Abstract
In recent years, a greater appreciation for the microbes inhabiting human body sites has emerged. In the female mammary gland, milk has been shown to contain bacterial species, ostensibly reaching the ducts from the skin. We decided to investigate whether there is a microbiome within the mammary tissue. Using 16S rRNA sequencing and culture, we analyzed breast tissue from 81 women with and without cancer in Canada and Ireland. A diverse population of bacteria was detected within tissue collected from sites all around the breast in women aged 18 to 90, not all of whom had a history of lactation. The principal phylum was Proteobacteria. The most abundant taxa in the Canadian samples were Bacillus (11.4%), Acinetobacter (10.0%), Enterobacteriaceae (8.3%), Pseudomonas (6.5%), Staphylococcus (6.5%), Propionibacterium (5.8%), Comamonadaceae (5.7%), Gammaproteobacteria (5.0%), and Prevotella (5.0%). In the Irish samples the most abundant taxa were Enterobacteriaceae (30.8%), Staphylococcus (12.7%), Listeria welshimeri (12.1%), Propionibacterium (10.1%), and Pseudomonas (5.3%). None of the subjects had signs or symptoms of infection, but the presence of viable bacteria was confirmed in some samples by culture. The extent to which these organisms play a role in health or disease remains to be determined.
INTRODUCTION
The human body is home to a large and diverse population of bacteria with properties that are both harmful and beneficial to health (1–6), and for this reason there has been a strong push in recent years to fully characterize the bacteria associated with different parts of the body under different health conditions. These studies have been made possible with the use of deep-sequencing technologies, and sites once thought of as sterile, such as the stomach, bladder, and lungs, have now been shown to harbor an indigenous microbiota (7–9). We hypothesized that microbes may also be present in breast tissue given the known presence of bacteria in human milk (10). This is not surprising considering that skin and oral bacteria have access to the mammary ducts through the nipple (11), with some recent studies suggesting their source to be from the mother's gastrointestinal tract (12). We rationalized that given the nutrient-rich fatty composition of the female breast, the widespread vasculature and lymphatics, and the diffuse location of the lobules and ducts leading from the nipple, bacteria would be widespread within the mammary glands, irrespective of lactation. Thus, the objective of the study was to determine, using culture and nonculture methods, whether breast tissue contains a microbiome. To ensure that the results obtained were not an artifact of a single demographic, tissue was collected from two distant countries, Canada and Ireland.
MATERIALS AND METHODS
Clinical samples and study design (Canadian samples).
Ethical approval for this study was obtained from Western Research Ethics Board and Lawson Health Research Institute, London, Ontario, Canada. Patients provided written consent for sample collection and subsequent analyses.
(i) Tissue collection and processing.
Breast tissue was collected from 43 women (aged 18 to 90) undergoing breast surgery at St. Joseph's Hospital in London, Ontario, Canada. Thirty-eight of those women underwent lumpectomies or mastectomies for either benign (n = 11) or cancerous (n = 27) tumors, and 5 women underwent breast reductions and had no history of breast cancer. For those women with tumors, the tissue obtained for analysis was collected outside the marginal zone, approximately 5 cm away from the tumor. After excision, the fresh tissue was immediately placed in a sterile vial on ice and homogenized within 30 min of collection. As an environmental control, a tube filled with sterile phosphate-buffered saline (PBS) was left open for the duration of the surgical procedure and then processed in parallel with the tissue samples.
Tissue samples were homogenized in sterile PBS using a PolyTron 2100 homogenizer at 28,000 rpm. The amount of PBS added was based on the weight of the tissue in order to obtain a final concentration of 0.4 g/ml. Fresh homogenate and the environmental controls were then plated on different agar plates for culture analysis and the remaining amount aliquoted and stored at −80°C until DNA extraction.
(ii) DNA isolation.
After tissue homogenates were thawed on ice, 400 μl (equivalent to 160 mg of tissue) was added to tubes containing 1.2 ml of ASL buffer (QIAamp DNA stool kit; Qiagen) and 400 mg of 0.1-mm-diameter zirconium-glass beads (BioSpec Products). Mechanical and chemical lyses were performed by bead beating at 4,800 rpm for 60 s at room temperature and then 60 s on ice (repeated twice) (Mini-Beadbeater-1; BioSpec Products), after which the suspension was incubated at 95°C for 5 min. Subsequent procedures were performed using the Qiagen QIAamp DNA stool kit according to the manufacturer's protocol, with the exception of the last step, in which the column was eluted with 120 μl of elution buffer. DNA was stored at −20°C until further use.
