Abstract
Vibrio anguillarum is an important pathogen in aquaculture, responsible for the disease vibriosis in many fish and invertebrate species. Disease control by antibiotics is a concern due to potential development and spread of antibiotic resistance. The use of bacteriophages to control the pathogen may offer a non-antibiotic-based approach to reduce vibriosis. A detailed understanding of the phage-host interaction is needed to evaluate the potential of phages to control the pathogen. In this study, we examined the diversity and interactions of 11 vibriophages, 24 V. anguillarum strains, and 13 Vibrio species strains. Together, the host ranges of the 11 phages covered all of the tested 37 Vibrio sp. host strains, which represented considerable temporal (20 years) and geographical (9 countries) differences in their origins of isolation. Thus, despite the occurrence of unique susceptibility patterns of the individual host isolates, key phenotypic properties related to phage susceptibility are distributed worldwide and maintained in the global Vibrio community for decades. The phage susceptibility pattern of the isolates did not show any relation to the physiological relationships obtained from Biolog GN2 profiles, demonstrating that similar phage susceptibility patterns occur across broad phylogenetic and physiological differences in Vibrio strains. Subsequent culture experiments with two phages and two V. anguillarum hosts demonstrated an initial strong lytic potential of the phages. However, rapid regrowth of both phage-resistant and phage-sensitive cells following the initial lysis suggested that several mechanisms of protection against phage infection had developed in the host populations.
INTRODUCTION
Vibriosis is one of the most prevalent and devastating diseases in marine aquaculture, causing substantial mortality and economic losses in both fish and shellfish cultures worldwide (1). The disease is caused by the marine pathogen Vibrio (Listonella) anguillarum, which infects >50 different species of fish and shellfish, with severe implications for the marine fish-rearing industry (1, 2). Vaccines against vibriosis have been developed and have been widely successful in grown fish (3). However, the larval stage is especially vulnerable to V. anguillarum infections, as the immune system is not fully developed and vaccines are not effective. Traditionally, antibiotics have been widely used in antimicrobial prophylaxis and treatment of vibriosis in aquaculture, despite concern about the development and dispersal of antibiotic resistance in the pathogen community. Tetracycline and quinolones are still the first drugs of choice, and multiple cases of antibiotic resistance have been reported for V. anguillarum in aquaculture environments (4, 5). Therefore, there is a need for the development of alternative methods to control and prevent V. anguillarum infection in aquaculture.
Bacteriophages are natural and abundant biological entities in the marine environment, playing important roles in, e.g., structuring bacterial diversity and succession in the ocean and promoting biogeochemical element cycling, and being key drivers of horizontal gene transfer (6). The strong lytic potential of bacteriophages against specific bacterial hosts has led to an increasing interest in the therapeutic use of lytic phages to control pathogenic bacterial infections in aquaculture. There has been a growing number of studies on phage therapy focusing on a variety of fish pathogens, including Aeromonas salmonicida in brook trout, Yersinia ruckeri in salmon, Lactococcus garvieae in yellowtail, Pseudomonas plecoglossicida in ayu, Flavobacterium psychrophilum in rainbow trout, V. harveyi in shrimp, and V. anguillarum in salmon (7–14). These studies have demonstrated the potential of specific phages to significantly control pathogen density and, in some cases, reduce fish mortality. Therefore, there is growing evidence that phages can be applied to control diseases in aquaculture.
The use of bacteriophages to control specific pathogens is complicated by the phenotypic and genotypic complexity of both phage and host communities, which may consist of multiple coexisting strains with different host range and susceptibility patterns (14, 15). In order to establish efficient phage control of a diverse host community, a large and well-characterized collection of phages is required which, in combination, covers a broad range of hosts.
In this study, a collection of Vibrio sp. strains and vibriophages was isolated from Danish aquaculture farms and characterized along with a collection of 20 V. anguillarum strains and one lytic V. anguillarum phage of different temporal and geographic origins. Patterns in phage and host phenotype, genotype, morphology, and phage-host ranges and interactions were studied to establish a collection of well-characterized vibriophages and to evaluate their potential for controlling the pathogen.
MATERIALS AND METHODS
Bacterial strains.
The collection consisted of 20 V. anguillarum strains provided by Technical University of Denmark (DTU, Denmark); V. anguillarum strain PF430-3, derived from strain PF4, which was originally isolated in Chile (16) but was reisolated after long-term (2 weeks) culturing; and 16 Vibrio sp. strains recently isolated from Danish aquaculture farms (Table 1). The 16 recent Danish isolates were obtained from 18 water samples collected from two fish farms (Venøsund and Maximus, Denmark) between May 2012 and August 2012. Aliquots of water samples (200 ml) were mixed with 200 ml 2× LB medium (12106; MO-BIO) and agitated at room temperature for 24 h. For isolation of Vibrio sp. strains, 100-μl subsamples from serial dilutions were plated on thiosulfate citrate bile salt sucrose agar (TCBS; 221872; Difco) incubated at 30°C for 24 h. Single colonies were then isolated and subsequently pure cultured by repeated streaking on TCBS agar. These isolates were then further characterized to determine if they were V. anguillarum (see below). All of the strains were kept at −80°C in LB medium with sterile glycerol (15%, vol/vol).
TABLE 1.
The 37 Vibrio sp. strains used in the study
| Numbera | Strain | Source | BLAST result (accession no.) | Homology (%) | API 20NE code | BIOLOG GN2c | 16S phylogenetic groupd | Reference or source |
|---|---|---|---|---|---|---|---|---|
| 1* | 87-9-117 | Rainbow trout, Finland | Vibrio anguillarum (CP006699.1) | 96 | 7577745 | 1 | 1 | 24, 49 |
| 2* | BA35 | Sockeye salmon, USA | Vibrio anguillarum (CP006699.1) | 95 | 7577745 | 1 | 1 | 49 |
| 3* | 51/82/2 | Rainbow trout, Germany | Vibrio anguillarum (CP006699.1) | 97 | 7577745 | 1 | 1 | 24, 49 |
| 4* | T265 | Salmon, UK | Vibrio anguillarum (CP006699.1) | 97 | 7577745 | 1 | 1 | 24, 49 |
| 5* | 91-8-178 | Turbot, Norway | Vibrio anguillarum (CP006699.1) | 97 | 7577745 | 1 | 1 | 24, 49 |
| 6* | 87-9-116 | Rainbow trout, Finland | Vibrio anguillarum (CP006699.1) | 97 | 7577745 | 1 | 1 | 24, 49 |
| 7* | 601/90 | Sea bass, Italy | Vibrio anguillarum (CP006699.1) | 96 | 7577745 | 1 | 1 | This study |
| 8* | 178/90 | Sea bass, Italy | Vibrio anguillarum (CP006699.1) | 96 | 7577745 | 1 | 1 | 24, 49 |
| 9* | 261/91 | Sea bass, Italy | Vibrio anguillarum (CP006699.1) | 97 | 7577645 | 1 | 1 | 24, 49 |
| 10b | PF430-3 | Salmon, Chile | Vibrio anguillarum (KF150778.1) | 96 | 7477745 | 1 | 1 | 17 |
| 11* | A023 | Turbot, Spain | Vibrio anguillarum (CP006699.1) | 95 | 7577345 | 1 | 1 | 49 |
| 12* | 4299 | Unknown | Vibrio anguillarum (KF150778.1) | 96 | 5576744 | 1 | 1 | 50 |
| 13* | LMG 12010 | Unknown | Vibrio anguillarum (CP006699.1) | 95 | 7577344 | 1 | 1 | 49 |
| 14* | 850610–1/6a | Rainbow trout, Denmark | Vibrio anguillarum (CP006699.1) | 97 | 7577745 | 1 | 1 | 49 |
| 15* | 90-11-287 | Water sample, Denmark | Vibrio anguillarum (CP006699.1) | 96 | 7577745 | 1 | 1 | 24, 51 |
| 16* | 91-7-154 | Trout, Denmark | Vibrio anguillarum (CP006699.1) | 97 | 7577745 | 1 | 1 | 49 |
| 17* | S2 2/9 | Rainbow trout, Denmark | Vibrio anguillarum (FJ824662.1) | 98 | 7577745 | 1 | 1 | 49 |
| 18* | 90-11-286 | Rainbow trout, Denmark | Vibrio anguillarum (KF150778.1) | 97 | 7576745 | 1 | 1 | 49 |
| 19* | HWU53 | Rainbow trout, Denmark | Vibrio anguillarum (CP006699.1) | 97 | 7576745 | 1 | 1 | 49 |
| 20* | 6018/1 | Rainbow trout, Denmark | Vibrio anguillarum (CP006699.1) | 97 | 7576745 | 1 | 1 | 49 |
| 21* | 9014/8 | Rainbow trout, Denmark | Vibrio anguillarum (CP006699.1) | 96 | 7577745 | 1 | 1 | 49 |
| 22 | VA1 | Salmon, Denmark | Vibrio anguillarum (KF150778.1) | 96 | 7577745 | 1 | 1 | This study |
| 23 | VA2 | Salmon, Denmark | Vibrio anguillarum (KF150778.1) | 95 | 7577745 | 1 | 1 | This study |
| 24 | VA3 | Salmon, Denmark | Vibrio anguillarum (KF150778.1) | 96 | 7576745 | 1 | 1 | This study |
| 25 | VSP1 | Salmon, Denmark | Vibrio splendidus (KC884584.1) | 96 | 7430044 | 2 | 2 | This study |
| 26 | VSP2 | Salmon, Denmark | Vibrio cyclitrophicus (KC185413.1) | 97 | 7520044 | 2 | 2 | This study |
| 27 | VSP3 | Salmon, Denmark | Vibrio cyclitrophicus (KF488567.1) | 97 | 7430044 | 2 | 2 | This study |
| 28 | VSP8 | Denmark | Vibrio cyclitrophicus (KF488567.1) | 95 | 7430044 | 2 | 2 | This study |
| 29 | VSP10 | Salmon, Denmark | Vibrio cyclitrophicus (KC185413.1) | 97 | 7430044 | 2 | 2 | This study |
| 30 | VSP12 | Denmark | Vibrio cyclitrophicus (KF488567.1) | 96 | 7430044 | 2 | 2 | This study |
| 31 | VSP4 | Sole, Denmark | Vibrio shilonii (JX999944.1) | 95 | 7420044 | 2 | 3 | This study |
| 32 | VSP5 | Sole, Denmark | Vibrio sp. (EU517619.1) | 97 | 7476744 | 3 | 3 | This study |
| 33 | VSP6 | Denmark | Vibrio sp. (FM957468.1) | 96 | 7476744 | 3 | 3 | This study |
| 34 | VSP7 | Denmark | Vibrio sp. (FM957468.1) | 97 | 7476744 | 3 | 3 | This study |
| 35 | VSP9 | Denmark | Vibrio sp. (KC178920.1) | 97 | 7476744 | 3 | 3 | This study |
| 36 | VSP11 | Denmark | Vibrio sp. (KC178920.1) | 97 | 7476744 | 3 | 3 | This study |
| 37 | VSP13 | Denmark | Vibrio sp. (FM957471.1) | 97 | 7476744 | 3 | 3 | This study |
Strains marked by an asterisk were provided by Technical University of Denmark (DTU).
