill the mesocosms, which were then placed in the excavated holes.
One of three treatments was randomly assigned to each mesocosm: control, water addition, or organic carbon/matter with water addition. Soil moisture was measured in the field using a HydroSense soil moisture probe (Campbell Scientific, Australia) calibrated for each salinity plot to determine the amounts of each treatment (water or leachate) to be added to mesocosms. For water treatments, sterile deionized (DI) water was added to the soil to reach a 10% final soil water content by weight. For organic matter treatments, a leachate (2,800 mg/liter dissolved organic carbon) was prepared from cyanobacterial mats collected along the lake edge by steeping in sterile DI water and filter sterilizing. As with the water treatments, organic matter treatments were added in volumes to bring the final soil water content to 10% by weight and to increase the soil organic carbon by 0.005%, an increase of approximately 1/10 of the total soil organic carbon. In addition to providing information regarding the alleviation of water stress on MDV soil communities, the water treatments also served as a control for the organic matter treatments with respect to the changes in soil ion solubility with the addition of water. Treatments were added using a sterile syringe and 10-cm needle by penetrating the soil at five points within each mesocosm five times over the 30-day incubation to maintain soil moisture at ∼10% by weight. After incubation, soils to be used for molecular analyses were preserved with an equal volume of sucrose lysis buffer (43), and all samples were immediately stored at −20°C.
Soil chemistry and microbial activity.
Soil pH, salinity, total nitrogen, and soil organic carbon were determined as described previously (17). Respiration rates were measured using a LI-COR 8100 closed gas exchange system with a LI-COR 8100-102 survey chamber (LI-COR Biosciences, Lincoln, NE) to detect CO2 flux. Extracellular enzyme activities were measured following the microplate methods of Zeglin et al. (20) for the potential activity of four enzymes, α-glucosidase (AG), β-glucosidase (BG), alkaline phosphatase (AP), and leucine aminopeptidase (LAP), optimized for the low organic matter content.
DNA extraction, sequencing, and sequence analysis.
Soils (0.7 g) from each of the 36 samples were extracted with the cetyltrimethylammonium bromide (CTAB) method adapted by Mitchell and Takacs-Vesbach (44). Bar-coded amplicon pyrosequencing of 16S rRNA genes was performed as described previously (17, 45, 46) using universal V6 bacterial primers 939F (5′-TTG ACG GGG GCC CGC ACA AG-3′) and 1492R (5′-GTT TAC CTT GTT ACG ACT T-3′) on a Roche 454 FLX instrument using Roche titanium reagents and titanium procedures at the Research and Testing Laboratory, Lubbock, TX.
The 16S rRNA gene sequences were quality filtered, denoised, screened for PCR errors, and chimera checked using the AmpliconNoise and Perseus programs (47). The Quantitative Insights into Microbial Ecology (QIIME) pipeline was used to analyze the 16S rRNA gene sequence data (48). Unique 16S rRNA gene sequences or operational taxonomic units (OTUs) were identified by use of the 97% DNA identity criterion. A representative sequence was picked from each OTU, aligned using the PyNAST aligner (49) and the Greengenes core set (50), and given taxonomic assignments using the Ribosomal Database Classifier program (51). The alpha diversity for each sample was assessed using Chao1 richness estimates of 1,000 randomly selected subsets of 1,421 sequences per sample to standardize for various sequencing efforts across samples. Measures of beta diversity to assess differences in community composition between sites and treatments were also performed with randomly selected subsets of sequences. Two phylogenetic methods were used to compare the community composition of the samples: the unweighted UniFrac method, which compares the unique branch length associated with each sample in a phylogenetic tree, and the weighted UniFrac method, which weights distances on the basis of the number of sequences associated with each branch (52). Additionally, we also used two nonphylogenetic distance methods (the Gower and Jaccard methods) shown to be effective in detecting differences in microbial communities (53).
Data analysis.
The within- and across-plot effects of water and organic matter additions on soil respiration and extracellular enzyme activities were assessed for significance using a two-way analysis of variance with Tukey's honestly significant difference (HSD) post hoc test in R (54). Weighted and unweighted UniFrac, Jaccard, and Gower distance matrices were imported into the Primer program (v6) (55), where the analysis of similarity (ANOSIM) function, which performs analysis of variance using similarity matrices, was used to test the overall (global) and pairwise dissimilarity of the community composition of samples from different plots (56). Groups were designated significantly different when the global test was significant (P < 0.05), the pairwise test was significant (P < 0.05), and the R statistic was greater than 0.40.
Accession numbers.
The individual standard flowgram format (.sff) files from this study were assigned accession numbers SAMN02259435 through SAMN02259470, and raw sequence data from this study are available through the NCBI Sequence Read Archive (SRA) as accession numbers SRX378286 through SRX378321 under Bioproject PRJNA228947.
RESULTS
Effects of water, organic matter, and salinity on microbial activity.
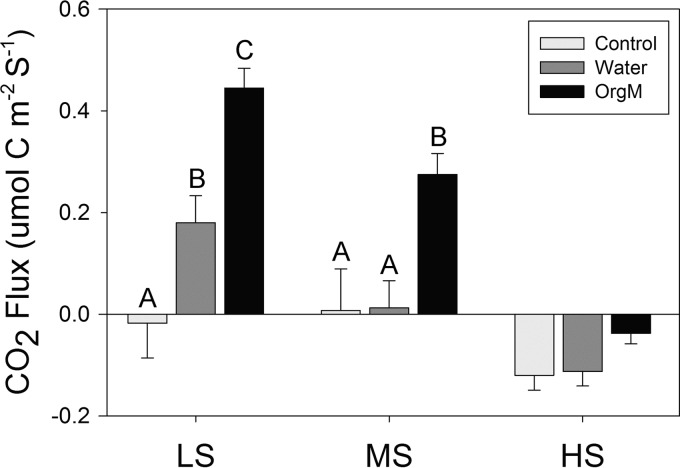
Microbial activity, measured by extracellular enzyme activities and soil respiration, varied widely among plots and treatments. Among plots, the soil respiration decreased significantly (P < 0.05) with increasing salinity for the water and organic matter additions, while significant differences between control samples were restricted to the moderate- and high-salinity plot comparisons (Fig. 1). Within plots, soil respiration varied in response to water and organic matter additions. In the low-salinity plot, water addition stimulated a significant (P < 0.05) increase in respiration, with an even greater significant increase being stimulated by organic matter addition (Fig. 1). In the moderate-salinity plot, only the organic matter addition significantly stimulated respiration, and in the high-salinity plot, neither organic matter nor water addition caused a significant respiration response (Fig. 1).