Clinical samples and study design (Irish samples).
Ethical approval for this study was obtained from the University College Cork Clinical Research Committee. Patients provided written consent for sample collection and subsequent analyses.
(i) Tissue collection.
Breast tissue was collected from 38 women (aged 20 to 85) undergoing breast surgery at Cork University Hospital or South Infirmary Victoria Hospital, Cork, Ireland. Thirty-three women underwent lumpectomies or mastectomies for cancerous tumors (taken at least 5 cm away from the primary tumor site), while 5 women underwent breast reductions and had no history of breast cancer. Once collected, the specimens were placed in sterile cryotubes, flash frozen in liquid nitrogen within 45 min of collection, and then stored at −80°C until DNA extraction.
(ii) DNA isolation.
Tissue samples were thawed on ice and the genomic DNA extracted using the Gene-Elute mammalian genomic DNA miniprep kit (Sigma-Aldrich) as per the manufacturer's protocol with the exception of the elution step, where the column was eluted with 70 μl of elution buffer.
Ion Torrent V6 16S rRNA sequencing. (i) PCR amplification.
At the London, Canada, lab, the genomic DNA isolated from Canadian and Irish clinical samples was amplified using the barcoded primers V6-LT and V6-RT (see Table S1 in the supplemental material), which amplify the V6 hypervariable region of the 16S rRNA gene. The PCR was carried out in a 40-μl reaction mixture containing 5 μl of DNA template (or nuclease-free water as a negative control), 1.5 mM MgCl2, 0.8 μM each primer, 4 μl of 10× PCR buffer (Invitrogen), 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.05U Taq polymerase (Invitrogen), and 0.15 μg/μl of bovine serum albumin. Thermal cycling was carried out in an Eppendorf Mastercyler under the following conditions: initial denaturation at 95°C for 2 min followed by 25 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. After amplification, the DNA concentration was measured with the Qubit 2.0 fluorometer (Invitrogen) using the broad-range assay. Equimolar amounts of each PCR product were then pooled and purified using the QIAquick PCR purification kit (Qiagen). The PCR-purified sample was then sent to the London Regional Genomics Center, London, Ontario, Canada, for V6 16S rRNA sequencing using the Ion Torrent platform as per the Center's standard operating procedure.
(ii) Sequence processing and taxonomic assignment.
Custom Perl and Bash scripts were used to demultiplex the reads and assign barcoded reads to individual samples. Reads were kept if the sequence included a perfect match to the barcode and the V6 16S rRNA gene primers and were within the length expected for the V6 variable region. Samples with more than 600 reads were kept, while those with fewer than 600 were discarded. Reads were then clustered by 97% identity into operational taxonomic units (OTUs) using UClust v. 3.0.617 (13). OTUs that represented ≥2% of the reads in at least one sample were kept, while those that did not meet the cutoff were discarded to account for the high error rate intrinsic to Ion Torrent sequencing. Taxonomic assignments for each OTU were made by the Ribosomal Database Project (RDP) SeqMatch tool (14). From the top 20 matches to the RDP named-isolate database, the full taxonomy was retained for matches with the highest S_ab score. For multiple top matches with equal scores, the highest common taxonomy was retained (e.g., genus level if multiple species matched equally well). Since the maximum number of matches displayed per sequence is 20, the RDP taxonomic assignments were verified by BLAST against the Greengenes named-isolate database with an output of 100 hits (15). Taxonomy was assigned based on hits with the highest percent identities and coverage. If multiple hits fulfilled this criterion, classification was reassigned to a higher common taxonomy. In instances where the highest percent identity/coverage yielded a single match, if this was less than 90% and the S_ab score from RDP was less than 0.7, taxonomy was assigned at the family level instead of at the genus level. The sequences for each OTU and the RDP/Greengenes classification are shown in Data Set S1 in the supplemental material. The Ion Torrent V6 16S rRNA sequences obtained in this study have been deposited in the Short Read Archive (SRA) at NCBI.
(iii) Data analysis.
