Abstract
Salmonella enterica rarely grows on healthy, undamaged plants, but its persistence is influenced by bacterial plant pathogens. The interactions between S. enterica, Xanthomonas perforans (a tomato bacterial spot pathogen), and tomato were characterized. We observed that virulent X. perforans, which establishes disease by suppressing pathogen-associated molecular pattern (PAMP)-triggered immunity that leads to effector-triggered susceptibility, created a conducive environment for persistence of S. enterica in the tomato phyllosphere, while activation of effector-triggered immunity by avirulent X. perforans resulted in a dramatic reduction in S. enterica populations. S. enterica populations persisted at ∼10 times higher levels in leaves coinoculated with virulent X. perforans than in those where S. enterica was applied alone. In contrast, S. enterica populations were ∼5 times smaller in leaves coinoculated with avirulent X. perforans than in leaves inoculated with S. enterica alone. Coinoculation with virulent X. perforans increased S. enterica aggregate formation; however, S. enterica was not found in mixed aggregates with X. perforans. Increased aggregate formation by S. enterica may serve as the mechanism of persistence on leaves cocolonized by virulent X. perforans. S. enterica association with stomata was altered by X. perforans; however, it did not result in appreciable populations of S. enterica in the apoplast even in the presence of large virulent X. perforans populations. Gene-for-gene resistance against X. perforans successively restricted S. enterica populations. Given the effect of this interaction, breeding for disease-resistant cultivars may be an effective strategy to limit both plant disease and S. enterica populations and, consequently, human illness.
INTRODUCTION
The occurrence of human pathogens, including Salmonella enterica, on raw produce is now responsible for the steady occurrence of food-borne bacterial illness in the United States (1). Nontyphoid salmonellosis outbreaks have occurred annually over the last decade due to the contamination of a variety of edible plants by various serovars of S. enterica. Outbreaks associated with S. enterica contamination of tomato consistently occur in agricultural areas where the pathogen is now considered endemic (2). S. enterica appears to establish reservoirs on plants and to use them as vectors to animals, a preferred host (3). What factors influence S. enterica persistence in plants is not well understood (4).
S. enterica demonstrates low epiphytic fitness on plants. The fitness of S. enterica on leaves is less than that of other common epiphytes, such as Pseudomonas syringae, Pseudomonas chlororaphis, and Pantoea agglomerans (5, 6). Numerous studies with undamaged plants have reported the lack of net S. enterica growth in the phyllosphere (5, 7–9). Given the poor fitness of S. enterica on plant surfaces, as measured by growth (9, 10), factors that increase S. enterica populations or cause increased persistence may play a significant role in maintaining populations of this human pathogen on agricultural plants, which could lead to human illness.
The colonization of plants by S. enterica has been shown to be influenced by bacterial plant pathogens (9, 11). S. enterica populations were higher on produce infected with bacterial soft rot than on produce that was mechanically damaged or affected by fungal soft rot (12). Brute force pathogens, such as those that cause soft rot, macerate tissue by overwhelming plant defenses once population levels reach a quorum (13); they can increase S. enterica survival on plants by liberating nutrients that the human pathogen cannot access on its own (4). Stealth plant pathogens (14), such as Xanthomonas spp. and Pseudomonas spp. that rely on a type 3 secretion system (T3SS) for pathogenicity and suppression of host defenses (13, 15) instead of plant degradation enzymes, have also been shown to increase S. enterica survival on tomato plants (9, 16). However, interactions between stealth phytobacterial and human bacterial pathogens remain to be well characterized.
Mechanisms that govern the interaction between stealth phytobacterial and human bacterial pathogens are poorly understood. Both pathogenicity and virulence factors of phytobacterial pathogens have been investigated as factors that affect human pathogen bacterial populations on leaves. The role of a functional phytopathogenic T3SS in S. enterica growth or survival on plants is complex. In leaves infiltrated with both S. enterica and P. syringae pv. tomato, a functional phytopathogenic T3SS was required for S. enterica growth (16). However, in leaves precolonized by P. syringae pv. syringae, S. enterica survival was independent of the phytopathogenic T3SS, even though the T3SS or effectors contributed to P. syringae fitness (10). Unlike previous work, we hypothesized that S. enterica survival in the phyllosphere is dependent on the pathogenicity of a cocolonizing stealth plant-pathogenic bacterium. Our approach was to examine the interaction of S. enterica and the plant pathogen X. perforans following inoculation of the two organisms onto the leaf surface. This approach requires bacterial survival on the leaf surface, migration and entry into the leaf apoplast via stomata, and suppression of plant defenses by the phytopathogen in the absence of abiotic leaf damage. We also hypothesized that effector-triggered immunity (ETI) induced against an avirulent phytopathogen reduces S. enterica survival in cocolonized leaves. To elucidate the mechanism that allows increased persistence, we also examined S. enterica colonization patterns and preferred niches.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Both S. enterica strains are clinical isolates from salmonellosis outbreaks caused by consumption of contaminated raw tomatoes. These S. enterica serovars are commonly associated with outbreaks of produce origin (serovar Baildon, tomato in 2010 and 1999; serovar Enteritidis, tomato in 2012 and 2005, pine nuts in 2011, and sprouts in 2011 [Centers for Disease Control and Prevention]). For studies other than microscopy, each S. enterica strain was transformed with plasmid pKT-Kan (17). For microscopy studies, each S. enterica strain was transformed with pWM1009, which constitutively expresses kanamycin resistance and the cyan fluorescent protein (CFP). X. perforans strains were transformed with pUFZ, which constitutively expresses kanamycin resistance and the green fluorescent protein (GFP) (Ptrp-GFP) (18). X. perforans strain 91-118, which contains avrXv3 and is recognized by the tomato cultivar NIL216 (avirulent), and an avrXv3 deletion mutant of X. perforans strain 91-118, which eludes recognition by cv. NIL216 (virulent), were used for plant studies. X. perforans was cultured on nutrient agar (NA) (Difco) and S. enterica on Luria-Bertani agar (19) amended with 50 μg/ml kanamycin (LB-kan).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| X. perforans | ||
| 91-118 | Tomato race 3; avirulent on tomato cv. NIL216; carries avrXv3; Rifr | 21 |
| 91-118 ΔavrXv3 | Tomato race 3; virulent on tomato cv. NIL216; marker exchange mutant; Rifr Kmr | 21 |
| 91-118 pUFZ75 | Transconjugant GFP constitutive; Rifr Kmr | This study |
| 91-118 ΔavrXv3 pUFZ75-Cm | Transconjugant GFP constitutive; Rifr Kmr Cmr | This study |
| S. enterica | ||
| 05x-02123 | Serovar Enteritidis | California State Department of Health (CSDH), Berkeley, California |
| 99A 23 | Serovar Baildon | 40 |
| 05x-02123 pWM1009 | CFP constitutive; Kmr | This study |
| 99A 23 pWM1009 | CFP constitutive; Kmr | This study |
| E. coli | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Bethesda Research Laboratories |
| Plasmids | ||
| pLAFR3 | Tcr rlx+ RK2 replicon; Tcr | 41 |
| pUFZ75 | pUFR034; ΔlacZV[(T1)4-Ptac-gfp-T1]; Kmr | 18 |
| pUFZ75-Cm | pUFR034; ΔlacZV[(T1)4-Ptac-gfp-Cmr-T1]; Kmr Cmr | This study |
| pWM1009 | pMW10 ΔlacZV[(T1)4-Pc-cfp-T1]; Kmr | 17 |
In planta S. enterica and X. perforans population dynamics.
