Abstract
Quantitative real-time PCR (qPCR) assays that target the human-associated HF183 bacterial cluster within members of the genus Bacteroides are among the most widely used methods for the characterization of human fecal pollution in ambient surface waters. In this study, we show that a current TaqMan HF183 qPCR assay (HF183/BFDrev) routinely forms nonspecific amplification products and introduce a modified TaqMan assay (HF183/BacR287) that alleviates this problem. The performance of each qPCR assay was compared in head-to-head experiments investigating limits of detection, analytical precision, predicted hybridization to 16S rRNA gene sequences from a reference database, and relative marker concentrations in fecal and sewage samples. The performance of the modified HF183/BacR287 assay is equal to or improves upon that of the original HF183/BFDrev assay. In addition, a qPCR chemistry designed to combat amplification inhibition and a multiplexed internal amplification control are included. In light of the expanding use of PCR-based methods that rely on the detection of extremely low concentrations of DNA template, such as qPCR and digital PCR, the new TaqMan HF183/BacR287 assay should provide more accurate estimations of human-derived fecal contaminants in ambient surface waters.
INTRODUCTION
Numerous host-associated indicators are used to identify human fecal pollution in ambient surface waters, with many relying on molecular methods such as quantitative real-time PCR (qPCR). The most widely used methods target the HF183 16S rRNA gene cluster of members of the genus Bacteroides initially identified in human fecal samples collected from the United States Pacific Northwest (1) and targeted by PCR to distinguish human-associated contamination (2). Over the past decade, researchers have developed (2–9) and implemented (10–15) many PCR-based methods targeting the HF183 cluster worldwide.
In most HF183 PCR applications, the forward primer targets the Bacteroides HF183 cluster, reported to include Bacteroides dorei (3). The reverse primer typically hybridizes to a wider diversity of Bacteroidales so as not to further restrict the range of targeted bacteria. Researchers have employed this strategy for endpoint PCR applications (2), as well as a variety of quantitative real-time PCR chemistries, such as the SYBR green (8, 16) and TaqMan (4, 5, 9, 17) chemistries.
In recent performance evaluation studies, PCR-based methods targeting the HF183 cluster, in particular, the TaqMan HF183/BFDrev assay (3), consistently outperformed other tested approaches in terms of specificity and sensitivity (18–21). TaqMan chemistry utilizes an internal oligonucleotide probe, greatly reducing the chance of false-positive results due to the accumulation of unintended amplification by-products (e.g., primer dimerization [PD] molecules). The TaqMan HF183/BFDrev qPCR assay was recently tested in a five-laboratory repeatability study and shown to be highly reproducible across laboratories when key components of the protocol were standardized (22). However, the current lack of a formal standardized method protocol for any HF183 method poses a large obstacle for integration into water management frameworks. In response, the United States Environmental Protection Agency (U.S. EPA) is conducting studies to further optimize the HF183 TaqMan qPCR assay in hopes of creating a new formal U.S. EPA standardized method for use in both marine water and freshwater.
In the optimization experiments presented herein, we found that the TaqMan HF183/BFDrev assay forms nonspecific amplification products with human fecal DNA composite dilutions, especially at low DNA target template concentrations. Elimination of spurious nonspecific amplification products was achieved by the development of a modified TaqMan multiplex qPCR assay (HF183/BacR287) and resulted in improved accuracy and limits of detection.
In addition to modifying the assay to eliminate amplification by-products, we incorporate new strategies and reagents to monitor amplification inhibition. Amplification inhibition can be widespread in environmental samples (23). However, the amplification inhibition reported during the qPCR enumeration of Enterococcus spp. in marine water samples was reduced by 91% when using reagents specifically tailored for the analysis of samples containing high levels of inhibitors (24). In addition, the widely accepted Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines state that to authenticate results, it is essential that an amplification control be utilized with each test sample reaction (25). We therefore optimized the new assay for use with reagents specifically developed to combat amplification inhibitors commonly associated with environmental samples and utilized a custom internal amplification control (IAC) designed to identify the presence of partial or complete inhibition in each test sample. Finally, we compared the performance of the modified TaqMan HF183/BacR287 assay to that of the current, top-performing TaqMan HF183/BFDrev assay via head-to-head experiments designed to measure limits of detection, analytical precision, and relative host distributions using reference fecal and sewage samples.
MATERIALS AND METHODS
Reference sample collections.
Fecal samples (n = 129) from individual animals were collected as previously described (19). Animal reference fecal samples represented nine different species, including Homo sapiens (human, n = 6), Felis catus (cat, n = 14), Gallus gallus (chicken, n = 15), Odocoileus virginianus (white-tail deer, n = 8), Canis familiaris (dog, n = 37), Bos taurus (unprocessed grain-fed cattle, n = 12), Bos taurus (grass-fed cattle, n = 10), Sus scrofa (pig, n = 9), Meleagris spp. (turkey, n = 8), and Antilocapra americana (pronghorn, n = 10). Additional chicken and dog samples were tested, as these animal sources are commonly reported to elicit false-positive results by HF183-based methods (19, 20). Wastewater treatment plant (WWTP) primary influent sewage samples from 58 different geographic locations across the United States (n = 54) and New Zealand (n = 4) were collected and filtered as previously described (26). Briefly, 500 ml influent was collected, stored on ice, and shipped to Cincinnati, OH. Twenty-five milliliters of each influent sample was filtered through a 0.2-μm-pore-size Supor-200 filter (Pall Co., Port Washington, NY) and stored in a 1.7-ml microcentrifuge tube at −80°C until the time of analysis. WWTP samples were from areas representing human population sizes ranging from 3,000 to 1.2 × 106 individuals, on the basis of the reported estimates of the population served (data not shown).
DNA extractions.