The QIIME pipeline (16) was used to (i) calculate weighted UniFrac distances and Shannon's diversity index (logarithms with base 2), (ii) summarize OTUs by different taxonomic levels, and (iii) generate unweighted-pair group method with arithmetic mean (UPGMA) trees representing hierarchical clustering of samples. The UniFrac distances were calculated by using a phylogenetic tree of OTU sequences built with FastTree (17) and based on an OTU sequence alignment with MUSCLE (18). Weighted UniFrac compares microbial profiles (presence/absence and abundance) between samples (i.e., beta diversity) (19), while Shannon's diversity index evaluates the microbial diversity within a sample (i.e., alpha diversity). The higher the Shannon's diversity index, the more diverse a sample is, and a value of zero indicates the presence of only one species (20). UPGMA trees allow one to visualize the distance matrix, and the robustness of what was observed was tested with jackknifing and bootstrapping. For beta-diversity analyses, the data set was rarified to the lowest read count/sample. Bar plots, box plots, and strip charts were all generated in R (21).
Culture analysis.
In an effort to prove that bacteria in the breast were viable, 100 μl of Canadian tissue homogenate from each of the 43 samples and 100 μl of each environmental control were plated on Columbia blood agar (CBA) plates and incubated aerobically or anaerobically at 37°C. Of note, the facilities to perform culture were not available at the Irish site. DNA from single colonies was extracted using the InstaGene Matrix (Bio-Rad) and then amplified using the eubacterial primers pA/pH, which amplify the 16S rRNA gene (see Table S1 in the supplemental material). The PCR was carried out in a 50-μl reaction mixture containing 10 μl of DNA template (or nuclease-free water as a negative control), 1.5 mM MgCl2, 1.0 μM each primer, 0.2 mM dNTP, 5 μl 10× PCR buffer (Invitrogen), and 0.05 Taq polymerase (Invitrogen). Thermal cycling was carried out in an Eppendorf Mastercycler under the following conditions: initial denaturation step at 95°C for 2 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. A final elongation step was performed at 72°C for 10 min. After running 10 μl of the PCR mixture on a 1% agarose gel to verify the presence of amplicons, 40 μl of the PCR mixture was then purified using the QIAquick PCR purification kit (Qiagen). The purified PCR products were then sent for Sanger sequencing to the London Regional Genomics Centre, London, Ontario, Canada. Sequences were analyzed using the GenBank 16S rRNA sequence database with the BLAST algorithm (22). Taxonomy was assigned based on the highest Max score.
Statistical analysis.
Statistical analysis was performed in R using the Kruskal-Wallis one-way analysis of variance followed by the Mann-Whitney U test with Bonferroni correction.
RESULTS
Tissue was obtained from various locations within the breast, from close to the nipple to as far back as the chest wall (Fig. 1). Regardless of the location sampled within the breast, presence/absence of breast malignancy, country of origin, age, history of pregnancy, and method of DNA preparation, a variety of bacteria were detected in breast tissue. (Fig. 2 and 3; see Data Set S2 in the supplemental material). Bacterial diversity within samples varied between individuals, with Shannon's diversity indices ranging from 0.8 to 5.2 with an average value of 3.6 (see Fig. S1 in the supplemental material). To put this into perspective, using the same index, oral and gut samples, which are known for their diverse bacterial communities, have values between 3.9 and 6.5 (23–26), while vaginal samples, of low bacterial diversity, generate values between 0.46 and 2.9 (27–29).
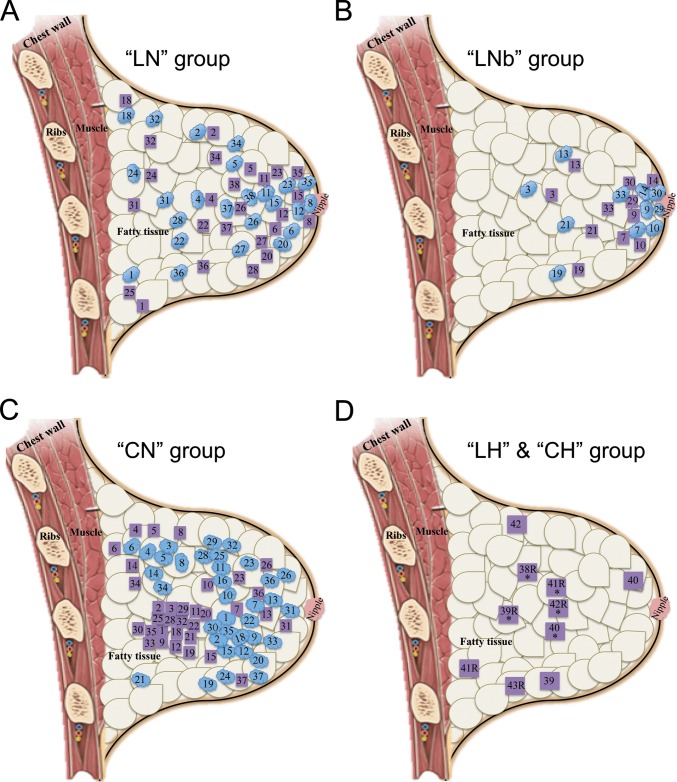
FIG 1.