FIG 1.
Mean soil respiration (CO2 flux in μmol cm−2 s−1) with standard deviations for the low-salinity (LS), moderate-salinity (MS), and high-salinity (HS) sites. Within-site significant differences (Tukey's HSD) are indicated with different letters. OrgM, organic matter.
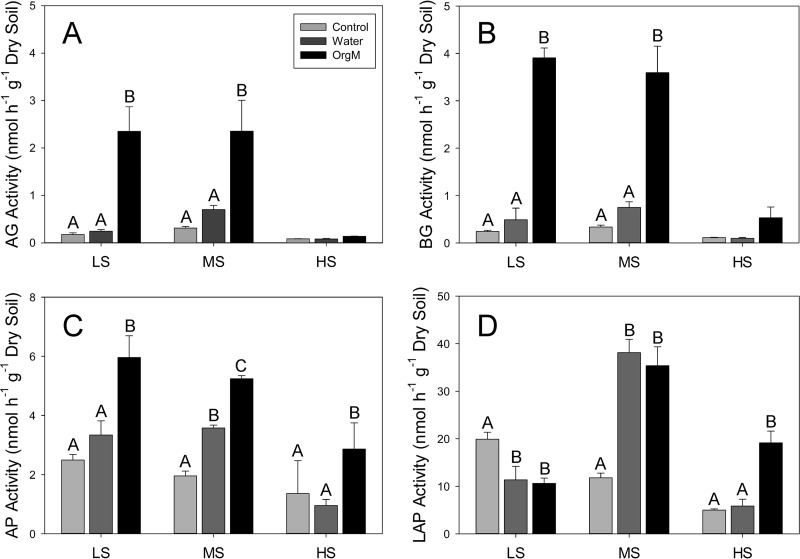
The responses of carbon-acquiring enzymes (α- and β-glucosidases) to water and organic matter additions were similar to those of soil respiration. Among plots, the only significant difference for the treatments was between the high-salinity plot (lower activity) and the low- and moderate-salinity plots (higher activity) (Fig. 2A and B). Within-plot comparisons showed no significant increase in response to water additions and a strong, significant (P < 0.05) increase in the organic matter addition treatments in both the low- and moderate-salinity plots. However, there was no response in the high-salinity plot (Fig. 2A and B).
FIG 2.
Mean extracellular enzyme activities (nmol h−1 g−1 dry soil) with standard deviations for the low-salinity (LS), moderate-salinity (MS), and high-salinity (HS) sites for α-glucosidase (AG) (A), β-glucosidase (BG) (B), alkaline phosphatase (AP) (C), and leucine aminopeptidase (LAP) (D). Within-site significant differences (Tukey's HSD) are indicated with different letters, while among-site differences are described in the text.
The responses of nitrogen (LAP)- and phosphorus (AP)-acquiring enzyme activities were varied. Among the three plots, AP activity in response to the addition of water and organic matter was significantly (P < 0.05) lower in the high-salinity plot than in the low- and moderate-salinity plots; however, the activity of the controls was not significantly different among the plots. Within plots, the activity of AP was significantly (P < 0.05) stimulated by organic matter additions in all three plots, while water addition resulted in a significant activity increase only in the moderate-salinity plot (Fig. 2C). Among the three plots, the activity of LAP was significantly different for all of the treatments, with the exception of the water additions in the low- and high-salinity plots (Fig. 2D). Within plots, LAP activity significantly decreased in response to both water and organic matter additions in the low-salinity plot, increased in response to both treatments in the moderate-salinity plot, and increased in response to only the organic matter addition in the high-salinity plot (Fig. 2D).
Bacterial community responses.
Pyrosequencing resulted in 213,176 high-quality 16S rRNA gene sequences from 34 samples (2 samples had an insufficient number of DNA sequences) after denoising and chimera removal. The number of sequences per sample ranged from 1,421 to 18,703, representing a total of 1,338 operational taxonomic units (OTUs; 97% sequence similarity). Chao1 richness estimates were used to assess differences in alpha diversity within and among sites by randomly selecting 1,421 sequences 1,000 times. The low-salinity site had the highest diversity, with an average of 296 ± 27, 333 ± 34, and 240 ± 55 OTUs found in the control, water addition, and organic matter addition treatments, respectively. The diversity of the control and water addition samples was significantly higher than that found with the addition of organic matter (P < 0.05). The moderate-salinity site had intermediate levels of alpha diversity, with an average of 186 ± 23, 191 ± 23, and 180 ± 30 OTUs in the control, water addition, and organic matter addition treatments, respectively, with no significant differences being found among treatments. The high-salinity site had the lowest diversity, with an average of 40 ± 9, 42 ± 16, and 18 ± 4 OTUs being found in the control, water addition, and organic matter addition treatments, respectively, and no significant differences being found among treatments. Among the plots, the average Chao1 richness values when all treatments were included were significantly different (P < 0.05), with dramatically lower diversity being detected at the high-salinity site (289 ± 57, 186 ± 29, and 33 ± 17 OTUs for the low-, moderate-, and high-salinity plots, respectively).