All DNA extractions of filtered sewage samples were performed with a DNA-EZ kit (GeneRite, North Brunswick, NJ) according to the manufacturer's instructions, as previously described (26). Prior to DNA extraction of fecal samples, approximately 1 g (wet weight) of fecal material was mixed with GITC buffer (5 M guanidine isothiocyanate, 100 mM EDTA [pH 8.0], 0.5% Sarkosyl) and vortexed until it was homogenized to create a fecal slurry. Eight hundred microliters of each fecal slurry was bead homogenized at 4.0 m/s for 30 s using a MP FastPrep-24 instrument (MP Biomedicals, LLC Solon, OH). Subsequent processing of these samples was performed in the same manner described for sewage samples. Extraction blanks were performed by adding 800 μl molecular-grade water to respective bead mill microcentrifuge tubes instead of fecal slurry and were included in each extraction preparation to monitor potential extraneous DNA contamination. Total DNA extraction yields were measured using a Quant-iT PicoGreen double-stranded DNA assay kit (Life Technologies, Carlsbad, CA) on a SpectraMax Paradigm multimode microplate detection platform (Molecular Devices, Sunnyvale, CA) according to the manufacturers' instructions. DNA extractions yielding less than 300 ng total DNA were discarded. Individual fecal DNA extracts were combined into composites for each animal source and population to estimate the concentration of HF183 genetic markers. Samples from a particular animal group were used to generate composites; an identical mass of total DNA from each extract was mixed to allow equivalent representation of each individual (0.5 ng μl−1 individual−1). Sewage DNA extracts were also prepared at a fixed amount (0.4 ng total DNA per reaction mixture) to standardize measurements. All individual DNA extracts and composites were stored at −20°C in GeneMate Slick low-adhesion microcentrifuge tubes (ISC BioExpress, Kaysville, UT) until the time of analysis (<3 months of storage time).
Oligonucleotides and plasmid DNA standards.
Primers, probes, and plasmid DNA constructs are reported in Table 1, and an alignment of HF183 assay oligonucleotides with a B. dorei reference sequence is provided in Fig. 1. The multiplex HF183/BFDrev and simplex Sketa22 qPCR methods are reported elsewhere (3). A new reverse primer (BacR287) and probes (BacP234MGB and BacP234IAC) were designed to develop the multiplex HF183/BacR287 qPCR assay. The BacR287 primer and BacP234MGB probe were identified on the basis of an alignment of Bacteroidetes 16S rRNA gene sequences with 100% similarity to the HF183 primer identified with the TestPrime (version 1.0) application (27) using the RefNR SILVA small subunit (SSU) rRNA gene database (release 115) (27, 28). Regions of similarity were identified by visual inspection with adjustment to optimal lengths and TaqMan chemistry with Primer Express (version 3.0) software (Life Technologies). The priming efficiency for BacR287 was estimated for each presumptive Bacteroides 16S rRNA gene target with the Oligo (version 6.71) program (Molecular Biology Insights, Cascade, CO) as previously described (3). The BacP234IAC probe sequence was generated via a rearrangement of the BacP234MGB probe sequence that continued to meet Primer Express (version 3.0) guidelines. A single plasmid DNA construct was developed to function as a reference standard for generation of both the HF183/BFDrev and HF183/BacR287 qPCR assay calibration curves (Integrated DNA Technologies, Coralville, IA). The same DNA construct was used for both HF183 qPCR assays to eliminate variability between methods arising from different reference DNA standard preparations and storage conditions. Competitive IAC constructs with sequences identical to those used for the respective reference DNA standards were generated for each HF183 qPCR assay, except that the probe hybridization region was replaced by the IAC probe sequence (Integrated DNA Technologies).
TABLE 1.
Primers, probes, and plasmid target constructs
| Oligonucleotide | Sequence (5′ → 3′) | Reference |
|---|---|---|
| HF183 | ATCATGAGTTCACATGTCCG | 2 |
| BFDRev | CGTAGGAGTTTGGACCGTGT | 3 |
| BFDFam | FAM-CTGAGAGGAAGGTCCCCCACATTGGA-TAMRAa | |
| BacR287 | CTTCCTCTCAGAACCCCTATCC | This study |
| BacP234MGB | FAM-CTAATGGAACGCATCCC-MGB | This study |
| BacP234IAC | VIC-AACACGCCGTTGCTACA-MGB | This study |
| Standard | ATCATGAGTTCACATGTCCGCATGATTAAAGGTATTTTCCGGTAGACGATGGGGATGCGTTCCATTAGCTCGAGATAGTAGGCGGGGTAACGGCCCACCTAGTCAACGATGGATAGGGGTTCTGAGAGGAAGGTCCCCCACATTGGAACTGAGACACGGTCCAAACTCCTACG | This study |
| HF183/BFDrev IAC | TCATGAGTTCACATGTCCGGGATAATTATTAAAGAATTTCGGTTGTCGATGGGGATGCGTTCCATTAGGCAGTTGGCGGGGTAACGGCCCACTCGAGCTACGATGGATAGGGGTTCCTGCCGTCTCGTGCTCCTCACCACATTGGAACTGAGACACGGTCCAAACTCCTACG | This study |
| HF183/BacR287 IAC | ATCATGAGTTCACATGTCCGCATGATTAAAGGTATTTTCCGGTAGACGATGTGTAGCAACGGCGTGTTATAGTAGGCGGGGTAACGGCCCACCTAGTCAACGATGGATAGGGGTTCTGAGAGGAAG | This study |
TAMRA, 6-carboxytetramethylrhodamine.
FIG 1.
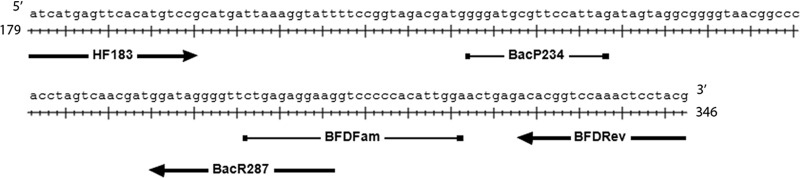
Sites of HF183/BFDrev and HF183/BacR287 oligonucleotide binding to a partial B. dorei 16S rRNA gene sequence (GenBank accession number AB242142; bases 179 to 346). The oligonucleotide with the rightward-pointing arrow indicates the HF183 forward primer. The oligonucleotides with the leftward-pointing arrows represent reverse primers (BFDRev and BacR287). Probe binding sites are indicated by bars (BFDFam and BacP234).
Oligonucleotide annealing predictions.
Primer sequences were used to query the SILVA RefNR SSU rRNA gene database (release 115) (27) by using the Primer Prospector program (29) and allowing zero, one, or two mismatches per oligonucleotide (no mismatches were allowed in the last 4 bp of the 3′ end). Predicted amplicon libraries were generated from matching sequences by trimming flanking regions outside the primer annealing regions. Primer Prospector was then used a second time to query the amplicon libraries for internal matches to each probe sequence, and zero mismatches were allowed. Sequence, alignment, and Pintail program qualities (30), as well as assigned taxonomies for sequences matching all primers and the probe for each assay, were obtained using the SILVA search tool. Sequences with Pintail qualities of less than 75% were assumed to be chimeric and removed from further analysis. Sequence and alignment qualities were greater than 90% for all sequences in the analysis.