Locations within the breast of tissue samples collected for bacterial analysis. Tissue samples were collected from 43 women from London, Canada, and 38 women from Cork, Ireland, undergoing different breast surgeries. (A and B) Locations of tissue collected from women in Canada undergoing lumpectomies or mastectomies for either malignant (A) or benign (B) tumors. Subject 25 underwent a prophylactic mastectomy due to previous cancer in the other breast. (C) Locations of tissue collected from women in Ireland undergoing lumpectomies or mastectomies for malignant tumors. Blue ovals represent the location of the tumor, and purple squares represent the location of the specimen obtained for bacterial analysis. The distances between the blue ovals and purple squares are approximate estimates of the distance between the tumor and the specimen, which was at least 5 cm away from the tumor. (D) Locations of tissue collected from women in both Canada and Ireland undergoing breast reduction surgery. Asterisks in the purple boxes underneath the subject number indicate samples from Ireland. All samples were a minimum of 1 cm deep into the skin, with the surgeons aiming for mid-deep rather than superficial locations. As shown in the diagram, specimens were obtained from a variety of locations within the breast.
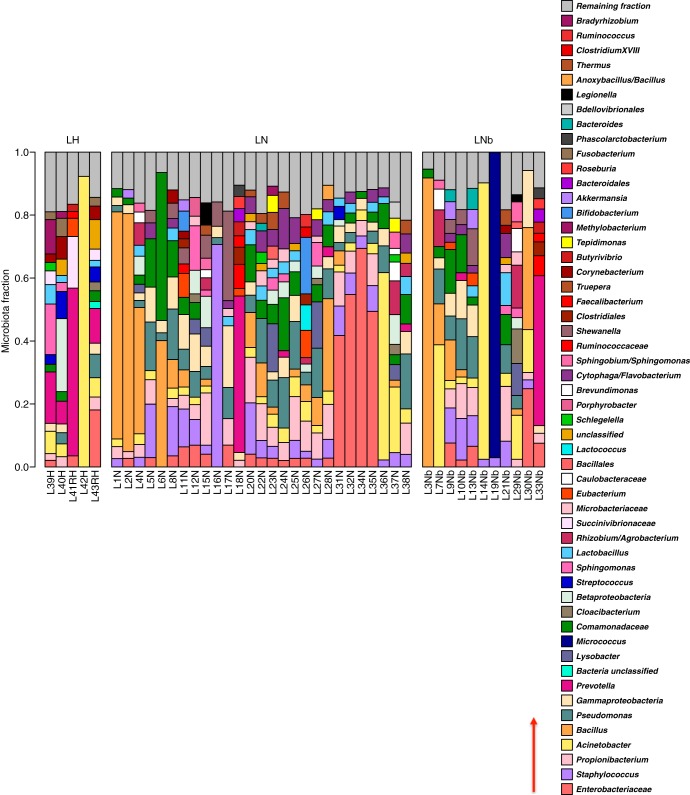
FIG 2.
Breast tissue microbiota in 43 Canadian women, identified by 16S rRNA sequencing. The relative abundances of bacterial taxa identified in different tissue samples were visualized by bar plots. Each bar represents a subject and each colored box a bacterial taxon. The height of a colored box represents the relative abundance of that organism within the sample. For example, sample L42H is dominated by Acinetobacter, whereas L19Nb is dominated by Micrococcus. Taxa present in less than 2% abundance in a given sample are displayed in the “Remaining fraction” at the top of the graph (gray boxes). As shown by the bar plots, a variety of bacteria were detected in breast tissue. The legend is read from bottom to top, as indicated by the red arrow, with the bottom organism on the legend corresponding to the bottom colored box on the bar plot. LH, tissue collected from women undergoing breast reductions; LN, nonmalignant tissue collected adjacent to cancerous tumors; LNb, nonmalignant tissue collected adjacent to benign tumors.