For plant inoculum preparation, overnight cultures of X. perforans and S. enterica from plates were resuspended in sterile tap water to an optical density at 600 nm of 0.3 for X. perforans and 0.2 for S. enterica, approximately 108 CFU/ml for both species. S. enterica strains were combined in a 1:1 ratio prior to inoculum dilution, and this cocktail was used for all experiments. Five-week-old tomato cultivar NIL216 plants containing the Xv3 resistance gene were coinoculated with avirulent X. perforans and S. enterica, virulent X. perforans and S. enterica, or S. enterica alone, with three replicates for each treatment. Inoculations were performed by dipping the plants for 30 s into a sterile tap water suspension of a 1:1 ratio of X. perforans (106 CFU/ml) and S. enterica (106 CFU/ml) and S. enterica alone (106 CFU/ml), amended with 0.025% (vol/vol) Silwet 77 (Fisher Scientific). Experiments were repeated at least twice.
For testing the effect of X. perforans populations on S. enterica survival, the initial inoculum concentration of virulent X. perforans was altered. Inoculations were performed by dipping plants for 30 s into a sterile tap water suspension of a 1:1 ratio of virulent X. perforans with two different concentrations (106 CFU/ml and 105 CFU/ml) and S. enterica (106 CFU/ml), amended with 0.025% (vol/vol) Silwet 77, with three replicates for each treatment. The inoculated plants were kept in a growth chamber at 25°C with a 12-h light-dark cycle at 95% humidity for 2 days, followed by 12 h high (>90%) humidity for the remainder of the experiment. Experiments were repeated at least twice.
Bacterial populations were determined every 48 h. To test the effect of leaf age on bacterial colonization, young, middle, and old leaves were assayed as shown in Fig. S1 in the supplemental material. A 2-cm2 area of leaf tissue was removed from each leaflet with a sterile cork borer. Using sterile forceps, two leaf disks were then placed into a sterile tube containing 1 ml sterile tap water and homogenized with a micro-Dremel tool (Dremel, PA). Standard 10-fold dilution plating of the homogenate was carried out, and suspensions were plated onto NA and LB-kan to estimate X. perforans and S. enterica populations, respectively, from each sample. The NA plates were incubated at 28°C for 3 days and the LB-kan plates at 37°C for 24 h. Colonies were counted, and bacterial populations were calculated as CFU per cm2 of leaf area.
CLSM.
Colonization of S. enterica in leaves cocolonized by X. perforans was assessed at 2 and 7 days postinoculation (dpi). Leaf samples (1 cm2) were mounted on a clean glass slide in 50% glycerol and covered with a coverslip that was held down tightly with clear plastic tape. The leaves were observed under confocal laser scanning microscopy (CLSM) (TCS-SP 5; Leica, Germany). Samples were scanned with a 458-nm excitation filter for CFP and a 488-nm filter for GFP, and a spectral detection window was selected at 460 to 540 nm (for CFP) and 505 to 550 nm (for GFP) for emission using a prism-based spectrometer. In order to avoid overlap between the two signals, samples were scanned with the “between-the-frames” scanning option. The images were acquired using LAS AF Lite version 3 software (Leica Microsystems, Germany). To count the stomata colonized by S. enterica, samples were observed under ×40 magnification, and stomata from 10 random fields of view per leaf were examined for the presence of S. enterica on one leaf taken from each of three plants. The experiment was repeated at least three times.
Statistical analysis.
Bacterial populations were log transformed to obtain normal distributions for analysis. A linear model (y = ax + b, where y is the S. enterica concentration in log CFU/cm2, x is the time [in days postinoculation], a is the slope of log CFU/cm2 · day−1, and b is the initial S. enterica concentration) was fitted to the transformed population data to describe the S. enterica population decline over time. To determine whether the average population of S. enterica differed between treatments or over time, analysis of covariance (ANCOVA) was used to test the potential effects of X. perforans cocolonization of leaves on S. enterica populations, with treatment, time (dpi), and leaf age as covariates. The same analysis was used to analyze X. perforans populations. Repetitions of the experiment were considered block factors. S. enterica populations (log CFU/cm2 leaf tissue) were compared between treatments at specific time points (dpi) using a two-sample t test. These statistical analyses were performed using R software (20). To determine whether the decline of the S. enterica population was different among treatments, a t test of the slopes was performed using SAS-JMP Pro10. To examine S. enterica leaf colonization, stomata in a single view were checked for the presence of S. enterica. The percentage of stomata colonized by S. enterica was calculated for each treatment. Differences among treatments were tested for significance with Duncan's multiple-range tests using SAS-Jump10.
RESULTS
Plant-phytopathogen interaction influences S. enterica survival.