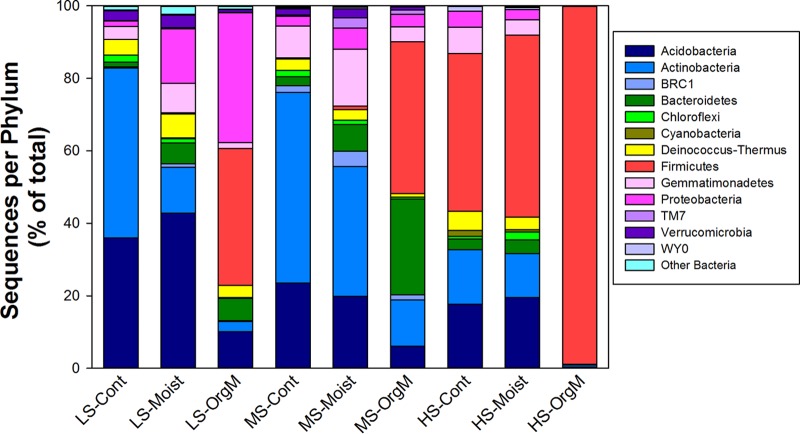
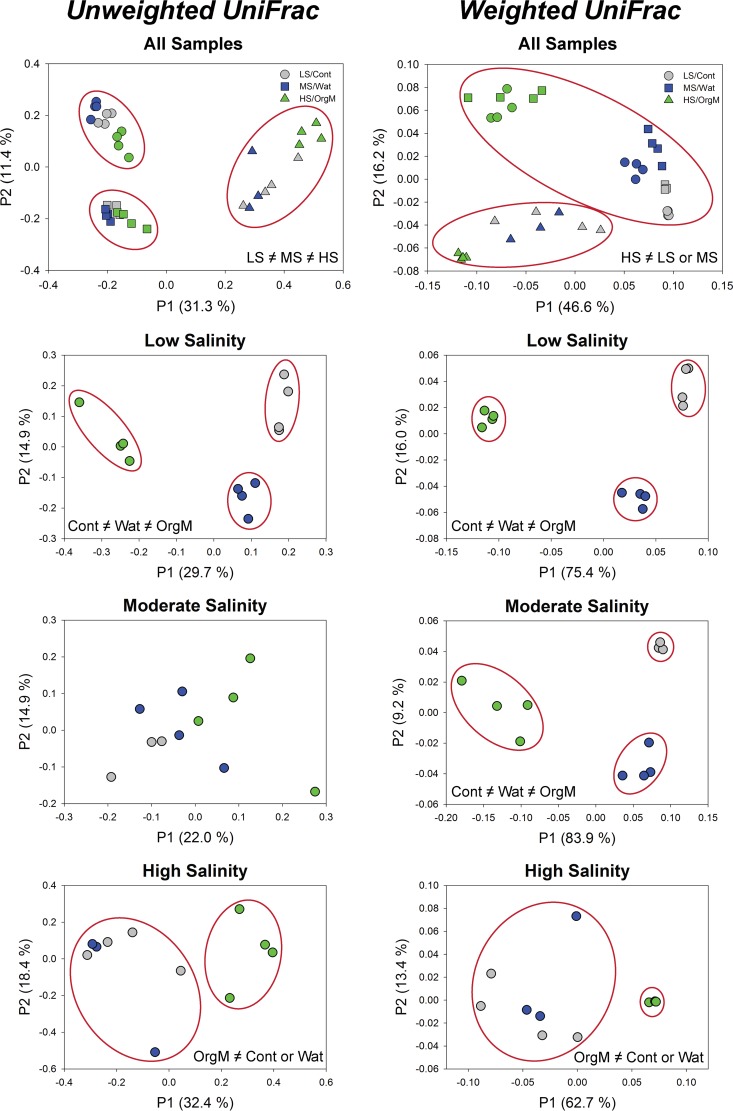
The community composition of the three plots differed in response to water and organic matter additions during the 30-day incubation period (Fig. 3). The taxonomic distribution in the low-salinity plot for the control treatment was dominated by Acidobacteria (26%) and Actinobacteria (47%), with Deinococcus-Thermus (4%), Gemmatimonadetes (4%), Verrucomicrobia (3%), and Chloroflexi (2%) making up the majority of the remaining taxa (Fig. 3). With the addition of water, Bacteroidetes (6%) and Proteobacteria (15%) replaced Actinobacteria (13%), with a further reduction of Actinobacteria and an increase in Proteobacteria (36%) and Firmicutes (38%) occurring with organic matter addition (Fig. 3). In the moderate-salinity control and water addition samples, Acidobacteria (∼20%) and Actinobacteria (∼35 to 50%) dominated, but these taxa decreased dramatically with the addition of organic matter as members of the Firmicutes (42%) and Bacteroidetes (26%) became prevalent (Fig. 3). In the high-salinity plot, Firmicutes (∼44%), Acidobacteria (∼18%), and Actinobacteria (∼15%) were the dominant members of the community in the control samples, with the frequency of all phyla other than Firmicutes (99%) decreasing with water and organic matter addition (Fig. 3). A single organism from the genus Paenisporosarcina found in all treatments was responsible for the majority of the increase in Firmicutes in the high-salinity plot. The ANOSIM analysis of both the phylogenetic and nonphylogenetic beta-diversity measures found strong support for global differences among plots and among treatments within plots; however, not all between-group comparisons (plots or treatments within plots) were significantly different (Fig. 4; see Table S1 and Fig. S2 in the supplemental material).
FIG 3.
Average phylum-level taxonomy for each treatment expressed as the percentage of the number of total sequences.
FIG 4.
Principal coordinate analyses of the unweighted UniFrac and weighted UniFrac distance matrices created from the OTU table (97% identity) for all samples combined and for each site individually. Control (Cont), water (Wat), and organic matter (OrgM) additions are represented by gray, blue, and green symbols, respectively, while low-salinity (LS), moderate-salinity (MS), and high-salinity (HS) sites are represented by circles, squares, and triangles, respectively. Groupings of samples that are significantly different from those for the other groups (ANOSIM, P < 0.05 and R > 0.40) are delineated with red ovals (see Table S1 in the supplemental material for detailed results).
DISCUSSION
Effects of a severity gradient on microbial communities.
The first objective of this study was to examine the effects of a salinity gradient on microbial communities in an environment that also experiences extreme cold and aridity. As salinity increased across sites, respiration rates and carbohydrate-degrading enzyme activity declined, findings that are consistent with those of other studies showing that salinity stress decreases rates of C cycling (57, 58). Additionally, as in other studies that have shown a salinity-related decline in hydrolytic and oxidative enzyme activities (59, 60), the activities of carbon- and nitrogen-acquiring enzymes in the control treatments were the lowest at the high-salinity site. However, this result may also be related to the higher SOM and total nitrogen found at the moderate- and high-salinity sites, as microbes regulate the production of enzymes to acquire limiting nutrients (61). Similar declines in microbial activity have been observed along natural salinity gradients (62, 63), in field sites that have experienced anthropogenic salinization (60, 64), and in salt additions in laboratory experiments (58, 65), suggesting that the negative impact of salinity on overall microbial community function is common.