The strain PF430-3 was provided by the Hellenic Centre for Marine Research.
Group numbers according to the BIOLOG GN2 dendrogram.
Group numbers according to 16S rRNA phylogenetic tree (see Fig. S1 in the supplemental material).
16S rRNA sequencing.
Bacterial genomic DNA was extracted using a commercial genomic DNA purification kit (Clontech). The 16S rRNA gene was amplified by universal primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′). PCR was performed in a 24-μl reaction mixture, which contained genomic DNA template, 10× PCR buffer, 2 mM deoxynucleoside triphosphates (dNTPs), primers (12.5 μM each), MgCl2 (25 mM), hot start polymerase, and water. PCR amplification was carried out on a thermal cycler (Applied Biosystems) for 5 min at 95°C for denaturation, followed by 35 cycles of 45 s at 95°C, 1 min at 51°C, and 2 min at 72°C and final extension at 72°C for 7 min. PCRs were verified using 1% agarose gels stained with ethidium bromide. PCR products were subsequently sequenced in both forward and reverse directions (Beijing Genomic Institute [BGI]).
A phylogenetic analysis based on neighbor joining was performed on 16S rRNA gene sequences. The original sequences were manually corrected, trimmed, and aligned using the BioEdit sequence alignment tool (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Each of the compiled sequences were compared to the available databases using the basic local alignment search tool (BLAST) to determine phylogenetic affiliations. Mega 5.1 software (http://www.megasoftware.net/) was used to align all 37 sequences. Overhanging ends were removed to allow direct comparison of sequences with the same length (695 bp). The phylogenetic relationship was bootstrapped 1,000 times, and the gaps were treated by pairwise deletion.
Phenotypic analysis of bacterial strains.
Biochemical tests were performed using an API 20NE system (bioMérieux) according to the instructions of the manufacturer. The inoculum was distributed into test strips which were then incubated at 30°C for 48 h. Biochemical reactions were read based on color development, translated into numerical codes, and interpreted with the API 20NE database.
The sensitivity of the strains to the vibriostatic compound agent 0129 (2,4-diamino-6,7-diisopropylpteridine phosphate) (Oxoid) was determined by the disk diffusion method (17) at both 10 and 150 μg per disk with incubation for 24 h at 30°C.
Biolog GN2 MicroPlates (Biolog) containing 95 different carbon sources were used to test the ability of the strains to use different carbon sources. Briefly, pure cultures of bacterial strains were grown on LB agar plates for 24 h at 30°C and subsequently transferred to 0.9% saline buffer (9 g NaCl in 1 liter distilled water) and washed twice to remove medium (8,000 × g for 10 min). From this bacterial suspension (ca. 108 CFU ml−1), 100-μl aliquots were inoculated into each well in duplicate plates and incubated in the dark at 30°C for 48 h. Data were analyzed based on visual inspection of tetrazolium violet development as an indicator of metabolic activity.
Bacteriophage isolation.
The water samples for bacterial isolation were also used for propagation of phages for subsequent isolation and purification of new vibriophages. Four different protocols were used (18). (i) A 100-ml water sample was sterile filtered (0.22 μm; Millipore) and mixed with 100 ml 2× LB medium containing a mixture of 5 random Vibrio strains (Table 1) to stimulate proliferation of phages specific to the added strains. After 24 h of incubation at room temperature, bacteria in the lysate were killed by chloroform (20 μl ml−1), followed by centrifugation (10,000 × g, 10 min) to remove bacterial debris. (ii) One-hundred-ml unfiltered water samples were mixed with 100 ml 2× LB broth and incubated at room temperature for 24 h to stimulate proliferation of phages specific to cooccurring Vibrio species host strains in the water sample. Aliquots of enrichment samples (50 ml) were then treated with chloroform and centrifuged as described above, and the supernatant was mixed with individual Vibrio strains (Table 1) in a second round of enrichment as described above. (iii) Five-ml water samples were filtered (0.22 μm pore size), and the filtrate was diluted and used directly for plaque assay (19) without previous enrichment. (iv) Two-hundred-ml water samples were filtered (0.22 μm), and phages were then concentrated using 30-kDa centrifugation filters (Amicon ultra 15 protein; Millipore) and stored at 4°C until further use.
The presence of Vibrio-specific phages in the water samples described above was tested against all 37 Vibrio sp. strains by spotting 5-μl samples on lawns of individual strains using the double-layer agar method (18). In the case of cell lysis in the spotted area, phages were scraped off the plate and transferred to SM buffer (50 mM Tris-Cl, pH 7.5, 99 mM NaCl, 8 mM MgSO4, 0.01% gelatin). These phage concentrates were then diluted, and the presence of phages was verified by plaque assay (19). For purification of individual phages, single plaques were isolated with a sterile pipet and transferred to SM buffer. This procedure was repeated at least three times to ensure purification of individual phages.
To obtain high-titer phage stocks, 5 ml SM buffer was added to plates with confluent lysis, and the top agar was shredded with an inoculation loop. The mixture was transferred to a sterile tube and incubated on a shaker for at least 2 h at room temperature to release phage particles and then centrifuged (10,000 × g, 10 min, 4°C). The supernatant was filtered through a 0.22-μm membrane (Millipore), and the phage stock was stored at 4°C in the dark. In addition to the phage isolates obtained from Danish aquaculture farms, a broad-host-range vibriophage (KVP40) (20) was kindly provided by the Hellenic Centre for Marine Research.
Phage host range and EOP.
The host range of each phage isolate was determined using a spot assay against all of the available Vibrio sp. strains (Table 1). Aliquots of 5 μl phage stock (phage titer between 106 and 109 PFU ml−1) were spotted on lawns of the potential host strain prepared as 4 ml top agar inoculated with 500 μl bacterial cultures in mid-log phase (ca. 108 CFU ml−1). After 24 h of incubation at 30°C, the spots were assessed by the clarity of plaques and scored as transparent, turbid, or no inhibition. To provide a quantitative measure of the phage lytic potential, the efficiency of plating (EOP) for each phage on each phage-sensitive strain was determined by measurement of the number of PFU produced on individual hosts from a given phage stock, as quantified by plaque assay (19).
Purification of bacteriophage genome DNA.
Aliquots (500 μl) of high-titer (ca. 109 PFU ml−1) bacteriophage stocks were further concentrated by centrifugation (1,000 × g, 25 min) in 30-kDa ultrafiltration Amicon Microcon spin devices (Millipore), and then the membrane was washed three times by 50 μl 100× Tris-EDTA buffer (TE buffer; 10 mmol Tris-Cl, 1 mmol EDTA, pH 8) (1,000 × g for 18 min each). After the washing, the phages were eluted with 20 μl 100× TE buffer by centrifugation of the inverted membrane into a new tube (300 × g for 5 min). To denature phage proteins and release the phage DNA, the concentrated phages were heated to 70°C for 10 min and then cooled on ice for subsequent determination of phage genome size. To confirm the concatemer phenomena of a number of phages (phage ΦH2, ΦH8, and ΦH20), additional extraction of DNA from these phages was performed using a QIAprep spin M13 kit (Qiagen) according to the manufacturer's instructions.