TaqMan and SYBR green qPCR amplification.
All qPCR amplifications and assays used the TaqMan chemistry unless it is explicitly stated that SYBR green chemistry was used. Both TaqMan and SYBR green amplifications were performed in a StepOne Plus real-time sequence detector (Life Technologies). The HF183/BFDrev assay was performed as a multiplex reaction, and the Sketa22 qPCR assay was run in simplex format (sewage and ambient water samples only) for use as a sample processing control (SPC), as previously described (3), except that TaqMan environmental master mix (version 2.0) was substituted for TaqMan fast mix PCR master mix (Life Technologies). For the HF183/BacR287 qPCR assay, the multiplex reaction mixtures contained 1× TaqMan environmental master mix (version 2.0), 0.2 mg/ml bovine serum albumin (Sigma-Aldrich Corp., St. Louis, MO), 1 μM each primer, 80 nM 6-carboxyfluorescein (FAM)-labeled probe, and 80 nM VIC-labeled probe. Multiplex reaction mixtures for both HF183 qPCR assays contained 10 to 1 × 105 copies of the IAC template combined with either 1 × 10−2 to 1 ng total DNA from sewage, 1 × 10−2 to 5 ng total DNA from fecal samples, or 10 to 1 × 105 target gene copies of reference plasmid DNA in a total reaction volume of 25 μl. The SYBR green reactions were performed in the same manner as the TaqMan reactions, except that the probe was excluded to prevent fluorescence interference with SYBR green dye and 0.1× SYBR green I dye (Life Technologies) was added to each reaction mixture. Fresh SYBR green dye was prepared daily. All reactions were performed in triplicate or more times in MicroAmp optical 96-well reaction plates with MicroAmp 96-well optical adhesive film (Life Technologies). Data were initially viewed with Sequence Detector software (version 2.3), and quantification cycle (Cq) values (0.03 threshold for all assays) were exported to Microsoft Excel.
Characterization of PD.
To characterize potential HF183/BFDrev and HF183/BacR287 PD or other spurious amplification products, SYBR green qPCR assays were performed with reaction mixtures containing 1 × 10−2 to 1 ng total DNA template from the human fecal DNA composite. Melt curve analysis of these reaction mixtures with a resolution of 0.3°C was used after thermal cycling to identify the melting temperature and intensity of spurious PCR products generated due to a lack of specificity and/or PD. Amplification plateau height fluorescence data (TaqMan reagents only) and maximum melt peak height data (SYBR green) were documented for the respective amplification conditions. Human fecal DNA composite dilution amplification products not containing SYBR green were visualized by electrophoresis with 2.0% GeneMate LE quick-dissolve agarose gels (ISC BioExpress) in 1× lithium borate buffer, and the results were recorded with a Kodak Gel Logic 100 digital imaging system (Carestream Health, Inc., Rochester, NY).
Optimization of multiplex qPCR for monitoring amplification interference.
Four multiplex calibration curves were generated for each HF183 assay to characterize the IAC range of quantification (ROQ) at five different IAC spike levels (10, 102, 103, 104, and 105 copies per reaction mixture). The IAC ROQ was defined as the range of plasmid reference DNA standard concentrations where there was less than a +1 Cq shift from the IAC mean Cq value at 10 copies (n = 12). Simple linear regression was used to assess the influence that IAC spike levels had on the reference DNA standard calibration curve ROQ (range of plasmid DNA standard concentrations where the calibration model was linear), amplification efficiency {E; equal to [10(−1/slope)] − 1}, slope, and y-intercept values.
Monitoring for extraneous DNA and amplification interference.
To monitor for false-positive results due to potential contaminants, no-template and extraction blanks were included, in which laboratory-grade water was substituted for test samples for each instrument run and sample batch. Substances inhibitory to qPCR amplification can persist after DNA purification. Therefore, a fixed amount of IAC plasmid constructs was spiked into each qPCR mixture (102 or 103 copies per reaction mixture) to evaluate the suitability of the isolated DNA for amplification. If amplification interference was present, confirmation of inhibition was achieved by comparison to a competition threshold. The competition threshold is the Cq value where the upper bound of the IAC ROQ intersects the respective multiplexed master calibration curve (31). Thus, evidence of inhibition occurred when the IAC Cq for a given test sample failed the interference criterion and the respective HF183 FAM test sample Cq did not exceed the competition threshold.
Calculations and statistics.
Master calibration curves and test sample concentration estimates were determined using a Monte Carlo Markov chain approach (32). Bayesian calculations were performed using the publicly available software WinBUGS (version 1.4.1). Linearity (R2), amplification efficiency (E), ROQ, precision (percent coefficient of variation [CV] of Cq values across all reference DNA standard concentrations), and the smallest amount of the log10 number of copies of the reference DNA standard per reaction mixture where at least 95% of replicates were detected (LOD95) were calculated from six independently generated calibration curves. Analysis of covariance (ANCOVA) and simple linear regression were used to compare data both within and between HF183/BFDrev and HF183/BacR287 assays. All reported statistical analyses were performed with SAS software (version 9.2; SAS Institute Inc., Cary, NC).
RESULTS
Extraneous DNA controls.
Quality assurance controls indicated the absence of contaminant HF183 markers in 100% of all amplifications (n = 464 reactions).
PD comparisons.
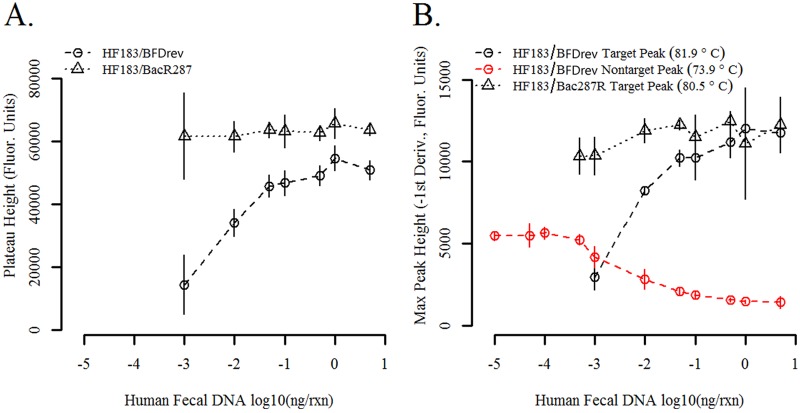
Characterization of amplification products indicated clear differences between the multiplex HF183/BFDrev and HF183/BacR287 assays: HF183/BFDrev amplifications consistently generated a primer dimerization (PD) product (∼50 bp), and HF183/BacR287 did not (Fig. 2). The amplification plateau height increased as human DNA template was added with the HF183/BFDRev assay but not with the HF183/BacR287 assay (Fig. 3A). A similar trend was observed in the SYBR green experiment: maximum melt peak heights increased as a function of the initial human DNA template concentration with HF183/BFDRev but not with HF183/BacR287 (Fig. 3B). In addition, the HF183/BFDrev reactions yielded a second, PD amplification product; primer dimer maximum peak heights appeared to be inversely proportional to the amount of DNA template in the reaction mixture and to target peak values (Fig. 3B).