To understand the influence of phytobacterial pathogenicity on the survival of S. enterica, we examined the effects of resistant and susceptible plant responses to X. perforans on S. enterica populations. Five-week-old tomato plants (cv. NIL216) were inoculated with S. enterica and virulent X. perforans, S. enterica and avirulent X. perforans, or S. enterica alone (the strains are listed in Table 1). Inoculation of the tomato cultivar NIL216 with virulent X. perforans triggers a susceptible plant response (water-soaked lesions), while inoculation with avirulent X. perforans triggers a resistant plant response (small necrotic lesions characteristic of a hypersensitive reaction) (see Fig. S2 in the supplemental material). Avirulent X. perforans translocates effector AvrXv3 via the T3SS into plant cells, which triggers this resistance response (ETI), restricting bacterial growth of avirulent X. perforans in the apoplast (21). No sign of disease appeared on the leaves inoculated with S. enterica alone. Both S. enterica and X. perforans populations were enumerated approximately every 2 days. No significant difference was found among experimental replications; data from two experiments are shown in Fig. 1 and 2. S. enterica populations were significantly different for each dpi (Fig. 1A) (P < 0.001). S. enterica population change over time (slope) was significantly different on plants inoculated with S. enterica and virulent X. perforans compared to the leaves without X. perforans or leaves inoculated with S. enterica and avirulent X. perforans (interaction of dpi and treatment [dpi:treatment]; P < 0.001). S. enterica populations were ∼10-fold higher by 14 days p.i. on leaves inoculated with S. enterica and virulent X. perforans than on the leaves inoculated with S. enterica alone (Fig. 1A). The average virulent X. perforans population surpassed 106 CFU/cm2 by 6 dpi (Fig. 1B), when S. enterica populations were ∼3- and 5-fold higher on leaves of plants inoculated with S. enterica and virulent X. perforans than on those inoculated with S. enterica alone and S. enterica and avirulent X. perforans, respectively (Fig. 1A). The avirulent X. perforans population did not reach beyond 106 CFU/cm2 (Fig. 1B), due to the restriction of avirulent X. perforans multiplication as a result of ETI. As early as 4 dpi, S. enterica populations were significantly higher on leaves coinoculated with virulent X. perforans than on leaves inoculated with avirulent X. perforans (P < 0.001). Thus, S. enterica survival in the phyllosphere was influenced by the host response to infection by virulent X. perforans. Interestingly, 14 dpi, S. enterica populations on the leaves with activated ETI (i.e., plants inoculated with avirulent X. perforans) were ∼3- and 10-fold lower than on the leaves inoculated with S. enterica alone and virulent X. perforans, respectively (Fig. 1A). In addition, when ETI was activated, the rate of decline of S. enterica populations was higher (a steeper slope) than that on leaves with S. enterica alone (P < 0.05). Thus, plant defense responses activated against avirulent X. perforans were also effective in compromising the survival of S. enterica in the tomato phyllosphere.
FIG 1.
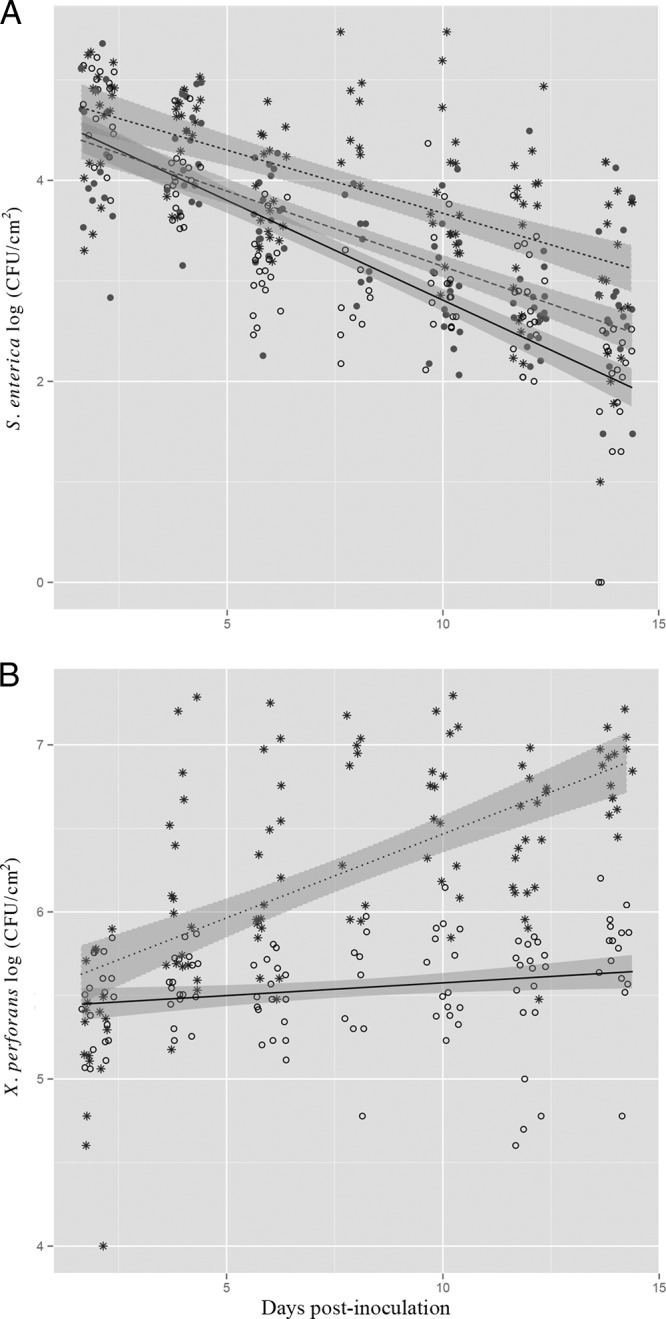
S. enterica and X. perforans population dynamics on tomato plants. S. enterica (A) and X. perforans (B) populations (log CFU/cm2) on tomato leaves were sampled every 2 days for 14 days postinoculation (dpi). Plants were inoculated with S. enterica and virulent X. perforans (asterisks and dotted lines), S. enterica and avirulent X. perforans (open circles and solid lines), or S. enterica alone (solid circles and dashed lines). The data represent two independent experiments combined, since no significant difference existed among experimental replicates (experiment:dpi:treatment; P = 0.1 [S. enterica] and P = 0.6 [X. perforans]). The lines correspond to a linear regression model and the shaded areas to their associated 95% confidence intervals.
FIG 2.