Phage genome size.
The bacteriophage genome sizes were determined by pulsed-field gel electrophoresis (PFGE) using the CHEF-DRIII pulsed-field electrophoresis system (Bio-Rad) (14). Phage DNA was run using 1% agarose gels in 0.5× Tris-borate-EDTA (TBE) buffer (10× TBE, 890 mM Tris, 20 mM EDTA, 890 mM boric acid, pH 8.3) with two different instrument settings for small and large genome sizes. For the large genome sizes (>50 kb), switch times were ramped from 50 to 90 s for 24 h and a voltage of 6 V/cm, whereas switch times ranged from 0.5 to 5 s and a total run time of 15 h for small genomes (<50 kb). An 8- to 48-kb molecular size DNA marker and a Lambda DNA ladder (Bio-Rad) were used as standards. The gels were stained with 0.01% (vol/vol) SYBR green I (Invitrogen) in 0.5× TBE for 1 h in the dark and then washed with distilled water for 30 min prior to visualization of the bands using the ChemiDoc MP imaging system (Bio-Rad).
Bacteriophage morphology by TEM.
For transmission electron microscopy (TEM) analysis of phage isolates, the phage stocks were further concentrated by ultracentrifugation (100,000 × g, 90 min, 20°C; 70.1Ti; Beckman), followed by resuspension of the pellet in 100 μl SM buffer. Formvar-carbon-coated copper grids were placed on top of the phage concentrates, allowing the phages to adsorb for 20 min. The grids were then stained with 10 μl 2% sodium phosphotungstate (pH 7.4; 0.02-μm-pore-size filters) for 2 min. Excess stain was removed by touching the edge of the grids with filter paper, and the grids were washed with several drops of distilled water and allowed to dry on the filter paper for 15 min. The grids were observed with a JEM-2100 transmission electron microscope (JEOL) operated at 80 kV.
One-step growth curve.
To characterize phage life cycle characteristics, one-step growth experiments were performed for two of the phage isolates using marine broth (MB; 2216; Difco) at a low multiplicity of infection (MOI; i.e., the initial ratio of phage particles to host cells), following a protocol modified from that of Ellis and Delbrück (21). For phage ΦH20, 1 ml of exponentially growing bacteria (108 CFU ml−1) was mixed with 100 μl lytic phage stock (108 PFU ml−1; MOI of 0.1) and incubated at room temperature for 20 min to allow the phages to adsorb to the host according to procedures described by Hidaka and Tokushige (22). Bacteria were then pelleted (7,000 × g, 10 min), and the nonadsorbed phages in the supernatant were discarded. The pellet was then resuspended in 1 ml MB and diluted to an appropriate phage titer (104 PFU ml−1). Two 100-μl subsamples of culture were collected at regular intervals for 60 min for enumeration. In one subsample, free phages were immediately quantified by plaque assay (18). The other subsample was pretreated with 10 μl chloroform to measure the release of intracellular phages and then centrifuged at 10,000 × g for 1 min prior to phage enumeration. For phage KVP40, the same procedure was applied, except that for this phage, the stock was diluted to 107 PFU ml−1, reducing the MOI to 0.01.
Susceptibility of V. anguillarum to phage infection at different MOIs.
The lytic potentials of two selected phages, ΦH20 and KVP40, against V. anguillarum were examined at MOIs of 0.1, 1, and 10 in liquid MB cultures during infection of V. anguillarum BA35 and PF430-3, respectively. Serial dilutions of the phage stocks (ca. 107 to 109 PFU ml−1) were inoculated in duplicate 50-ml mid-log-phase cultures of host bacteria in MB at a density of ∼107 CFU ml−1. For each experiment, duplicate control cultures without phage addition were also established. The effects of phage lysis on host cell density were monitored by regular measurements of the optical density at 600 nm (OD600) during the 24-h incubations. Phage abundance was quantified in one of the experiments (MOI, 0.1) from the number of PFU obtained on lawns of host strains by plaque assay (19) periodically over 24 h. At various time points, individual colonies were isolated from the cultures on TCBS plates and subsequently purified through 2 to 3 rounds of reisolation of single colonies to determine changes in phage susceptibility in the host community during incubation.
Statistical analysis.
For all of the dendrograms, UPGMA (unweighted-pair group method with arithmetic mean) construction experiments and Dice coefficients were calculated between pairs of each strain or phage and grouped by UPGMA and 100 bootstrap replicates using the dendrogram construction utility DendroUPGMA (Biochemistry and Biotechnology Department, Rovira i Virgili University, Tarragona, Spain) (http://genomes.urv.cat/UPGMA/index.php).
RESULTS
Isolation of Vibrio strains.
A total of 16 Vibrio sp. strains isolated from Danish aquaculture were susceptible to phage KVP40; therefore, these 16 strains were tentatively classified as closely related Vibrio sp. strains. Of the total of 37 strains examined, 35 were sensitive to the vibriostatic agent O129 at both 10 and 150 μg, whereas two isolates (VSP8 and VSP9) were resistant to O129 at concentrations of up to 150 μg. The 37 Vibrio sp. strains used in this study covered a considerable spatial variability, with isolates originating from the Mediterranean Sea (Spain and Italy), the Baltic Sea (Finland and Germany), the North Sea (Norway and United Kingdom), the Atlantic Ocean (United States), the south Pacific Ocean (Chile), and inner Danish waters; the range in time of isolation covered ∼20 years (23).
Identification of Vibrio strains.
Based on 16S rRNA gene similarity, 24 of the isolates were presumptively identified as V. anguillarum according to NCBI BLAST analysis (NCBI BLAST; http://www.ncbi.nlm.nih.gov/BLAST/) (Table 1), including the entire DTU collection, the Chilean strain (PF430-3), and 3 Danish water isolates (VA1, VA2, and VA3), whereas the remaining 13 Danish isolates yielded identification as V. cyclitrophicus, V. slendidus, V. shilonii, and Vibrio spp. (Table 1). A phylogenetic tree based on the 16S rRNA gene sequence analysis showed three main groups: group 1 (V. anguillarum), containing 20 DTU strains, strain PF430-3, and three Danish V. anguillarum isolates (VA1, VA2, and VA3); group 2 (V. splendidus and V. cyclitrophicus), containing six Danish water isolates (VSP1, VSP2, VSP3, VSP8, VSP10, and VSP12); and group 3 (V. shilonii and Vibrio spp.), containing the remaining strains (VSP4, VSP5, VSP6, VSP7, VSP9, VSP11, and VSP13) (see Fig. S1 in the supplemental material).
API 20NE fingerprinting.
Eleven different identities (Table 1) were generated from the 37 strains using API 20NE test. For all of the strains, positive results were obtained for potassium nitrate, d-glucose, esculin ferric citrate, 4-nitrophenyl β-d-galactopyranoside, malic acid, and oxidase reactions, and negative reactions were found for urea, capric acid, adipic acid, and phenylacetic acid, whereas the remaining 11 biochemical tests gave various results (data not shown).
Biolog fingerprint.
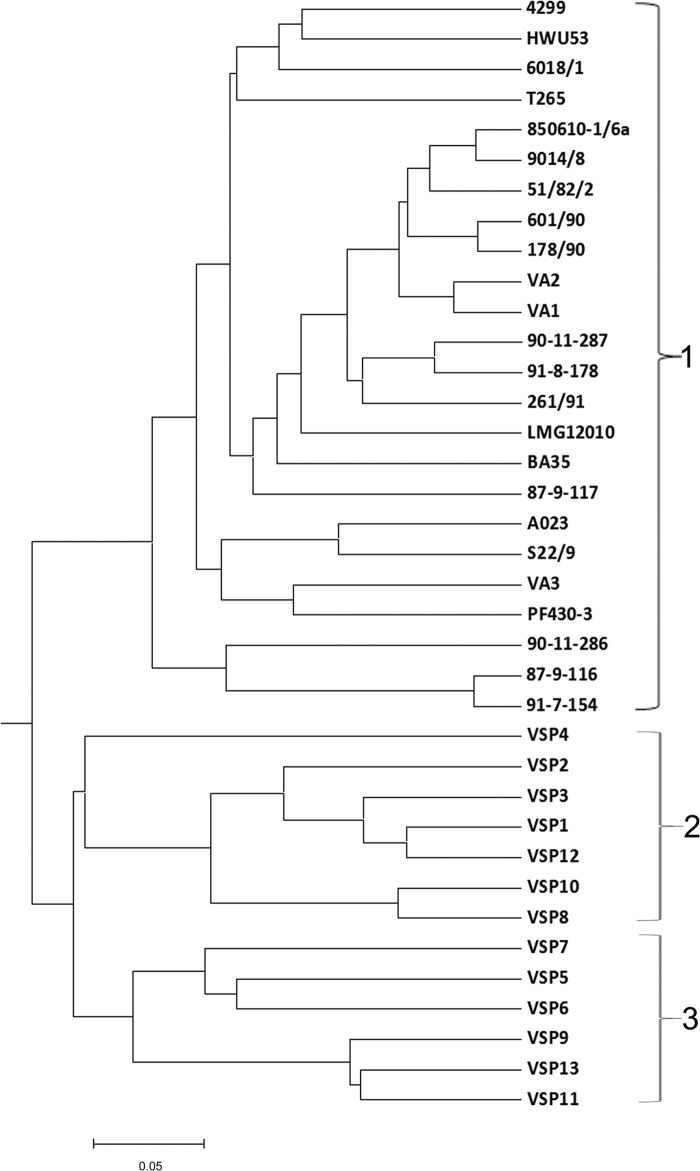
The phenotypic diversity of and relationship between the 37 strains were explored by studying the differences in carbon resource utilization, which showed a clustering of the strains into three primary phenotypic groups (Fig. 1). Cluster 1 contained 24 strains, including all of the strains in the DTU collection, the PF430-3 strain, and three of the Danish isolates (VA1, VA2, and VA3), whereas the remaining 13 Danish isolates were grouped into clusters 2 and 3 (Fig. 1). Overall, the number of substrates used varied from 54 (strain BA35) to 22 (strain VSP10) of the 95 tested substrates, with the highest number of available substrates found in cluster 1 (33 to 54) and the lowest in cluster 2 (22 to 38). All of the 22 substrates used by strain VSP10 could also be used by all other strains, suggesting that the utilization of these 22 substrates was a fundamental property of Vibrio strains.
