Abstract
Bacterial multicomponent monooxygenase gene targets in Pseudonocardia dioxanivorans CB1190 were evaluated for their use as biomarkers to identify the potential for 1,4-dioxane biodegradation in pure cultures and environmental samples. Our studies using laboratory pure cultures and industrial activated sludge samples suggest that the presence of genes associated with dioxane monooxygenase, propane monooxygenase, alcohol dehydrogenase, and aldehyde dehydrogenase are promising indicators of 1,4-dioxane biotransformation; however, gene abundance was insufficient to predict actual biodegradation. A time course gene expression analysis of dioxane and propane monooxygenases in Pseudonocardia dioxanivorans CB1190 and mixed communities in wastewater samples revealed important associations with the rates of 1,4-dioxane removal. In addition, transcripts of alcohol dehydrogenase and aldehyde dehydrogenase genes were upregulated during biodegradation, although only the aldehyde dehydrogenase was significantly correlated with 1,4-dioxane concentrations. Expression of the propane monooxygenase demonstrated a time-dependent relationship with 1,4-dioxane biodegradation in P. dioxanivorans CB1190, with increased expression occurring after over 50% of the 1,4-dioxane had been removed. While the fraction of P. dioxanivorans CB1190-like bacteria among the total bacterial population significantly increased with decrease in 1,4-dioxane concentrations in wastewater treatment samples undergoing active biodegradation, the abundance and expression of monooxygenase-based biomarkers were better predictors of 1,4-dioxane degradation than taxonomic 16S rRNA genes. This study illustrates that specific bacterial monooxygenase and dehydrogenase gene targets together can serve as effective biomarkers for 1,4-dioxane biodegradation in the environment.
INTRODUCTION
Bacterial multicomponent monooxygenases (BMMs), also known as soluble di-iron monooxygenases (SDIMOs), are an important group of enzymes that play essential roles in the biotransformation of many environmental pollutants (1, 2). The components of a BMM usually consist of a multiple-subunit hydroxylase, a reductase, and a protein B (3) with nucleotide sequences that typically associate into six distinct groups. As such, many studies have focused on the identification of biomarkers within this functional group of enzymes (4–8). Biomarkers can serve as indicators of cellular or physiological variation in a biological system and have been used in a wide range of applications, from clinical diagnostics to microbial source tracking of pollutants in impacted water bodies. Indeed, BMMs have been used to study the biodegradation of toluene, tetrahydrofuran (THF), and phenol, among other contaminants of concern (3). The transformation of environmental pollutants by natural microbial processes is an attractive approach for the remediation of recalcitrant compounds because significant cost savings might be achieved compared to alternative physical-chemical treatment strategies.
Genetic biomarkers are powerful tools that can be used as indicators for potential contaminant biodegradation. Biomarkers targeting conserved regions of the 16S rRNA gene to identify the presence or absence of microbial organisms are useful when biodegradation is dependent on a specific microbial strain. This approach has been effective in studying reductive dechlorination of chlorinated solvents by Dehalococcoides spp. (9). Alternatively, functional genes can be targeted for processes that involve BMMs with broad substrate specificities, such as the soluble (sMMO) and particulate (pMMO) methane monooxygenase enzymes (10). The pmoA gene codes for the alpha subunit of pMMO and was previously shown to be expressed by a mixed community of methanotrophs in soil capable of biodegrading trichloroethylene (TCE) (11). Consequently, biomarkers are becoming increasingly important in bioremediation and monitoring applications for environmental contaminants, with the development of several assays that target specific genes involved in compound degradation (8, 12, 13). For example, quantification of the cytochrome P450 monooxygenase-encoding gene, ethB, was used as an indicator for the microbial transformation of methyl tert-butyl ether (MTBE) in samples with low levels of contamination (14). Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) genes associated with BMMs also play an important role in the biotransformation of recalcitrant compounds. For example, the degradation of THF breakdown products by Pseudonocardia tetrahydrofuranoxydans strain K1 involves aldehyde and semialdehyde dehydrogenase genes located within the gene cluster associated with THF degradation (15), suggesting these dehydrogenase genes should be investigated during biodegradation of recalcitrant compounds to gain insights into the complete oxidation of organic contaminants. The quantification of important microbial populations and identification of enzymes responsible for degradation processes would greatly improve the design of biological treatment for recalcitrant compounds in complex environmental samples. In conjunction with temporal monitoring of 1,4-dioxane concentrations, analysis of biomarkers provides valuable information to assess whether changes in contaminant concentration are occurring through physical, chemical, or biological processes. Thus, biomarkers are important tools for advanced characterization to determine the biodegradation potential of contaminants in wastewater, sediments, soils, and groundwater.
Biotransformation of emerging contaminants is a significant challenge because of our constantly evolving understanding of degradation kinetics and pathways for these compounds in the environment. 1,4-Dioxane (hereafter “dioxane”) is a water contaminant of emerging concern with the potential for transformation by a selected group of microorganisms (16–20). Dioxane is classified as a likely human carcinogen (21) and has been detected at several hazardous waste sites (22) due to its widespread use as a stabilizer for chlorinated solvents (mainly 1,1,1-trichloroethane) and improper storage and disposal of dioxane-containing solvents (23). In polyester manufacturing processes, dioxane is a by-product during esterification of dicarboxylic acid (i.e., terephthalic acid) and dihydroxy alcohol (i.e., ethylene glycol) (23, 24); thus, dioxane is also an important contaminant in industrial waste streams. Dioxane is found at trace levels in personal care products as it is an impurity in ethoxylated surfactants and emulsifiers (25). As a result, dioxane can eventually reach municipal wastewater treatment plants (26). This compound is generally resistant to conventional water and wastewater treatment strategies and may ultimately be discharged into the environment. Currently available water and wastewater treatment processes for dioxane typically require energy-intensive advanced oxidation processes. However, a recent study presented several lines of evidence that microbial biodegradation of dioxane was naturally occurring at a contaminated site (27), suggesting the potential for widespread application of biologically mediated dioxane degradation processes. Thus, it is essential to evaluate intrinsic and enhanced bioremediation as an effective approach for removing dioxane from contaminated waste streams and water resources.
Previous research efforts have determined that the microbial transformation of dioxane involves monooxygenase enzymes for both metabolic and cometabolic degradation pathways (28, 29). Metabolic degradation is less common, with only a few microorganisms being able to use dioxane as the sole source for carbon and energy (18, 30–32). Cometabolic pathways include several monooxygenase enzymes required for dioxane degradation, including the methane (MMO), propane (PrMO), phenol (PHE), tetrahydrofuran (THFMO), and toluene (TOL, T4MO, or RMO), monooxygenases (19, 20, 28, 33, 34). THFMO was proven to catalyze THF as well as dioxane biodegradation in Pseudonocardia sp. strain ENV478 (19). In addition, the recently completed genomic assembly and annotation of Pseudonocardia dioxanivorans CB1190 (hereafter “CB1190”) (35), a bacterium able to use dioxane as the sole source of carbon and energy (18, 32), facilitated a comprehensive in silico analysis of genes encoding several multicomponent monooxygenase clusters (35), including a plasmid-encoded region containing the entire THF degradation gene cluster from Pseudonocardia sp. strain ENV478. Indeed, further study of THFMO in CB1190 has shown this BMM was upregulated during dioxane biodegradation (29). Consequently, this gene cluster within CB1190 is also referred to as the dioxane monooxygenase (DXMO).
The present study extends from previous research on metabolic and cometabolic dioxane biodegradation by studying gene abundance and expression of select monooxygenases and dehydrogenases in CB1190 and in a consortium of microorganisms in industrial activated sludge actively transforming dioxane. A time series approach was used to determine the correlation between candidate biomarkers and dioxane biodegradation rates. To our knowledge, this is the first article to evaluate the expression of monooxygenase and dehydrogenase genes as biomarkers for the biotransformation of dioxane in complex environmental samples.