FIG 2.
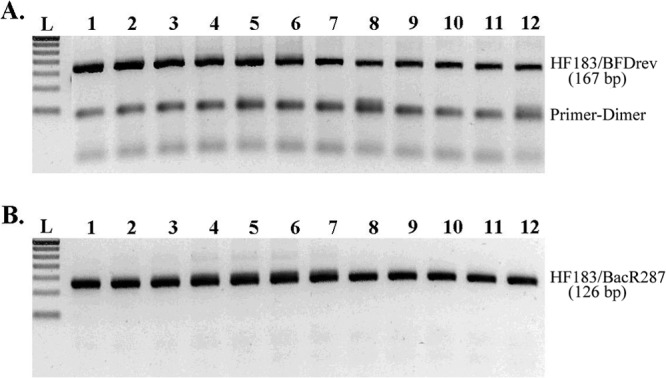
Gel electrophoresis of TaqMan qPCR products showing PD products present in HF183/BFDrev products (A) and absent in HF183/BacR287 amplification products (B). Lanes 1 to 11, amplicons generated from 5 to 1 × 105 ng per reaction mixture of human composite DNA template; lanes 12, no-template control (the DNA target product was visible due to amplification of the IAC spike); lanes L, molecular size ladder. The lowest band on the molecular ladder indicates 50 bp.
FIG 3.
Reduction of amplification performance at low DNA template concentrations due to the formation of spurious PD products in HF183/BFDrev products but not in HF183/BacR287 products. (A) Reduction of the amplification plateau height for the HF183/BFDrev TaqMan assay but not for the HF183/BacR287 TaqMan assay as the concentration of DNA template decreases. (B) SYBR green maximum peak heights at different human fecal DNA composite template concentrations indicating the formation of low-melting-temperature products at low template concentrations with HF183/BFDrev but not HF183/BacR287. Error bars represent 2 standard deviations from the mean (n = 4). Amplification curves that did not fit a 7-parameter sigmoidal model with an R2 value of ≥0.999, which was typically the result of no or poor amplification, were omitted from the analysis. rxn, reaction mixture; −1st Deriv., negative first derivative.
Comparison of calibration model performance.
Master calibration curve equations, associated performance metrics, and competition threshold values are reported in Table 2. HF183 assays indicated R2 values of ≥0.995, E values of ≥0.993, and ROQ values spanning 1 to 5 log10 copies of the reference DNA standard (the entire range tested in this study). LOD95 was 10-fold lower for the HF183/BacR287 assay. ANCOVA indicated a significant difference in the y-intercept values (P < 0.0001) but no difference in slopes (P = 0.1042) between the two HF183 TaqMan assays. Precision averaged a 1.0% lower CV in HF183/BacR287 than HF183/BFDrev (data not shown).
TABLE 2.
Calibration model performance parametersa
| Assay | Slope | y intercept | R2 | E | ROQ | CV (%) |
LOD95 (no. of copies) | |
|---|---|---|---|---|---|---|---|---|
| 10 copies | 100 copies | |||||||
| HF183/BFDrev | −3.279 | 36.07 | 0.995 | 1.018 | 1 to 5 | 3.56 | 2.59 | 100 |
| HF183/BacR287 | −3.338 | 36.74 | 0.996 | 0.993 | 1 to 5 | 2.45 | 1.19 | 10 |
R2, coefficient of determination; E, amplification efficiency, calculated as [10(−1/slope)] − 1; ROQ, range of quantification, based on the log10 number of copies per reaction mixture; CV, coefficient of variation (results for 10 copies and 100 copies of the plasmid standard calculated from 34 to 36 replicate test reactions are shown); LOD95, log10 number of copies of the target per reaction mixture that is detected in at least 95% of replicates (n = 36).
Comparison of serial dilutions of two common human fecal pollution sources.
To evaluate the ability of each HF183 assay to detect low concentrations of human fecal pollution sources, parallel experiments were conducted with serial dilutions of the composite human fecal DNA extracts and sewage DNA extracts (Table 3). Linearity (R2) and LOD95 values were very similar between assays with human fecal DNA but clearly different with sewage template DNA. Values from a 100-copy IAC spike for each HF183 assay indicated the absence of inhibition in all fecal composite DNA extracts.
TABLE 3.
Comparison of results obtained with serial dilutions of two human fecal pollution sourcesa
| DNA target type | Assay | Slope | y intercept | R2 | E | LOD95 (pg) |
|---|---|---|---|---|---|---|
| Human fecal DNA | HF183/BFDrev | −3.415 | 24.5 | 0.993 | 0.992 | 1 |
| HF183/BacR287 | −3.432 | 24.6 | 0.992 | 0.956 | 1 | |
| Sewage DNA | HF183/BFDrev | −3.473 | 26.2 | 0.941 | 0.941 | 50 |
| HF183/BacR287 | −3.455 | 26.2 | 0.997 | 0.947 | 5 |
R2, coefficient of determination; E, amplification efficiency, calculated as [10(−1/slope)] − 1; LOD95, mass of DNA per reaction mixture that is detected in 95% of replicates (n = 8 for both human fecal DNA and sewage DNA).
Performance of custom IAC multiplex qPCR.
Calibration models and associated performance metrics for HF183 assays multiplexed with different IAC spike concentrations are reported in Table 4. In both assays, R2 values were ≥0.987. However, E values ranged from 1.03 to 0.71 for HF183/BFDrev and 0.99 to 0.86 for HF183/BacR287. ANCOVA comparing 10, 102, and 103 IAC spike levels per reaction mixture by assay (n = 87) indicated that both slope and y-intercept values significantly shifted for HF183/BFDrev (P < 0.003) but did not change for HF183/BacR287 (P > 0.0884).