FIG 1.
Phenotype dendrogram derived by UPGMA cluster analysis of all 37 strains in the Biolog GN2 profiles. The data were scored and compared by using Dice coefficient algorithm. The scale bar indicates the dissimilarity.
Bacteriophage enrichment and isolation.
Ten different bacteriophage isolates (Table 2) were obtained from Danish waters using different enrichment protocols. Phages ΦH20, ΦH8, ΦS4-7, and ΦS4-18 were isolated by following protocol I, using a mixture of Vibrio sp. strains as hosts for enrichment, whereas phages ΦH1, ΦH2, ΦH4, ΦH5, ΦH7, and Φ2E-1 were isolated using a single Vibrio sp. strain for enrichment (protocol II).
TABLE 2.
Method of isolation, morphological characteristics, and genome size of vibriophages isolated from Danish aquaculture farms and phage KVP40 (Japan)
| Family | Phage | Isolation protocola | Genome size (kb) | Host | Head diam (nm) | Head length (nm) | Tail diam (nm) | Tail length (nm) | Reference or source |
|---|---|---|---|---|---|---|---|---|---|
| Myoviridae | KVP40 | Unknown | 242 | PF430-3 | 76 | 120 | 30 | 120 | 32 |
| Myoviridae | ΦH1 | II | 194 | 4299 | 107 | 129 | 19 | 233 | This study |
| Myoviridae | ΦH7 | II | 194 | 90-11-286 | 117 | 129 | 22 | 226 | This study |
| Myoviridae | ΦS4-7 | I | 145 | VSP10 | 81 | 80 | 26 | 58 | This study |
| Myoviridae | ΦH4 | II | 95 | 90-11-287 | 72 | 80 | 22 | 229 | This study |
| Myoviridae | ΦH5 | II | 95 | 51/82/2 | 63 | 77 | 24 | 222 | This study |
| Siphoviridae | ΦH8 | I | 50 | T265 | 114 | 127 | 19 | 237 | This study |
| Siphoviridae | ΦH20 | I | 50 | BA35 | 54 | 65 | 12 | 144 | This study |
| Podoviridae | ΦS4-18 | I | 45 | VA1 | 63 | 64 | 12 | 13 | This study |
| Podoviridae | Φ2E-1 | II | 42 | VSP8 | 53 | 46 | 8 | 10 | This study |
| Podoviridae | ΦH2 | II | 11 | A023 | 89 | 82 | 15 | 32 | This study |
I, phage isolation method using a single enrichment step with a mixture of potential host strains; II, phage isolation method using two steps of enrichments with a single strain. For further information, see Materials and Methods.
Phage genome sizes, as determined by PFGE, showed large differences among the 11 phages and covered a 20-fold range, from 11 kb to 244 kb (Table 2). An interesting phenomenon was the systematic observation of several distinct bands appearing on the PFGE gels for phage ΦH20, ΦH8, and ΦH2 despite up to 5 rounds of phage purification from a single plaque. For example, PFGE analysis of phage ΦH20 revealed seven bands, ranging from 50 to 291 kb, with each band size increasing by 50 kb, and phage ΦH8, for which two bands of 50 kb and 100 kb, respectively, were observed (Fig. 2; also see Fig. S2 in the supplemental material).
FIG 2.
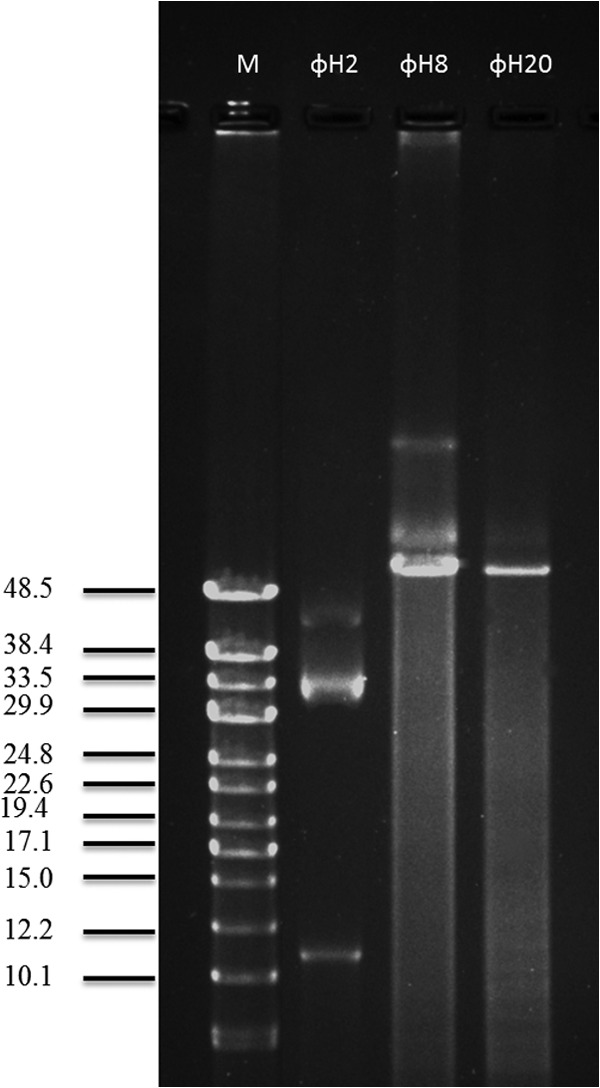
PFGE analysis of selected phage genomes with concatemers (ΦH2, ΦH8, and ΦH20), with switch times ranging from 0.5 to 5 s and a total run time of 15 h.
Host range and EOP.
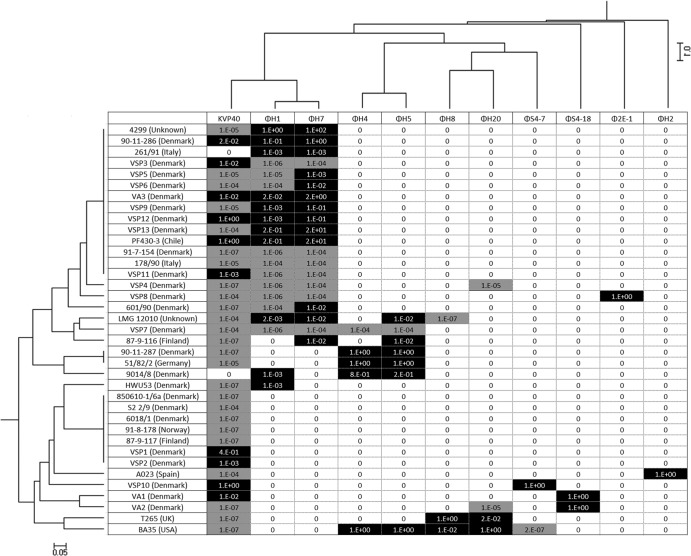
The host range and lytic potential of the 11 vibriophages were tested against 24 V. anguillarum and 13 Vibrio sp. strains by spotting 5 μl of a serially diluted phage stock onto top agar containing the potential host strain. Together, the collection of phages was able to infect all 37 strains; however, there were large differences in host range pattern and lytic efficiency among the phages (Fig. 3). Six phages (ΦH2, ΦH8, ΦH20, ΦS4-7, ΦS4-18, and Φ2E-1) exhibited a narrow host range, infecting only the host used for its isolation and perhaps one or two additional strains. Phages ΦH1, ΦH7, ΦH4, ΦH5, and KVP40, on the other hand, were characterized by broader host ranges covering both local host isolates and isolates obtained from geographically distant locations. Conversion of the phage host range pattern into a dendrogram using the Dice coefficients comparing algorithm showed some distinct clusters of the phages and bacterial strains (Fig. 3). Generally, phages with the same genome size had similar host ranges, for example, the ΦH1 and ΦH7 pair and the ΦH4 and ΦH5 pair, which had pairwise identical genome sizes and also clustered together according to host range (Fig. 3). Interestingly, the phages with the largest genomes, i.e., ΦH1, ΦH7 (194 kb), and KVP40 (244 kb), also had the largest host range, infecting many of the same hosts, despite the fact that they were isolated from Danish (ΦH1 and ΦH7) and Japanese (KVP40) locations (24). Phage susceptibility pattern, on the other hand, did not show obvious connection to the other genotypic or phenotypic relationships.
