Abstract
Biological nitrogen fixation is the primary supply of N to most ecosystems, yet there is considerable uncertainty about how N-fixing bacteria will respond to global change factors such as increasing atmospheric CO2 and N deposition. Using the nifH gene as a molecular marker, we studied how the community structure of N-fixing soil bacteria from temperate pine, aspen, and sweet gum stands and a brackish tidal marsh responded to multiyear elevated CO2 conditions. We also examined how N availability, specifically, N fertilization, interacted with elevated CO2 to affect these communities in the temperate pine forest. Based on data from Sanger sequencing and quantitative PCR, the soil nifH composition in the three forest systems was dominated by species in the Geobacteraceae and, to a lesser extent, Alphaproteobacteria. The N-fixing-bacterial-community structure was subtly altered after 10 or more years of elevated atmospheric CO2, and the observed shifts differed in each biome. In the pine forest, N fertilization had a stronger effect on nifH community structure than elevated CO2 and suppressed the diversity and abundance of N-fixing bacteria under elevated atmospheric CO2 conditions. These results indicate that N-fixing bacteria have complex, interacting responses that will be important for understanding ecosystem productivity in a changing climate.
INTRODUCTION
Nitrogen is the most common nutrient limiting productivity in terrestrial ecosystems and enters ecosystems predominantly through bacterial fixation. However, we lack a clear understanding of how N-fixing bacteria respond to climate change drivers or how conserved those responses might be across biomes in a geographic region. Nonagricultural biomes in the eastern United States that experience elevated atmospheric CO2 and increasing N deposition include hardwood forests to the north, pine forests to the south, and brackish marsh areas along the eastern seaboard. Rising atmospheric CO2 concentrations and shifting patterns of N deposition can interact and affect N fixation processes in soil (1). To determine if populations of N-fixing bacteria in soils of different biomes showed similarities in composition and in responses to elevated CO2, we conducted a systematic survey of soil N-fixing bacterial communities across four biomes in the eastern United States, utilizing long-term, free-air CO2 enrichment (FACE) experiments (2). One of these field experiments combined elevated CO2 and N fertilization treatments, allowing us to determine their interactive effects on the N-fixing community in a pine forest in the southeastern United States.
Progressive N limitation theory proposes that ecosystems become more N limited with rising CO2, which suggests that the continued sequestration of CO2 in terrestrial biomass will require greater N fixation inputs (3, 4). The increased ecosystem demand for N under elevated CO2 has been documented after several years of whole-forest CO2 enrichment (5, 6). N-fixing bacteria, which span many taxonomic groups with high levels of endemism and exhibit complex responses to CO2, are thought to contribute ∼97% of the N input into unmanaged terrestrial ecosystems (1). Because many biomes lack large N inputs from symbiotic N fixation, an increase in N fixation demand will most likely be met by free-living, N-fixing bacteria (1).
Nitrogen-fixing bacteria are capable of responding to climate change drivers through alterations in diversity, abundance, and fixation rates. Elevated CO2 typically increases N demand through mechanisms such as increased C/N ratio of plant inputs to soils, which favors conditions for N fixation (7). However, N fertilization or other additions often lower fixation rates, since free-living N-fixing bacteria are typically facultative and can potentially suppress N fixation when N is relatively abundant (1). The community diversity of soil N-fixing bacteria shifts quickly and is a strong predictor of N fixation activity (8, 9). Based on these factors, we hypothesized that long-term elevated atmospheric CO2 would increase the abundance of N-fixing bacteria and alter bacterial community composition but that increased inorganic N supply through fertilization would suppress the CO2 enhancement of N-fixing-bacterial abundance.
MATERIALS AND METHODS
Soil collection.
Soil cores were collected from three free-air CO2 enrichment (FACE) field research sites and one open-top chamber (OTC) site. The FACE field sites were as follows: (i) a sweet gum (Liquidambar styraciflua) plantation, Oak Ridge, TN (TNO); (ii) a loblolly pine (Pinus taeda L.) plantation, Orange County, NC (NCD); and (iii) an aspen plantation forested with Populus tremuloides Michx. (trembling aspen), Rhinelander, WI (WIR). The OTC site was a brackish marsh in the high intertidal zone within a subestuary of the Chesapeake Bay, in Maryland, with a patchy plant cover comprised of Spartina patens (Ait.) Muhl, Scirpus olneyi Gray, Distichlis spicata (L.) Greene, Typha angustifolia L., and Iva fructescens L (MDE). Following collection, soil samples from all sites were immediately placed on dry ice for transport to the laboratory and stored at −70°C. Further descriptions of these field sites and soil characteristics are available in references 2 and 10 and at http://public.ornl.gov/face/. Complete information on soil chemical characteristics can be found in Table S2 of reference 2.
Soil at the aspen plantation site (WIR) is a Padus sandy loam. Beginning in 1998, 30-m-diameter FACE rings at the WIR site were fumigated with an additional 397.4 ± 14.7 ppm CO2 (elevated) or 84.0 ± 16.6 ppm CO2 (ambient) during daylight hours over the course of each growing season (sampling for this study occurred during the 10th season of CO2 fumigation). Five soil cores (2.5- by 5-cm cores, 0- to 10-cm depth) were collected in July 2007 from the aspen stands in each of three elevated CO2 rings and three ambient CO2 rings after removal of loose litter from the forest floor (i.e., Oi horizon). Soil cores from each ring (n = 5) were pooled to yield a single, composite 0- to 10-cm soil sample from each ring (three elevated and three ambient soil composites).
Soil at the loblolly pine plantation (NCD) is an acidic clay loam of the Enon series of moderately low fertility (6). Since 1996, three 30-m-diameter FACE rings have been fumigated with elevated (ambient plus 200 ppm) CO2 and another three rings with ambient (∼370 ppm) levels of CO2. Fumigation was applied during daylight hours when the ambient temperature was greater than 5°C and average wind speed above the canopy was less than 5 m s−1. Beginning in 2005, fertilization treatment (NH4NO3 pellets broadcast by hand once or twice yearly, 11.2 g N m−2 year−1) was applied to two quadrants (one half) of the existing elevated- and ambient-CO2 rings (http://face.env.duke.edu/). Triplicate soil cores (2.5- by 5-cm cores, 0- to 10-cm depth) were randomly collected and then pooled from the fertilized and unfertilized quadrants of three elevated CO2 rings and three ambient CO2 rings in June 2007 (24 composite soil samples). Following DNA extraction (see below), equimolar quantities of DNA from fertilized or unfertilized quadrants of a given ring were pooled, yielding three DNA samples from each: (i) elevated CO2 without fertilizer, (ii) elevated CO2 with fertilizer, (iii) ambient CO2 minus fertilizer, and (iv) ambient CO2 with fertilizer.
Soil at the sweet gum plantation (TNO) is a slightly acidic silty clay loam classified as Wolftever series (http://public.ornl.gov/face/ORNL/ornl_site_characteristics.shtml). Carbon dioxide fumigation was initiated for three 25-m-diameter FACE rings in 1998 and has been performed yearly, 24 h day−1 from April to November. Between 1998 and 2007, CO2 concentrations within the airspace of the elevated rings and ambient rings ranged from 528 to 559 ppm and 384 to 402 ppm, respectively. Triplicate soil cores (2.5- by 5-cm cores, 0- to 10-cm depth) were randomly collected from two elevated CO2 rings and two ambient CO2 rings. The triplicate soil samples from each ring were pooled to yield a single, composite 0- to 10-cm soil sample for each ring (two elevated and two ambient CO2 composite soil samples).