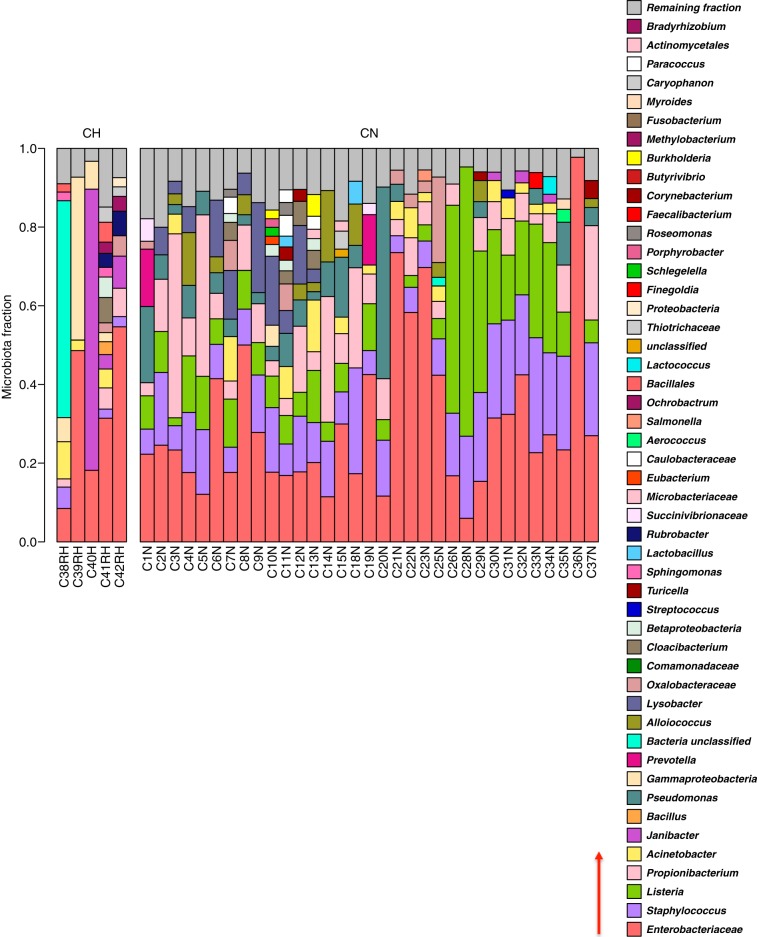
FIG 3.
Breast tissue microbiota in 38 Irish women identified, by 16S rRNA sequencing. The relative abundances of bacterial taxa identified in different tissue samples were visualized by bar plots. Each bar represents a subject and each colored box a bacterial taxon. The height of a colored box represents the relative abundance of that organism within the sample. For example, sample C40H is dominated by Janibacter, whereas C36N is dominated by Enterobacteraceae. Taxa present in less than 2% abundance in a given sample are displayed in the “Remaining fraction” at the top of the graph (gray boxes). As shown by the bar plots, a variety of bacteria were detected in breast tissue. The legend is read from bottom to top, as indicated by the red arrow, with the bottom organism on the legend corresponding to the bottom colored box on the bar plot. CH, tissue collected from women undergoing breast reductions; CN, nonmalignant tissue collected adjacent to cancerous tumors.