TABLE 4.
Comparison of HF183/BFDrev and HF183/BacR287 multiplex assay performancea
| IAC spike | HF183/BFDrev assay |
HF183/BacR287 assay |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calibration equation | R2 | E | ROQ | IAC ROQ | CT | Calibration equation | R2 | E | ROQ | IAC ROQ | CT | |
| 10 | y = −3.29x + 36.4 | 0.998 | 1.01 | 1 to 5 | 1 to 2 | 29.85 | y = −3.33 x + 36.5 | 0.999 | 0.99 | 1 to 5 | 1 to 2 | 29.80 |
| 102 | y = −3.26 x + 36.2 | 0.995 | 1.03 | 1 to 5 | 1 to 3 | 26.47 | y = −3.40 x + 36.8 | 0.992 | 0.97 | 1 to 5 | 1 to 3 | 26.58 |
| 103 | y = −3.47 x + 37.0 | 0.993 | 0.94 | 1 to 5 | 1 to 4 | 23.13 | y = −3.58 x + 37.5 | 0.987 | 0.90 | 1 to 5 | 1 to 4 | 23.17 |
| 104 | y = −3.50 x + 37.2 | 0.997 | 0.93 | 2 to 5 | 1 to 5 | 19.74 | y = −3.44 x + 36.9 | 0.996 | 0.95 | 2 to 5 | 1 to 5 | 19.76 |
| 105 | y = −4.28 x + 40.6 | 0.987 | 0.71 | 2 to 5 | 1 to 5 | 19.74 | y = −3.71 x + 38.2 | 0.988 | 0.86 | 3 to 5 | 1 to 5 | 19.76 |
IAC spike, number copies of the internal amplification control in each qPCR; R2, coefficient of determination; E, amplification efficiency, calculated as [10(−1/slope)] − 1; ROQ, range of quantification, based on the standard log10 number of copies per reaction mixture; IAC ROQ, internal amplification control range of concentration, reported as the standard log10 number of copies per reaction mixture; CT, competition threshold.
Primer and probe hybridization predictions.
In silico prediction of the hybridization of HF183 primers and probes to all 16S rRNA reference gene sequences available in the SILVA RefNR database (release 115) (27) is provided in Table 5. Minor differences in the targeted diversity between the assays were predicted. Regardless of the mismatch allowances tested, the HF183/BacR287 oligonucleotides were predicted to hybridize to a slightly larger number of Bacteroidetes, Bacteroides, and uncultivated Bacteroides sequences than the HF183/BFDrev oligonucleotides. Sequences predicted to amplify under any condition were originally obtained from human sources, except for four sequences obtained from mouse cecal contents (GenBank accession numbers GQ491336, GQ492074, GQ493896, and GQ493974), which were predicted targets of both assays under all tested mismatch conditions. Sixty-four sequences (28.8%) were removed from the data set due to Pintail scores of <75% (data not shown) (30).
TABLE 5.
In silico prediction of HF183/BFDrev and HF183/BacR287 assay amplification by querying the SILVA 16S rRNA gene reference databasea
| Taxonomy | No. of sequencesb |
||||||
|---|---|---|---|---|---|---|---|
| HF183/BacR287 assay |
HF183/BFDrev assay |
Total | |||||
| 0 MM | 1 MM | 2 MM | 0 MM | 1 MM | 2 MM | ||
| Bacteroidetes | 158 | 167 | 167 | 156 | 165 | 166 | 40,923 |
| Bacteroides | 158 | 167 | 167 | 156 | 165 | 166 | 5,396 |
| B. dorei | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Uncultivated Bacteroides | 155 | 164 | 164 | 153 | 162 | 163 | —c |
The SILVA 16S rRNA gene reference database (RefNR, release 115) was queried with the Primer Prospector program.
The number of sequences within each taxonomy that are predicted to be amplified and detected under each mismatch condition. Total, the number of sequences in the database for each category; MM, number of mismatches (none were allowed in the last 4 bp of the 3′ ends of the primers).
—, the number of uncultivated Bacteroides sequences in the database could not be easily identified because of the incomplete annotation of many sequences.
Distribution of genetic targets in target and nontarget pollution sources.
The abundance of HF183 genetic markers in human and nontarget fecal sources is reported in Table 6. Despite differences in PD formation and predicted hybridization targets, the abundance of HF183 genetic markers in human and nonhuman fecal composites was almost identical between assays. Low marker concentrations were observed in one chicken and turkey population, both of which originated from the same agricultural facility. Cq values from a 100-copy IAC spike for each HF183 assay indicated the absence of inhibition in all fecal composite DNA extracts (data not shown).
TABLE 6.
Estimated log10 concentration of genetic target in reference fecal source composites
| Animal | Localitya | HF183/BFDrev assayb |
HF183/BacR287 assay |
||
|---|---|---|---|---|---|
| n | Mean log10 concn ± SD | n | Mean log10 concn ± SD | ||
| Chicken | CA (6) | ||||
| KY (9) | 4 | 0.89 ± 0.017 | 4 | 0.95 ± 0.018 | |
| Turkey | KY (8) | 4 | 1.26 ± 0.021 | 4 | 1.19 ± 0.020 |
| Dog | CA (8) | ||||
| OH (10) | |||||
| FL (9) | |||||
| WY (10) | |||||
| Cat | OH (14) | ||||
| Deer | FL (8) | ||||
| Pronghorn | WY (10) | ||||
| Swine | GA (9) | ||||
| Cow | OH (12) | ||||
| WY (10) | |||||
| Humanc | OH (6) | 4 | 4.71 ± 0.023 | 4 | 4.79 ± 0.024 |
Values in parentheses refer to the number of individual reference fecal samples used to prepare respective composites.
n, number of replicate measurements used to estimate target concentrations; mean, estimated log10 number of copies per 1 ng of total DNA from fecal sample; standard deviations are for four qPCR replicates.
Human fecal sample composite, not sewage DNA.
Fifty-eight untreated sewage samples were used to estimate the distribution of HF183 genetic markers in human populations. Both assays detected genetic markers in all sewage samples (data not shown). Simple linear regression of the estimated log10 number of copies per reaction mixture for each assay suggested a high degree of correlation (R2 = 0.91), even though 89.3% of the sewage samples yielded higher genetic marker concentration estimates for the HF183/BacR287 assay. A two-way analysis of variance comparison of sewage data indicated a significant difference (P < 0.001) between TaqMan HF183 assay genetic marker concentration estimates, even though the average difference was minimal (∼0.5 Cq).