The brackish tidal marsh site (MDE) is located in an occasionally flooded region of the Rhode Island River, a subestuary of the Chesapeake Bay. Salinity at this site ranges from 0 to 18 ppt (11), and the site floods during roughly 28% of high tides, though the peat soil remains nearly constantly saturated (Patrick Megonigal, personal communication). The MDE site has a pH of 6 to 6.5 and carbonates in sediments, but the elevated CO2 treatment has been shown to raise both air and sediment inorganic carbon concentrations (12). A CO2 fumigation experiment with a randomized block design was initiated in 1987, consisting of five replicate open-top chambers fumigated with 686 ± 30 μl liter−1 CO2 (elevated) and another five replicate chambers fumigated with 350 ± 22 μl liter−1 CO2 (ambient) (13). Triplicate soil cores (2.5- by 5-cm cores, 0 to 10 cm depth) were collected from different compass points within each chamber in September 2008 and pooled to yield a single, composite 0- to 10-cm soil sample for each chamber (10 composite soil samples total). At the time of sampling, S. olneyi was the dominant plant in the open-top chambers.
DNA extraction.
DNA was extracted from composite soil samples (0.5 g; except TNO samples, 0.25 g) using the FastDNA spin kit for soil (MP Biomedicals) following the manufacturer's instructions. Large, visually obvious plant debris (roots, leaves, and needles) was removed prior to DNA extraction. DNA extracts were examined qualitatively on 1.2% agarose gels in 0.5× Tris-borate-EDTA (TBE) with ethidium bromide, quantified using the Quant-It PicoGreen double-strand DNA (dsDNA) assay kit (Invitrogen), and stored at −70°C until further analysis. DNA yields were as follows: WIR, 48.5 ± 5.9 μg/g soil; NCD, 63.8 ± 17.7 μg/g soil; TNO, 33.4 ± 11.0 μg/g soil; MDE, 34.4 ± 5.2 μg/g soil.
PCR, cloning, and sequencing of the bacterial nifH gene.
An ∼506- to 521-bp portion of the nifH gene, which encodes the reductase subunit of dinitrogenase, was amplified from soil DNA extracts using the degenerate primers 19F (5′-GCIWTYTAYGGIAARGGIGG) (14) and nifH3 (5′-ATRTTRTTNGCNGCRTA) (15). Each reaction was carried out in triplicate with a 50-μl volume containing the following: 1× PCR buffer, 1.5 mM MgCl2 (Applied Biosystems), 31 pmol each primer, 0.8 mM concentrations of deoxynucleoside triphosphates (dNTPs) (Applied Biosystems), 10 μg bovine serum albumin (BSA), 2.5 U Ampli-Taq LD polymerase (Applied Biosystems), and 2 μl template DNA (either a 1:10 or 1:100 dilution of the soil extract DNA). The thermal cycle profile consisted of: 95°C for 5 min; 35 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 60 s; and a 72°C final extension step for 10 min. PCR product length was confirmed by agarose gel electrophoresis. Triplicate reactions were pooled and gel purified using a 1.2% agarose gel and the Qiagen QIAquick PCR purification kit. Purified nifH amplicons were cloned using the TOPO-TA pCR 2.1 kit (Invitrogen) according to the manufacturer's directions. Clones were sequenced bidirectionally with M13 primers using Sanger technology.
Sequence processing and analysis.
Sequences were assembled using Fincon (Los Alamos National Laboratory program; courtesy of Cliff Han), and short sequences, chimeric sequences, and sequences containing ambiguous bases were discarded. Translations and phylogenetic analysis of amino acid sequences (161 characters) were performed with MEGA software version 5.05 (16). Paralogues of nifH exhibiting similarity to the protochlorophyllide reductase (bchX) and chlorophyllide reductase (bchL) genes were excluded from further analysis.
Amino acid sequences were aligned using the Protdist program within the Phylip v. 3.69 software package (17). The resulting alignment was used to generate operational taxonomic units (OTUs) in the mothur software package version 1.25.1 (18). Simpson's reciprocal index of OTU diversity (rarefied for even sampling), Venn diagrams, and principal coordinate analyses (PCoA) were generated using the mothur software suite. Simpson's reciprocal index was chosen since it is less sensitive to sample size than other diversity indexes; when combined with rarefaction, this index is ideal for comparing diversity across disparate sites with different sampling intensities (19). For the PCoA analyses, values between samples were determined using the Bray-Curtis dissimilarity index. Sequences were pooled by site, from both ambient and elevated CO2 plots, for a cross-site comparison yielding 779, 241, 288, and 292 total nifH sequences for MDE, NCD, TNO, and WIR, respectively. Rarefaction curves were generated using OTU cutoff values of 90, 95, and 99% for translated nifH sequences (158 amino acids) (see Fig. S1 in the supplemental material). At the 99% OTU level, coverage ranged from 77 to 79% for sites MDE, NCD, and TNO and was 88% for WIR (Table 1). Coverage was high for libraries from each site (92 to 99%) when OTUs were binned at ≥95% similarity.
TABLE 1.
Intrasite diversity of translated nifH amino acid sequences at different OTU cutoff valuesa
| OTU similarity (%) | Site | No. of sequences | No. of OTUs | 1/Db | Coverage |
|---|---|---|---|---|---|
| 99 | MDE | 791 | 295 | 48.7 a | 0.77 |
| NCD | 242 | 97 | 27.7 a | 0.77 | |
| TNO | 287 | 102 | 30.8 a | 0.79 | |
| WIR | 292 | 63 | 5.39 b | 0.88 | |
| 95 | MDE | 791 | 143 | 25.6 a | 0.93 |
| NCD | 242 | 36 | 5.47 b | 0.92 | |
| TNO | 287 | 35 | 5.51 b | 0.94 | |
| WIR | 292 | 29 | 4.31 b | 0.96 | |
| 90 | MDE | 791 | 69 | 8.02 a | 0.96 |
| NCD | 242 | 17 | 3.33 b | 0.97 | |
| TNO | 287 | 20 | 3.49 b | 0.98 | |
| WIR | 292 | 12 | 3.04 b | 0.99 |
Temperate brackish tidal marsh (MDE), n = 5; warm temperate pine plantation (NCD), n = 3; warm temperate deciduous plantation (TNO), n = 2; cool temperate deciduous plantation (WIR), n = 3. See Materials and Methods for detailed site and soil information.
Reciprocal Simpson's index, derived from rarefied data to ensure equal sampling across sites. Values with different letters are significantly different (P < 0.05).
Quantitative PCR (qPCR) of the nifH gene.
Quantitative PCR was performed targeting the nifH gene using the PolF/PolR primer pair (5′-TGCGAYCCSAARGCBGACTC-3′ and 5′-ATSGCCATCATYTCRCCGGA-3′) and techniques identical to those described by Gaby and Buckley (20, 21). This primer pair was selected for its wide coverage of N-fixing bacterial groups and optimal fragment size (∼360) for qPCR (17). Standard curves for qPCR were constructed from a 10-fold dilution series of PCR products amplified from genomic extracts of a nifH-containing strain of Klebsiella pneumoniae. PCR amplicons for the standard curve were purified from a 1% agarose gel and quantified for mass via PicoGreen fluorimetry using the Quant-IT kit (Invitrogen) to determine nifH copy number.