The decline in bacterial richness along the salinity gradient in this study suggests that, as with soil metazoans in the MDV (26, 66), salinity poses a significant stress for the majority of bacterial species found there. The effects of salinity on the diversity and community composition of soil microbial communities are less well studied than the effects of salinity on microbial activity, though some data for comparison do exist. In a study of soils and lake sediments associated with a hypersaline lake in Texas, moderate declines in richness were observed along a salinity gradient (67). However, increasing salinity has more commonly been shown to either have no effect on total richness (68, 69) or decrease overall phylogenetic diversity while promoting higher diversity within a few specific lineages (29, 70, 71). Therefore, our results are contrary to the results found in most other studies, suggesting that microbial diversity may be significantly constrained by high salinity only when communities also experience other stressors or resource limitations.
Bacterial community composition was significantly different among the three sites with various salinities, in spite of numerous shared attributes: close proximity (within 2 km) and similar aspect and climate. The phylum-level community composition in the control treatments at the low- and moderate-salinity sites was similar to that of other oligotrophic desert communities (72): samples contained a high abundance of Actinobacteria and Acidobacteria, with contributions from the Proteobacteria and Bacteroidetes. However, compared to other deserts (72), the Proteobacteria and Bacteroidetes were less abundant and the Verrucomicrobia and Deinococcus-Thermus phyla were more abundant in the MDV soils. The composition of the high-salinity site was strikingly different from that of other desert ecosystems, with Firmicutes largely replacing Acidobacteria and Actinobacteria. The dominance of Firmicutes observed here may in part be due to their Gram-positive cell walls and spore-forming ability, which make them resistant to desiccation stress and harsh environmental conditions (73, 74). In recent studies in the MDV, Firmicutes were found to be abundant at high-elevation, low-SOM, and low-soil-moisture sites (17) and increased in response to resource additions (25).
Effects of resource amendments on microbial activity and community composition.
The second and third goals of this study were to assess the responsiveness of microbial communities to the alleviation of two primary limitations, water and organic matter, and the role of salinity in mediating community responses. While water is a limiting resource in these soils, rapid water additions may also cause dilution stress for the microbes in these soils, as they must rapidly discharge or utilize osmolytes to avoid cell lysis (74). The effects of resource addition and the associated stressors on bacterial activity and community composition were expected to vary along the salinity gradient, as resistance and resilience to disturbance decline with decreases in diversity (7, 75) and along gradients of increasing stress (76, 77).
At the low-salinity site, the addition of water or water plus dilute native organic matter resulted in a substantial increase in microbial activity: bulk soil respiration and also extracellular enzyme activities increased significantly. Similar responses, particularly responses resulting from organic matter additions, have been observed in soils from a variety of other temperate/mesic biomes (78, 79), suggesting that low-salinity soils in the MDV have overall functions similar to those found in other ecosystems. The muted respiration and carbon-acquiring enzyme activity responses to additions at the high-salinity site indicate that these microbial communities had a reduced capacity to respond to increased resource availability. The negative CO2 flux values observed at the high-salinity site are common in the MDV and are attributed to abiotic dissolution of CO2 in soil water rather than autotrophy; negative CO2 fluxes can be considered to reflect soil respiration below the detection level (19, 23, 80).
The effects of water and organic matter additions on microbial community composition and structure were rapid. However, as with the activity response, water addition alone had less of an effect at the moderate- and high-salinity sites. At the low-salinity site, water addition resulted in a dramatic decline of Actinobacteria, while Proteobacteria and Bacteroidetes increased to relative abundances similar to those in other desert soils (72). At all three sites, the addition of organic matter resulted in a sharp decrease in the relative abundance of Acidobacteria, as has been reported in other organic matter addition studies (78, 81, 82) and high-carbon environments (83), providing further confirmation of the oligotrophic designation of this phylum (83). The organic matter-related increase in Proteobacteria, specifically, the Gammaproteobacteria and Betaproteobacteria subphyla, observed at the low-salinity site also agrees with the copiotrophic classification of these groups from other organic matter enrichment studies (78, 79, 82), as does the increase in Bacteroidetes at the moderate-salinity site (83). The organic matter-related increase in Firmicutes at all sites, with a nearly complete dominance at the high-salinity site, was striking, suggesting that the community compositions of the most extreme soil communities in the MDV have limited resistance to enrichment. The single organism from the genus Paenisporosarcina responsible for the majority of this increase in Firmicutes at the high-salinity site is a close relative of an organism isolated from a cyanobacterial mat from the MDV (84), indicating that some MDV bacteria are highly competitive under enriched conditions. Interestingly, a recent multiyear experiment in the MDV in which a mummified seal was moved from one location to another found that Firmicutes were abundant in soils impacted by the seal carcass (25), suggesting that organisms from this phylum respond predictably to resource addition in the MDV.
The community composition and function of the MDV soil microbial communities will likely be altered by the landscape-scale change currently occurring in this region (13) due to the apparent responsiveness of these bacterial communities to altered resources; however, these responses are expected to be complex. Increased water availability from the melting of massive buried ice, permafrost thaw, and increased stream flow will alleviate water stress in some areas and is likely to stimulate the in situ primary production which has been observed in MDV soils with consistent soil moisture inputs (17, 85). If increased moisture is persistent, organic matter inputs will further stimulate microbial activity and result in a shift in community composition to copiotrophic organisms. However, these responses will likely vary across the MDV habitats, which demonstrate significant heterogeneity. For example, transient increases in soil moisture in high-salinity areas may act to dilute and mobilize salts, increasing diversity and shifting the community from a dominance by Firmicutes toward a typical desert soil community.
Implications for the resistance and function of microbial communities in extreme environments.