FIG 3.
Host ranges of 11 phages (columns) against 37 Vibrio sp. strains (rows). Black boxes indicate clear plaques, gray boxes indicate turbid plaques, and white boxes indicate no infection. Numbers express the efficiency of plating, i.e., the fraction of infectivity of the phage stock on a given host compared to the infectivity on the original host of isolation (isolation efficiency of the host was 100%). Dendrograms were derived by UPGMA cluster analysis. The data were scored and compared by using the Dice coefficient algorithm. The scale bar indicates the dissimilarity.
Phage morphology.
Phage morphology was examined by TEM for all 11 phages (Table 2 and Fig. 4). According to the International Committee on Taxonomy of Viruses (ICTV), three morphological groups of phages were represented by the 11 phages: Siphoviridae (ΦH8 and ΦH20), with flexible tails, Podoviridae (ΦH2, Φ2E-1, and ΦS4-18), with short tails, and Myoviridae (ΦH1, ΦH4, ΦH5, ΦH7, ΦS4-7, and KVP40), possessing an icosahedral head and contractile tails with tail fibers and relatively large genomes (25) (Fig. 4 and Table 2). Interestingly, some vibriophages with different genome sizes, e.g., ΦH2 and ΦS4-18, belong to the same family.
FIG 4.
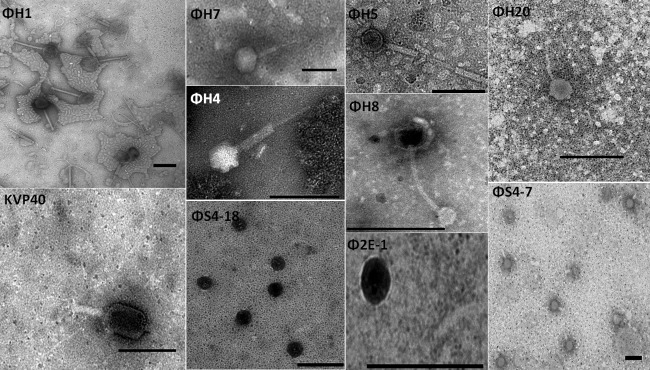
TEM pictures of selected phages. Phages were stained with 2% sodium phosphotungstate. Scale bar, 200 nm.
Phage-host interactions.
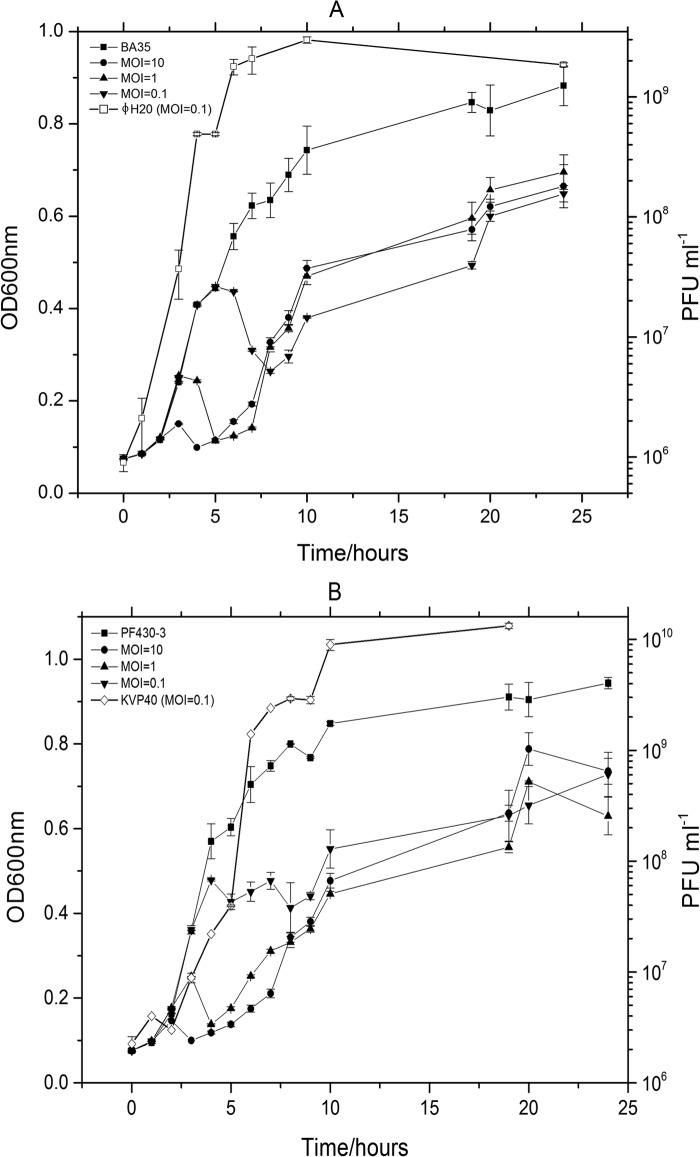
Life cycle characteristics were determined for phages ΦH20 and KVP40 from one-step growth curves during incubation with their host strains BA35 and PF430-3, respectively. Despite different morphologies and genome sizes, phages ΦH20 and KVP40 had quite similar burst sizes (70 and 80 PFU per cell, respectively) and identical eclipse (20 min) and latent periods (25 min).
The parallel long-term (24 h) infection experiments with phages ΦH20 and KVP40 and the hosts BA35 and PF430-3, respectively, at different MOIs also showed similar lytic potential of the two phages, with a strong dependence on the initial MOI for phage control of the host population (Fig. 5). In batch cultures at an MOI of 0.01, both phage KVP40 and ΦH20 showed rapid propagation during the first 10 h of incubation, resulting in phage titers stabilizing around 1010 and 109 PFU ml−1, respectively (Fig. 5). The host growth rates in the control cultures were 0.241 ± 0.002 h−1 and 0.231 ± 0.007 h−1 for PF430-3 and BA35, respectively, and both strains reached stationary phase after 10 h. In the phage-amended cultures, phages were able to control growth of the host for 5 to 6 h followed by regrowth of the host population.
FIG 5.
(A) Optical density (OD600) in cultures of V. anguillarum BA35 amended with phage ΦH20 at MOIs of 0 (control), 0.1, 1, and 10 and a corresponding abundance of PFU ml−1 in the treatment at an MOI of 0.1. (B) Optical density (OD600) in cultures of V. anguillarum PF430-3 amended with phage KVP40 at MOIs of 0 (control), 0.1, 1, and 10 and the corresponding PFU abundance in the treatment at an MOI of 0.1. Error bars represent standard deviations from all experiments carried out in duplicate.
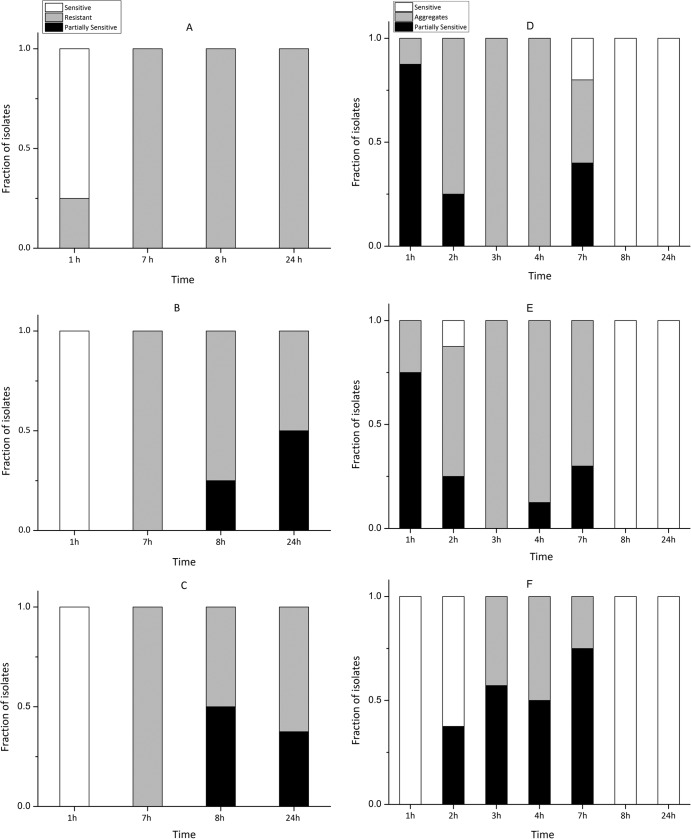
Accordingly, decreasing the initial MOI resulted in a delayed and less pronounced decrease in the OD600 (Fig. 6). Regrowth occurred in all cultures, and after 20 h the OD600 stabilized at 0.6 to 0.7, corresponding to 65 to 75% of the final OD600 in the control cultures. In addition to the OD600 measurements, bacterial isolates were obtained during the incubations for further analysis of changes in phage susceptibility in the host population (Fig. 6). In the BA35-plus-ΦH20 cultures, the occurrence of the bacteriophage-resistant isolates varied considerably over time and between treatments. According to the spot test results, the emergence of the bacteriophage-resistant bacteria was faster at high than at low MOI. Within the first hour after incubation, resistant strains were isolated at an MOI of 10, whereas resistant phenotypes were not detected at MOIs of 1 and 0.1. At high MOI, only resistant isolates were obtained after 7 h of incubation. At the lower MOIs, resistant isolates dominated during the regrowth phase (7 h). However, partially sensitive bacteria (i.e., reduced sensitivity compared to the sensitive wild-type strain) were isolated from 8 h onwards and constituted 40 to 50% of the isolates after 24 h. All of the resistant and partially sensitive strains isolated from the phage-amended BA35 culture after 24 h formed smaller colonies with lower growth rates on TCBS plates than those seen in the control cultures without phages. For the partially sensitive and resistant BA35-derived isolates, a second round of purification and subsequent susceptibility test were carried out. Interestingly, all of the partially sensitive strains returned to their sensitive phenotype after purification in the absence of phages, whereas all of the completely resistant isolates remained resistant.