MATERIALS AND METHODS
In silico analysis of P. dioxanivorans CB1190 to identify candidate biomarkers.
Identification of candidate biomarkers was focused on monooxygenase and dehydrogenase genes based on earlier work describing the dioxane degradation pathway for monooxygenase-expressing bacteria (29, 36) and expression of dehydrogenase genes in bacteria during utilization of monooxygenase-inducing substrates (37–39). To facilitate gene selection, an in silico analysis was performed to identify all annotated monooxygenase groups within CB1190 and its associated plasmids (35). Candidate targets were selected from this comprehensive list based on their functional roles in the biological transformation of recalcitrant compounds. Dehydrogenase gene targets were also selected based on sequence similarity to alcohol and aldehyde dehydrogenase genes previously shown to be associated with BMMs (15, 29) or involved in the oxidation of primary substrates in monooxygenase-expressing bacteria (38, 39).
Bacterial strains and culture conditions.
Pure cultures of CB1190 were used to study gene abundance and expression during metabolic biodegradation of dioxane. CB1190 was initially cultured in sterile 100-ml bottles containing 20 ml of ammonia mineral salt (AMS) medium as previously described (18, 32), with the exception that 20 mM glucose was used as the carbon source. After 5 days of growth on glucose, 60 ml of CB1190 cells was filtered onto 0.2-μm-pore cellulose filters (Nalgene, Rochester, NY) and washed three times with phosphate-buffered saline (PBS) solution to remove glucose. Cells were collected from the filter and resuspended in 5 ml of AMS medium to an average concentration of 3.2 × 108 cells/liter (standard deviation [SD], 4.8 × 107 cells/liter). For dioxane biodegradation experiments, 1 ml of concentrated CB1190 was used to seed triplicate 100-ml screw-cap bottles containing a total volume of 20 ml AMS medium supplemented with 100 mg/liter dioxane. Bacterial cultures were incubated at 30°C with 150 rpm of agitation.
Acetylene inhibition of monooxygenase activity.
An acetylene exposure experiment was performed as previously described (28) to study the effects of a monooxygenase-inhibiting compound on gene abundance and expression in pure cultures. CB1190 was cultured in 250-ml Boston round screw-cap bottles equipped with Mininert valves (Alltech Associates, Inc., Deerfield, IL) containing 50 ml of AMS medium supplemented with 100 mg/liter dioxane. Cultures were maintained for three cycles of complete dioxane degradation prior to acetylene exposure. Cultures were exposed to 5% acetylene gas in the headspace while shaking for 30 min, followed by flushing with nitrogen gas for 5 min. Unexposed controls were treated in a similar fashion without the addition of acetylene. Dioxane was added to 100 mg/liter, and the cultures were incubated at 30°C with 150 rpm of agitation. Cultures were monitored every 8 h, and 0.5-ml samples were collected for gene abundance and expression analysis until dioxane concentrations fell below the detection limit.
Biodegradation of dioxane by a microbial consortium from industrial activated sludge.
Activated sludge samples were collected from an industrial wastewater treatment plant with dioxane concentrations ranging between 50 and 100 mg/liter. Fifty milliliters of unamended activated sludge samples was incubated in triplicate sterile 250-ml Boston round bottles equipped with Mininert valves at 30°C with 150 rpm of agitation. Bottles were maintained under aerobic conditions and monitored until dioxane concentrations reached below the detection limit. At each time point, a 0.5-ml sample was collected for subsequent gene abundance and expression analyses.
Extraction of nucleic acids and cDNA synthesis.
Total nucleic acids were extracted from samples using a modified phenol-chloroform extraction method (40). Briefly, bacterial cells were centrifuged for 3 min at 13,000 × g, and the supernatant was removed. Cells were resuspended in 250 μl of lysis buffer (50 mM sodium acetate, 10 mM EDTA [pH 5.1]), 100 μl 10% sodium dodecyl sulfate, and 1.0 ml pH 8.0 buffer-equilibrated phenol. One gram of 100-μm-diameter zirconia-silica beads (Biospec Products, Bartlesville, OK) were added to the samples, and cells were lysed by being heated to 65°C for 2 min, undergoing bead beating for 2 min with a Mini bead beater (Biospec Products, Bartlesville, OK), and being incubated for 8 min at 65°C, followed by an additional bead beating for 2 min. Samples were centrifuged for 5 min at 13,000 × g, and the aqueous lysate was transferred to a new DNase- and RNase-free microcentrifuge tube. Nucleic acids were extracted twice with 1 volume of phenol-chloroform-isoamyl alcohol (25:24:1) and once with 1 volume of chloroform-isoamyl alcohol (24:1). Isolated nucleic acids were precipitated from solution at −20°C overnight with the addition of 0.1 volume of 3 M sodium acetate and 1 volume of isopropanol. The precipitate was collected by centrifugation at 4°C for 30 min at 20,000 × g, washed with 70% ethanol, and resuspended in 100 μl DNase- and RNase-free water. The concentration and purity of the DNA and RNA were measured using a Nanodrop 2000C spectrophotometer (Thermo Scientific, Wilmington, DE). All extracts were stored at −80°C.
DNA was removed from samples by treatment with DNase I (Turbo DNase, Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Briefly, 4 U of Turbo DNase and 1× Turbo DNase buffer were added to each nucleic acid extract. Samples were incubated at 37°C for 30 min, and DNase was removed by extraction with phenol-chloroform-isoamyl alcohol solution (25:24:1). RNA concentrations were quantified on a Nanodrop 2000C (Thermo Scientific), and extracts were stored at −80°C until ready for use.
Reverse transcription was performed on RNA extracts using 50 ng of total RNA, 2.5 μM random hexamers, 0.5 mM each deoxynucleoside triphosphate, 1× First Strand buffer (Life Technologies, Carlsbad, CA), 10 mM dithiothreitol (DTT), 40 U RNaseOUT recombinant RNase inhibitor, and 200 U Moloney murine leukemia virus (MMLV) reverse transcriptase in a total volume of 20 μl per reaction. Control reactions consisted of a reverse transcriptase-free control to identify potential contamination by residual genomic DNA and a no-template control. Samples were incubated at 37°C for 1 h and then transferred to 70°C for 15 min to inactivate the reverse transcription reaction. The synthesized cDNA was stored at −80°C until ready for use.
qPCR.
Candidate biomarkers were amplified by quantitative PCR (qPCR) using the primers listed in Table 1. A temperature gradient was performed to identify the optimal annealing temperature for the primers. All qPCRs were performed in a total volume of 20 μl containing: 1× Maxima Sybr green master mix (Thermo Scientific, Waltham, MA), 0.3 μM each primer (IDTDNA, Coralville, IA), 0.2 μg/ml bovine serum albumin (BSA), and 2 μl template DNA or cDNA. BSA was included because it was previously shown to mitigate inhibition in environmental samples (41, 42). The cycling parameters for qPCR of all targets, with the exception of the universal 16S rRNA gene primers, included 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and annealing at 60°C for 45 s. Universal 16S rRNA gene primers followed a similar cycling profile, except annealing was performed at 50°C as previously described by Harms and coworkers (43). A melt curve analysis accompanied each qPCR to confirm the specificity of the amplification plots. All reactions were performed and analyzed with an Applied Biosystems StepOnePlus (Life Technologies, Carlsbad, CA) real-time PCR system. Standard curves for gene abundance analysis were generated from genomic DNA extracted from pure cultures of CB1190. The minimum detection limit for all primer sets used in this study was 2 × 106 copies/liter.
TABLE 1.