Statistical analyses.
Effects of elevated CO2 on OTU diversity (Simpson's reciprocal index) across biomes were tested using a general linear model (ordinary least squares) with site and CO2 treatment as main effects. Diversity index values were pooled by replicated CO2 plot (i.e., by rings and treatments) for analysis (MDE, n = 5; NCD, n = 3; TNO, n = 2; WIR, n = 3). For NCD site data, general linear models were used to test the effects of CO2, N fertilization, and their interaction on Simpson's reciprocal index and nifH copy number pooled by N treatment within replicated CO2 treatment (n = 3 for each factorial CO2 and N fertilization combination). All statistical analyses were conducting using PROC GLM in SAS 9.3 (SAS Institute, Cary, NC, USA).
Nucleotide sequence accession numbers.
The nifH sequences obtained in this study were deposited in GenBank under accession numbers KF846581 through KF848170.
RESULTS
Diversity of nifH across biomes.
The diversity of nifH sequences was greatest for the MDE wetland system, regardless of the OTU level assessed, exhibiting Simpson's diversity index values (1/D) that were 1.6- to 9-fold greater than those of the other sites (Table 1). When examined at the 99% OTU level, though, MDE diversity was not statistically distinguishable from that of the loblolly pine site at NCD and the sweet gum site at TNO (Table 1). In contrast, nifH diversity at the aspen site at WIR was lower than at the other sites at each OTU level examined. The analysis indicated that levels of nifH diversity at sites NCD and TNO were remarkably similar (1/D values were with 10% of each other at each OTU level tested).
A consistent difference in nifH diversity between ambient and elevated CO2 soils was not observed (Table 2). Elevated CO2 was found to significantly raise diversity (ANOVA; P < 0.05) only at the NCD site at the 99% OTU level. Regardless of CO2 treatment, the MDE site exhibited the highest N-fixing-bacterial diversity, particularly at the lower OTU cutoff values (P < 0.05, 95% and 90% OTU levels [Table 2]).
TABLE 2.
Numbers of translated nifH sequences and OTUs, diversity, and coverage at different amino acid sequence similarity levels in ambient and elevated CO2 plots across sitesa
| OTU similarity (%) | Site | No. of sequences |
No. of OTUs |
1/D |
Coverage |
||||
|---|---|---|---|---|---|---|---|---|---|
| Amb | Elv | Amb | Elv | Amb | Elv | Amb | Elv | ||
| 99 | MDE | 383 | 408 | 170 | 185 | 40.9 | 53.9 | 0.71 | 0.71 |
| NCD | 121 | 121 | 53 | 64 | 20.0 | 35.5b | 0.79 | 0.65 | |
| TNO | 116 | 171 | 60 | 66 | 41.1 | 20.5 | 0.69 | 0.78 | |
| WIR | 133 | 159 | 36 | 40 | 5.33 | 5.46 | 0.83 | 0.88 | |
| 95 | MDE | 383 | 408 | 95 | 102 | 22.8 | 27.5 | 0.87 | 0.88 |
| NCD | 121 | 121 | 17 | 29 | 6.02 | 4.92 | 0.96 | 0.84 | |
| TNO | 116 | 171 | 18 | 27 | 6.78 | 4.23 | 0.95 | 0.92 | |
| WIR | 133 | 159 | 19 | 19 | 4.57 | 4.05 | 0.95 | 0.97 | |
| 90 | MDE | 383 | 408 | 47 | 48 | 6.12 | 9.29 | 0.94 | 0.96 |
| NCD | 121 | 121 | 9 | 14 | 3.70 | 2.97 | 0.98 | 0.93 | |
| TNO | 116 | 171 | 11 | 16 | 3.38 | 3.59 | 0.97 | 0.96 | |
| WIR | 133 | 159 | 10 | 10 | 3.00 | 3.10 | 0.99 | 0.99 | |
Amb, ambient; Elv, elevated. Temperate brackish tidal marsh (MDE), n = 5; warm temperate pine plantation (NCD), n = 3; warm temperate deciduous plantation (TNO), n = 2; cool temperate deciduous plantation (WIR), n = 3. See Materials and Methods for detailed site and soil information.
Significant effect of elevated CO2 (P < 0.05).
Phylogenetic composition of nifH genes across four biomes.
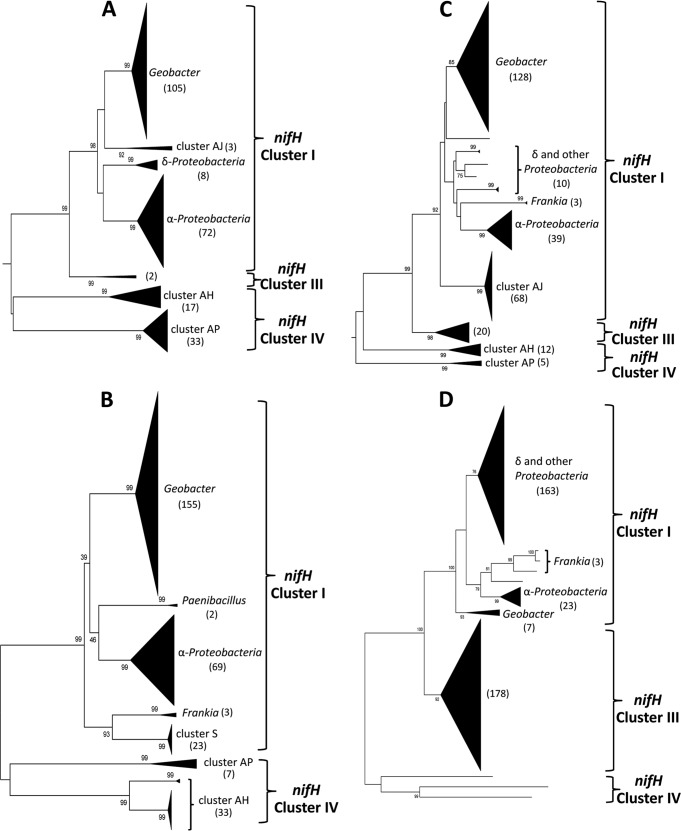
Phylogenetic trees were generated for the 1,600 translated nifH sequences (161 amino acids) collected from the four sites (Fig. 1). From each of the forested sites (NCD, TNO, and WIR), sequences belonging to the canonical nifH cluster I comprised ≥80% of the total. In each case, the most abundant sequences were most similar to those of various Geobacter spp. and related genera (Pedobacter and Anaeromyxobacter) (Fig. 1A to C). Sequences most similar to those from the Alphaproteobacteria were the other dominant clade at the forested sites, except for TNO, where subcluster AJ comprised the second most abundant cluster of sequences. Subcluster AJ sequences fall squarely into nifH cluster I and are 96 to 99% similar to translated sequences from clones retrieved from mangrove and paddy soils/sediments (22); however, they cannot be firmly assigned to a specific phylum at this time (80 to 85% similar to multiple phyla).
FIG 1.