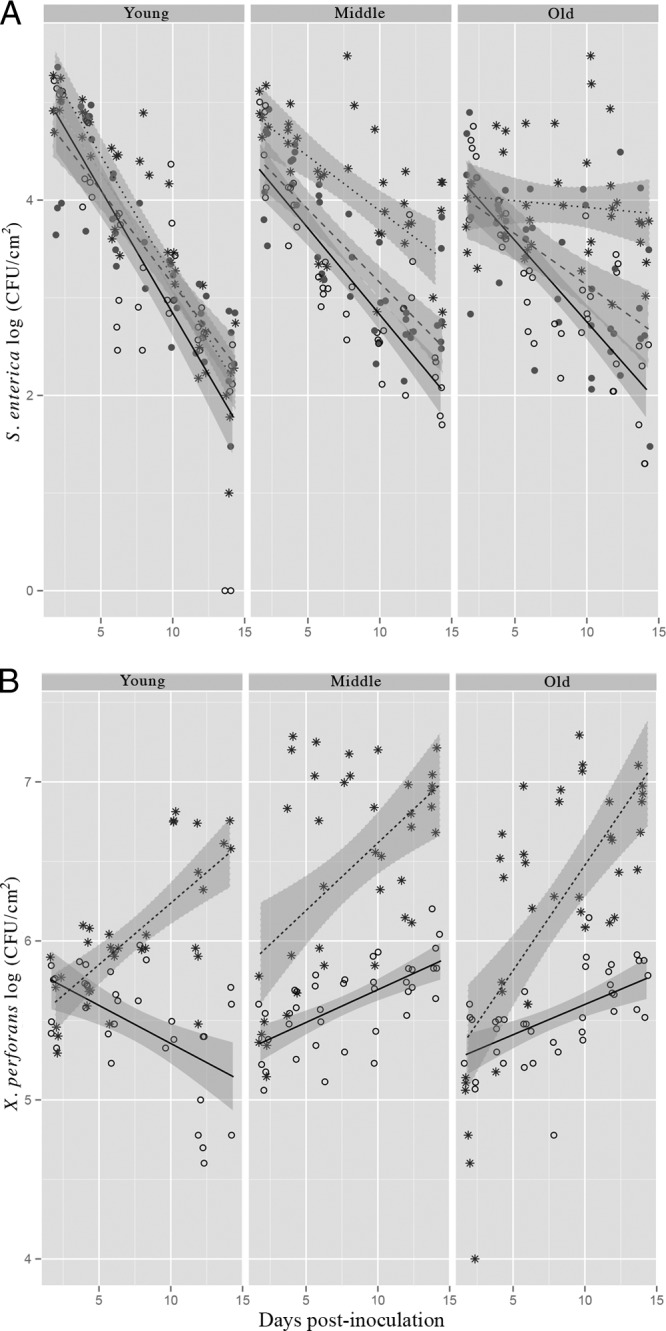
Effect of leaf age on S. enterica and X. perforans population dynamics on tomato plants. Shown is separation of the data in Fig. 1 by leaf age. S. enterica (A) and X. perforans (B) populations (log CFU/cm2) on tomato leaves were sampled every 2 days for 14 days postinoculation (dpi). Plants were inoculated with S. enterica and virulent X. perforans (asterisks and dotted lines), S. enterica and avirulent X. perforans (open circles and solid lines), or S. enterica alone (solid circles and dashed lines). The data represent two independent experiments combined, since no significant difference existed among experimental replicates (experiment:dpi:treatment:age; P = 0.1 [S. enterica] and P = 0.5 [X. perforans]). The lines correspond to a linear regression model and the shaded areas to their associated 95% confidence intervals.
Leaf age influences S. enterica and X. perforans populations.
To understand the role of leaf age in persistence of S. enterica in the phyllosphere, we examined different leaf age categories in which S. enterica was applied alone or was coinoculated with virulent or avirulent X. perforans. Leaves were labeled as old, middle, and young at the time of inoculation (see Fig. S1 in the supplemental material). Water-soaked lesions (a symptom of bacterial spot disease, a result of suppression of pathogen-associated molecular pattern [PAMP]-triggered immunity [PTI]) were observed initially on middle leaves inoculated with S. enterica and virulent X. perforans at 4 dpi, followed by old and young leaves by 6 dpi (data not shown). No water-soaked lesions appeared on leaves inoculated with S. enterica and avirulent X. perforans; however, small necrotic lesions typical of a hypersensitive reaction (22) that did not spread or expand over time (a sign of plant resistance) appeared 5 dpi (see Fig. S2 in the supplemental material). The rates of S. enterica population decline were significantly different among different-aged leaves (Fig. 2A) (dpi:treatment:leaf age; P < 0.001). Leaf age also affected X. perforans populations (Fig. 2B), most apparently for avirulent X. perforans in young leaves. Population trends were similar between middle and old leaves for both S. enterica and X. perforans. Interestingly, S. enterica populations on young leaves were similar regardless of treatment, even though the rates of growth of virulent X. perforans were similar between different-aged leaves (Fig. 2B). Thus, X. perforans virulence and leaf age both influenced S. enterica population dynamics in the phyllosphere.
S. enterica persistence is dependent on X. perforans pathogenicity but independent of the X. perforans population level.
In the experiments discussed above, we observed a sharp decline in S. enterica populations on leaves treated with avirulent X. perforans but not on those treated with virulent X. perforans. We speculated that higher populations of virulent X. perforans as opposed to pathogenicity per se may be important in S. enterica persistence. For these experiments, we applied S. enterica alone or in combination with the virulent X. perforans at two concentrations, 105 CFU/ml and 106 CFU/ml, or the avirulent X. perforans at 106 CFU/ml. The lower concentration of virulent X. perforans was selected so that the resulting virulent X. perforans populations would be close to the avirulent X. perforans concentration for a few days following inoculation and would provide information as to the importance of X. perforans populations for S. enterica persistence. We chose a single leaf age category (middle) for further experimentation. The lower virulent X. perforans inoculum level delayed bacterial spot development by 3 or 4 days (lesions appeared at 10 dpi) (data not shown) compared to the higher inoculum density (lesions appeared at 6 dpi). Although similar X. perforans populations in leaves were attained for avirulent and virulent X. perforans (at the low inoculum) by 4 dpi (Fig. 3B), S. enterica populations were 5-fold higher by 6 dpi and 10-fold higher by 14 dpi on leaves treated with virulent X. perforans (low inoculum) than on leaves treated with avirulent X. perforans (Fig. 3A) (dpi:treatment; P < 0.01). These results suggest that virulence of X. perforans, and thus manipulation of host defenses, is more important for the persistence of S. enterica in the phyllosphere than X. perforans population densities in the phyllosphere. This population difference also complements our findings in earlier experiments (Fig. 1) that ETI activated on the leaves with avirulent X. perforans is important in restricting survival of S. enterica in the phyllosphere.
FIG 3.