Soil microbial communities in even the most hospitable MDV soils have alpha-diversity values one-third to half (17) of those found in other soil ecosystems (40). Other studies and reviews have demonstrated that the species composition of microbial communities is sensitive to change (86, 87). However, the response of the communities to environmental change (over a 30-day period) was more rapid than that which has been shown to occur in other soil communities (86). Additionally, the high-salinity site, which had the lowest initial diversity, was the most responsive to organic matter addition, resulting in a nearly complete dominance by a single organism. The speed with which these changes took place suggests that the microbial community composition in low-diversity and environmentally extreme ecosystems such as soils in the MDV has limited resistance to environmental change. These results are supported by previous studies of longer duration in the MDV which have also found that soil communities are responsive to altered conditions: soil nematodes responded dramatically to a discrete flood event, with changes persisting for multiple years (88), and a multiyear study involving the transplant of a mummified seal carcass showed a shift in the bacterial community composition and an increase in bacterial biomass after 3 years in the new location (25).
The high diversity in soil microbial communities (89) has promoted the view that while community composition may shift in response to disturbances or altered conditions, these shifts may be of limited functional importance due to redundancy between species. While this may be accurate for very broad level processes, such as soil respiration, other more specialized functions are likely more susceptible to shifts in community composition (90, but see reference 91). The shift in the broad, phylum-level community composition observed in this study may significantly impact the functioning of these communities, given recent lines of evidence suggesting that many microbial functions, including carbon utilization and nutrient processing, are phylogenetically linked (81, 83, 92–95). In fact, altered function in the form of respiration and enzyme activity was observed to change very rapidly in this study. More detailed metagenomic and metatranscriptomic studies are needed to determine the specific functions that have been affected.
Conclusions.
The MDV are experiencing climate-induced changes, including increased moisture and organic matter availability, which are likely to intensify in the near future. The results from this experimental work suggest that the microbial communities in this cold, arid, and oligotrophic environment may change in response to altered resources, with an altered bacterial community composition and increased carbon and nutrient cycling occurring. Investigating these effects along a gradient of habitat severity induced by salinity demonstrated the differential responses of communities under various levels of stress and suggests that the communities in the harshest habitats are the least resistant to disturbance.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by NSF OPP grants 0838879 to J.E.B., M.N.G., and C.D.T.-V. and 1142102 to C.D.T.-V. and D.J.V.H. Additional support was provided by the McMurdo LTER, NSF grant 1115245.
Footnotes
Published ahead of print 7 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03414-13.
REFERENCES
- 1.Engelen B, Ziegelmueller K, Wolf L, Koepke B, Gittel A, Cypionka H, Treude T, Nakagawa S, Inagaki F, Lever MA, Steinsbu BO. 2008. Fluids from the oceanic crust support microbial activities within the deep biosphere. Geomicrobiol. J. 25:56–66. 10.1080/01490450701829006 [DOI] [Google Scholar]
- 2.Lynch RC, King AJ, Farías ME, Sowell P, Vitry C, Schmidt SK. 2012. The potential for microbial life in the highest-elevation (>6000 m.a.s.l.) mineral soils of the Atacama region. J. Geophys. Res. 117:G02028. 10.1029/2012JG001961 [DOI] [Google Scholar]
- 3.Rosnes JT, Torsvik T, Lien T. 1991. Spore-forming thermophilic sulfate-reducing bacteria isolated from North Sea oil field waters. Appl. Environ. Microbiol. 57:2302–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou W, Wang S, Dong H, Jiang H, Briggs BR, Peacock JP, Huang Q, Huang L, Wu G, Zhi X. 2013. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS One 8:e53350. 10.1371/journal.pone.0053350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neilson JW, Quade J, Ortiz M, Nelson WM, Legatzki A, Tian F, LaComb M, Betancourt JL, Wing RA, Soderlund CA, Maier RM. 2012. Life at the hyperarid margin: novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles 16:553–566. 10.1007/s00792-012-0454-z [DOI] [PubMed] [Google Scholar]
- 6.Takacs-Vesbach C, Inskeep WP, Jay ZJ, Herrgard MJ, Rusch DB, Tringe SG, Kozubal MA, Hamamura N, Macur RE, Fouke BW. 2013. Metagenome sequence analysis of filamentous microbial communities obtained from geochemically distinct geothermal channels reveals specialization of three Aquificales lineages. Front. Microbiol. 4:84. 10.3389/fmicb.2013.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tilman D. 1996. Biodiversity: population versus ecosystem stability. Ecology 77:350–363. 10.2307/2265614 [DOI] [Google Scholar]
- 8.Bamforth SS, Wall DH, Virginia RA. 2005. Distribution and diversity of soil protozoa in the McMurdo Dry Valleys of Antarctica. Polar Biol. 28:756–762. 10.1007/s00300-005-0006-4 [DOI] [Google Scholar]
- 9.Fell JW, Scorzetti G, Connell L, Craig S. 2006. Biodiversity of micro-eukaryotes in Antarctic Dry Valley soils with <5% soil moisture. Soil Biol. Biochem. 38:3107–3119. 10.1016/j.soilbio.2006.01.014 [DOI] [Google Scholar]