Neighbor-joining trees constructed from translated nifH sequences (161 amino acids) from NCD (A), WIR (B), TNO (C), and MDE (D). Brackets indicate canonical nifH clusters: cluster I, typical Mo-Fe nitrogenases from the Cyanobacteria, most Proteobacteria, and a limited number of Firmicutes and Actinobacteria; cluster III, Mo-Fe nitrogenases primarily from anaerobic organisms, including clostridia, acetogenic bacteria, methanogens, spirochetes, green sulfur bacteria, and sulfate-reducing bacteria; cluster IV, uncharacterized, divergent nifH paralogues mainly from archaea and some anoxygenic photosynthetic bacteria (as defined in reference 26). Values in parentheses indicate the number of sequences in each subcluster.
Subcluster S sequences, which also fall within nifH cluster I, were found only at the WIR site. These sequences are very similar to numerous sequences from soil samples collected from the Qin Zhang plateau of the Tibetan region of China (23), from ephemerally wetted soils of the Antarctic Dry Valleys, and from soils of a moderately burned, northern New Mexico conifer forest (11, 24, 25). These nifH sequences appear to form a distinct cluster that is most similar to Frankia nifH; however, they cannot be confidently linked to a specific taxon, even at the phylum level.
The majority of the remaining nifH sequences from the forested sites could be assigned to nifH cluster IV, in subcluster AH or AP. Subcluster AH sequences are very similar (>97%) to a second nifH copy in Geobacter daltonii. Subcluster AP sequences are novel, sharing less than 62% similarity with nifH sequences in GenBank's nonredundant protein database (all nonredundant GenBank CDS translations plus entries in PDB, Swiss-Prot, PIR, and PRF, excluding environmental samples from whole-genome sequencing projects).
The MDE nifH libraries were dominated by nifH cluster I and III sequences (26). The sequences from the MDE estuary sediments belonging to nifH cluster III were similar to those from anaerobic Deltaproteobacteria. Most nifH cluster I sequences from the MDE samples were most similar to Gammaproteobacteria and Betaproteobacteria, including purple sulfur bacteria involved in sulfur oxidation/reduction, iron-oxidizing bacteria, and methanotrophs. A small number of nifH cluster I sequences from MDE were very similar to nifH sequences from Alphaproteobacteria or Geobacter spp., similar to those identified in the forest soil samples.
Community composition across biomes.
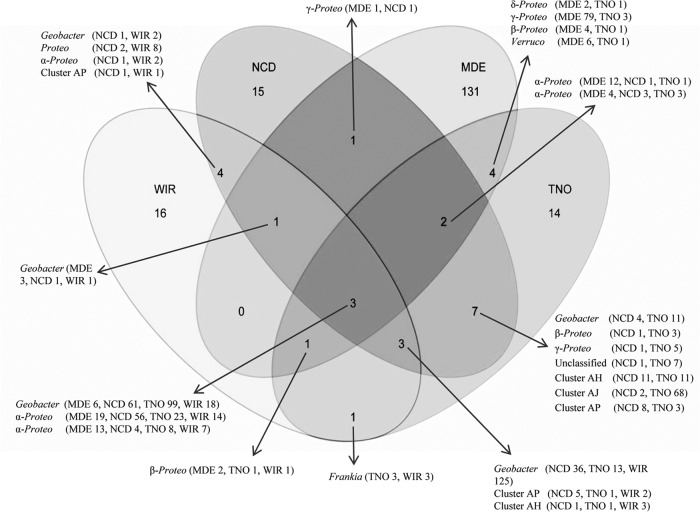
Principal coordinate analysis across sites showed strong differences in composition of N-fixing bacterial communities across the four biomes (Fig. 2). N-fixing-bacterial-community composition was distinct in the brackish tidal marsh (MDE) compared to the forest biomes. The two warm (Tennessee and North Carolina) temperate forest communities (NCD and TNO) were more similar to each other than the cold (Wisconsin) temperate forest community (WIR). This similarity was striking, since the NCD site was dominated by loblolly pine (Pinus taeda) and the TNO and WIR sites were both deciduous hardwood plantations. Analysis of shared OTUs with 95% similarity (OTU95s) revealed that there was a very high level of endemism, with the majority of unique OTU95s belonging to the estuary (MDE) site; however, some of the MDE OTU95s were also present in low abundance across the forest sites (Fig. 3). Seventy-nine percent of the nifH sequences from the forest sites belonged to OTU95s that were identified in at least two of the forest sites. In contrast, only 19% of the nifH sequences from the estuary sediments belonged to OTU95s that were also found in the forest sites.
FIG 2.
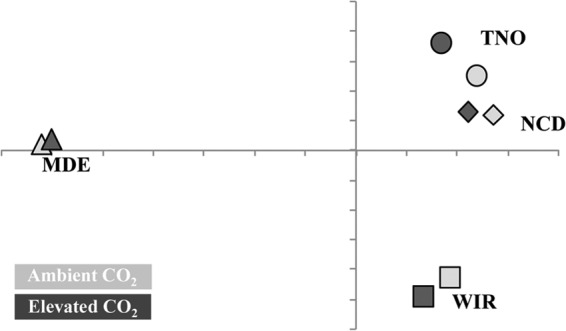
Principal coordinate analysis of nifH translated amino acid sequences at the 95% similarity OTU threshold.
FIG 3.
Venn diagram displaying the distribution of nifH OTU95s across the four field sites (WIR, NCD, TNO, and MDE). Numbers within the diagram show the number of OTU95s shared between sites or unique to a given site. Arrows point to the taxonomic identity of the OTU95s, and the accompanying values in parenthesis indicate the number of sequences from each site comprising each OTU95.
Interaction of elevated atmospheric CO2 and N fertilization in the pine forest.
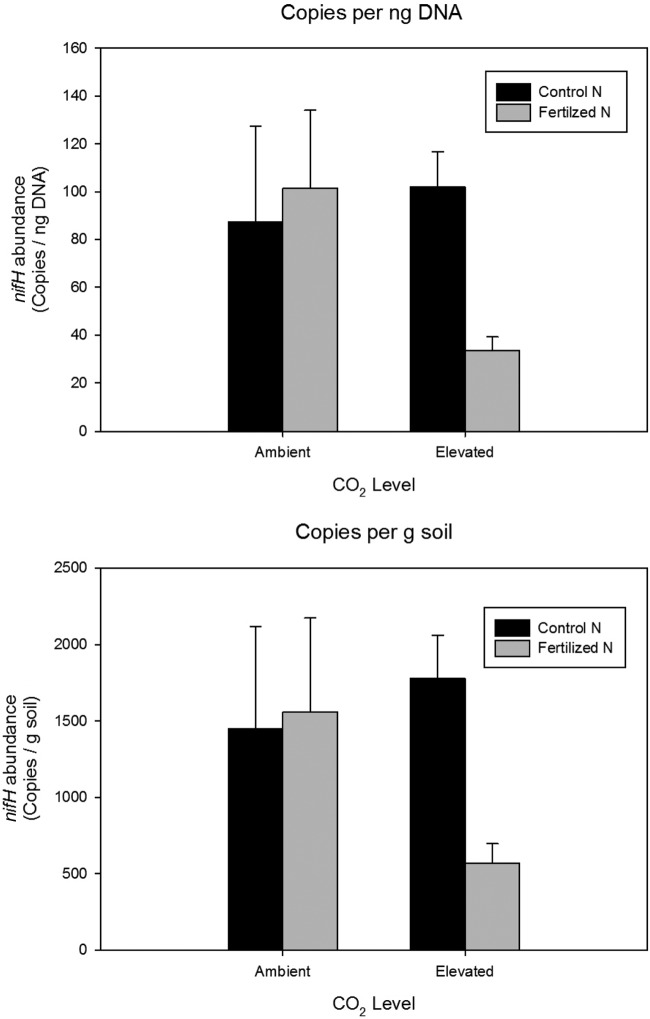
The interaction between elevated CO2 and N fertilization on the soil N-fixing bacterial community was examined at the NCD site, which at the time of sampling had received 10 years of CO2 and 2 years of N fertilization in a factorial design (6). N fertilization had no effect on relative abundance of nifH genes (Fig. 4) under ambient CO2 conditions. In contrast, N fertilization reduced the abundance of nifH gene copies 3-fold (ANOVA; P < 0.05) under elevated CO2 (Fig. 4). This decrease was evident in both nifH gene copy number per nanogram of DNA (Fig. 4, top) and copy number per gram of soil (Fig. 4, bottom).