FIG 6.
Phage susceptibility properties of V. anguillarum isolates obtained from cultures enriched with phages at different MOIs. (A to C) V. anguillarum strain BA35 plus phage ΦH20 at an MOI of 10 (A), 1 (B), or 0.1 (C). (D to F) V. anguillarum strain PF430-3 plus phage KVP40 at an MOI of 10 (D), 1 (E), or 0.1 (F). “Sensitive” indicates that isolates were fully sensitive to the phage. “Resistant” indicates that isolates were not susceptible to phage ΦH20 infection in the spot test. “Partially resistant” indicates that isolates had a reduced susceptibility to the phage. “Aggregates” indicates that isolates were forming aggregates, which protected against phage infection.
In the parallel experiment with V. anguillarum strain PF430-3 and phage KVP40, no completely resistant isolates were obtained from any of the treatments during the incubation, and the colony morphology did not change over time. Instead, all of the isolates were sensitive or partially sensitive strains, and after 8 h all obtained isolates were fully sensitive to KVP40. In contrast to the control cultures, visible biofilm was formed in the KVP40-amended cultures, and some isolates formed aggregates when subsequently grown in liquid culture. For the aggregating and partially sensitive PF430-3-derived isolates, a second and third round of purification and subsequent spot assay showed that all strains returned to the original phenotype and were fully sensitive to phage KVP40.
DISCUSSION
Successful application of phage therapy in the treatment of vibriosis requires detailed knowledge of the diversity and distribution of the phage susceptibility properties of Vibrio sp. pathogens associated with vibriosis as well as a collection of well-characterized phages that covers host diversity. In this study, we characterized 11 vibriophages and 37 Vibrio sp. strains (Table 1 and 2), covering considerable temporal and spatial variability. Further, two V. anguillarum strains, BA35 and PF430-3, and the specific phages ΦH20 and KVP40 were selected for more detailed studies of phage lytic potential and phage-host interactions.
Diversity of V. anguillarum strains.
The V. anguillarum strains examined in the present study (Table 1) showed close similarity according to 16S rRNA gene sequences irrespective of time or place of isolation. Thirty-seven of the isolates which grouped together covered a period of isolations of >20 years and a wide range of geographical locations (Chile, Spain, Italy, Germany, United Kingdom, Norway, Finland, Denmark, and the United States) and fish species (turbot, trout, sea bass, and salmon). This suggests that the 16S rRNA gene phylogeny of V. anguillarum is highly uniform and stable in time and space. Thus, the method is unable to discriminate between potentially pathogenic and nonpathogenic V. anguillarum strains or resolve differences in V. anguillarum communities across spatial and temporal scales. Similarly, genetic fingerprinting methods (enterobacterial repetitive-element intergenic consensus sequence [ERIC] PCR) showed that Vibrio sp. isolates from coastal British Columbia did not group according to geography, suggesting genetic homogeneity among the environmental strains (26). Previous studies have, however, revealed a large degree of microheterogeneity in 16S rRNA genes within Vibrio sp. strains (27), limiting the use of 16S rRNA genes for phylogenetic studies in Vibrionaceae. Higher taxonomic resolution has been obtained using multilocus sequence analyses (MLSA) of six protein-coding genes (28); however, this method also focuses on variations in housekeeping genes and is applicable mainly for identification of Vibrio strains at the species level.
The V. anguillarum isolates clustered in distinct groups based on the Biolog GN2 profiles and differed from the other identified clusters of Vibrio strains. In contrast, the phage susceptibility pattern of the V. anguillarum isolates did not show any connection to the species or physiological relationships obtained from Biolog profiles. In fact, phage susceptibility patterns did not even group according to the species clusters defined for the 37 strains. This suggests that phenotypic traits related to sensitivity to phages are not resolved by the methods used in this study. This supports previous studies of Cellulophaga baltica (29), V. parahaemolyticus (15), F. psychrophilum (14), and Escherichia coli (30), which have all demonstrated a high degree of phenotypic diversity in host susceptibility at the strain level that was uncoupled from the overall genetic relationships among the host isolates. Thus, our results confirm highly variable patterns in phage sensitivity even within relatively narrow phylogenetic and phenotypic groups of bacteria, and at the same time they demonstrate similar susceptibility patterns across a broad phylogenetic scale in Vibrio spp., emphasizing the complexity of phage-host interactions in this group.
As observed for the physiological fingerprints, there was overlap in the phage susceptibility patterns for V. anguillarum strains isolated across the large temporal and spatial scales and from different fish species represented in the current study. For example, the strains 90-11-286 and VA3, which were isolated from Danish rainbow trout and salmon farms, respectively, with 20 years between isolations, were both infected by the same 3 phages (ΦH1, ΦH7, and KVP40). Similarly, strains VIB88 and BA35, isolated from rainbow trout farms and salmon farms in Germany and the United States, respectively, both were infected by phages ΦH4 and ΦH5, isolated from a Danish turbot farm 20 years later. This demonstrates that despite the occurrence of unique susceptibility patterns of individual isolates, key phenotypic properties related to phage susceptibility are distributed worldwide and maintained in the global V. anguillarum community for decades.
Phage isolation and diversity.
The 10 vibriophages isolated from Danish fish farms all had unique host ranges when tested against the collection of 37 Vibrio strains. All three major morphological families, Myoviridae, Podoviridae, and Siphoviridae, were represented, and the phages ranged in genome size from 11 to 194 kb, distributed in 6 distinct groups (Table 2). The phage isolates showed large differences in host specificity, and together the Danish phages were able to infect and lyse 30 of the 37 strains, covering most of the phylogenetic, geographical, and temporal variation that characterized the host isolates. This is interesting, as it suggests that vibriophage infective properties are not restricted to local cooccurring host communities but rather widely distributed across large spatial and temporal scales, as also suggested by previous observations of an ocean-scale distribution of genetically related vibriophages (31).
The two broad-host-range phages isolated in this study (ΦH1 and ΦH7) both belonged to the Myoviridae family and had relatively large genomes (194 kb). This is consistent with the host range pattern observed for F. psychrophilum phages, where the large-genome phages (>90 kb) had substantially broader host ranges than small-genome phages (<12 kb) (29). In accordance with this, the previously isolated large-genome vibriophages KVP40 (244 kb) (32) and φpp2 (246 kb) (33) also have very broad host ranges covering several species of Vibrio. KVP40 is known to cause infection through the common outer membrane protein OmpK and has previously been shown to infect eight Vibrio species, including V. anguillarum, V. parahaemolyticus, and V. cholerae (20). The presence of several copies of genes encoding proteins associated with phage tail or tail fibers in the KVP40 genome suggested an increased flexibility in host range adaptation, increasing host range (32). Despite the fact that KVP40 was originally isolated in Japan >20 years ago (24), it still infected approximately 40% of the Vibrio sp. isolates obtained from Danish aquaculture, confirming its cross-species host range, and it maintained infective properties across large temporal and spatial scales. Thus, the characteristic of the new phage isolates ΦH1 and ΦH7 adds to the emerging view of a global distribution of large-genome, broad-host-range vibriophages.
Generally, vibriophages were widespread in Danish aquaculture, as phages were isolated from all of the investigated farms independently of outbreaks of vibriosis. However, the fact that phages could not be isolated without previous propagation in enrichment cultures indicated that densities of specific vibriophages were relatively low in the fish farms. Some samples from which Vibrio sp. hosts were isolated did not harbor their corresponding phages, suggesting that specific hosts and lytic phages did not always coexist in the same environment. However, it should be noted that the filtration of the water samples and chloroform treatment prior to phage isolation may have resulted in the loss of phages (18, 34).