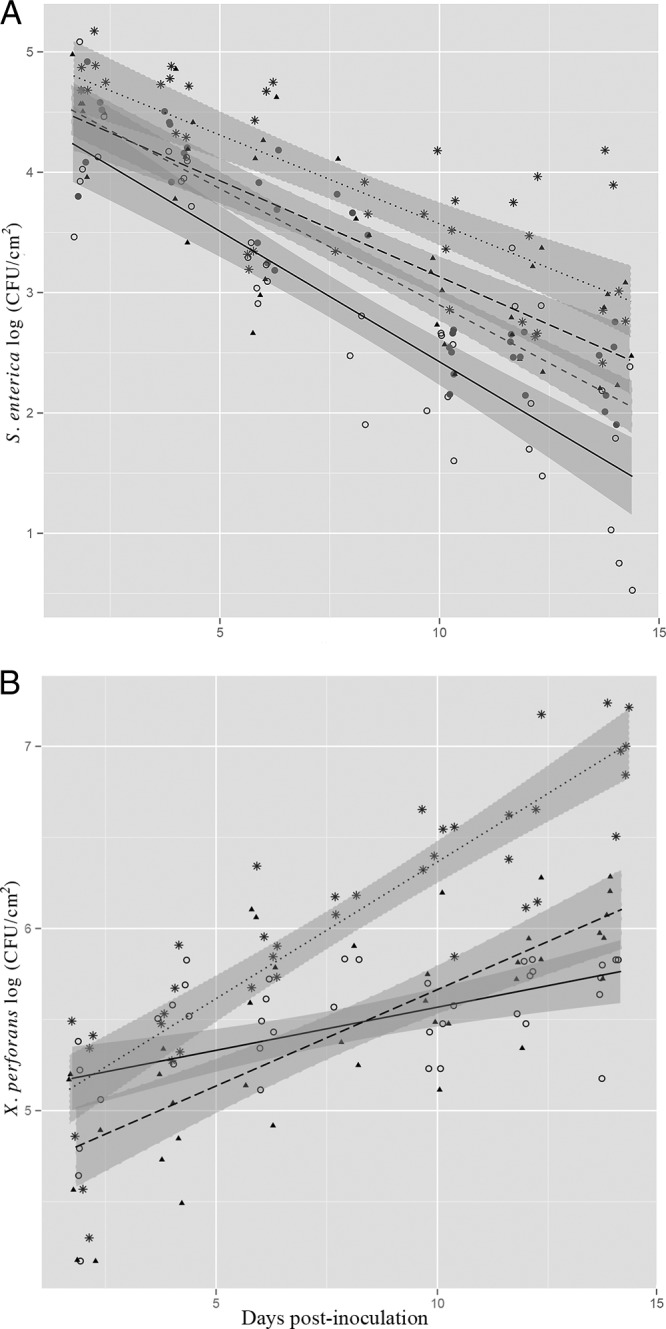
S. enterica populations are influenced by pathogenicity of X. perforans. S. enterica (A) and X. perforans (B) populations (log CFU/cm2) on tomato leaves were sampled every 2 days for 14 days postinoculation (dpi). Plants were inoculated with S. enterica and virulent X. perforans (106 CFU/ml, asterisks and dotted lines; 105 CFU/ml, triangles and long-dashed lines), S. enterica and avirulent X. perforans (open circles and solid lines), or S. enterica alone (solid circles and short-dashed line). The data represent two independent experiments combined, since no significant difference existed among experimental replicates (experiment:dpi:treatment; P = 0.5 [S. enterica] and P = 0.1 [X. perforans]). The lines correspond to a linear regression model and the shaded areas to their associated 95% confidence intervals.
Virulent X. perforans influences S. enterica aggregation on leaves.
To investigate the effect of X. perforans on S. enterica leaf colonization patterns, plants were inoculated with S. enterica and virulent X. perforans, S. enterica and avirulent X. perforans, or S. enterica alone and examined at 2 and 7 dpi with confocal laser scanning microscopy. These time points were chosen to compare the initial X. perforans infection period (2 dpi) and the time of established disease (7 dpi). At 2 dpi, S. enterica had colonized depressions between epidermal cells, areas close to veins, a few stomata (data not shown), and trichomes, as previously described (23, 24). This spatial distribution over the entire leaf surface occurred in the absence of X. perforans, as well as on leaves cocolonized by X. perforans. By 7 dpi, virulent X. perforans formed large aggregates in the apoplast (see Fig. S3 in the supplemental material) while avirulent X. perforans was limited to small aggregates on the leaf surface (data not shown). When inoculated alone, S. enterica was observed as aggregates in 20% of the fields showing small aggregates of 2 to 10 cells (data not shown). In contrast, S. enterica formed aggregates of more than two cells in close proximity (observed in the same field of view) to virulent X. perforans aggregates in 95% of the fields examined (Fig. 4). However, mixed aggregates of S. enterica and X. perforans, which would appear as a mixture of blue and green cells, were rarely observed, even near bacterial spot lesions. S. enterica was found in aggregates of more than two cells on the leaves colonized by avirulent X. perforans in 40% of the fields examined, usually in the substomatal chambers. S. enterica was rarely observed in the apoplast, even in the presence of virulent X. perforans, indicating that surface colonization in aggregates is a strategy adapted by S. enterica on the leaves with bacterial spot disease (see Fig. S3 in the supplemental material). Colonization in the form of aggregates might allow S. enterica to survive for longer periods by providing protection from environmental stress, in contrast to the solitary cells, as previously described (25).
FIG 4.
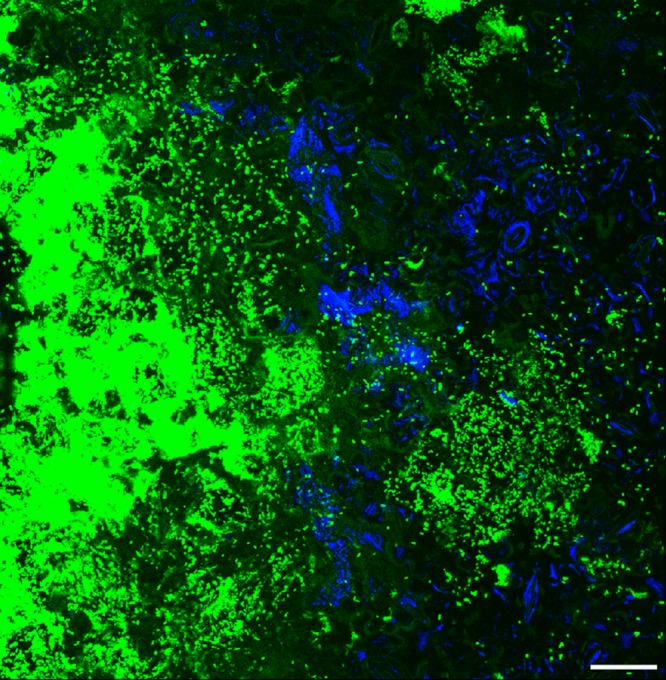
S. enterica and virulent X. perforans tomato leaf colonization. Plants were inoculated with S. enterica and virulent X. perforans, S. enterica and avirulent X. perforans, or S. enterica alone, and middle leaves were sampled for confocal microscopy 7 days postinoculation. Shown is a representative overlapping z-stack from confocal scanning laser microscopy of a middle leaf from a plant inoculated with S. enterica and virulent X. perforans (z-stack spanning 27-μm depth at ×40 magnification). S. enterica cells appear blue, and X. perforans appears green. Scale bar = 25 μm.
Since X. perforans enters the leaf apoplast via stomata, we examined S. enterica association with substomatal chambers or stomatal guard cells (Fig. 5). We found that at 2 dpi, when X. perforans initially enters the leaf apoplast via stomata (18), S. enterica associations with stomata were similar among treatments (Fig. 6) (P = 0.66). In leaves coinoculated with S. enterica and virulent X. perforans, bacterial spot symptoms were present at 7 dpi, and S. enterica was found in association with stomata almost twice as often as on leaves inoculated with S. enterica alone (P < 0.05). However, there was no difference in S. enterica association with stomata between plants coinoculated with virulent X. perforans and S. enterica or with avirulent X. perforans and S. enterica (P = 0.52). These data, coupled with our finding that the S. enterica populations on leaves coinoculated with S. enterica and virulent X. perforans are 30-fold larger on middle leaves than on leaves coinoculated with S. enterica and avirulent X. perforans (Fig. 2A), suggest S. enterica association with stomata plays little role in persistence. Furthermore, S. enterica fails to aggregate in the apoplast to appreciable populations even in the presence of large aggregations of virulent X. perforans (see Fig. S3 in the supplemental material). S. enterica aggregates are seen in sections near the surface of the leaf, up to 15 μm from the lower leaf surface, where stomata are still visible. In contrast, in sections greater than 16.5 μm from the leaf surface (the apoplast), S. enterica is rarely seen.