FIG 4.
Interactive effects of elevated CO2 and N fertilization on nifH abundance at the NCD site. (Top) Copies of nifH per nanogram of DNA analyzed; (bottom) copies of nifH per gram of soil. Data in both panels show that N fertilization reduced abundance of nifH only under elevated CO2 (P < 0.05).
A similar pattern was observed in the diversity of cluster I nifH OTU95s from the loblolly pine forest at the NCD site. Under ambient CO2, diversity of nifH OTUs did not differ between N-fertilized and unfertilized soils (Fig. 5). In contrast, under elevated CO2, N fertilization reduced diversity at the 95% OTU similarity level to half that of the unfertilized soils (ANOVA; P < 0.05) (Fig. 5). Elevated CO2 alone doubled the diversity of nifH OTUs in soils with no N addition (ANOVA; P < 0.05) but had no significant effect when soils were fertilized with N (Fig. 5).
FIG 5.
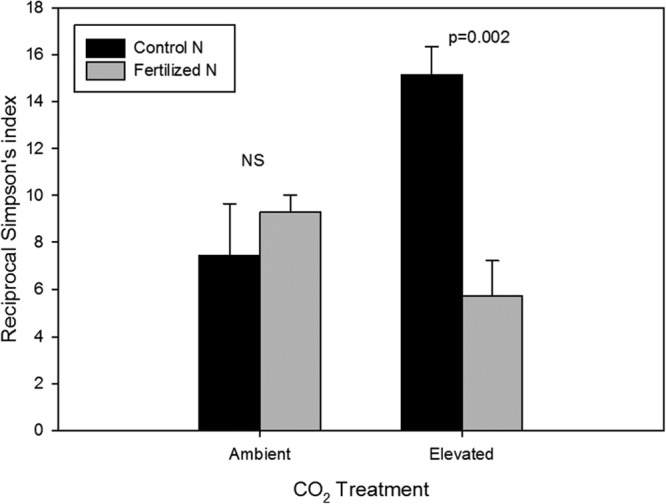
Interactive effects of elevated CO2 and N fertilization on nifH diversity index at the NCD site. Similar to abundance of nifH, diversity of nifH OTU95s decreased with N fertilization only under elevated CO2 (P < 0.05). Under control N conditions (no N added), diversity was significantly greater under elevated CO2 (P < 0.05).
DISCUSSION
Here, we present a nifH gene sequencing survey (1,600 sequences) (Table 1) of soils from three temperate forest biomes and a brackish tidal marsh that had been exposed to several years of CO2 enrichment or a combination of elevated CO2 and N fertilization. This data set significantly increases the taxonomic coverage of high-quality, publicly available sequences for this important biogeochemical function.
Using qPCR and sequencing approaches, we demonstrated the effects of interacting global change drivers (elevated CO2 and N deposition) on N-fixing bacteria in a forest system. The nifH libraries from the NCD, TNO, and WIR forest soils were dominated by cluster I sequences very similar to nifH sequences from Alphaproteobacteria and Geobacteraceae (Deltaproteobacteria). Assuming that nifH cluster IV sequences are not involved in nitrogen fixation (27, 28), sequences from Alphaproteobacteria and Geobacteraceae comprised 93%, 62%, and 89% of the functional nifH in the NCD, TNO, and WIR libraries, respectively. These results are consistent with those from a recent pyrosequencing survey of four terrestrial soils (Alaska boreal forest/taiga, Florida subtropical/dry forest, Hawaii subtropical/lower montane wet forest, and Utah grassland/shrubland), where 65 to 80% of the nifH sequences were identified as belonging to either Alphaproteobacteria or Deltaproteobacteria (29).
The presumptive alphaproteobacterial nifH sequences could be clustered into four predominant OTU95s (see Table S1 in the supplemental material) and were most closely matched to nifH sequences from well-known N-fixing soil genera, such as Bradyrhizobium, Azospirillum, Hyphomicrobium, and Gluconacetobacter. The most common Alphabacteria nifH OTU detected in this study (OTU 1) (see Table S1) was the most common nifH sequence type detected in the recent cross-soil pyrosequencing soil study by Wang et al. (29). Additionally, the second most common OTU detected in this study (OTU 2 in Table S1) was previously found to be the fourth most frequent OTU95 in a global survey of nifH sequences (the three OTUs that were observed more frequently were primarily from marine environments) (30). These results confirm the ubiquity of genera such as Azospirillum and Bradyrhizobium, not only as symbiotic N-fixing bacteria, but also potentially as free-living soil diazotrophs (31, 32).
A large proportion (45 to 62%) of the functional nifH homologs retrieved from the three temperate forest sites examined in this study appear to belong to the Geobacteraceae. Sequences very similar to nifH from Geobacter have been identified in previous soil surveys (25, 26, 33, 34). This sequence cluster, defined as subcluster 1A by Zehr et al. (26), is common in nifH soil libraries and typically contributes <15% of the total sequence number. Gaby and Buckley (30) found that it comprised 14% of all soil nifH sequences (n = 5,748) in their 2010 census of multiple public databases. However, subcluster 1A was found to comprise ≥50% of the nifH sequences obtained from roots and surrounding soil of the perennial grass Molinia caerulea in a littoral meadow in Switzerland (34), a conifer forest in northern New Mexico (25), a maize field in Clinton County, New York (8), an 8-year ice-free section of the forefield of the Damma glacier in Switzerland (35), and the temperate forest soils of this study. Until recently, the only nifH homolog from a cultured representative within subcluster 1A belonged to Geobacter metallireducens; thus, it was difficult to determine the cluster's phylogenetic breadth. However, recent whole-genome sequencing efforts have revealed that nifH from related genera within the family Geobacteraceae (i.e., Pelobacter, Desulfuromonas, and Anaeromyxobacter) fall within subcluster 1A and clearly cluster with all Geobacter-related sequences recovered in this study (see Fig. S2 in the supplemental material).
The Geobacteraceae have largely been categorized as subsurface, facultative anaerobes that are important agents of bioremediation [oxidation of organics and certain metals coupled to the dissimilatory reduction of Fe(III) sulfate] due to their advanced capacity for syntrophic metabolism and extracellular electron transfer (36, 37). It has been posited that their ability to fix N2 provides a selective advantage in the nutrient-depleted subsurface (37). In addition, members of this clade are also found in surface soil habitats with fluctuating oxic/anoxic conditions like Antarctic hyporheic zones/lake margins and the forefields of alpine glaciers, where the metabolic flexibility of the Geobacteraceae would be favored (24, 35). Our results indicate that whatever selective advantage leads to the numeric dominance of Geobacteraceae among free-living, N-fixing populations in such exotic soils could also be at play in less extreme soil systems such as temperate forests, as we observed.