Sequences of oligonucleotide primers used in this studya
| Target gene/target enzyme | Primer | Sequence (5′→3′) |
|---|---|---|
| dxmB/dioxane monooxygenase (DXMO) | DXMO-For | CCAAACGGGCGTCAGTCAT |
| DXMO-Rev | AGAACGTGCGCTCCCAAAG | |
| poxD/phenol-2-monooxygenase (PHE) | PHE-For | TGGTCCGGCGAGCCCTTGTA |
| PHE-Rev | CACACCTGCTCCGACGGCTG | |
| prm1A/propane monooxygenase (PrMO) | PrMO-For | GAAGAGTCGTGGAAGCAGATC |
| PrMO-Rev | GTACTTGTACTCGAACCACTCG | |
| sad/aldehyde dehydrogenase (ALDH) | ALDH-For | GCCGACGCTTTTAGCAGATG |
| ALDH-Rev | TCATTAACGGCAGCAAACGC | |
| adh/alcohol dehydrogenase (ADH) | ADH-For | ACCAAGGACCTCACCTCGTA |
| ADH-Rev | AACGGATGCGCGTTGTTC | |
| CB1190-like 16S rRNA | CB119016S-For | ACGGTCTCGCAGCCCTCTGT |
| CB119016S-Rev | AGCGGGTTATGCCGGGGACT | |
| Universal 16S rRNA | Uni16S-For | ATGGCTGTCGTCAGCT |
| Uni16S-Rev | ACGGGCGGTGTGTAC |
Analytical techniques.
A gas chromatograph equipped with a flame ionization detector (GC-FID) was used to measure dioxane concentrations. Fifty-microliter liquid samples were collected and filtered through 0.2-μm-pore nylon syringe filters. Two microliters of the filtered liquid samples was directly injected into a Hewlett-Packard 6890 GC-FID (Hewlett-Packard, Atlanta, GA) with a Restek Stabilwax-DB capillary column (30-m by 0.53-mm inside diameter by 1 μm; Restek, Bellefonte, PA). The injector and detector were maintained at 220°C and 250°C, respectively. The oven was programmed to ramp from 80°C to 140°C at 20°C/min and held for 1 min at 140°C. The retention time of dioxane was 3.7 min. The minimum detection limit was determined to be 0.80 mg/liter, and the accuracy of measurement was better than ±4% with this method.
Gene expression results were calculated using the comparative threshold cycle (CT) method as previously described (44). The efficiency of qPCR amplification for each target gene was determined through analysis of a serial dilution of cDNA derived from CB1190 actively transforming dioxane. For time course experiments, data were first normalized to the rpoD gene as described by Grostern and coworkers (29) because this gene produced stable expression across different treatments. A second normalization to the initial transcription levels was also performed to quantify increases in expression relative to time zero. In addition, dioxane degradation rates were calculated during consecutive intervals as the change in dioxane concentration divided by time for the mixed microbial community in activated sludge.
RESULTS
Phylogenetic analysis of DNA sequences in CB1190 encoding select monooxygenase genes.
A comprehensive evaluation of monooxygenase-associated genes in CB1190 is presented in Fig. S1 in the supplemental material. Monooxygenase homologs in CB1190 were selected from this list based on previous laboratory studies demonstrating the ability of bacteria harboring these enzymes to transform dioxane or other similarly recalcitrant compounds (19, 20, 28). We evaluated DXMO, PHE, PrMO, alcohol dehydrogenase (ADH), and aldehyde dehydrogenase (ALDH) as potential gene targets that could specifically indicate dioxane degradation in environmental samples. A Basic Local Alignment Search Tool (BLAST) analysis of these selected genes was conducted to compare sequence similarities to other bacteria and to identify regions for qPCR primer development that would only quantify gene targets from known dioxane-degrading bacteria. Based on the phylogenetic analysis, the DXMO, ADH, and ALDH gene sequences were the most specific targets for bacteria that were able to degrade dioxane by metabolic or cometabolic pathways. The PrMO and PHE gene targets show similarity to bacteria that are able to cometabolically transform dioxane; however, these gene targets were also found in a number of bacteria whose ability to biodegrade dioxane has not yet been determined.
Time course analysis of monooxygenase gene abundance and expression in pure cultures.
Gene abundance and expression were monitored during dioxane biodegradation by pure cultures of CB1190. This strain was initially cultured on AMS medium supplemented with 20 mM glucose to increase biomass and to determine whether monooxygenase gene expression was induced in the presence of dioxane. Cells were cultured until a biomass of a 40- to 60-mg/liter total protein concentration was achieved. Cells were washed then transferred into fresh AMS medium supplemented with 1 mM dioxane. This approach allowed a time course evaluation of gene abundance and expression during biotransformation of dioxane (Fig. 1 and 2, respectively). Glucose-grown CB1190 demonstrated a 3-day lag period prior to biodegradation of dioxane. All gene targets were present and abundant in DNA extracts of CB1190 ranging from 1.1 × 107 to 1.0 × 1012 gene copies per liter. Over 95% of the dioxane was removed in just 5 days. In addition, gene copies increased an average of 4 orders of magnitude over the course of 7 days. A Pearson's correlation indicated a statistically significant inverse relationship between the concentration of monooxygenase gene targets and dioxane over the course of this study (Table 2). Dioxane decreased to below the method detection limit after 6 days, which corresponded to a decrease in all gene targets. However, gene abundance for all gene targets increased at day 7 in the absence of dioxane.
FIG 1.
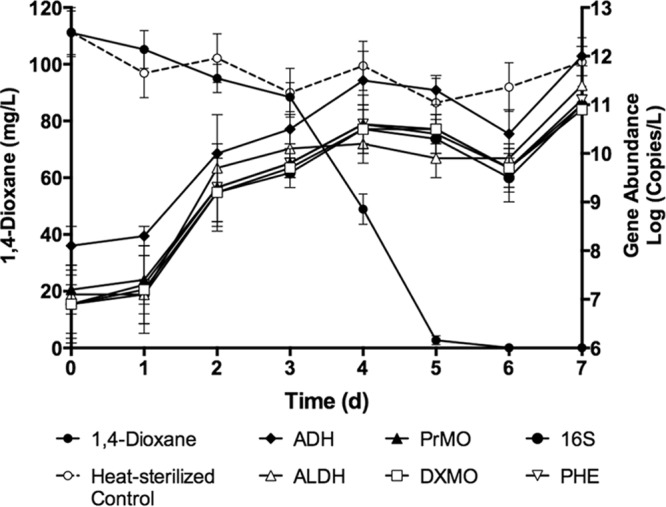
Time course analysis of gene abundance for 16S rRNA, dioxane monooxygenase (DXMO), propane monooxygenase (PrMO), alcohol dehydrogenase (ADH), and aldehyde dehydrogenase (ALDH) gene targets during biotransformation of 1,4-dioxane by pure cultures of Pseudonocardia dioxanivorans CB1190 following enrichment on AMS medium supplemented with 20 mM glucose. Enrichment on glucose increased the lag time before 1,4-dioxane biodegradation was observed. The abundance of all gene targets increased due to increase in CB1190 populations. This analysis also revealed a lag phase before and after 1,4-dioxane biodegradation, which suggests growth on a 1,4-dioxane breakdown product. Error bars represent the standard deviations of three replicates.
FIG 2.