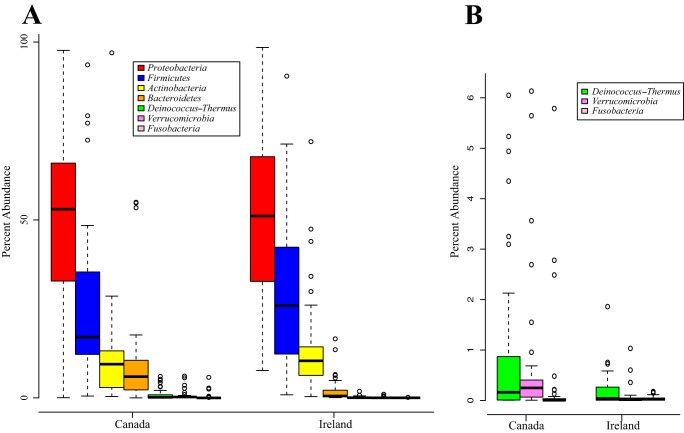
The bacteria identified in tissue were grouped into 121 operational taxonomic units (OTUs) based on 97% sequence similarity (see Data Set S3 in the supplemental material). These OTUs belonged to 7 different phyla, Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, Deinococcus-Thermus, Verrucomicrobia, and Fusobacteria, with Proteobacteria being the most abundant, followed by Firmicutes (specifically from the class Bacilli) (Fig. 4). Of the 121 OTUs identified, 57 could be classified at the genus level and 25 at the species level (see Data Set S3 in the supplemental material). The most abundant taxa in the Canadian samples were Bacillus (11.4%), Acinetobacter (10.0%), unclassified Enterobacteriaceae (8.3%), Pseudomonas (6.5%), Staphylococcus (6.5%), Propionibacterium (5.8%), unclassified Comamonadaceae (5.7%), unclassified Gammaproteobacteria (5.0%), and Prevotella (5.0%). In the Irish samples the most abundant taxa were unclassified Enterobacteriaceae (30.8%), Staphylococcus (12.7%), Listeria welshimeri (12.1%), Propionibacterium (10.1%), and Pseudomonas (5.3%).
FIG 4.
Percent abundances of different bacterial phyla in breast tissue identified by 16S rRNA sequencing. (A) Box plots of the seven different phyla identified in breast tissue from the Canadian and Irish samples. The box signifies the 75% (upper) and 25% (lower) quartiles and thus shows where 50% of the samples lie. The black line inside the box represents the median. The whiskers represent the lowest datum still within 1.5 interquartile range (IQR) of the lower quartile and the highest datum still within 1.5 IQR of the upper quartile. Outliers are shown with open circles. (B) The least-abundant phyla shown in panel A were plotted on another graph with a smaller scale to allow for better visualization of percentage. In samples from both countries Proteobacteria was the most abundant phylum, followed by Firmicutes (Kruskal-Wallis test/Mann-Whitney U test with Bonferroni correction, P ≤ 0.001).
Of the 18 environmental samples that passed quality control, 13 of them had lower read counts than their respective tissue samples. Of those with higher reads, weighted UniFrac UPGMA hierarchical clustering showed that microbial profiles differed between the control and its respective tissue sample (see Fig. S2 in the supplemental material). The 7 lowest-read counts overall belonged to the environmental controls (<600 reads).
Bacteria were able to be cultured from all 43 of the Canadian samples (culture analysis was not performed on the Irish samples), with amounts ranging from 75 to 2,000 CFU/gram of tissue, depending on the sample. Collectively, eight different strains were identified: Bacillus sp., Micrococcus luteus, Propionibacterium acnes, Propionibacterium granulosum, Staphylococcus sp., Staphylococcus saprophyticus, Streptococcus oralis, and Streptococcus agalactiae. No bacteria were isolated from the environmental controls.
DISCUSSION
We first suggested the potential for a breast microbiome in 2012 (10), and this has now been confirmed in the present study, and recently by Xuan et al. (30), who examined breast tumor and normal adjacent tissue also by using next-generation sequencing techniques. In our study, none of the 81 women recruited had any clinical signs or symptoms of breast infection, yet bacteria were still detected in all Canadian samples, which were analyzed by both culture and molecular techniques, and Irish samples, which were analyzed just by molecular techniques. While only 8 different strains were able to be cultured, we only used Columbia blood agar for isolation, and some fastidious bacteria, such as Prevotella and Bacteroides, require more nutrient-rich conditions for growth. Regardless, all those bacteria that were cultured were also detected by 16S rRNA sequencing, supporting the argument that the DNA amplimers were from viable bacteria and not solely from remnant bacterial DNA.
Proteobacteria was the most abundant phylum in breast tissue, unlike in the vagina, oral cavity, bladder, skin, and gastrointestinal tract, where members of this phylum make up only a small proportion of the overall bacterial community (5, 8, 31–34). This finding is similar to what was observed by Xuan et al. (30) and suggests that breast tissue may have a unique microbiota, distinct from that found at other body sites, which is consistent with published studies showing that bacterial communities vary across body habitats (5, 35). The higher abundance of Proteobacteria and Firmicutes (specifically the class Bacilli) compared with other taxonomic groups may be a result of host microbial adaptation to the fatty acid environment in the tissue. A recent study documented a positive correlation between Proteobacteria and Bacilli and the metabolic by-products of fatty acid metabolism, as well as host-derived genes involved in fatty acid biosynthesis (36). It has also been documented that wild-type mice fed a high-fat diet have a gut microbiota enriched in Firmicutes (37). Of interest, Proteobacteria is also the principal phylum in human milk (38), with many of the same bacteria that we detected in tissue, raising the possibility that the tissue microbiota could be a source of bacterial inocula for babies.