Soil chemistry at the four sites differed markedly, with dramatically higher values for soil organic matter (SOM) and cations in the MDE brackish marsh than the forest sites (see Table 5 in reference 2). The diversity of translated nifH sequences was markedly greater in sediments from the MDE brackish tidal marsh than the forest soils (Tables 1 and 2). The diversity of the MDE nifH libraries was largely driven by nifH cluster III sequences that are most closely aligned with those from anaerobic microorganisms, primarily sulfate reducers (Desulfovibrio, Desulfatibacillum, Desulfobacca, etc.), spirochetes involved in H2-CO2 acetogenesis (Treponema) and fermentative syntrophs (Syntrophobacter). These results are consistent with those of Gaby and Buckley (30), who used a meta-analysis of global nifH sequences and found that nifH cluster III contained the greatest diversity among the canonical nifH clusters. They also support the idea that sulfate-reducing bacteria are key N2 fixers in marine and estuarine sediments (38).
Among the three forest sites, the cold-climate aspen forest soils were 20- to 100-fold higher in nitrate and phosphorus than the warm-climate pine or sweet gum forests (see Table 5 in reference 2). Thus, N-fixing bacteria differences among the forest sites appeared to correlate more with geographic location (warm versus cold climates) or soil chemical conditions than with tree type (hardwoods versus pine) (Fig. 2). In addition, the pine-dominated site (NCD) had been a mixed hardwood stand prior to clearing and planting of pine 20 years prior to the initiation of CO2 fumigation. The consistency among warm-climate soil communities is interesting, since it suggests that long-term effects of geography, climate, and soil history and chemistry are more important to the composition of N-fixing bacterial communities than current forest type. This agrees with other research that suggests that plant species can alter soil microbial community composition but that these plant effects are constrained by soil impacts (39, 40, 41). A review by Berg and Smalla (42) suggests that plants can alter microbial communities, but only by increasing or reducing the abundance of microbial groups already present within the soil reservoir. Plants certainly exert an influence on N-fixing-bacterial composition in the rhizosphere, but it appears that climate and soil type may exert a stronger control on community composition of free-living, N-fixing bacteria in the bulk soil.
The effects of CO2 treatment on the diversity of the N-fixing bacterial community were relatively small compared to the differences across biomes. This result was contrary to our initial hypothesis but is consistent with other studies that observed elevated CO2-induced shifts in microbial communities were driven by functional groups that did not necessarily possess nifH (2). These groups are presumably more sensitive to elevated CO2 than those containing nifH genes identified in this study. Taken together, the results of this study and previous work suggest potentially important effects of elevated CO2 on soil microbial communities, but those differences are likely confined to specific phylogenetic or functional groups and certain biomes.
Our analysis of the interaction of CO2 and N fertilization on N-fixing bacteria showed that N fertilization decreased diversity and abundance of nifH under elevated CO2. Previous studies of N fixation rates at the NCD found no difference in N fixation rates with elevated CO2 (43); however, N fixation process rates have not been measured at the site since N fertilization began. Furthermore, the method that was used to previously assess N fixation rates in soils at the NCD site (e.g., acetylene block [43]) provides a gross measure of the process and may not be capable of detecting subtle shifts in N fixation. Beyond forest ecosystems, a meta-analysis of the effects of elevated CO2 on N fixation rates was not able to detect an effect of soil N fertilization on N fixation rates (44). As both diversity and abundance of nitrogenase genes are positively correlated with rates of N fixation (8, 9), our molecular analysis provides novel insight into the interactive effects of CO2 and N fertilization on potential N fixation rates.
The interacting effects of CO2 and N fertilization (Fig. 4 and 5) also imply that elevated CO2 can create an N-fixing community that is more sensitive to N additions than communities with ambient CO2. Suppression of N fixation by N addition under elevated CO2 has been documented in systems with symbiotic N-fixing legumes (45, 46) but has not been studied in forest systems that lack legumes. At the pine forest site, this suppressive effect of N fertilization on N-fixing bacteria under elevated CO2 is potentially explained by observed changes in tree root exudation and its response to N fertilization under elevated CO2 (47, 48). Root exudation at this site also increases with elevated CO2, but this increase is then suppressed by N fertilization, which mirrors our results on nifH diversity and abundance (48). The connection between root exudation and N fixation has been observed previously (49, 50). Our results suggest that changes in root exudation could impact potential N-fixing bacterial communities and rates of N fixation more strongly in a future higher-CO2 environment.
Although the sequencing depth for each sample is small compared to that achievable with newer sequencing technologies, the nifH Sanger sequencing data are very high quality (bidirectional sequences), and the relatively long reads (474 nucleotides; 158 amino acids) allowed the higher taxonomic resolution we required for this comparative survey (∼100 to 200 bp). These sequences span key amino acid residues important for NifH structure and function, including Lys15, Ser16, Cys38, Cys85, Cys97, Arg100, Thr104, Asp125, Asp129, and Cys132, that are required for accurate functional prediction (51, 52, 53). It is important to note that biases in PCR, especially at high cycle numbers such as those used in this study (i.e., 35 cycles), can result in a skewed representation of the relative abundance of nifH phylotypes. Indeed, significant differences have been noted in the relative abundance of nifH phylotypes in a sample assessed using qPCR versus clone library representation (24, 54). Yet, the primary goal of this study was to evaluate nifH composition across treatments (CO2 versus ambient and interactions with N fertilization) and sites using a single standardized assay, where shifts in N-fixing-bacterial-community composition could be attributed to location or treatment rather than experimental bias.
In conclusion, this study provides key new information about an important microbial functional group that mediates N additions to soils and about how that group responds to global change drivers. The cross-biome approach supplied valuable insights into similarities and contrasts among ecosystem types. For example, regional effects (warm temperate versus cool temperate) were larger than differences across forest types (hardwood versus pine) for N-fixing bacterial communities. Despite these biome contrasts, roughly half of the sequences for nifH came from the same family (Geobacteraceae). Our approach also highlighted important interactions between C and N cycles in N-fixing bacterial communities. Furthermore, N deposition may have a strong interactive and suppressive effect on N fixation in higher CO2 scenarios. Future research should consider multiple interacting drivers and biome-specific factors to accurately predict N fixation under changing climate conditions.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to many people at the four field research sites for access to the field sites and assistance with soil sample collection.
This study was supported by the U.S. Department of Energy, Biological and Environmental Research Division, through a Science Focus Area grant to C.R.K. (2009LANLF260). Sanger sequencing was conducted at Los Alamos National Laboratory by the U.S. DOE Joint Genome Institute. The four field research sites in this study were supported by the U.S. Department of Energy Climate Program. R.B.J. acknowledges support from the Department of Energy (DE-FG02-95ER62083) and the National Science Foundation (DEB-02-35425). S.T.B. was supported by Agriculture and Food Research Initiative competitive grant no. 2012-67012-19816 from the USDA National Institute of Food and Agriculture.
Footnotes
Published ahead of print 7 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04034-13.