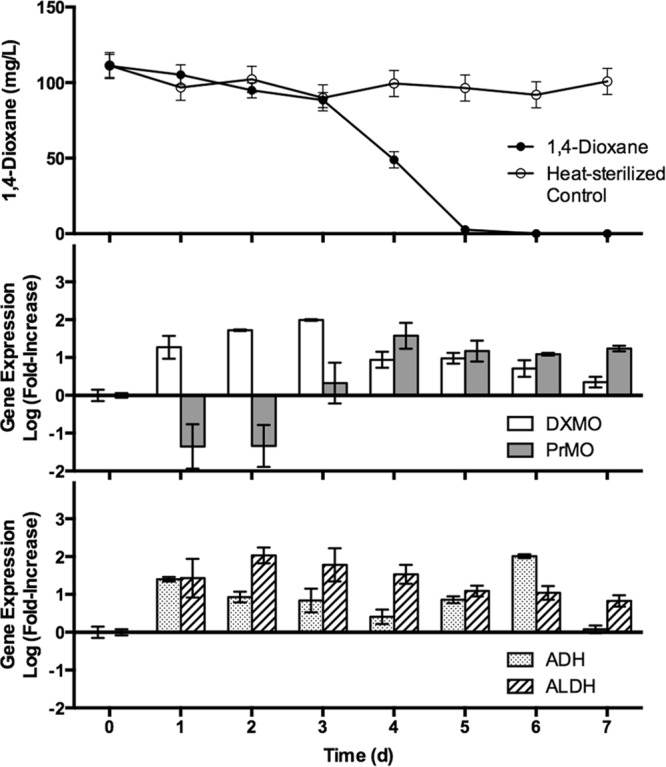
Time course gene expression analysis of select monooxygenase and dehydrogenase gene targets in pure cultures of Pseudonocardia dioxanivorans CB1190 during biodegradation of 1,4-dioxane following enrichment on AMS medium supplemented with 20 mM glucose. The transcripts of DXMO and ALDH together varied with 1,4-dioxane, whereas transcripts of PrMO were high only when 1,4-dioxane was undergoing active transformation at lower concentrations below 50%. PHE is not displayed because transcripts from each sampling point were below the detection limit. Error bars represent the standard deviations of three replicates.
TABLE 2.
Correlations between concentration of gene targets and 1,4-dioxane during biodegradation by pure cultures of Pseudonocardia dioxanivorans CB1190
| Target enzyme | Correlation witha: |
|
|---|---|---|
| 1,4-Dioxane | 16S rRNA | |
| DXMO | −0.786* | 0.998** |
| PrMO | −0.770* | 0.999** |
| PHE | −0.773* | 0.999** |
| ADH | −0.781* | 0.997** |
| ALDH | −0.714* | 0.972** |
All gene targets were significantly correlated to biodegradation of 1,4-dioxane by CB1190. *, P < 0.05; **, P < 0.01.
Monooxygenase and dehydrogenase gene expression analyses for pure cultures of CB1190 in the presence of dioxane are presented in Fig. 2. DXMO, PrMO, ADH, and ALDH showed increased gene expression during biodegradation of dioxane. Interestingly, the DXMO, ADH, and ALDH gene targets were highly upregulated after only 1 day of incubation in AMS supplemented with 100 mg/liter dioxane, and this increased expression occurred prior to biotransformation of dioxane. Expression of DXMO and ALDH gene targets was significantly correlated (r = 0.92, P < 0.01). Both DXMO and ALDH genes had the highest levels of expression when dioxane was present in high concentrations. After 3 days, expression of DXMO and ALDH steadily decreased.
ADH was initially upregulated; however, this increased expression steadily dropped until 50% of the dioxane remained. Surprisingly, ADH expression increased on days 5 and 6, when dioxane was nearly completely removed. PrMO expression remained below initial levels until day 3, just prior to significant dioxane removal, at which point expression increased to day 0 levels. Subsequently, expression of PrMO peaked at 1.57 orders of magnitude on day 4, when approximately 50% of the dioxane had been utilized, and remained high even in the absence of measureable concentrations of dioxane. Relative gene expression of PHE was also investigated; however, copy numbers measured at all time points were below the detection limit of the method.
Application of biomarkers to industrial activated sludge.
Figure 3 illustrates the biodegradation of dioxane and the fluctuations of selected gene targets as dioxane was transformed by a consortium of microorganisms within the activated sludge of an industrial wastewater treatment plant. Average concentrations of dioxane in these samples were measured at 81.97 mg/liter (SD, 1.56 mg/liter). Microorganisms in the sludge were able to remove dioxane to levels below the detection limit in 10 days (Fig. 3) after an initial lag period of nearly 5 days.
FIG 3.
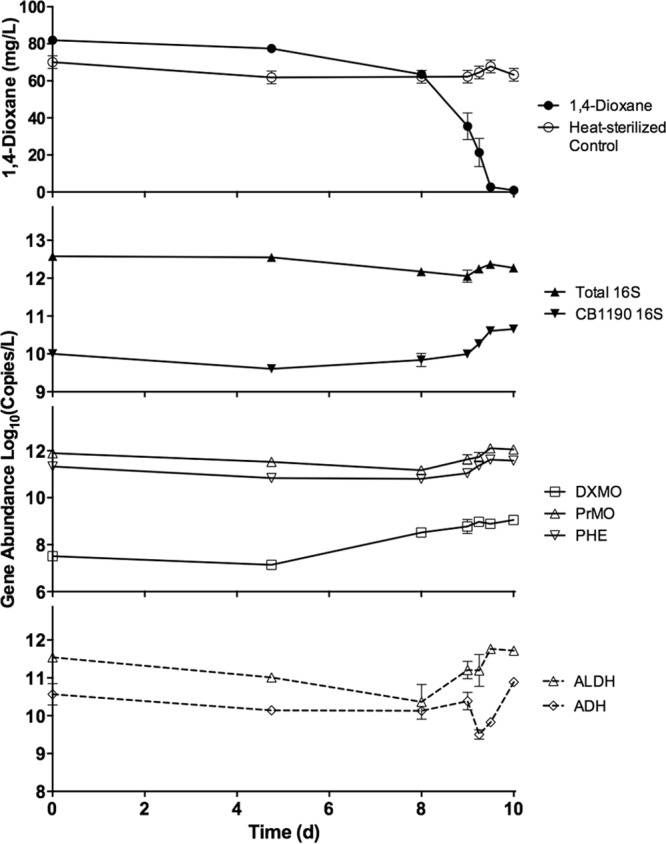
Gene abundance analysis and biodegradation of 1,4-dioxane by a microbial consortium in industrial activated sludge. Total bacterial populations experienced relatively small changes compared to bacterial populations containing the DXMO or CB1190 16S rRNA genes. DXMO and CB1190 16S rRNA genes were the only targets significantly correlated with 1,4-dioxane concentrations. Error bars represent the standard deviations of three replicates.
The concentration of total bacteria in activated sludge did not vary greatly over the course of the experiment and ranged between 1.1 × 1012 and 3.8 × 1012 copies/liter (Fig. 3). Copies of the DXMO gene demonstrated the largest increase of all targets monitored, with an increase of nearly 100-fold, while dioxane was degraded after day 5. A Pearson's correlation analysis of gene abundance and dioxane concentrations resulted in statistically significant correlations for the CB1190 16S rRNA gene (r = −0.90, P < 0.01) and DXMO gene (r = −0.88, P < 0.01) targets, supporting these genes as promising biomarkers for dioxane degradation. The copy numbers of ADH (P = 0.81), ALDH (P = 0.21), PHE (P = 0.06), and universal 16S rRNA (P = 0.28) gene targets were not significantly correlated with dioxane concentrations.
The dioxane degradation rate was calculated as the change in dioxane divided by the change in time elapsed during active degradation. Activated sludge samples were able to remove dioxane at a maximum rate of 97.0 mg/liter/day, which occurred during a 5-h interval from day 9.3 to day 9.5 (Fig. 4). In addition, the percentage of CB1190-like bacteria quantified using the concentration of 16S rRNA gene copies specific to sequences associated with CB1190 and other closely related Pseudonocardia strains within the total bacterial population demonstrated a statistically significant correlation with decreasing dioxane concentrations (r = −0.94, P < 0.01).