- 10.Freckman DW, Virginia RA. 1997. Low-diversity Antarctic soil nematode communities: distribution and response to disturbance. Ecology 78:363–369. 10.1890/0012-9658(1997)078[0363:LDASNC]2.0.CO;2 [DOI] [Google Scholar]
- 11.Freckman DW, Virginia RA. 1998. Soil biodiversity and community structure in the McMurdo Dry Valleys, Antarctica. Antarctic Res. Ser. 72:323–335 [Google Scholar]
- 12.Treonis AM, Wall DH, Virginia RA. 1999. Invertebrate biodiversity in Antarctic Dry Valley soils and sediments. Ecosystems 2:482–492. 10.1007/s100219900096 [DOI] [Google Scholar]
- 13.Fountain AG, Levy JS, Van Horn DJ, Gooseff MN. The McMurdo Dry Valleys: a landscape on the threshold of change. Geomorphology, in press [Google Scholar]
- 14.Doran PT, McKay CP, Clow GD, Dana GL, Fountain AG, Nylen T, Lyons WB. 2002. Valley floor climate observations from the McMurdo Dry Valleys, Antarctica, 1986-2000. J. Geophys. Res. 107:4772. 10.1029/2001JD002045 [DOI] [Google Scholar]
- 15.Simmons BL, Wall DH, Adams BJ, Ayres E, Barrett JE, Virginia RA. 2009. Long-term experimental warming reduces soil nematode populations in the McMurdo Dry Valleys, Antarctica. Soil Biol. Biochem. 41:2052–2060. 10.1016/j.soilbio.2009.07.009 [DOI] [Google Scholar]
- 16.Lee CK, Barbier BA, Bottos EM, McDonald IR, Cary SC. 2012. The inter-valley soil comparative survey: the ecology of Dry Valley edaphic microbial communities. ISME J. 6:1046–1057. 10.1038/ismej.2011.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Horn DJ, Van Horn ML, Barrett JE, Gooseff MN, Altrichter AE, Geyer KM, Zeglin LH, Takas-Vesbach CD. 2013. Factors controlling soil microbial biomass and bacterial diversity and community composition in a cold desert ecosystem: role of geographic scale. PLoS One 8:e66103. 10.1371/journal.pone.0066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregorich EG, Hopkins DW, Elberling B, Sparrow AD, Novis P, Greenfield LG, Rochette P. 2006. Emission of CO2, CH4 and N2O from lakeshore soils in an Antarctic dry valley. Soil Biol. Biochem. 38:3120–3129. 10.1016/j.soilbio.2006.01.015 [DOI] [Google Scholar]
- 19.Shanhun FL, Almond PC, Clough TJ, Smith CMS. 2012. Abiotic processes dominate CO2 fluxes in Antarctic soils. Soil Biol. Biochem. 53:99–111. 10.1016/j.soilbio.2012.04.027 [DOI] [Google Scholar]
- 20.Zeglin LH, Sinsabaugh RL, Barrett JE, Gooseff MN, Takacs-Vesbach CD. 2009. Landscape distribution of microbial activity in the McMurdo Dry Valleys: linked biotic processes, hydrology, and geochemistry in a cold desert ecosystem. Ecosystems 12:562–573. 10.1007/s10021-009-9242-8 [DOI] [Google Scholar]
- 21.Cowan DA, Casanueva A. 2007. Stability of ATP in Antarctic mineral soils. Polar Biol. 30:1599–1603. 10.1007/s00300-007-0324-9 [DOI] [Google Scholar]
- 22.Cowan DA, Russell NJ, Mamais A, Sheppard DM. 2002. Antarctic dry valley mineral soils contain unexpectedly high levels of microbial biomass. Extremophiles 6:431–436. 10.1007/s00792-002-0276-5 [DOI] [PubMed] [Google Scholar]
- 23.Ball BA, Virginia RA, Barrett JE, Parsons AN, Wall DH. 2009. Interactions between physical and biotic factors influence CO2 flux in Antarctic dry valley soils. Soil Biol. Biochem. 41:1510–1517. 10.1016/j.soilbio.2009.04.011 [DOI] [Google Scholar]
- 24.Sparrow AD, Gregorich EG, Hopkins DW, Novis P, Elberling B, Greenfield LG. 2011. Resource limitations on soil microbial activity in an Antarctic dry valley. Soil Sci. Soc. Am. J. 75:2188–2197. 10.2136/sssaj2010.0303 [DOI] [Google Scholar]
- 25.Tiao G, Lee CK, McDonald IR, Cowan DA, Cary SC. 2012. Rapid microbial response to the presence of an ancient relic in the Antarctic Dry Valleys. Nat. Commun. 3:660. 10.1038/ncomms1645 [DOI] [PubMed] [Google Scholar]
- 26.Poage MA, Barrettt JE, Virginia RA, Wall DH. 2008. The influence of soil geochemistry on nematode distribution, McMurdo Dry Valleys, Antarctica. Arct. Antarct. Alp. Res. 40:119–128. 10.1657/1523-0430(06-051)[POAGE]2.0.CO;2 [DOI] [Google Scholar]
- 27.Zahran HH. 1997. Diversity, adaptation and activity of the bacterial flora in saline environments. Biol. Fertil. Soils 25:211–223. 10.1007/s003740050306 [DOI] [Google Scholar]
- 28.Lozupone CA, Knight R. 2007. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U. S. A. 104:11436–11440. 10.1073/pnas.0611525104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Yang D, Zhang Y, Shen J, van der Gast C, Hahn MW, Wu Q. 2011. Do patterns of bacterial diversity along salinity gradients differ from those observed for macroorganisms? PLoS One 6:e27597. 10.1371/journal.pone.0027597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fountain AG, Nylen TH, Monaghan A, Basagic HJ, Bromwich D. 2010. Snow in the McMurdo Dry Valleys, Antarctica. Int. J. Climatol. 30:633–642. 10.1002/joc.1933 [DOI] [Google Scholar]
- 31.Clow G, McKay C, Simmons GJ, Wharton RJ. 1988. Climatological observations and predicted sublimation rates at Lake Hoare, Antarctica. J. Clim. 1:715–728. [DOI] [PubMed] [Google Scholar]
- 32.Fountain AG, Lyons WB, Burkins MB, Dana GL, Doran PT, Lewis KJ, McKnight DM, Moorhead DL, Parsons AN, Priscu JC. 1999. Physical controls on the Taylor Valley ecosystem, Antarctica. Bioscience 49:961–971. 10.2307/1313730 [DOI] [Google Scholar]
- 33.Bockheim J, McLeod M. 2008. Soil distribution in the McMurdo Dry Valleys, Antarctica. Geoderma 144:43–49. 10.1016/j.geoderma.2007.10.015 [DOI] [Google Scholar]
- 34.Hall BL, Denton GH, Hendy CH. 2000. Evidence from Taylor Valley for a grounded ice sheet in the Ross Sea, Antarctica. Geogr. Ann. Ser. A Phys. Geogr. 82:275–303. 10.1111/j.0435-3676.2000.00126.x [DOI] [Google Scholar]