REFERENCES
- 1.Reed SC, Cleveland CC, Townsend AR. 2011. Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annu. Rev. Ecol. Evol. Syst. 42:489–512. 10.1146/annurev-ecolsys-102710-145034 [DOI] [Google Scholar]
- 2.Dunbar J, Eichorst SA, Gallegos-Graves LV, Silva S, Xie G, Hengartner NW, Evans RD, Hungate BA, Jackson RB, Megonigal JP, Schadt CW, Vilgalys R, Zak DR, Kuske CR. 2012. Common bacterial responses in six ecosystems exposed to 10 years of elevated atmospheric carbon dioxide. Environ. Microbiol. 14:1145–1158. 10.1111/j.1462-2920.2011.02695.x [DOI] [PubMed] [Google Scholar]
- 3.Finzi AC, Moore DJP, DeLucia EH, Lichter J, Hofmockel KS, Jackson RB, Kim HS, Matamala R, McCarthy HR, Oren R, Pippen JS, Schlesinger WH. 2006. Progressive nitrogen limitation of ecosystem processes under elevated CO2 in a warm-temperate forest. Ecology 87:15–25. 10.1890/04-1748 [DOI] [PubMed] [Google Scholar]
- 4.Kelley AM, Fay PA, Polley HW, Gill RA, Jackson RB. 2011. Atmospheric CO2 and soil extracellular enzyme activity: a meta-analysis and CO2 gradient experiment. Ecosphere 2:art96. 10.1890/ES11-00117.1 [DOI] [Google Scholar]
- 5.Hofmockel KS, Gallet-Budynek A, McCarthy HR, Currie WS, Jackson RB, Finzi AC. 2011. Sources of increased N uptake in forest trees growing under elevated CO2: results of a large-scale 15N study. Glob. Change Biol. 17:3338–3350. 10.1111/j.1365-2486.2011.02465.x [DOI] [Google Scholar]
- 6.McCarthy HR, Oren R, Johnsen KH, Gallet-Budynek A, Pritchard SG, Cook CW, LaDeau SL, Jackson RB, Finzi AC. 2010. Re-assessment of plant carbon dynamics at the Duke free-air CO2 enrichment site: interactions of atmospheric CO2 with nitrogen and water availability over stand development. New Phytol. 185:514–528. 10.1111/j.1469-8137.2009.03078.x [DOI] [PubMed] [Google Scholar]
- 7.Luo Y, Su B, Currie WS, Dukes JS, Finzi A, Hartwig U, Hungate B, McMurtrie RE, Oren R, Parton WJ, Pataki DE, Shaw MR, Zak DR, Field CB. 2004. Progressive nitrogen limitation of ecosystem responses to rising atmospheric carbon dioxide. Bioscience 54:731–739. 10.1641/0006-3568(2004)054[0731:PNLOER]2.0.CO;2 [DOI] [Google Scholar]
- 8.Hsu S-F, Buckley DH. 2009. Evidence for the functional significance of diazotroph community structure in soil. ISME J. 3:124–136. 10.1038/ismej.2008.82 [DOI] [PubMed] [Google Scholar]
- 9.Reed SC, Townsend AR, Cleveland CC, Nemergut DR. 2010. Microbial community shifts influence patterns in tropical forest nitrogen fixation. Oecologia 164:521–531. 10.1007/s00442-010-1649-6 [DOI] [PubMed] [Google Scholar]
- 10.Weber CF, Zak DR, Hungate BA, Jackson RB, Vilgalys R, Evans RD, Schadt CW, Megonigal JP, Kuske CR. 2011. Responses of soil cellulolytic fungal communities to elevated atmospheric CO2 are complex and variable across five ecosystems. Environ. Microbiol. 13:2778–2793. 10.1111/j.1462-2920.2011.02548.x [DOI] [PubMed] [Google Scholar]
- 11.Jordan TE, Correll DL. 1991. Continuous automated sampling of tidal exchanges of nutrients by brackish marshes. Estuar. Coast. Shelf Sci. 32:527–545. 10.1016/0272-7714(91)90073-K [DOI] [Google Scholar]
- 12.Marsh AS, Rasse DP, Drake BG, Megonigal JP. 2005. Effect of elevated CO2 on carbon pools and fluxes in a brackish marsh. Estuaries 28:694–704. 10.1007/BF02732908 [DOI] [Google Scholar]
- 13.Curtis PS, Drake BG, Leadley PW, Arp WJ, Whigham DF. 1989. Growth and senescence in plant communities exposed to elevated CO2 concentrations on an estuarine marsh. Oecologia 78:20–26. 10.1007/BF00377193 [DOI] [PubMed] [Google Scholar]
- 14.Ueda T, Suga Y, Yahiro N, Matsuguchi T. 1995. Phylogeny of Sym plasmids of rhizobia by PCR-based sequencing of a nodC segment. J. Bacteriol. 177:468–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zani S, Mellon MT, Collier JL, Zehr JP. 2000. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl. Environ. Microbiol. 66:3119–3124. 10.1128/AEM.66.7.3119-3124.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 18.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Horn DJV, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soetaert K, Heip C. 1990. Sample-size dependence of diversity indices and the determination of sufficient sample size in a high-diversity deep-sea environment. Mar. Ecol. Prog. Ser. 59:305–307. 10.3354/meps059305 [DOI] [Google Scholar]
- 20.Gaby JC, Buckley DH. 2012. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7:e42149. 10.1371/journal.pone.0042149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poly F, Monrozier LJ, Bally R. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152:95–103. 10.1016/S0923-2508(00)01172-4 [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Peng M, Li Y. 2012. Phylogenetic diversity of nitrogen-fixing bacteria and the nifH gene from mangrove rhizosphere soil. Can. J. Microbiol. 58:531–539. 10.1139/w2012-016 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Li D, Wang H, Xiao Q, Liu X. 2006. Molecular diversity of nitrogen-fixing bacteria from the Tibetan Plateau, China. FEMS Microbiol. Lett. 260:134–142. 10.1111/j.1574-6968.2006.00317.x [DOI] [PubMed] [Google Scholar]
- 24.Niederberger TD, Sohm JA, Tirindelli J, Gunderson T, Capone DG, Carpenter EJ, Cary SC. 2012. Diverse and highly active diazotrophic assemblages inhabit ephemerally wetted soils of the Antarctic Dry Valleys. FEMS Microbiol. Ecol. 82:376–390. 10.1111/j.1574-6941.2012.01390.x [DOI] [PubMed] [Google Scholar]
- 25.Yeager CM, Northup DE, Grow CC, Barns SM, Kuske CR. 2005. Changes in nitrogen-fixing and ammonia-oxidizing bacterial communities in soil of a mixed conifer forest after wildfire. Appl. Environ. Microbiol. 71:2713–2722. 10.1128/AEM.71.5.2713-2722.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehr JP, Jenkins BD, Short SM, Steward GF. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539–554. 10.1046/j.1462-2920.2003.00451.x [DOI] [PubMed] [Google Scholar]
- 27.Dos Santos P, Fang Z, Mason S, Setubal J, Dixon R. 2012. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. 10.1186/1471-2164-13-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raymond J, Siefert J, Staples C, Blankenship R. 2004. The natural history of nitrogen fixation. Mol. Biol. Evol. 21:541–554. 10.1093/molbev/msh047 [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Quensen JF, Fish JA, Lee TK, Sun Y, Tiedje JM, Cole JR. 2013. Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio 4:e00592-13. 10.1128/mBio.00592-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaby JC, Buckley DH. 2011. A global census of nitrogenase diversity. Environ. Microbiol. 13:1790–1799. 10.1111/j.1462-2920.2011.02488.x [DOI] [PubMed] [Google Scholar]