FIG 4.
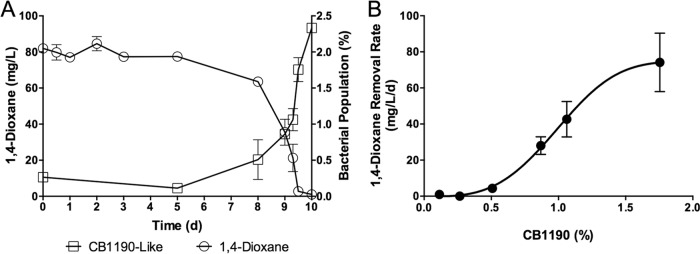
The relationship between 1,4-dioxane concentrations and abundance of CB1190-like bacteria in activated sludge samples. (A) The percentage of CB1190-like bacteria within the total bacterial population was a good indicator of 1,4-dioxane biodegradation. (B) 1,4-Dioxane degradation rates increased with increases in the fraction of CB1190-like bacteria within the total bacterial population. Error bars represent the standard deviations of three replicates.
Expression of monooxygenase gene targets in activated sludge samples is presented in Fig. 5. DXMO and PrMO gene targets demonstrated increased gene transcription during biodegradation of dioxane, whereas PHE was not related to dioxane concentrations. DXMO had the highest relative expression; however, increased levels of expression were delayed until day 8, when dioxane reached 63.55 mg/liter. PrMO experienced increased expression on the 5th day, and expression remained high for the duration of the experiment. ADH and ALDH gene expression data are presented in Fig. 5. Both dehydrogenase targets had increased expression over the course of dioxane biodegradation in activated sludge. The highest levels of expression for ADH and ALDH were achieved when 70% of the dioxane had been utilized.
FIG 5.
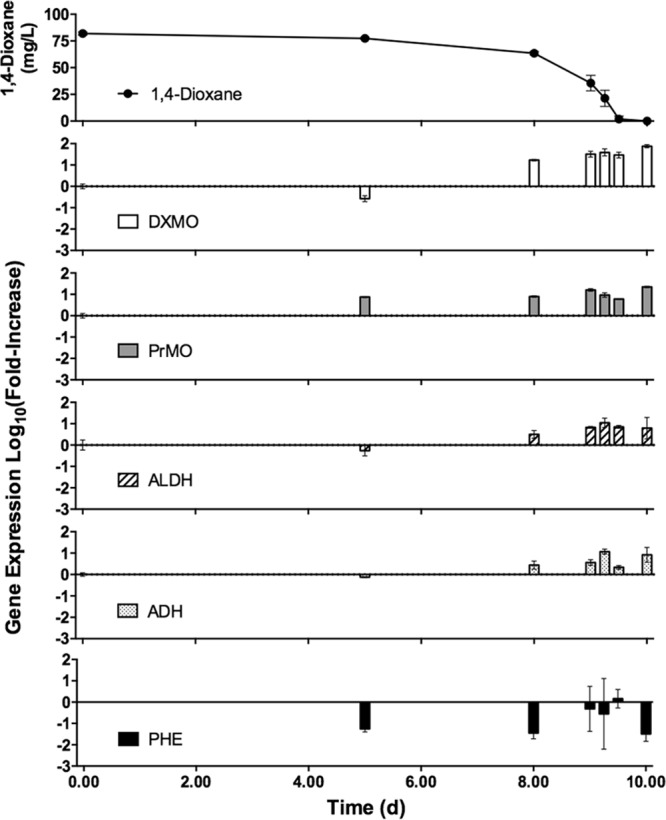
Time-dependent relationship between gene expression of selected monooxygenase and dehydrogenase targets and concentrations of 1,4-dioxane in industrial activated sludge. Biodegradation of 1,4-dioxane accompanied increased expression of the DXMO, ALDH, and PrMO gene targets. Error bars represent the standard deviations of three replicates.
Inhibition of monooxygenase activity by acetylene exposure.
Pure cultures of CB1190 grown using dioxane as the sole carbon source were exposed to acetylene gas to inhibit monooxygenase activity during biodegradation of dioxane (Fig. 6A and B). Bacteria exposed to acetylene exhibited a 24-h period of inhibition followed by recovery leading to complete degradation of dioxane in 48 h (Fig. 6B). This resulted in an approximately 40-fold (1.6 orders of magnitude) increase of DXMO at 8 h. DXMO expression remained elevated in the presence of dioxane. However, biodegradation resumed, and DXMO was repressed when dioxane concentrations were below the detection limit at 48 h. In comparison, DXMO expression increased a maximum of 2-fold (0.3 order of magnitude) greater than initial concentrations when less than 20% of the dioxane remained in the unexposed control cultures.
FIG 6.
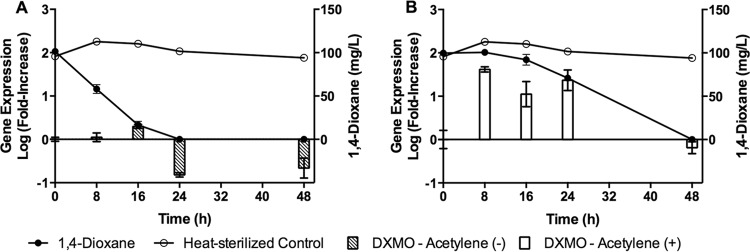
1,4-Dioxane biodegradation and gene expression of DXMO in control (A) and acetylene-exposed (B) pure cultures of Pseudonocardia dioxanivorans CB1190. Expression of DXMO in unexposed cells was relatively low because samples were collected from a culture that was actively using 1,4-dioxane as the sole carbon source. Expression of DXMO was elevated in acetylene-exposed cells because 1,4-dioxane concentrations remained high for 24 h, confirming that biodegradation was inhibited due to inactivation of the monooxygenase protein by exposure to acetylene. Error bars represent the standard deviations of three replicates.
DISCUSSION
Our study has shown that gene abundance and expression analysis of BMM targets has the capacity to indicate the potential for dioxane biodegradation in pure cultures and complex microbial communities, such as those found within activated sludge. Quantification of monooxygenase and dehydrogenase genes in pure cultures of CB1190 clearly demonstrated the effectiveness of qPCR to monitor growth of bacterial cultures in relation to dioxane biodegradation (Fig. 1). Gene concentrations are representative of cell copies, with CB1190 containing 3 copies of the 16S rRNA gene and single copies of the selected monooxygenase and dehydrogenase genes (35). All targets experienced an initial lag phase, exponential growth phase, and subsequent stationary phase until day 6, when dioxane was removed to levels below the detection limit. Surprisingly, all gene targets showed an increase of over 10-fold on day 7 in the absence of dioxane. This result could be a combination of endogenous respiration on cell lysates or utilization of a secondary substrate that is likely a breakdown product of dioxane. While biphasic growth has not been confirmed in CB1190, diauxie in other actinomycetes, such as several Streptomyces spp., has been widely documented (45, 46). The results of our gene abundance analysis for DXMO, PHE, and ALDH in pure cultures of CB1190 demonstrate that these genes can be used as taxonomic identifiers of CB1190 and other dioxane-degrading organisms because of their specificity, as indicated by screening against sequences in the National Center for Biotechnology Information (NCBI) GenBank databases.