- 35.Stuiver M, Denton GH, Hughes TJ, Fastook JL. 1981. History of the marine ice sheet in West Antarctica during the last glaciation: a working hypothesis, p 319–436 In Denton GH, Hughes TJ. (ed), The last great ice sheets. Wiley-Interscience, New York, NY [Google Scholar]
- 36.Burkins MB, Virginia RA, Chamberlain CP, Wall DH. 2000. Origin and distribution of soil organic matter in Taylor Valley, Antarctica. Ecology 81:2377–2391. 10.1890/0012-9658(2000)081[2377:OADOSO]2.0.CO;2 [DOI] [Google Scholar]
- 37.Burkins MB, Virginia RA, Wall DH. 2001. Organic carbon cycling in Taylor Valley, Antarctica: quantifying soil reservoirs and soil respiration. Global Change Biol. 7:113–125. 10.1046/j.1365-2486.2001.00393.x [DOI] [Google Scholar]
- 38.Barrett JE, Virginia RA, Wall DH, Parsons AN, Powers LE, Burkins MB. 2004. Variation in biogeochemistry and soil biodiversity across spatial scales in a polar desert ecosystem. Ecology 85:3105–3118. 10.1890/03-0213 [DOI] [Google Scholar]
- 39.Barrett JE, Gooseff MN, Takacs-Vesbach C. 2009. Spatial variation in soil active-layer geochemistry across hydrologic margins in polar desert ecosystems. Hydrol. Earth Syst. Sci. 6:3725–3751. 10.5194/hessd-6-3725-2009 [DOI] [Google Scholar]
- 40.Chu HY, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P. 2010. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12:2998–3006. 10.1111/j.1462-2920.2010.02277.x [DOI] [PubMed] [Google Scholar]
- 41.Connell L, Redman R, Craig S, Rodriguez R. 2006. Distribution and abundance of fungi in the soils of Taylor Valley, Antarctica. Soil Biol. Biochem. 38:3083–3094. 10.1016/j.soilbio.2006.02.016 [DOI] [Google Scholar]
- 42.Rao S, Chan Y, Lacap DC, Hyde KD, Pointing SB, Farrell RL. 2012. Low-diversity fungal assemblage in an Antarctic Dry Valleys soil. Polar Biol. 35:567–574. 10.1007/s00300-011-1102-2 [DOI] [Google Scholar]
- 43.Giovannoni SJ, DeLong EF, Schmidt TM, Pace NR. 1990. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl. Environ. Microbiol. 56:2572–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell KR, Takacs-Vesbach CD. 2008. A comparison of methods for total community DNA preservation and extraction from various thermal environments. J. Ind. Microbiol. Biotechnol. 35:1139–1147. 10.1007/s10295-008-0393-y [DOI] [PubMed] [Google Scholar]
- 45.Andreotti R, Pérez de León AA, Dowd SE, Guerrero FD, Bendele KG, Scoles GA. 2011. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 11:6. 10.1186/1471-2180-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dowd SF, Sun Y, Wolcott RD, Domingo A, Carroll JA. 2008. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog. Dis. 5:459–472. 10.1089/fpd.2008.0107 [DOI] [PubMed] [Google Scholar]
- 47.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. 10.1186/1471-2105-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R. 2010. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat. Methods 7:813–819. 10.1038/nmeth.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Development Core Team R. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 55.Clarke K, Gorley R. 2006. Primer v6: user manual/tutorial. Primer-E, Plymouth, United Kingdom [Google Scholar]
- 56.Clarke KR. 1993. Nonparametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117–143. 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- 57.Garcia C, Hernandez T. 1996. Influence of salinity on the biological and biochemical activity of a calciorthird soil. Plant Soil 178:255–263. 10.1007/BF00011591 [DOI] [Google Scholar]
- 58.Ghollarata M, Raiesi F. 2007. The adverse effects of soil salinization on the growth of Trifolium alexandrinum L. and associated microbial and biochemical properties in a soil from Iran. Soil Biol. Biochem. 39:1699–1702. 10.1016/j.soilbio.2007.01.024 [DOI] [Google Scholar]
- 59.Frankenberger WT, Bingham FT. 1982. Influence of salinity on soil enzyme activities. Soil Sci. Soc. Am. J. 46:1173–1177. 10.2136/sssaj1982.03615995004600060011x [DOI] [Google Scholar]
- 60.Rietz DN, Haynes RJ. 2003. Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biol. Biochem. 35:845–854. 10.1016/S0038-0717(03)00125-1 [DOI] [Google Scholar]
- 61.Hill BH, Elonen CM, Seifert LR, May AA, Tarquinio E. 2012. Microbial enzyme stoichiometry and nutrient limitation in US streams and rivers. Ecol. Indic. 18:540–551. 10.1016/j.ecolind.2012.01.007 [DOI] [Google Scholar]
- 62.Shah SA, Shah Z. 2011. Changes in soil microbial characteristics with elevated salinity. Sarhad J. Agric. 27:233–244 [Google Scholar]
- 63.Yuan B-C, Li Z-Z, Liu H, Gao M, Zhang Y-Y. 2007. Microbial biomass and activity in salt affected soils under arid conditions. Appl. Soil Ecol. 35:319–328. 10.1016/j.apsoil.2006.07.004 [DOI] [Google Scholar]
- 64.Tripathi S, Kumari S, Chakraborty A, Gupta A, Chakrabarti K, Bandyapadhyay BK. 2006. Microbial biomass and its activities in salt-affected coastal soils. Biol. Fertil. Soils 42:273–277. 10.1007/s00374-005-0037-6 [DOI] [Google Scholar]
- 65.Chowdhury N, Marschner P, Burns RG. 2011. Soil microbial activity and community composition: impact of changes in matric and osmotic potential. Soil Biol. Biochem. 43:1229–1236. 10.1016/j.soilbio.2011.02.012 [DOI] [Google Scholar]
- 66.Nkem J, Virginia R, Barrett J, Wall D, Li G. 2006. Salt tolerance and survival thresholds for two species of Antarctic soil nematodes. Polar Biol. 29:643–651. 10.1007/s00300-005-0101-6 [DOI] [Google Scholar]
- 67.Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, Gentry TJ. 2010. Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J. 4:829–838. 10.1038/ismej.2010.3 [DOI] [PubMed] [Google Scholar]
- 68.Andronov E, Petrova S, Pinaev A, Pershina E, Rakhimgalieva SZ, Akhmedenov K, Gorobets A, Sergaliev NK. 2012. Analysis of the structure of microbial community in soils with different degrees of salinization using T-RFLP and real-time PCR techniques. Eurasian Soil Sci. 45:147–156. 10.1134/S1064229312020044 [DOI] [Google Scholar]