Some of the organisms detected in breast tissue have been found at other body sites, such as Lactobacillus iners and Prevotella (vagina), Enterobacteriaceae (gastrointestinal tract), Fusobacterium and Streptococcus (oral cavity), Propionibacterium and Micrococcus (skin), and Pseudomonas (respiratory tract). Species known for their health-conferring properties, such as Lactobacillus and Bifidobacterium, were detected, as well as taxa known for pathogenesis at other sites, such as Enterobacteriaceae, Pseudomonas, and Streptococcus agalactiae. We can only speculate why potentially pathogenic bacteria able to metabolize fat that is present in abundance in the breast do not multiply and induce infections. One concept that has been proposed is that it is due to a host microbial sensory system aligned with antibacterial gene expression (30).
Of interest, in the study by Xuan et al. (30), differential abundance was observed with the genera Sphingomonas and Methylbacterium between paired normal and tumor tissue, with the authors suggesting a potential role in tumor development. While we also detected these two genera in our tissue samples, further studies, with a larger sample size of healthy women, would be needed to determine whether differential abundance between healthy and normal adjacent tissue also exists. While a comparison between tissue from cancer patients and healthy women was not the focus of this study, we did notice a higher abundance of Escherichia coli in women with cancer than in healthy controls, with this species known for its cancer-promoting activity (39). However, it is premature to suggest a cause-and-effect relationship before more work is done in this area.
One limitation of the study was the inability to make direct comparisons between the two countries due to the different collection and DNA extraction techniques used. Thus, no claims can be made as to whether bacteria detected at one site differ significantly from those detected at the other.
This research shows that breast tissue is not sterile but contains a diverse community of bacteria, adding to the literature that body sites once believed to be sterile do indeed have an endogenous microbiome. It has been proposed that the breast microbiome contributes to maintenance of healthy breast tissue by stimulating resident immune cells (30), but the type of bacteria and their metabolic activity, such as the ability to degrade carcinogens, may also contribute. Further studies are warranted to determine how this breast microbiome is established, why no infections accompanied colonization, what impact these organisms have on the host, and whether external factors such as diet, antibiotics, and illness affect this bacterial community and the subsequent consequences for the woman.
Supplementary Material
ACKNOWLEDGMENTS
C.U was the recipient of a CIHR Strategic Training Program in Cancer Research and Technology Transfer (CaRTT) scholarship from CIHR and the Translational Breast Cancer Studentship from the London Regional Cancer Program. We acknowledge support relevant to this study from University College Cork School of Medicine TRAP and the Irish Health Research Board (HRA_POR 2012/99).
We thank Bill Bennett and Marian Manning (Pathology Department, CUH, Cork, Ireland) for facilitating sample collection and Robert Richards (St. Joseph's Hospital, London, Canada) and Sean O'Sullivan (SIVUH, Cork, Ireland) for collecting tissue samples from breast reduction patients. We are also grateful to David Carter (London Genomics Research Center, London, Canada) and Michelle Angelini and all the clinical and administrative staff at London Health Sciences Centre (London, Canada) and Cork University Hospital (Cork, Ireland) for their help and support. We are also indebted to all the women who participated in the study.
Footnotes
Published ahead of print 7 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00242-14.