The results of the time course expression analysis of monooxygenase and dehydrogenase gene targets in CB1190 during dioxane degradation illustrate the transcription of DXMO and PrMO genes as well as both dehydrogenase targets analyzed. Although PHE was abundant in pure culture DNA extracts, this target was not an effective indicator of actual dioxane biotransformation because the relative expression of this gene was below the detection limit at all time points evaluated (Fig. 2). DXMO and ALDH are homologs of the tetrahydrofuran oxygenase beta subunit (thmB) and the aldehyde dehydrogenase (thmH) of Pseudonocardia tetrahydrofuranoxydans strain K1, respectively (15, 47). These targets have been shown to be involved in the metabolic and cometabolic biodegradation of tetrahydrofuran and dioxane (19, 29). Grostern and coworkers (29) identified these genes as being differentially upregulated in CB1190 during growth on dioxane compared to pyruvate/glyoxylate, which is supported by our time course characterization of these genes in CB1190. However, our results for PrMO gene expression differ from those in the previous study, where it was determined that only DXMO-related genes among the different BMMs within the CB1190 genome were significantly expressed during growth on dioxane. We observed the relative expression of PrMO to be much lower than day 0 levels, until approximately 50% of the dioxane was removed at day 4. Furthermore, gene expression remained high through the termination of the experiment after 7 days. The contrasting results may be explained by differences in preculture carbon sources and the time-dependent relationship revealed by our gene expression analysis. Our study utilized glucose to increase biomass prior to the time course experiment and to investigate inducible gene expression. Several studies have shown that glucose can impart a more defined diauxic effect compared to alternative sugars (48, 49), which can potentially influence gene expression.
Toluene, a monooxygenase-inducing substrate, was evaluated to determine whether biomarkers for dioxane biodegradation would be expressed during growth on this alternative carbon source (see Fig. S2 in the supplemental material). DXMO was the only gene target that did not experience higher levels of expression throughout growth relative to the initial time of inoculation. Interestingly, expression of PrMO and PHE was nearly 2 orders of magnitude higher than the initial levels (see Fig. S2). These results highlight the importance of analyzing multiple gene targets involved in the dioxane degradation pathway to specifically identify the potential for dioxane biodegradation in the environment.
The influence of sample complexity on gene abundance and expression emphasizes the benefits of quantifying genes and transcripts involved in contaminant biodegradation. Our study used industrial activated sludge as a model for mixed microbial communities because these samples demonstrated active dioxane-degrading capabilities within a complex engineered environment. Gene expression analysis of PHE in these samples suggests this target is a poor indicator of active contaminant biodegradation (Fig. 5). Rather, the gene abundance of PHE served as a more appropriate indicator of dioxane biodegradation (Fig. 3), suggesting the PHE primer set is a good taxonomic indicator of CB1190 and other very closely related microorganisms. This finding is attributed to the high homology of the PHE primer sequences to CB1190 and further supported by the significant positive correlation (r = 0.93, P < 0.01) between CB1190 16S rRNA genes and PHE abundance in activated sludge. DXMO and ALDH expression profiles were very similar, which was expected since these targets are located within the same gene cluster associated with dioxane or tetrahydrofuran degradation in CB1190 (35), Pseudonocardia tetrahydrofuranoxydans K1 (15), Pseudonocardia sp. strain ENV478 (19), and Rhodococcus sp. strain YYL (50). The time course gene expression results in activated sludge samples lend further support to the inducible nature of DXMO. Dioxane degradation lagged in activated sludge samples for a period of 5 days, and DXMO expression was repressed.
Expression of biomarker genes demonstrated temporal differences in pure cultures compared to activated sludge samples. Pure cultures of CB1190 showed increased expression of DXMO, ADH, and ALDH targets relative to time zero before dioxane degradation was observed, whereas expression of these targets increased during observed dioxane degradation in activated sludge samples. Decreased levels of expression for DXMO, ALDH, and ADH in the presence of dioxane are attributed to inhibition or toxicity of CB1190-like bacteria, as evidenced by lower concentrations of CB1190 16S rRNA genes in activated sludge samples (Fig. 3). CB1190-like bacterial populations decreased nearly 60%, from 1.00 × 1010 cells/liter on day 0 to 4.04 × 109 cells/liter, while total bacterial populations were relatively unchanged during the same time period, indicating that the expression of genes involved in dioxane degradation would be much lower with respect to the rpoD reference gene.
In addition, PrMO demonstrated higher levels of expression after more than 50% of the dioxane was degraded by pure cultures. However, expression of PrMO was increased before biodegradation of dioxane had occurred in activated sludge samples (Fig. 5). It is possible that a PrMO-inducing carbon source was present in the activated sludge samples given the high concentrations of these genes (Fig. 3) and transcripts. Indeed, waste streams from the polyester manufacturing process often contain dioxane and other aromatic compounds (23, 51, 52). Based on our pure culture results using toluene as the sole carbon source (see Fig. S2 in the supplemental material), it is reasonable for PrMO expression to be induced by toluene or another aromatic compound in this industrial activated sludge sample. In addition, other contaminants, like acetone and phenol, have been determined to increase expression of the prmA gene of the propane monooxygenase in Mycobacterium goodii (53). While the broad specificity of our PrMO target may limit its effectiveness as a biomarker, multiple studies have demonstrated the ability of propanotrophs to cometabolize dioxane, supporting the use of this target as a potential biomarker for dioxane biodegradation (20, 28, 54).
Interestingly, with the exception of PHE, all monooxygenase and dehydrogenase genes evaluated in this study maintained increased gene expression in activated sludge samples, even in the absence of dioxane. This was also the case for all gene targets, except PHE and ADH, in pure cultures of CB1190. Despite the increased expression after dioxane was removed below the detection limit, our study determined statistically significant inverse relationships between dioxane concentrations and both DXMO and ALDH (Table 3). A number of factors may contribute to the observed expression of DXMO and ALDH in the absence of dioxane, including induction by daughter products of dioxane metabolism as determined in previous studies (29, 55). Continued expression upon depletion of inducing substrate has also been shown for reductive dehalogenase-encoding genes. A previous study determined that the expression of tceA or vcrA remained elevated for more than 48 h after the disappearance of TCE (12). A more recent study on expression of the benzylsuccinate synthase gene, bssA, identified a rapid decline of mRNA transcripts by over 2 orders of magnitude upon depletion of nitrate (56). However, similar to our study showing maintained expression levels of DXMO and ALDH, bssA transcripts stabilized and maintained greater than 1.0 × 104 copies/ml for over 3 weeks after nitrate was removed (56).
TABLE 3.
Correlation matrix for 1,4-dioxane concentrations and transcripts of select monooxygenase and dehydrogenase gene targets at different times during biodegradation of 1,4-dioxane in industrial activated sludge
| Target enzyme | Correlation witha: |
||||
|---|---|---|---|---|---|
| PrMO | ALDH | PHE | ADH | 1,4-Dioxane | |
| DXMO | 0.617 | 0.961** | −0.002 | 0.873** | −0.845* |
| PrMO | 0.556 | −0.524 | 0.634 | −0.614 | |
| ALDH | 0.171 | 0.876** | −0.852* | ||
| PHE | −0.178 | −0.103 | |||
| ADH | −0.737 | ||||
Expression of DXMO and ALDH was significantly correlated (boldface) with 1,4-dioxane biodegradation. *, P < 0.05; **, P < 0.01.
It should be recognized that a genetic biomarker approach relies upon the indirect association between gene transcripts and enzyme activity (Fig. 6). Our results demonstrated transient inhibition of dioxane biodegradation in pure cultures by exposure to acetylene, a potent inhibitor of monooxygenase activity (57, 58). Acetylene binds to specific protein regions, resulting in inactivation of the enzyme (57) with no effect on gene transcription and translation. This mechanism of inhibition complicates the interpretation of biomarker analysis (Fig. 6). In cultures exposed to acetylene, dioxane biodegradation was temporarily suspended for 24 h; however, DXMO expression was elevated. DXMO expression remained high until cultures resumed biodegradation of dioxane, suggesting a potential false-positive expression analysis of the DXMO biomarker. The increased expression of DXMO could be explained by the inducible monooxygenase. While dioxane was present, transcription of DXMO was induced, and the proteins were synthesized. Acetylene exposure did not influence transcription; therefore, expression of DXMO was not downregulated. Ultimately, CB1190 was able to completely degrade dioxane below the method detection limit in 48 h, suggesting that comprehensive characterization of environmental samples for potential inhibitors of enzyme activity is critical for the effective application of biomarkers utilizing the abundance and expression of catabolic genes.