In all isolate-based studies of phage diversity, the results are biased by the choice of host strains used for isolation, as each individual strain will only target a subset of the infective phages present in the sample. In the current study, broad-host-range phages ΦH1 and ΦH7 were isolated using strains from the global Vibrio species collection, whereas the more host-specific phages, such as Φ2E-1, ΦS4-18, and ΦS4-7, were isolated using a cooccurring Vibrio sp. host obtained from the same sample. As the evolution of a broad host range in phages is believed to be accompanied by a fitness cost, i.e., a reduced infective efficiency in its original hosts (35), it is likely that the observed differences in phage isolation pattern reflect different phage properties. Therefore, we speculate that highly specific phages are efficient, opportunistic phages which propagate rapidly in response to the growth of their specific hosts and will dominate in samples where their host is present. Consequently, these phages are the most likely to be isolated using cooccurring hosts as target strains. Broad-host-range phages, on the other hand, may follow a K-strategy (36), maintaining lower densities but for longer periods; therefore, they may be more likely to be pulled out from a sample using non-cooccurring or rare hosts. Due to these differences in isolation pattern, we do not know to what extent the isolated phages are representative of the local phage community.
PFGE provides a good discrimination for phage genome sizes and clearly demonstrates a large variability in genome size within the group of vibriophages. Several phage genomes, including ΦH20, ΦH8, and ΦH2, yielded several bands in PFGE analysis, representing multiplications of its own genome size. These bands may represent concatemers of the phage genome produced during DNA replication, as previously demonstrated in λ phage (37). Also, concatemer formation of phage DNA has been observed as a result of DNA cleavage during heating and subsequent reassembly at ambient temperature (38). The possibility of coinfection of other phages still cannot be ruled out; however, we did not see different phage morphotypes during TEM analysis of single isolates. A previous report of different phage morphotypes associated with multiple bands on a PFGE gel, however, suggested that coinfection of more than one phage may be an explanation for the occurrence of several genome sizes on a PFGE gel of a purified phage (29). Further genomic analyses are required to elucidate the origin of the multiple bands obtained in the PFGE analyses.
Phage-host interactions.
The in vitro bacterial challenge assay with V. anguillarum strains BA35 and PF430-3 demonstrated significant but temporary phage control of both strains which was strongly dependent on the initial MOI. Even at the highest MOI, the host bacteria were not completely eliminated and regrowth of cells following phage infection suggested the emergence of phage-resistant strains after 5 to 6 h of incubation. Lytic bacteriophages have previously been shown to select for the emergence of resistant strains in culture studies with marine heterotrophic bacteria (39–41), where phage-resistant strains rapidly replaced the sensitive wild type following exposure to phages.
Isolation and characterization of bacterial isolates during the experiments confirmed that completely resistant strains, and strains with reduced sensitivity, dominated the bacterial population after 7 h in the culture containing strain BA35 and phage ΦH20. Interestingly, however, phage-resistant strains were not isolated after exposure of strain PF430-3 to phage KVP40, as has also been observed in other phage infection studies with hosts such as Salmonella enterica (42), S. enterica serovar Oranienburg (43), and E. coli O157:H7 (44). In the current experiment with strain PF430-3, phage exposure led to the formation of bacterial aggregates, and all of the isolated colonies had maintained full sensitivity to the phage after purification. These results suggest that different mechanisms were responsible for the regrowth of cells in the presence of phages following lysis of the initial sensitive population. The BA35 and phage ΦH20 interaction results in a succession toward phage-resistant strains, whereas the strain PF430-3 apparently is protected against phage KVP40 infection by aggregation. The underlying mechanism providing bacterial protection against phages via aggregation, allowing the coexistence of lytic phages and sensitive strains, is not revealed in the present study. However, previous studies have shown that phage-induced lysis can promote the formation of bacterial biofilms (45), which may offer protection against phage infection via the exopolysaccharide (EPS) matrix covering the biofilm (46) or the presence of bacterial refuges in the spatially heterogeneous environment (47). More recently, Høyland-Kroghsbo et al. (48) found that N-acyl-l-homoserine lactone (AHL) quorum-sensing signals in E. coli caused a transient reduction in the phage adsorption rate and suggested this as a mechanism allowing coexistence of lytic phages and sensitive hosts. Such mechanisms may also have contributed to the observed coexistence of sensitive strains of PF430-3 and the lytic phage KVP40 in the current study.
Potential for phage therapy in aquaculture.
One of the main challenges in using bacteriophages to control pathogens in aquaculture is to cover the diversity and temporal and spatial variations in pathogen composition, as well as the phage resistance that may rapidly develop. A detailed characterization of phage properties and understanding of phage-host interactions are essential requirements for the successful application of phage-based pathogen control. The local occurrence of both specific and broad-host-range phages which, in combination, were able to infect all of the tested host strains, covering large geographical and temporal scales, suggested that vibriophage-host systems are ubiquitously distributed. In a phage therapy context this is promising, as it suggests the possibility of establishing a phage library with broad application against V. anguillarum communities worldwide. However, the fast development of phage resistance during exposure to specific phages and the indication that different mechanisms provided resistance in different hosts also emphasized that the susceptibility to phages is a dynamic property in the Vibrio sp. hosts. Consequently, phage resistance mechanisms and dynamics and the implications for host properties are important to address in future exploration of phage control of V. anguillarum in aquaculture.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Danish Strategic Research Council (ProAqua; project 12-132390), by a grant from the Danish Council for Independent Research (FNU 09-072829), and by the EU-IRSES-funded project AQUAPHAGE.
We thank Pantelis Katharios, Hellenic Centre for Marine Research, for kindly providing V. anguillarum PF430-3 and vibriophage KVP40 and Jette Melchiorsen for excellent technical support.
Footnotes
Published ahead of print 7 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03544-13.
REFERENCES
- 1.Frans I, Michiels CW, Bossier P, Willems K, Lievens B, Rediers H. 2011. Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J. Fish Dis. 34:643–661. 10.1111/j.1365-2761.2011.01279.x [DOI] [PubMed] [Google Scholar]
- 2.Ransom DP. 1978. Bacteriologic, immunologic and pathogenic studies of Vibrio spp. pathologic to salmonids. Ph.D. dissertation Oregon State University, Corvallis, OR [Google Scholar]
- 3.Joosten PHM, Tiemersma E, Threels A, Caumartin Dhieux C, Rombout JHWM. 1997. Oral vaccination of fish against Vibrio anguillarum using alginate microparticles. Fish Shellfish Immunol. 7:471–485. 10.1006/fsim.1997.0100 [DOI] [Google Scholar]
- 4.Aoki T, Satoh T, Kitao T. 1987. New tetracycline resistance determinant on R plasmids from Vibrio anguillarum. Antimicrob. Agents Chemother. 31:1446. 10.1128/AAC.31.9.1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stamm JM. 1989. In vitro resistance by fish pathogens to aquacultural antibacterials, including the quinolones difloxacin (A-56619) and sarafloxacin (A-56620). J. Aquat. Anim. Health 1:135–141. [DOI] [Google Scholar]
- 6.Middelboe M. 2008. Microbial disease in the sea: effects of viruses on carbon and nutrient cycling. Princeton University Press, Princeton, NJ [Google Scholar]
- 7.Park SC, Nakai T. 2003. Bacteriophage control of Pseudomonas plecoglossicida infection in ayu Plecoglossus altivelis. Dis. Aquat. Org. 53:33–39. 10.3354/dao053033 [DOI] [PubMed] [Google Scholar]
- 8.Castillo D, Higuera G, Villa M, Middelboe M, Dalsgaard I, Madsen L, Espejo R. 2012. Diversity of Flavobacterium psychrophilum and the potential use of its phages for protection against bacterial cold water disease in salmonids. J. Fish Dis. 35:193–201. 10.1111/j.1365-2761.2011.01336.x [DOI] [PubMed] [Google Scholar]
- 9.Nakai T, Sugimoto R, Park K, Matsuoka S, Mori K, Nishioka T, Maruyama K. 1999. Protective effects of bacteriophage on experimental Lactococcus garvieae infection in yellowtail. Dis. Aquat. Org. 37:33–41. 10.3354/dao037033 [DOI] [PubMed] [Google Scholar]