FIG 5.
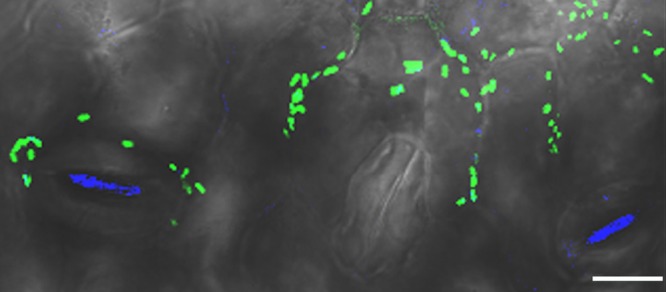
S. enterica colonization of stomata in the presence of X. perforans. Plants were inoculated with S. enterica and virulent X. perforans, S. enterica and avirulent X. perforans, or S. enterica alone, and middle leaves were sampled for confocal microscopy at 2 days postinoculation (dpi) and 7 dpi. Shown is a representative confocal laser scanning micrograph of a middle leaf from a plant inoculated with S. enterica and virulent X. perforans sampled at 7 dpi. S. enterica cells appear blue, and X. perforans appears green. Magnification, ×40. Scale bar = 25 μm.
FIG 6.
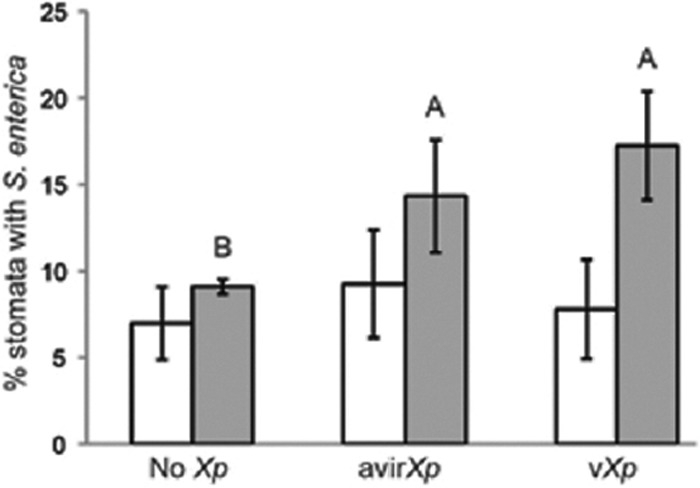
S. enterica association with stomata in the presence of X. perforans is independent of phytobacterial pathogenicity. Plants were inoculated with S. enterica and virulent X. perforans (vXp), S. enterica and avirulent X. perforans (avirXp), or S. enterica alone (No Xp), and middle leaves were sampled for confocal microscopy at 2 days postinoculation and 7 dpi. Stomata from 10 random fields of view per leaf at ×40 magnification were examined for the presence of S. enterica from one leaf taken from each of three plants. The experiment was repeated at least three times. Average percentages of stomata colonized by S. enterica are shown for 2 dpi (white bars) and 7 dpi (gray bars). The error bars indicate standard deviations.
DISCUSSION
With a goal of reducing human illness caused by consumption of contaminated raw produce, it is important to understand how plant-pathogenic bacteria influence populations of the plant symbiont S. enterica. It is recognized that brute force pathogens macerate plant tissue with cell wall-degrading enzymes during infection and thereby influence the growth of human pathogens in affected tissue (26, 27). Previous work in our laboratory and others has shown that S. enterica populations also increase on leaves coinoculated with stealth phytobacterial pathogens (9, 16). In this study, we found that survival of S. enterica on leaves is directly influenced by phytobacterial pathogen virulence. As early as 4 dpi, S. enterica populations were significantly larger in the phyllosphere coinoculated with S. enterica and virulent X. perforans than on leaves coinoculated with S. enterica and avirulent X. perforans or S. enterica alone. During the compatible interaction, virulent X. perforans delivers an array of type III effectors, some of which are responsible for suppressing PTI during early events of disease establishment and some of which are involved in imparting effector-triggered susceptibility (28). The suppression of PTI by virulent X. perforans during the establishment of disease helps S. enterica to persist in the phyllosphere. These results complement the findings of Meng et al. (16) in that S. enterica benefits from a functional phytobacterial T3SS. However, by infiltrating S. enterica into leaves, the work of Meng et al. (16) was based on two assumptions: (i) S. enterica can generally access the apoplast, presumably via stomata, and (ii) S. enterica can reach or maintain large populations in the apoplast. In our study, we inoculated plants in a manner that mimics natural contamination of field-grown produce by S. enterica via irrigation water or pesticide diluent and found that S. enterica rarely colonizes the apoplast in significant populations, even in the presence of a virulent phytopathogen, X. perforans. Persistence of S. enterica was observed on the leaf surface as opposed to in the apoplast.
Bacterial cell aggregation is a mechanism for survival on plants (25). In this study, we observed that S. enterica rarely formed cell aggregates on leaves inoculated with S. enterica alone. Previous findings that solitary cells are more susceptible to leaf surface stresses and die first (25) may explain the restricted survival of S. enterica in the phyllosphere when inoculated alone. Furthermore, S. enterica that was unable to produce cellulose and thus aggregate had reduced survival on leaves (J. D. Barak, unpublished data). A net increase in S. enterica populations over the course of this study was not observed, even in the presence of the virulent X. perforans. However, large aggregates of S. enterica were observed on leaves cocolonized by virulent X. perforans, suggesting local replication, perhaps with simultaneous death of cells (presumably single cells) elsewhere, as opposed to cell migration from great distances to form large aggregates. If the former is true, then S. enterica may utilize limited nutrients liberated by X. perforans in a compatible interaction. During the infection process, phytopathogens manipulate plant metabolism to benefit their ability to colonize, both by suppressing plant defenses (compatible interaction) and by reallocating photoassimilates to supply the pathogen with nutrients (29). The compatible interaction increases apoplastic carbohydrate levels in infected leaves by converting the leaves from sources to sinks (30). In addition to changes due to infection, the physical and chemical composition of leaves also changes due to age (31, 32), which can affect the associated bacterial community, including members such as S. enterica and E. coli. We found that the rates of S. enterica population decline were significantly different among leaf ages in the presence of virulent X. perforans, with the most rapid decline on young leaves and with old and middle leaves maintaining higher S. enterica populations. Our data suggest that the higher populations on leaves coinoculated with virulent X. perforans is most likely due to the ability of S. enterica to form aggregates on these leaves. Aggregated S. enterica survives for a longer period by gaining protection against environmental stress, as has been described previously for phyllosphere symbionts (33). Even without a net population increase, persistence of S. enterica in the phyllosphere increases the risk of fruit contamination (24, 34) or direct consumption of an infectious dose in plants with edible leaves.