- 31.Kahindi JHP, Woomer P, George T, de Souza Moreira FM, Karanja NK, Giller KE. 1997. Agricultural intensification, soil biodiversity and ecosystem function in the tropics: the role of nitrogen-fixing bacteria. Appl. Soil Ecol. 6:55–76. 10.1016/S0929-1393(96)00151-5 [DOI] [Google Scholar]
- 32.Okubo T, Tsukui T, Maita H, Okamoto S, Oshima K, Fujisawa T, Saito A, Futamata H, Hattori R, Shimomura Y, Haruta S, Morimoto S, Wang Y, Sakai Y, Hattori M, Aizawa S, Nagashima KV, Masuda S, Hattori T, Yamashita A, Bao Z, Hayatsu M, Kajiya-Kanegae H, Yoshinaga I, Sakamoto K, Toyota K, Nakao M, Kohara M, Anda M, Niwa R, Jung-Hwan P, Sameshima-Saito R, Tokuda S, Yamamoto S, Yokoyama T, Akutsu T, Nakamura Y, Nakahira-Yanaka Y, Takada Hoshino Y, Hirakawa H, Mitsui H, Terasawa K, Itakura M, Sato S, Ikeda-Ohtsubo W, Sakakura N, Kaminuma E, Minamisawa K. 2012. Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microbes Environ. 27:306–315. 10.1264/jsme2.ME11321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bürgmann H, Widmer F, Von Sigler W, Zeyer J. 2004. New molecular screening tools for analysis of free-living diazotrophs in soil. Appl. Environ. Microbiol. 70:240–247. 10.1128/AEM.70.1.240-247.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamelin J, Fromin N, Tarnawski S, Teyssier-Cuvelle S, Aragno M. 2002. nifH gene diversity in the bacterial community associated with the rhizosphere of Molinia coerulea [sic], an oligonitrophilic perennial grass. Environ. Microbiol. 4:477–481. 10.1046/j.1462-2920.2002.00319.x [DOI] [PubMed] [Google Scholar]
- 35.Duc L, Noll M, Meier B, Bürgmann H, Zeyer J. 2009. High diversity of diazotrophs in the forefield of a receding alpine glacier. Microb. Ecol. 57:179–190. 10.1007/s00248-008-9408-5 [DOI] [PubMed] [Google Scholar]
- 36.Butler J, Young N, Lovley D. 2010. Evolution of electron transfer out of the cell: comparative genomics of six Geobacter genomes. BMC Genomics 11:40. 10.1186/1471-2164-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes DE, Nevin KP, Lovley DR. 2004. Comparison of 16S rRNA, nifD, recA, gyrB, rpoB and fusA genes within the family Geobacteraceae fam. nov. Int. J. Syst. Bacteriol. 54:1591–1599. 10.1099/ijs.0.02958-0 [DOI] [PubMed] [Google Scholar]
- 38.Bertics VJ, Sohm JA, Treude T, Chow C-ET, Capone DC, Fuhrman JA, Ziebis W. 2010. Burrowing deeper into benthic nitrogen cycling: the impact of bioturbation on nitrogen fixation coupled to sulfate reduction. Mar. Ecol. Prog. Ser. 409:1–15. 10.3354/meps08639 [DOI] [Google Scholar]
- 39.Grayston SJ, Prescott CE. 2005. Microbial communities in forest floors under four tree species in coastal British Columbia. Soil Biol. Biochem. 37:1157–1167. 10.1016/j.soilbio.2004.11.014 [DOI] [Google Scholar]
- 40.Marschner P, Crowley D, Yang CH. 2004. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 261:199–208. 10.1023/B:PLSO.0000035569.80747.c5 [DOI] [Google Scholar]
- 41.Marschner P, Yang CH, Lieberei R, Crowley DE. 2001. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33:1437–1445. 10.1016/S0038-0717(01)00052-9 [DOI] [Google Scholar]
- 42.Berg G, Smalla K. 2009. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68:1–13. 10.1111/j.1574-6941.2009.00654.x [DOI] [PubMed] [Google Scholar]
- 43.Hofmockel KS, Schlesinger WH. 2007. Carbon dioxide effects on heterotrophic dinitrogen fixation in a temperate pine forest. Soil Sci. Soc. Am. J. 71:140–144. 10.2136/sssaj2006.110 [DOI] [Google Scholar]
- 44.van Groenigen K-J, Six J, Hungate BA, de Graaff M-A, van Breemen N, van Kessel C. 2006. Element interactions limit soil carbon storage. Proc. Natl. Acad. Sci. U. S. A. 103:6571–6574. 10.1073/pnas.0509038103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lüscher A, Hartwig UA, Suter D, Nösberger J. 2000. Direct evidence that symbiotic N2 fixation in fertile grassland is an important trait for a strong response of plants to elevated atmospheric CO2. Glob. Change Biol. 6:655–662. 10.1046/j.1365-2486.2000.00345.x [DOI] [Google Scholar]
- 46.Zanetti S, Hartwig UA, Luscher A, Hebeisen T, Frehner M, Fischer BU, Hendrey GR, Blum H, Nosberger J. 1996. Stimulation of symbiotic N2 fixation in Trifolium repens L. under elevated atmospheric pCO2 in a grassland ecosystem. Plant Physiol. 112:575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson RB, Cook CW, Pippen JS, Palmer SM. 2009. Increased belowground biomass and soil CO2 fluxes after a decade of carbon dioxide enrichment in a warm-temperate forest. Ecology 90:3352–3366. 10.1890/08-1609.1 [DOI] [PubMed] [Google Scholar]
- 48.Phillips RP, Finzi AC, Bernhardt ES. 2011. Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long-term CO2 fumigation. Ecol. Lett. 14:187–194. 10.1111/j.1461-0248.2010.01570.x [DOI] [PubMed] [Google Scholar]
- 49.Dommergues Y, Balandreau J, Rinaudo G, Weinhard P. 1973. Non-symbiotic nitrogen fixation in the rhizospheres of rice, maize and different tropical grasses. Soil Biol. Biochem. 5:83–89. 10.1016/0038-0717(73)90094-1 [DOI] [Google Scholar]
- 50.Jones DL, Farrar J, Giller KE. 2003. Associative nitrogen fixation and root exudation—what is theoretically possible in the rhizosphere? Symbiosis 35:19–38 [Google Scholar]
- 51.Dang H, Luan X, Zhao J, Li J. 2009. Diverse and novel nifH and nifH-like gene sequences in the deep-sea methane seep sediments of the Okhotsk Sea. Appl. Environ. Microbiol. 75:2238–2245. 10.1128/AEM.02556-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovell CR, Friez MJ, Longshore JW, Bagwell CE. 2001. Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67:5308–5314. 10.1128/AEM.67.11.5308-5314.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. 1997. Structure of ADP·AIF4−-stabilized nitrogenase complex and its implications for signal transduction. Nature 387:370–376. 10.1038/387370a0 [DOI] [PubMed] [Google Scholar]
- 54.Turk AK, Rees AP, Zehr JP, Pereira N, Swift P, Shelley R, Lohan M, Woodward EMS, Gilbert J. 2011. Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic. ISME J. 5:1201–1212. 10.1038/ismej.2010.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.