- 69.Ikenaga M, Guevara R, Dean AL, Pisani C, Boyer JN. 2010. Changes in community structure of sediment bacteria along the Florida coastal Everglades marsh-mangrove-seagrass salinity gradient. Microb. Ecol. 59:284–295. 10.1007/s00248-009-9572-2 [DOI] [PubMed] [Google Scholar]
- 70.Benlloch S, Lopez-Lopez A, Casamayor EO, Ovreas L, Goddard V, Daae FL, Smerdon G, Massana R, Joint I, Thingstad F, Pedros-Alio C, Rodriguez-Valera F. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349–360. 10.1046/j.1462-2920.2002.00306.x [DOI] [PubMed] [Google Scholar]
- 71.Foti MJ, Sorokin DY, Zacharova EE, Pimenov NV, Kuenen JG, Muyzer G. 2008. Bacterial diversity and activity along a salinity gradient in soda lakes of the Kulunda Steppe (Altai, Russia). Extremophiles 12:133–145. 10.1007/s00792-007-0117-7 [DOI] [PubMed] [Google Scholar]
- 72.Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. 2009. Global patterns in belowground communities. Ecol. Lett. 12:1238–1249. 10.1111/j.1461-0248.2009.01360.x [DOI] [PubMed] [Google Scholar]
- 73.Onyenwoke RU, Brill JA, Farahi K, Wiegel J. 2004. Sporulation genes in members of the low G+C Gram-type-positive phylogenetic branch (Firmicutes). Arch. Microbiol. 182:182–192. 10.1007/s00203-004-0696-y [DOI] [PubMed] [Google Scholar]
- 74.Schimel J, Balser TC, Wallenstein M. 2007. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88:1386–1394. 10.1890/06-0219 [DOI] [PubMed] [Google Scholar]
- 75.Tilman D, Reich PB, Knops JMH. 2006. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441:629–632. 10.1038/nature04742 [DOI] [PubMed] [Google Scholar]
- 76.Crain CM, Albertson LK, Bertness MD. 2008. Secondary succession dynamics in estuarine marshes across landscape-scale salinity gradients. Ecology 89:2889–2899. 10.1890/07-1527.1 [DOI] [PubMed] [Google Scholar]
- 77.Slocum MG, Mendelssohn IA. 2008. Use of experimental disturbances to assess resilience along a known stress gradient. Ecol. Indic. 8:181–190. 10.1016/j.ecolind.2007.01.011 [DOI] [Google Scholar]
- 78.Cleveland CC, Nemergut DR, Schmidt SK, Townsend AR. 2007. Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 82:229–240. 10.1007/s10533-006-9065-z [DOI] [Google Scholar]
- 79.Eilers KG, Lauber CL, Knight R, Fierer N. 2010. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42:896–903. 10.1016/j.soilbio.2010.02.003 [DOI] [Google Scholar]
- 80.Parsons AN, Barrett JE, Wall DH, Virginia RA. 2004. Soil carbon dioxide flux in Antarctic dry valley ecosystems. Ecosystems 7:286–295. 10.1007/s10021-003-0132-1 [DOI] [Google Scholar]
- 81.Goldfarb KC, Karaoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK, Wallenstein MD, Brodie EL. 2011. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2:94. 10.3389/fmicb.2011.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nemergut DR, Cleveland CC, Wieder WR, Washenberger CL, Townsend AR. 2010. Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol. Biochem. 42:2153–2160. 10.1016/j.soilbio.2010.08.011 [DOI] [Google Scholar]
- 83.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. 10.1890/05-1839 [DOI] [PubMed] [Google Scholar]
- 84.Krishnamurthi S, Bhattacharya A, Mayilraj S, Saha P, Schumann P, Chakrabarti T. 2009. Description of Paenisporosarcina quisquiliarum gen. nov., sp. nov., and reclassification of Sporosarcina macmurdoensis Reddy et al. 2003 as Paenisporosarcina macmurdoensis comb. nov. Int. J. Syst. Evol. Microbiol. 59:1364–1370. 10.1099/ijs.0.65130-0 [DOI] [PubMed] [Google Scholar]
- 85.Geyer KM, Altrichter AE, Van Horn DJ, Takacs-Vesbach CD, Gooseff MN, Barrett JE. 2013. Environmental controls over bacterial communities in polar desert soils. Ecosphere 4:art127. 10.1890/ES13-00048.1 [DOI] [Google Scholar]
- 86.Allison SD, Martiny JBH. 2008. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105:11512–11519. 10.1073/pnas.0801925105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JB. 2012. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3:417. 10.3389/fmicb.2012.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barrett J, Virginia R, Wall D, Doran P, Fountain A, Welch K, Lyons W. 2008. Persistent effects of a discrete warming event on a polar desert ecosystem. Global Change Biol. 14:2249–2261. 10.1111/j.1365-2486.2008.01641.x [DOI] [Google Scholar]
- 89.Curtis TP, Sloan WT, Scannell JW. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U. S. A. 99:10494–10499. 10.1073/pnas.142680199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schimel J, Schaeffer SM. 2012. Microbial control over carbon cycling in soil. Front. Microbiol. 3:348. 10.3389/fmicb.2012.00348#sthash.ZVoptcde.dpuf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wertz S, Degrange V, Prosser JI, Poly F, Commeaux C, Guillaumaud N, Le Roux X. 2007. Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ. Microbiol. 9:2211–2219. 10.1111/j.1462-2920.2007.01335.x [DOI] [PubMed] [Google Scholar]
- 92.Lennon JT, Aanderud ZT, Lehmkuhl BK, Schoolmaster DR., Jr 2012. Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 93:1867–1879. 10.1890/11-1745.1 [DOI] [PubMed] [Google Scholar]
- 93.Martiny AC, Treseder K, Pusch G. 2013. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 7:830–838. 10.1038/ismej.2012.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB, Hallin S. 2010. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 8:523–529. 10.1038/nrmicro2367 [DOI] [PubMed] [Google Scholar]
- 95.Placella SA, Brodie EL, Firestone MK. 2012. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc. Natl. Acad. Sci. U. S. A. 109:10931–10936. 10.1073/pnas.1204306109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.