Differential amplification interference trends in reference sewage samples.
A distinct trend emerged during the comparison of IAC spike Cq values from untreated sewage DNA extracts. For both HF183 assays, a 103 IAC spike was used to monitor for amplification interference in sewage DNA extracts (n = 58). A 103 IAC spike was suitable on the basis of comparisons of IAC ROQ values (Table 4) and no significant difference in calibration curve slope and y-intercept parameters (P ≥ 0.132). The amplification interference and competition thresholds for HF183/BacR287 were 28.5 Cqs and 22.7 Cqs, respectively. Evaluation of HF183/BacR287 data indicated the absence of amplification interference in all sewage DNA extracts. However, 30 samples (51.7%) exhibited inhibition when tested with the HF183/BFDrev assay (interference threshold = 28.7 Cqs; competition threshold = 23.4 Cqs).
DISCUSSION
Influence of PD on HF183 assay performance.
Characterization of potential spurious PCR products generated by the HF183/BFDrev and HF183/BacR287 assays illustrates the effects that PD can have on TaqMan chemistry qPCR assay performance. PD can occur when a short region of complementary bases is shared between oligonucleotides in the same amplification reaction, resulting in the accumulation of a second product that can compete for limited reaction reagents. Our data suggest that the generation of PD molecules and HF183/BFDrev target amplicons is interrelated and dependent on initial target template concentrations. PD formation was most prevalent when initial target template concentrations were low (−5 to −3 log10 ng/reaction mixture of human fecal DNA) and then gradually decreased as initial template quantities increased. This trend suggests that the availability of primers and perhaps other regents influences PD formation in the absence of high initial DNA template concentrations (>−1 log10 ng/reaction mixture of human fecal DNA) where reagents are readily available for PD formation, but as initial template concentrations increase, PD formation becomes limited. The addition of sequence-specific probes, such as those used in TaqMan chemistry, can minimize the contribution of PD molecules to the overall fluorescence measurement used for a quantification estimate but does not prevent the accumulation of PD molecules from influencing the availability of limiting reagents. As a result, the presence of PD molecules reduced the precision (percent CV) and analytical sensitivity (LOD95) in HF183/BFDrev experiments and could potentially interfere with the measurement of IAC multiplex products. Thus, the elimination of PD molecule formation with the HF183/BacR287 assay represents an important advantage over the widely used HF183/BFDrev qPCR assay.
This study evaluated performance only within the qPCR ROQ for these assays (10 to 106 copies per reaction mixture). It is unclear how other quantification strategies that rely on the detection of smaller initial template quantities (<10 copies per reaction mixture), such as most probable number PCR and digital PCR, will perform with the HF183/BFDrev assay. It is likely that the deleterious effects of PD molecules generated from the HF183/BFDrev assay will be greater when analyzing data from samples with extremely low levels of HF183 template DNA. The use of HF183/BFDrev with most probable number PCR, digital PCR, and high numbers of thermal cycle applications (>40 cycles) may suffer from the systematic underestimation of test sample template detection if PD formation persists with these strategies. Additional research is necessary to confirm the suitability of the HF183/BFDrev assay and the predicted advantage of the new HF183/BacR287 method when target sequences are at such low levels.
Assays have similar marker distributions among hosts.
To ensure that modifications to the HF183/BFDrev assay did not have detrimental effects on the specificity of the assay, three strategies were used to evaluate the distribution of HF183 genetic markers in fecal and other samples. In silico predictions suggested that oligonucleotides from both assays hybridize to a similar, but not identical, group of Bacteroidetes 16S rRNA gene sequences. These results should be interpreted cautiously because in silico predictions are highly dependent on the quality of database sequences and may be sensitive to biased representation from particular sources (e.g., human and murine microbiota). Despite these limitations, testing of fecal and sewage DNAs supported in silico predictions well: both assays provided similar estimates of marker concentrations in human, chicken, and turkey fecal DNA and only a small difference in marker concentrations in sewage DNA extracts. Considering the narrow range of diversity targeted by HF183 compared to that targeted by either reverse primer, these results are not unexpected.
New tools to combat amplification inhibition.
The potential for amplification inhibition is a significant challenge when using qPCR to quantify genetic markers isolated from environmental samples (33). Since the presence of inhibitors can decrease the accuracy and precision of qPCR measurements and in extreme circumstances can lead to false-negative results (34), the inclusion of an inhibition control with each test sample helps ensure the integrity of the findings and promote consistency between laboratories (25). Because different qPCR assays may not have equal susceptibilities to inhibitors (23, 35), our strategy was to mimic the reaction conditions of the IAC so that they were as close as possible to those of the target amplification. This competitive amplification strategy utilizes a composite primer technique (36) where the target and the IAC are amplified with a common set of primers and under the same conditions in the same reaction. In this strategy, it is important to identify the IAC spike concentration range where multiplex amplifications are reproducible because simultaneous amplification of the target and IAC can result in inhibition or facilitation, depending on the molar ratio, the amplicon length, and/or the sequence secondary structure of each fragment (37, 38). In some instances, a competitive IAC strategy does not work (33); however, careful testing of HF183 multiplex qPCR assays in this study indicated that IAC spikes do not always significantly change calibration model parameters (P > 0.05) over a broad range of initial DNA template concentrations (Table 4), making the IAC competitive strategy ideal for fecal source identification applications.
When HF183/BFDrev and HF183/BacR287 IAC amplification trends were compared, we found that the HF183/BFDrev assay gave significantly different (P < 0.05) estimates of target DNA concentration in test samples when IAC spike levels (10, 102, or 103 copies per reaction mixture) were altered, suggesting that factors such as the presence of PD molecules and/or amplicon length (167 bp) influence performance. In contrast, the HF183/BacR287 assay generated similar results with different IAC spike levels, further supporting the notion that the presence of PD molecules and/or a shorter amplicon length (126 bp) may play an important role in the accurate measurement of an IAC spike. The ability to use different IAC spike levels with the HF183/BacR287 assay will provide researchers with more flexibility, allowing the optimization of experiments on the basis of the anticipated target DNA concentrations in environmental samples.
Recommendations for HF183/BacR287 application.