- 10.Vinod M, Shivu M, Umesha K, Rajeeva B, Krohn G, Karunasagar I, Karunasagar I. 2006. Isolation of Vibrio harveyi bacteriophage with a potential for biocontrol of luminous vibriosis in hatchery environments. Aquaculture 255:117–124. 10.1016/j.aquaculture.2005.12.003 [DOI] [Google Scholar]
- 11.Imbeault S, Parent S, Lagacé M, Uhland CF, Blais J-F. 2006. Using bacteriophages to prevent furunculosis caused by Aeromonas salmonicida in farmed brook trout. J. Aquat. Anim. Health 18:203–214. 10.1577/H06-019.1 [DOI] [Google Scholar]
- 12.Stevenson R, Airdrie D. 1984. Isolation of Yersinia ruckeri bacteriophages. Appl. Environ. Microbiol. 47:1201–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higuera G, Bastías R, Tsertsvadze G, Romero J, Espejo RT. 2013. Recently discovered Vibrio anguillarum phages can protect against experimentally induced vibriosis in Atlantic salmon, Salmo salar. Aquaculture 392-395:128–133. 10.1016/j.aquaculture.2013.02.013 [DOI] [Google Scholar]
- 14.Stenholm AR, Dalsgaard I, Middelboe M. 2008. Isolation and characterization of bacteriophages infecting the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 74:4070–4078. 10.1128/AEM.00428-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comeau AM, Chan AM, Suttle CA. 2006. Genetic richness of vibriophages isolated in a coastal environment. Environ. Microbiol. 8:1164–1176. 10.1111/j.1462-2920.2006.01006.x [DOI] [PubMed] [Google Scholar]
- 16.Silva-Rubio A, Avendano-Herrera R, Jaureguiberry B, Toranzo AE, Magarinos B. 2008. First description of serotype O3 in Vibrio anguillarum strains isolated from salmonids in Chile. J. Fish Dis. 31:235–239. 10.1111/j.1365-2761.2007.00878.x [DOI] [PubMed] [Google Scholar]
- 17.Lorian V. 2005. Antibiotics in laboratory medicine. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 18.Middelboe M, Chan AM, Bertelsen SK. 2010. Isolation and life cycle characterization of lytic viruses infecting heterotrophic bacteria and cyanobacteria, p 118–133 Wilhelm SW, Weinbauer MG, Suttle CA. (ed), Manual of aquatic viral ecology. ASLO, Waco, TX [Google Scholar]
- 19.Adams MH. 1959. Bacteriophages. Interscience Publishers, New York, NY [Google Scholar]
- 20.Inoue T, Matsuzaki S, Tanaka S. 1995. A 26-kDa outer membrane protein, OmpK, common to Vibrio species is the receptor for a broad-host-range vibriophage, KVP40. FEMS Microbiol. Lett. 125:101–105. 10.1111/j.1574-6968.1995.tb07342.x [DOI] [PubMed] [Google Scholar]
- 21.Ellis EL, Delbrück M. 1939. The growth of bacteriophage. J. Gen. Physiol. 22:365–384. 10.1085/jgp.22.3.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hidaka T, Tokushige A. 1978. Isolation and characterization of Vibrio parahaemolyticus bacteriophages in sea water. Mem. Fac. Fish. Kagoshima Univ. 27:79–90 [Google Scholar]
- 23.Skov MN, Pedersen K, Larsen JL. 1995. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl. Environ. Microbiol. 61:1540–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuzaki S, Tanaka S, Koga T, Kawata T. 1992. A broad-host-range vibriophage, KVP40, isolated from sea water. Microbiol. Immunol. 36:93–97. 10.1111/j.1348-0421.1992.tb01645.x [DOI] [PubMed] [Google Scholar]
- 25.Comeau AM, Tremblay D, Moineau S, Rattei T, Kushkina AI, Tovkach FI, Krisch HM, Ackermann HW. 2012. Phage morphology recapitulates phylogeny: the comparative genomics of a new group of myoviruses. PLoS One 7:e40102. 10.1371/journal.pone.0040102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comeau AM, Suttle CA. 2007. Distribution, genetic richness and phage sensitivity of Vibrio spp. from coastal British Columbia. Environ. Microbiol. 9:1790–1800. 10.1111/j.1462-2920.2007.01299.x [DOI] [PubMed] [Google Scholar]
- 27.Jensen S, Frost P, Torsvik VL. 2009. The nonrandom microheterogeneity of 16S rRNA genes in Vibrio splendidus may reflect adaptation to versatile lifestyles. FEMS Microbiol. Lett. 294:207–215. 10.1111/j.1574-6968.2009.01567.x [DOI] [PubMed] [Google Scholar]
- 28.Pascual J, Macián MC, Arahal DR, Garay E, Pujalte MJ. 2010. Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol. 60:154–165. 10.1099/ijs.0.010702-0 [DOI] [PubMed] [Google Scholar]
- 29.Holmfeldt K, Middelboe M, Nybroe O, Riemann L. 2007. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl. Environ. Microbiol. 73:6730–6739. 10.1128/AEM.01399-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer CR, Yoichi M, Unno H, Tanji Y. 2004. The coexistence of Escherichia coli serotype O157:H7 and its specific bacteriophage in continuous culture. FEMS Microbiol. Lett. 241:171–177. 10.1016/j.femsle.2004.10.017 [DOI] [PubMed] [Google Scholar]
- 31.Kellogg CA, Rose JB, Jiang SC, Thurmond JM, Paul JH. 1995. Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar. Ecol. Prog. Ser. 120:89–98. 10.3354/meps120089 [DOI] [Google Scholar]
- 32.Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Microbiol. 185:5220–5233. 10.1128/JB.185.17.5220-5233.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YR, Lin CS. 2012. Genome-wide characterization of Vibrio phage Φpp2 with unique arrangements of the mob-like genes. BMC Genomics 13:224. 10.1186/1471-2164-13-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts LM, Dunker AK. 1993. Structural changes accompanying chloroform-induced contraction of the filamentous phage fd. Biochemistry 32:10479–10488. 10.1021/bi00090a026 [DOI] [PubMed] [Google Scholar]
- 35.Duffy S, Turner PE, Burch CL. 2006. Pleiotropic costs of niche expansion in the RNA bacteriophage Φ6. Genetics 172:751–757. 10.1534/genetics.105.051136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Southwood T, May R, Hassell M, Conway G. 1974. Ecological strategies and population parameters. Am. Nat. 108:791–804. 10.1086/282955 [DOI] [Google Scholar]
- 37.Waterbury PG, Lane MJ. 1987. Generation of lambda phage concatemers for use as pulsed field electrophoresis size markers. Nucleic Acids Res. 15:3930–3930. 10.1093/nar/15.9.3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun M, Louie D, Serwer P. 1999. Single-event analysis of the packaging of bacteriophage T7 DNA concatemers in vitro. Biophys. J. 77:1627–1637. 10.1016/S0006-3495(99)77011-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Middelboe M. 2000. Bacterial growth rate and marine virus-host dynamics. Microb. Ecol. 40:114–124. 10.1007/s002480000050 [DOI] [PubMed] [Google Scholar]
- 40.Riemann L, Grossart HP. 2008. Elevated lytic phage production as a consequence of particle colonization by a marine Flavobacterium (Cellulophaga sp.). Microb. Ecol. 56:505–512. 10.1007/s00248-008-9369-8 [DOI] [PubMed] [Google Scholar]
- 41.Middelboe M, Holmfeldt K, Riemann L, Nybroe O, Haaber J. 2009. Bacteriophages drive strain diversification in a marine Flavobacterium: implications for phage resistance and physiological properties. Environ. Microbiol. 11:1971–1982. 10.1111/j.1462-2920.2009.01920.x [DOI] [PubMed] [Google Scholar]
- 42.Kim M, Ryu S. 2011. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 77:2042–2050. 10.1128/AEM.02504-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kocharunchitt C, Ross T, McNeil D. 2009. Use of bacteriophages as biocontrol agents to control Salmonella associated with seed sprouts. Int. J. Food Microbiol. 128:453–459. 10.1016/j.ijfoodmicro.2008.10.014 [DOI] [PubMed] [Google Scholar]
- 44.Sheng H, Knecht HJ, Kudva IT, Hovde CJ. 2006. Application of bacteriophages to control intestinal Escherichia coli O157:H7 levels in ruminants. Appl. Environ. Microbiol. 72:5359–5366. 10.1128/AEM.00099-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gödeke J, Paul K, Lassak J, Thormann KM. 2011. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 5:613–626. 10.1038/ismej.2010.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327. 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- 47.Heilmann S, Sneppen K, Krishna S. 2012. Coexistence of phage and bacteria on the boundary of self-organized refuges. Proc. Natl. Acad. Sci. U. S. A. 109:12828–12833. 10.1073/pnas.1200771109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Høyland-Kroghsbo NM, Mærkedahl RB, Svenningsen SL. 2013. A quorum-sensing-induced bacteriophage defense mechanism. mBio 4:e00362-12. 10.1128/mBio.00362-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen K, Gram L, Austin D, Austin B. 1997. Pathogenicity of Vibrio anguillarum serogroup O1 strains compared to plasmids, outer membrane protein profiles and siderophore production. J. Appl. Microbiol. 82:365–371. 10.1046/j.1365-2672.1997.00373.x [DOI] [PubMed] [Google Scholar]
- 50.Schrøder MB, Ellingsen T, Mikkelsen H, Norderhus EA, Lund V. 2009. Comparison of antibody responses in Atlantic cod (Gadus morhua L.) to Vibrio anguillarum, Aeromonas salmonicida and Francisella sp. Fish Shellfish Immunol. 27:112–119. 10.1016/j.fsi.2008.11.016 [DOI] [PubMed] [Google Scholar]
- 51.Hjelm M, Bergh Ø, Riaza A, Nielsen J, Melchiorsen J, Jensen S, Duncan H, Ahrens P, Birkbeck H, Gram L. 2004. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 27:360–371. 10.1078/0723-2020-00256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.