The ability of mixed microbial communities to degrade dioxane was determined by several studies conducted using wastewater, groundwater, and soil samples (24, 27, 59). Sei and colleagues (59) investigated the ability of activated sludge from different wastewater treatment plants to biodegrade dioxane and found that only one-third of examined sludge samples were able to degrade dioxane. Furthermore, this activity was conferred only with the addition of tetrahydrofuran as the primary substrate. A recent study by Li and coworkers (60) investigating the distribution of different multicomponent monooxygenase genes in a dioxane-contaminated aquifer identified that the dxmA gene, which encodes the alpha subunit of the dioxane hydroxylase, was present at the source, middle, and edge of the plume. The study reported that dxmA was highly enriched at the source zone compared to all other locations, which supports the dxmA gene as a promising biomarker target (60). However, this target was also found in background samples collected in uncontaminated areas. These findings suggest that while BMMs may be widely distributed, their presence alone is insufficient to indicate biodegradation.
Biomarkers predicting the potential for dioxane biodegradation are currently unavailable to environmental professionals and researchers. Our research builds upon previous studies that have identified the differential expression of monooxygenase gene targets during growth on dioxane or tetrahydrofuran relative to growth on alternative carbon sources (19, 29) through the development of a time-dependent gene expression profile before, during, and throughout dioxane utilization by pure cultures and mixed microbial communities in a complex environment. This approach provides a level of characterization that addresses whether dioxane-degrading bacteria are not only present but active through an evaluation of the abundance as well as expression of potential biomarker candidates. Biomarkers targeting functional genes involved in the biodegradation of dioxane would greatly benefit practitioners in identifying the most appropriate microbes and biogeochemical conditions for the removal of dioxane contamination in waste streams and other environments.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Air Force Civil Engineer Center award FA8903-11-C-8005. P. Pornwongthong was awarded a Royal Thai Government Scholarship through the Ministry of Science and Technology, Thailand.
Linda Tseng, Michelle Myers, Thomas Folker, and Conor Carr assisted with sample collection and analysis. We acknowledge Adria Bodour, Janet Anderson, and Richard H. Anderson for comments during the preparation of the manuscript.
Footnotes
Published ahead of print 14 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04162-13.
REFERENCES
- 1.Notomista E, Lahm A, Di Donato A, Tramontano A. 2003. Evolution of bacterial and archaeal multicomponent monooxygenases. J. Mol. Evol. 56:435–445. 10.1007/s00239-002-2414-1 [DOI] [PubMed] [Google Scholar]
- 2.Leahy JG, Batchelor PJ, Morcomb SM. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 27:449–479. 10.1016/S0168-6445(03)00023-8 [DOI] [PubMed] [Google Scholar]
- 3.Holmes AJ. 2009. The diversity of soluble di-iron monooxygenases with bioremediation applications, p 91–102 In Singh A, Kuhad RC, Ward OP. (ed), Advances in applied bioremediation. Springer-Verlag, Berlin, Germany [Google Scholar]
- 4.Nebe J, Baldwin BR, Kassab RL, Nies L, Nakatsu CH. 2009. Quantification of aromatic oxygenase genes to evaluate enhanced bioremediation by oxygen releasing materials at a gasoline-contaminated site. Environ. Sci. Technol. 43:2029–2034. 10.1021/es900146f [DOI] [PubMed] [Google Scholar]
- 5.Dionisi HM, Layton AC, Harms G, Gregory IR, Robinson KG, Sayler GS. 2002. Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl. Environ. Microbiol. 68:245–253. 10.1128/AEM.68.1.245-253.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman NV, Bui NB, Holmes AJ. 2006. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ. Microbiol. 8:1228–1239. 10.1111/j.1462-2920.2006.01015.x [DOI] [PubMed] [Google Scholar]
- 7.Dominguez RF, da Silva MLB, McGuire TM, Adamson D, Newell CJ, Alvarez PJJ. 2008. Aerobic bioremediation of chlorobenzene source-zone soil in flow-through columns: performance assessment using quantitative PCR. Biodegradation 19:545–553. 10.1007/s10532-007-9160-4 [DOI] [PubMed] [Google Scholar]
- 8.Yagi JM, Suflita JM, Gieg LM, DeRito CM, Jeon CO, Madsen EL. 2010. Subsurface cycling of nitrogen and anaerobic aromatic hydrocarbon biodegradation revealed by nucleic acid and metabolic biomarkers. Appl. Environ. Microbiol. 76:3124–3134. 10.1128/AEM.00172-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee PKH, Macbeth TW, Sorenson KS, Deeb RA, Alvarez-Cohen L. 2008. Quantifying genes and transcripts to assess the in situ physiology of “Dehalococcoides” spp. in a trichloroethene-contaminated groundwater site. Appl. Environ. Microbiol. 74:2728–2739. 10.1128/AEM.02199-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald IR, Bodrossy L, Chen Y, Murrell JC. 2008. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 74:1305–1315. 10.1128/AEM.02233-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shukla AK, Vishwakarma P, Upadhyay SN, Tripathi AK, Prasana HC, Dubey SK. 2009. Biodegradation of trichloroethylene (TCE) by methanotrophic community. Bioresour. Technol. 100:2469–2474. 10.1016/j.biortech.2008.12.022 [DOI] [PubMed] [Google Scholar]
- 12.Lee PKH, Johnson DR, Holmes VF, He JZ, Alvarez-Cohen L. 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl. Environ. Microbiol. 72:6161–6168. 10.1128/AEM.01070-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monard C, Martin-Laurent F, Lima O, Devers-Lamrani M, Binet F. 2012. Estimating the biodegradation of pesticide in soils by monitoring pesticide-degrading gene expression. Biodegradation 24:203–213. 10.1007/s10532-012-9574-5 [DOI] [PubMed] [Google Scholar]
- 14.Jechalke S, Rosell M, Martinez-Lavanchy PM, Perez-Leiva P, Rohwerder T, Vogt C, Richnow HH. 2011. Linking low-level stable isotope fractionation to expression of the cytochrome P450 monooxygenase-encoding ethB gene for elucidation of methyl tert-butyl ether biodegradation in aerated treatment pond systems. Appl. Environ. Microbiol. 77:1086–1096. 10.1128/AEM.01698-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiemer B, Andreesen JR, Schrader T. 2003. Cloning and characterization of a gene cluster involved in tetrahydrofuran degradation in Pseudonocardia sp. strain K1. Arch. Microbiol. 179:266–277. 10.1007/s00203-003-0526-7 [DOI] [PubMed] [Google Scholar]
- 16.Kim YM, Jeon JR, Murugesan K, Kim EJ, Chang YS. 2009. Biodegradation of 1,4-dioxane and transformation of related cyclic compounds by a newly isolated Mycobacterium sp. PH-06. Biodegradation 20:511–519. 10.1007/s10532-008-9240-0 [DOI] [PubMed] [Google Scholar]