S. enterica leaf colonization was altered by X. perforans.
In this study, S. enterica did not form mixed aggregates with X. perforans and S. enterica was found twice as often in substomatal chambers or in association with stomata guard cells in plants coinoculated with X. perforans as in plants where only S. enterica was applied to the leaf surface. This result suggests that S. enterica can colonize the protected niche of the substomatal chamber in leaves cocolonized by X. perforans regardless of X. perforans pathogenicity. Previously, S. enterica was reported to use flagellum-mediated motility to locate stomata and chemotaxis to access the leaf apoplast via stomata (35). Since that study examined detached leaf pieces, it was unknown whether similar mechanisms would cause a niche switch from cell junctions and glandular trichomes, as we and others previously reported (10, 23, 24), to substomatal chambers in live plants cocolonized with a human pathogen and phytopathogen (this study). Although substomatal chambers shelter bacteria from ultraviolet light exposure and desiccation on the leaf surface, we conclude that association with stomata alone cannot account for the larger S. enterica populations seen in the presence of virulent X. perforans, since the incidences of S. enterica colonization of stomata were similar between leaves coinoculated with virulent or avirulent X. perforans. While it is known that X. perforans manipulates stomatal closure (36), our finding suggests X. perforans accesses substomatal chambers prior to pathogen recognition by the plant. Cumulatively, these data support the conclusion that the benefit gained by S. enterica from the virulent phytopathogen occurs in the absence of S. enterica reaching large populations in the apoplast.
ETI, which is induced during incompatible plant-pathogen interactions, is accompanied by programmed cell death in the form of a strong hypersensitive response that restricts the multiplication and spread of an avirulent pathogen. Our study shows that the ETI host responses elicited against an avirulent phytobacterial pathogen negatively impacted colonization of the phyllosphere by the symbiont S. enterica. During incompatible interactions (in this case, wild-type X. perforans on NIL216), suppression of PTI is overridden by a strong activation of ETI, which results in stronger, quicker, more prolonged and robust plant immune responses (28, 37). Since S. enterica populations differed among treatments (coinoculation with virulent X. perforans or avirulent X. perforans) prior to bacterial spot appearance (day 4), events prior to bacterial spot disease development may be important for survival of S. enterica. The rate of decline of S. enterica on ETI-induced leaves was steeper than on leaves with S. enterica alone. Thus, the gene-for-gene resistance against X. perforans, which is triggered soon after phytobacterial entry into stomata, also proved to be effective in restricting S. enterica populations on the leaf surface.
The discovery that colonization of tomato plants by X. perforans coincides with longer persistence of S. enterica is alarming, as the opportunity to contaminate fruit rises with lingering S. enterica populations on leaves (24, 34, 38). Currently, the produce industry lacks an effective control method to ensure complete removal or killing of food-borne pathogens in fresh or minimally processed fruits and vegetables. The contamination incidences at the preharvest level indicate the need to design new intervention strategies for ensuring the safety of fresh produce, possibly at the level of the field. Tomato production areas where S. enterica is endemic (2) are also plagued by bacterial speck and bacterial spot of tomato, caused by P. syringae pv. tomato and xanthomonads, respectively (39). Our studies indicate that plant disease caused by stealth plant pathogens, e.g., X. perforans, is likely a risk factor for increased persistence of human pathogens, such as S. enterica, on leaves closest to developing fruit or those directly consumed (middle and older leaves), thereby increasing the potential for human illness. Resistance against bacterial plant disease appears to be effective in minimizing S. enterica populations, indicating a possible strategy of deploying resistance against phytobacterial pathogens to reduce preharvest incidences of S. enterica contamination.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the USDA-NIFA (grant no. 2011-670137-30166) and the Food Research Institute, University of Wisconsin—Madison.
We thank the Interdisciplinary Center for Biotechnology Research Flow Cytometry Laboratory at the University of Florida for technical support with scanning laser microscopy.
Footnotes
Published ahead of print 14 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00345-14.
REFERENCES
- 1.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 19:407–415. 10.3201/eid1903.111866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ottesen A, Pena A, White J, Pettengill J, Li C, Allard S, Rideout S, Allard M, Hill T, Evans P, Strain E, Musser S, Knight R, Brown E. 2013. Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 13:114. 10.1186/1471-2180-13-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak JD, Schroeder BK. 2012. Interrelationships of food safety and plant pathology: the life cycle of human pathogens on plants. Annu. Rev. Phytopathol. 50:241–266. 10.1146/annurev-phyto-081211-172936 [DOI] [PubMed] [Google Scholar]
- 4.Teplitski M, Barak JD, Schneider KR. 2009. Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr. Opin. Biotechnol. 20:166–171. 10.1016/j.copbio.2009.03.002 [DOI] [PubMed] [Google Scholar]
- 5.O'Brien RD, Lindow SE. 1989. Effect of plant species and environmental conditions on epiphytic population sizes of Pseudomonas syringae and other bacteria. Phytopathology 79:619–627. 10.1094/Phyto-79-619 [DOI] [Google Scholar]
- 6.Brandl MT, Mandrell RE. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68:3614–3621. 10.1128/AEM.68.7.3614-3621.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam M, Morgan J, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl. Environ. Microbiol. 70:2497–2502. 10.1128/AEM.70.4.2497-2502.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Islam M, Morgan J, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog. Dis. 1:27–35. 10.1089/153531404772914437 [DOI] [PubMed] [Google Scholar]
- 9.Barak JD, Liang AS. 2008. Role of soil, crop debris, and a plant pathogen in Salmonella enterica contamination of tomato plants. PLoS One 3:e1657. 10.1371/journal.pone.0001657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poza-Carrion C, Suslow T, Lindow S. 2013. Resident bacteria on leaves enhance survival of immigrant cells of Salmonella enterica. Phytopathology 103:341–351. 10.1094/PHYTO-09-12-0221-FI [DOI] [PubMed] [Google Scholar]