A systematic comparison of two HF183 qPCR assays provided important insights regarding the application of these methods to characterize human fecal pollution in ambient surface water samples. Comparisons of PD formation, calibration curve parameters, LOD95, precision (percent CV), multiplexed IAC performance, and the DNA target distribution in untreated sewage samples all showed that the new HF183/BacR287 assay is superior to the HF183/BFDrev assay. The changes in the reverse primer and probe for the HF183/BacR287 assay do not appear to have altered the occurrence or magnitude of cross-reactivity when the assay was tested against nonhuman fecal DNA composites reported to elicit false-positive results with other HF183 protocols. However, specificity should still be confirmed by testing reference fecal pollution source material collected directly from the local watershed of interest before application. Depending on which fecal sources are present and the degree of specificity observed, other human-associated methods, such as those involving HumM2 (39) or gyrB (40), may be used in place of or in addition to HF183/BacR287. The addition of a second assay may decrease the likelihood of overestimating the impacts of human contaminants in areas where dogs, chickens, turkeys, or perhaps untested fecal sources that can elicit a false-positive result are thought to contribute to the overall fecal load in a water body.
Although the HF183/BacR287 assay can be used with different IAC spike concentrations, it is desirable to use the lowest possible spike level that allows accurate measurement of the IAC at the target DNA concentrations anticipated in ambient surface water test samples. Although we did not directly test the performance of either assay in the presence of PCR inhibitors, the incorporation of the TaqMan environmental master mix will likely reduce the incidence of amplification inhibition in ambient water samples, as was observed previously with enterococcal qPCR protocols (24). Other considerations are the source, preparation, and handling of the reference DNA material used to generate calibration models or IAC spike controls. Previous studies have emphasized the importance of a centralized source for all reference materials in order to achieve high levels of reproducibility across laboratories (22, 41). Thus, for a group of laboratories that use the HF183/BacR287 protocol and routinely compare results, a single, centralized source for all reference DNA materials is ideal.
The HF183/BacR287 protocol reported here was specifically designed and optimized for the characterization of HF183 16S rRNA gene cluster DNA targets from ambient surface water samples at volumes of ≤100 ml. It remains unclear how assay performance will be influenced by changes in protocol, such as filtration of larger volumes of water, testing of higher concentrations of DNA template, use of different thermal cycling instrumentation, interpretation of Cq values beyond the defined lower limit of quantification (10 copies per reaction mixture), changing of qPCR reagent brands, running of reactions as a simplex rather than a multiplex format, as well as testing of other environmental matrices (i.e., soils, groundwater, etc.) besides ambient surface waters, sewage, and fecal samples. It is likely that the HF183/BacR287 assay will perform adequately under other conditions, but additional work will be required to confirm compatibility.
ACKNOWLEDGMENTS
Information has been subjected to U.S. EPA peer and administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the U.S. EPA. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print 7 March 2014
REFERENCES
- 1.Bernhard AE, Field KG. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587–1594. 10.1128/AEM.66.4.1587-1594.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571–4574. 10.1128/AEM.66.10.4571-4574.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haugland RA, Varma M, Kelty CA, Peed L, Sivaganesan M, Shanks OC. 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by real-time PCR. Syst. Appl. Microbiol. 33:348–357. 10.1016/j.syapm.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 4.Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701–3715. 10.1016/j.watres.2007.06.037 [DOI] [PubMed] [Google Scholar]
- 5.Layton A, McKay L, Williams D, Garrett V, Gentry R, Sayler G. 2006. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72:4214–4224. 10.1128/AEM.01036-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okabe S, Okayama N, Savichtcheva O, Ito T. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890–901. 10.1007/s00253-006-0714-x [DOI] [PubMed] [Google Scholar]
- 7.Reischer GH, Kasper DC, Steinborn R, Farnleitner AH, Mach RL. 2007. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area. Lett. Appl. Microbiol. 44:351–356. 10.1111/j.1472-765X.2006.02094.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seurinck S, Defoirdt T, Verstraete W, Siciliano S. 2005. Detection and quantification of human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol. 7:249–259. 10.1111/j.1462-2920.2004.00702.x [DOI] [PubMed] [Google Scholar]
- 9.Lee D-Y, Wier SC, Trevors JT. 2010. Quantitative identification of fecal water pollution sources by TaqMan real-time PCR assays using Bacteroidales 16S rRNA genetic markers. Appl. Microbiol. Biotechnol. 88:1373–1383. 10.1007/s00253-010-2880-0 [DOI] [PubMed] [Google Scholar]
- 10.Ahmed W, Kirs M, Gilpin B. 2011. Source tracking in Australia and New Zealand: case studies, p 485–513 In Hagedorn C, Blanch AR, Harwood VJ. (ed), Microbial source tracking: methods, applications, and case studies. Springer, New York, NY [Google Scholar]
- 11.Ahmed W, Stewart JR, Powell D, Gardner T. 2008. Evaluation of Bacteroides markers for the detection of human faecal pollution. Lett. Appl. Microbiol. 46:237–242. 10.1111/j.1472-765X.2007.02287.x [DOI] [PubMed] [Google Scholar]
- 12.Ahmed W, Yusuf R, Hasan I, Goonetilleke A, Gardner T. 2010. Quantitative PCR assay of sewage-associated Bacteroides markers to assess sewage pollution in an urban lake in Dhaka, Bangladesh. Can. J. Microbiol. 56:838–845. 10.1139/W10-070 [DOI] [PubMed] [Google Scholar]
- 13.Gawler A, Beecher JE, Brandoa J, Carroll NM, Falcao L, Gourmelon M, Masterson B, Nunes B, Porter J, Rince A, Rodrigues R, Thorp M, Walters JM, Meijer WG. 2007. Validation of host-specific Bacteroidales 16S rRNA genes as markers to determine the origin of faecal pollution in Atlantic Rim countries of the European Union. Water Res. 41:3780–3784. 10.1016/j.watres.2007.01.028 [DOI] [PubMed] [Google Scholar]