REFERENCES
- 1.Hooper LV. 2004. Bacterial contributions to mammalian gut development. Trends Microbiol. 12:129–134. 10.1016/j.tim.2004.01.001 [DOI] [PubMed] [Google Scholar]
- 2.Macpherson AJ, Harris NL. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4:478–485. 10.1038/nri1373 [DOI] [PubMed] [Google Scholar]
- 3.Serino M, Luche E, Chabo C, Amar J, Burcelin R. 2009. Intestinal microflora and metabolic diseases. Diabetes Metab. 35:262–272. 10.1016/j.diabet.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 4.Backhed F. 2012. Host responses to the human microbiome. Nutr. Rev. 70(Suppl 1):S14–S17. 10.1111/j.1753-4887.2012.00496.x [DOI] [PubMed] [Google Scholar]
- 5.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Q, Gao R, Wu W, Qin H. 2013. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 34:1285–1300. 10.1007/s13277-013-0684-4 [DOI] [PubMed] [Google Scholar]
- 7.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U. S. A. 103:732–737. 10.1073/pnas.0506655103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, Fitzgerald M, Mueller ER, Schreckenberger P, Dong Q, Nelson DE, Brubaker L. 2012. Evidence of uncultivated bacteria in the adult female bladder. J. Clin. Microbiol. 50:1376–1383. 10.1128/JCM.05852-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck JM, Young VB, Huffnagle GB. 2012. The microbiome of the lung. Transl. Res. 160:258–266. 10.1016/j.trsl.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urbaniak C, Burton JP, Reid G. 2012. Breast, milk and microbes: a complex relationship that does not end with lactation. Womens Health (Lond. Engl.). 8:385–398. 10.2217/whe.12.23 [DOI] [PubMed] [Google Scholar]
- 11.Ramsay DT, Kent JC, Owens RA, Hartmann PE. 2004. Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 113:361–367. 10.1542/peds.113.2.361 [DOI] [PubMed] [Google Scholar]
- 12.Donnet-Hughes A, Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, Serrant P, Segura-Roggero I, Schiffrin EJ. 2010. Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc. Nutr. Soc. 69:407–415. 10.1017/S0029665110001898 [DOI] [PubMed] [Google Scholar]
- 13.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 14.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill TC, Walsh KA, Harris JA, Moffett BF. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1–11. 10.1111/j.1574-6941.2003.tb01040.x [DOI] [PubMed] [Google Scholar]
- 21.R Development Core Team. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 23.Li K, Bihan M, Yooseph S, Methe BA. 2012. Analyses of the microbial diversity across the human microbiome. PLoS One 7:e32118. 10.1371/journal.pone.0032118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA. 2010. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 4:962–974. 10.1038/ismej.2010.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4554–4561. 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4680–4687. 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim TK, Thomas SM, Ho M, Sharma S, Reich CI, Frank JA, Yeater KM, Biggs DR, Nakamura N, Stumpf R, Leigh SR, Tapping RI, Blanke SR, Slauch JM, Gaskins HR, Weisbaum JS, Olsen GJ, Hoyer LL, Wilson BA. 2009. Heterogeneity of vaginal microbial communities within individuals. J. Clin. Microbiol. 47:1181–1189. 10.1128/JCM.00854-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. 2008. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74:4898–4909. 10.1128/AEM.02884-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xuan C, Shamonki JM, Chung A, Dinome ML, Chung M, Sieling PA, Lee DJ. 2014. Microbial dysbiosis is associated with human breast cancer. PLoS One 9:e83744. 10.1371/journal.pone.0083744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hummelen R, Fernandes AD, Macklaim JM, Dickson RJ, Changalucha J, Gloor GB, Reid G. 2010. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One 5:e12078. 10.1371/journal.pone.0012078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017. 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wade WG. 2013. The oral microbiome in health and disease. Pharmacol. Res. 69:137–143. 10.1016/j.phrs.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 34.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Comparative Sequencing Program NISC. Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Aidy S, Derrien M, Merrifield CA, Levenez F, Dore J, Boekschoten MV, Dekker J, Holmes E, Zoetendal EG, van Baarlen P, Claus SP, Kleerebezem M. 2013. Gut bacteria-host metabolic interplay during conventionalisation of the mouse germfree colon. ISME J. 7:743–755. 10.1038/ismej.2012.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy EF, Cotter PD, Healy S, Marques TM, O'Sullivan O, Fouhy F, Clarke SF, O'Toole PW, Quigley EM, Stanton C, Ross PR, O'Doherty RM, Shanahan F. 2010. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 59:1635–1642. 10.1136/gut.2010.215665 [DOI] [PubMed] [Google Scholar]
- 38.Ward TL, Hosid S, Ioshikhes I, Altosaar I. 2013. Human milk metagenome: a functional capacity analysis. BMC Microbiol. 13:116. 10.1186/1471-2180-13-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis CP, Cohen MS, Hackett RL, Anderson MD, Warren MM. 1991. Urothelial hyperplasia and neoplasia. III. Detection of nitrosamine production with different bacterial genera in chronic urinary tract infections of rats. J. Urol. 145:875–880 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.