- 11.Wells JM, Butterfield JE. 1997. Salmonella contamination associated with bacterial soft rot of fresh fruits and vegetables in the marketplace. Plant Dis. 81:867–872. 10.1094/PDIS.1997.81.8.867 [DOI] [PubMed] [Google Scholar]
- 12.Wells JM, Butterfield JE. 1999. Incidence of Salmonella on fresh fruits and vegetables affected by fungal rots or physical injury. Plant Dis. 83:722–726. 10.1094/PDIS.1999.83.8.722 [DOI] [PubMed] [Google Scholar]
- 13.Toth IK, Birch PRJ. 2005. Rotting softly and stealthily. Curr. Opin. Plant Biol. 8:424–429. 10.1016/j.pbi.2005.04.001 [DOI] [PubMed] [Google Scholar]
- 14.Rico A, Jones R, Preston GM. 2009. Adaptation to the plant apoplast by plant pathogenic bacteria, p 63–89 In Jackson RW. (ed), Plant pathogenic bacteria: genomics and molecular biology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 15.Alfano JR, Collmer A. 1996. Bacterial pathogens in plants: life up against the wall. Plant Cell 8:1683–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng F, Altier C, Martin GB. 2013. Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ. Microbiol. 15:2418–2430. 10.1111/1462-2920.12113 [DOI] [PubMed] [Google Scholar]
- 17.Miller WG, Leveau JH, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243–1250. 10.1094/MPMI.2000.13.11.1243 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Callaway E, Jones J, Wilson M. 2009. Visualisation of hrp gene expression in Xanthomonas euvesicatoria in the tomato phyllosphere. Eur. J. Plant Pathol. 124:379–390. 10.1007/s10658-008-9423-x [DOI] [Google Scholar]
- 19.Miller JH. 1972. Experiments in molecular genetics, p 466 Cold Spring Harbor Laboratory. Cold Spring Harbor, NY [Google Scholar]
- 20.R Development Core Team. 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- 21.Astua-Monge G, Minsavage GV, Stall RE, Davis MJ, Bonas U, Jones JB. 2000. Resistance of tomato and pepper to T3 strains of Xanthomonas campestris pv. vesicatoria is specified by a plant-inducible avirulence gene. Mol. Plant Microbe Interact. 13:911–921. 10.1094/MPMI.2000.13.9.911 [DOI] [PubMed] [Google Scholar]
- 22.Somodi GC, Jones JB, Scott JW. 1991. Populations of Xanthomonas campestris pv. vesicatoria in lesions of susceptible and resistant tomato genotypes. Plant Dis. 75:357–360. 10.1094/PD-75-0357 [DOI] [Google Scholar]
- 23.Cevallos-Cevallos JM, Gu G, Danyluk MD, van Bruggen AHC. 2012. Adhesion and splash dispersal of Salmonella enterica Typhimurium on tomato leaflets: effects of rdar morphotype and trichome density. Int. J. Food Microbiol. 160:58–64. 10.1016/j.ijfoodmicro.2012.09.021 [DOI] [PubMed] [Google Scholar]
- 24.Barak JD, Kramer LC, Hao L. 2011. Plant cultivar alters Salmonella enterica colonization of tomato and type I trichomes are preferential colonization sites. Appl. Environ. Microbiol. 77:498–504. 10.1128/AEM.01661-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monier JM, Lindow SE. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. U. S. A. 100:15977–15982. 10.1073/pnas.2436560100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamazaki A, Li J, Hutchins WC, Wang L, Ma J, Ibekwe AM, Yang C-H. 2011. Commensal effect of pectate lyases secreted from Dickeya dadantii on proliferation of Escherichia coli O157:H7 EDL933 on lettuce leaves. Appl. Environ. Microbiol. 77:156–162. 10.1128/AEM.01079-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwan G, Charkowski AO, Barak JD. 2013. Salmonella enterica suppresses Pectobacterium carotovorum subsp. carotovorum population and soft rot progression by acidifying the microaerophilic environment. mBio 4:e00557–12. 10.1128/mBio.00557-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones JDG, Dangl JL. 2006. The plant immune system. Nature 444:323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 29.Sonnewald S, Priller JPR, Schuster J, Glickmann E, Hajirezaei M-R, Siebig S, Mudgett MB, Sonnewald U. 2012. Regulation of cell wall-bound invertase in pepper leaves by Xanthomonas campestris pv. vesicatoria type three effectors. PLoS One 7:e51763. 10.1371/journal.pone.0051763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biemelt S, Sonnewald U. 2006. Plant-microbe interactions to probe regulation of plant carbon metabolism. J. Plant Physiol. 163:307–318. 10.1016/j.jplph.2005.10.011 [DOI] [PubMed] [Google Scholar]
- 31.Humphries EC, Wheeler AW. 1963. The physiology of leaf growth. Annu. Rev. Plant Physiol. 14:385–410. 10.1146/annurev.pp.14.060163.002125 [DOI] [Google Scholar]
- 32.Abou Hadid AF, El-Beltagy AS, Smith AR, Hall MA. 1986. The influence of leaf position and age upon the production of stress ethylene in tomatoes. Acta Hort. 190:397–404 [Google Scholar]
- 33.Monier JM, Lindow SE. 2005. Spatial organization of dual-species bacterial aggregates on leaf surfaces. Appl. Environ. Microbiol. 71:5484–5493. 10.1128/AEM.71.9.5484-5493.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu G, Cevallos-Cevallos JM, Vallad GE, van Bruggen AHC. 2013. Organically managed soils reduce internal colonization of tomato plants by Salmonella enterica serovar Typhimurium. Phytopathology 103:381–388. 10.1094/PHYTO-04-12-0072-FI [DOI] [PubMed] [Google Scholar]
- 35.Kroupitski Y, Golberg D, Belausov E, Pinto R, Swartzberg D, Granot D, Sela S. 2009. Internalization of Salmonella in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 75:6076–6086. 10.1128/AEM.01084-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gudesblat GE, Torres PS, Vojnov AA. 2009. Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiol. 149:1017–1027. 10.1104/pp.108.126870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomma BPHJ, Nurnberger T, Joosten MHAJ. 2011. Of PAMPS and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23:4–15. 10.1105/tpc.110.082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu G, Hu J, Cevallos-Cevallos JM, Richardson SM, Bartz JA, van Bruggen AHC. 2011. Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS One 6:e27340. 10.1371/journal.pone.0027340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graves AS, Alexander SA. 2002. Managing bacterial speck and spot of tomato with acibenzolar-S-methyl in Virginia. Plant Health Prog.. 10.1094/PHP-2002-0220-01-RS [DOI] [Google Scholar]
- 40.Cummings K, Barrett E, Mohle-Boetani JC, Brooks JT, Farrar J, Hunt T, Fiore A, Komatsu K, Werner SB, Slutsker L. 2001. A multistate outbreak of Salmonella enterica serotype Baildon associated with domestic raw tomatoes. Emerg. Infect. Dis. 7:1046–1048. 10.3201/eid0706.010625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staskawicz BJ, Dahlbeck D, Keen N, Napoli C. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