- 14.Jenkins MW, Tiwari S, Lorente M, Gichaba CM, Wuertz S. 2009. Identifying human and livestock sources of fecal contamination in Kenya with host-specific Bacteroidales assays. Water Res. 43:4956–4966. 10.1016/j.watres.2009.07.028 [DOI] [PubMed] [Google Scholar]
- 15.Shanks OC, Nietch C, Simonich MT, Younger M, Reynolds D, Field KG. 2006. Basin-wide analysis of the dynamics of fecal contamination and fecal source identification in Tillamook Bay, Oregon. Appl. Environ. Microbiol. 72:5537–5546. 10.1128/AEM.03059-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green H, Shanks OC, Sivaganesan M, Haugland R, Field K. 2011. Differential decay of human faecal Bacteroides in marine and freshwater. Environ. Microbiol. 13:3235–3249. 10.1111/j.1462-2920.2011.02549.x [DOI] [PubMed] [Google Scholar]
- 17.Haugland RA, Siefring S, Lavender J, Varma M. 2012. Influences of sample interference and interference controls on quantification of enterococci fecal indicator bacteria in surface water samples by the qPCR method. Water Res. 46:5989–6001. 10.1016/j.watres.2012.08.017 [DOI] [PubMed] [Google Scholar]
- 18.Boehm AB, Van De Werfhorst L, Griffith JF, Holden P, Jay J, Shanks OC, Wang D, Weisberg SB. 2013. Performance of forty-one microbial source tracking methods: a twenty-seven lab evaluation study. Water Res. 47:6812–6828. 10.1016/j.watres.2012.12.046 [DOI] [PubMed] [Google Scholar]
- 19.Shanks OC, White K, Kelty CA, Sivaganesan M, Blannon J, Meckes M, Varma M, Haugland RA. 2010. Performance of PCR-based assays targeting Bacteroidales genetic markers of human fecal pollution in sewage and fecal samples. Environ. Sci. Technol. 44:6281–6288. 10.1021/es100311n [DOI] [PubMed] [Google Scholar]
- 20.Layton B, Cao Y, Ebentier DL, Hanley KT, Van De Werfhorst L, Wang D, Madi T, Whitman RL, Byappanahalli MN, Balleste E, Meijier W, Schriewer A, Wuertz S, Converse RR, Noble RT, Srinivasan S, Rose JB, Lee CS, Lee J, Shields J, Stewart JR, Reischer G, Farnleitner A, Sinigalliano CD, Rodrigues R, Lozach S, Gourmelon M, Peed L, Shanks OC, Jay J, Holden P, Boehm AB, Griffith JF. 2013. Performance of human fecal anaerobe-associated PCR-based assays in a multi-laboratory method evaluation study. Water Res. 47:6897–6908. 10.1016/j.watres.2013.05.060 [DOI] [PubMed] [Google Scholar]
- 21.Staley C, Gordon KV, Schoen ME, Harwood VJ. 2012. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl. Environ. Microbiol. 78:7317–7326. 10.1128/AEM.01430-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebentier DL, Hanley KT, Cao Y, Badgley BD, Boehm AB, Ervin JS, Goodwin KD, Gourmelon M, Griffith JF, Holden PA, Kelty CA, Lozach S, McGee C, Peed LA, Raith M, Ryu H, Sadowsky MJ, Scott EA, Santo Domingo J, Schriewer A, Sinigalliano CD, Shanks OC, Van De Werfhorst LC, Wang D, Wuertz S, Jay JA. 2013. Evaluation of the repeatability and reproducibility of a suite of qPCR-based microbial source tracking methods. Water Res. 47:6839–6848. 10.1016/j.watres.2013.01.060 [DOI] [PubMed] [Google Scholar]
- 23.Green HC, Field KG. 2012. Sensitive detection of sample interference in environmental qPCR. Water Res. 46:3251–3260. 10.1016/j.watres.2012.03.041 [DOI] [PubMed] [Google Scholar]
- 24.Cao Y, Griffith JF, Dorevitch S, Weisberg SB. 2012. Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters. J. Appl. Microbiol. 113:66–75. 10.1111/j.1365-2672.2012.05305.x [DOI] [PubMed] [Google Scholar]
- 25.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 26.Kelty CA, Varma M, Sivaganesan M, Haugland R, Shanks OC. 2012. Distribution of genetic marker concentrations for fecal indicator bacteria in sewage and animal feces. Appl. Environ. Microbiol. 78:4225–4232. 10.1128/AEM.07819-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:188–196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. 2011. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27:1159–1161. 10.1093/bioinformatics/btr087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashelford KE, Khuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736. 10.1128/AEM.71.12.7724-7736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peed LA, Nietch CT, Kelty CA, Meckes M, Mooney T, Sivaganesan M, Shanks OC. 2011. Combining land use information and small stream sampling with PCR-based methods for better characterization of diffuse sources of human fecal pollution. Environ. Sci. Technol. 45:5652–5659. 10.1021/es2003167 [DOI] [PubMed] [Google Scholar]
- 32.Sivaganesan M, Seifring S, Varma M, Haugland RA, Shanks OC. 2008. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinformatics 9:120. 10.1186/1471-2105-9-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatt JK, Ritalahti KM, Ogles DM, Lebrón CA, Löffler FE. 2013. Design and application of an internal amplification control to improve Dehalococcoides mccartyi 16S rRNA gene enumeration by qPCR. Environ. Sci. Technol. 47:11131–11138. 10.1021/es4019817 [DOI] [PubMed] [Google Scholar]
- 34.Gibson KE, Schwab KJ, Spencer SK, Borchardt MA. 2012. Measuring the mitigating inhibition during quantitative real time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res. 46:4281–4291. 10.1016/j.watres.2012.04.030 [DOI] [PubMed] [Google Scholar]
- 35.Huggett J, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, Zumla A. 2008. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognized phenomenon. BMC Res. Notes 28:70. 10.1186/1756-0500-1-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siebert PD, Larrick JW. 1992. Competitive PCR. Nature 359:557–558. 10.1038/359557a0 [DOI] [PubMed] [Google Scholar]
- 37.Hoorfar J, Malorny B, Abdulmawjood A, Cook N, Wagner M, Fach P. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863–1868. 10.1128/JCM.42.5.1863-1868.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki MT, Giovannoni SJ. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA. 2009. Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol. 75:5507–5513. 10.1128/AEM.00305-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee CS, Lee J. 2010. Evaluation of new gyrB-based real-time PCR system for the detection of B. fragilis as an indicator of human-specific fecal contamination. J. Microbiol. Methods 82:311–318. 10.1016/j.mimet.2010.07.012 [DOI] [PubMed] [Google Scholar]
- 41.Shanks OC, Sivaganesan M, Peed L, Kelty CA, Noble RT, Blackwood AD, Bushon RN, Stelzer EA, Kinzelman J, Anan'eva T, Sinigalliano CD, Wanless D, Griffith JF, Cao Y, Weisberg SB, Harwood VJ, Staley C, Oshima KH, Varma M, Haugland R. 2012. Inter-laboratory comparison of real-time PCR methods for quantification of general fecal indicator bacteria. Environ. Sci. Technol. 46:945–953. 10.1021/es2031455 [DOI] [PubMed] [Google Scholar]