- 17.Li MY, Fiorenza S, Chatham JR, Mahendra S, Alvarez PJJ. 2010. 1,4-Dioxane biodegradation at low temperatures in Arctic groundwater samples. Water Res. 44:2894–2900. 10.1016/j.watres.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 18.Mahendra S, Alvarez-Cohen L. 2005. Pseudonocardia dioxanivorans sp. nov., a novel actinomycete that grows on 1,4-dioxane. Int. J. Syst. Evol. Microbiol. 55:593–598. 10.1099/ijs.0.63085-0 [DOI] [PubMed] [Google Scholar]
- 19.Masuda H, McClay K, Steffan RJ, Zylstra GJ. 2012. Biodegradation of tetrahydrofuran and 1,4-dioxane by soluble diiron monooxygenase in Pseudonocardia sp. strain ENV478. J. Mol. Microbiol. Biotechnol. 22:312–316. 10.1159/000343817 [DOI] [PubMed] [Google Scholar]
- 20.Vainberg S, McClay K, Masuda H, Root D, Condee C, Zylstra GJ, Steffan RJ. 2006. Biodegradation of ether pollutants by Pseudonocardia sp. strain ENV478. Appl. Environ. Microbiol. 72:5218–5224. 10.1128/AEM.00160-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.US Environmental Protection Agency. 2012. Drinking water standards and health advisories. Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 22.Anderson RH, Anderson JK, Bower PA. 2012. Co-occurrence of 1,4-dioxane with trichloroethylene in chlorinated solvent groundwater plumes at US Air Force installations: fact or fiction. Integr. Environ. Assess. Manag. 8:731–737. 10.1002/ieam.1306 [DOI] [PubMed] [Google Scholar]
- 23.Mohr T, Stickney J, Diguiseppi B. 2010. Environmental investigation and remediation: 1,4-dioxane and other solvent stabilizers. CRC Press, Boca Raton, FL [Google Scholar]
- 24.Han TH, Han JS, So MH, Seo JW, Ahn CM, Min DH, Yoo YS, Cha DK, Kim CG. 2012. The removal of 1,4-dioxane from polyester manufacturing process wastewater using an up-flow biological aerated filter (UBAF) packed with tire chips. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 47:117–129. 10.1080/10934529.2012.630291 [DOI] [PubMed] [Google Scholar]
- 25.Tanabe A, Kawata K. 2008. Determination of 1,4-dioxane in household detergents and cleaners. J. AOAC Int. 91:439–444 http://aoac.publisher.ingentaconnect.com/content/aoac/jaoac/2008/00000091/00000002/art00024 [PubMed] [Google Scholar]
- 26.Zenker MJ, Borden RC, Barlaz MA. 2003. Occurrence and treatment of 1,4-dioxane in aqueous environments. Environ. Eng. Sci. 20:423–432. 10.1089/109287503768335913 [DOI] [Google Scholar]
- 27.Chiang SY, Mora R, Diguiseppi WH, Davis G, Sublette K, Gedalanga P, Mahendra S. 2012. Characterizing the intrinsic bioremediation potential of 1,4-dioxane and trichloroethene using innovative environmental diagnostic tools. J. Environ. Monit. 14:2317–2326. 10.1039/c2em30358b [DOI] [PubMed] [Google Scholar]
- 28.Mahendra S, Alvarez-Cohen L. 2006. Kinetics of 1,4-dioxane biodegradation by monooxygenase-expressing bacteria. Environ. Sci. Technol. 40:5435–5442. 10.1021/es060714v [DOI] [PubMed] [Google Scholar]
- 29.Grostern A, Sales CM, Zhuang WQ, Erbilgin O, Alvarez-Cohen L. 2012. Glyoxylate metabolism is a key feature of the metabolic degradation of 1,4-dioxane by Pseudonocardia dioxanivorans strain CB1190. Appl. Environ. Microbiol. 78:3298–3308. 10.1128/AEM.00067-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernhardt D, Diekmann H. 1991. Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Appl. Microbiol. Biotechnol. 36:120–123. 10.1007/BF00164711 [DOI] [PubMed] [Google Scholar]
- 31.Nakamiya K, Hashimoto S, Ito H, Edmonds JS, Morita M. 2005. Degradation of 1,4-dioxane and cyclic ethers by an isolated fungus. Appl. Environ. Microbiol. 71:1254–1258. 10.1128/AEM.71.3.1254-1258.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parales RE, Adamus JE, White N, May HD. 1994. Degradation of 1,4-dioxane by an actinomycete in pure culture. Appl. Environ. Microbiol. 60:4527–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan MP, Pembroke JT, Adley CC. 2007. Ralstonia pickettii in environmental biotechnology: potential and applications. J. Appl. Microbiol. 103:754–764. 10.1111/j.1365-2672.2007.03361.x [DOI] [PubMed] [Google Scholar]
- 34.Fishman A, Tao Y, Wood TK. 2004. Toluene 3-monooxygenase of Ralstonia pickettii PKO1 is a para-hydroxylating enzyme. J. Bacteriol. 186:3117–3123. 10.1128/JB.186.10.3117-3123.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sales CM, Mahendra S, Grostern A, Parales RE, Goodwin LA, Woyke T, Nolan M, Lapidus A, Chertkov O, Ovchinnikova G, Sczyrba A, Alvarez-Cohen L. 2011. Genome sequence of the 1,4-dioxane-degrading Pseudonocardia dioxanivorans strain CB1190. J. Bacteriol. 193:4549–4550. 10.1128/JB.00415-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahendra S, Petzold CJ, Baidoo EE, Keasling JD, Alvarez-Cohen L. 2007. Identification of the intermediates of in vivo oxidation of 1,4-dioxane by monooxygenase-containing bacteria. Environ. Sci. Technol. 41:7330–7336. 10.1021/es0705745 [DOI] [PubMed] [Google Scholar]
- 37.Vangnai AS, Arp DJ. 2001. An inducible 1-butanol dehydrogenase, a quinohaemoprotein, is involved in the oxidation of butane by ‘Pseudomonas butanovora.’ Microbiology 147:745–756 http://mic.sgmjournals.org/content/147/3/745.short [DOI] [PubMed] [Google Scholar]
- 38.Smith CA, O'Reilly KT, Hyman MR. 2003. Characterization of the initial reactions during the cometabolic oxidation of methyl tert-butyl ether by propane-grown Mycobacterium vaccae JOB5. Appl. Environ. Microbiol. 69:796–804. 10.1128/AEM.69.2.796-804.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotani T, Yamamoto T, Yurimoto H, Sakai Y, Kato N. 2003. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J. Bacteriol. 185:7120–7128. 10.1128/JB.185.24.7120-7128.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson DR, Lee PKH, Holmes VF, Alvarez-Cohen L. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 71:3866–3871. 10.1128/AEM.71.7.3866-3871.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang MY, Olson BH, Chang JS. 2007. Improving PCR and qPCR detection of hydrogenase A (hydA) associated with clostridia in pure cultures and environmental sludges using bovine serum albumin. Appl. Microbiol. Biotechnol. 77:645–656. 10.1007/s00253-007-1196-1 [DOI] [PubMed] [Google Scholar]
- 42.Kreader CA. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harms G, Layton AC, Dionisi HM, Gregory IR, Garrett VM, Hawkins SA, Robinson KG, Sayler GS. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343–351. 10.1021/es0257164 [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 45.Robbins PW, Overbye K, Albright C, Benfield B, Pero J. 1992. Cloning and high-level expression of chitinase-encoding gene of Streptomyces plicatus. Gene 111:69–76. 10.1016/0378-1119(92)90604-N [DOI] [PubMed] [Google Scholar]
- 46.Novotna J, Vohradsky J, Berndt P, Gramajo H, Langen H, Li XM, Minas W, Orsaria L, Roeder D, Thompson CJ. 2003. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol. Microbiol. 48:1289–1303. 10.1046/j.1365-2958.2003.03529.x [DOI] [PubMed] [Google Scholar]
- 47.Kampfer P, Kohlweyer U, Thiemer B, Andreesen JR. 2006. Pseudonocardia tetrahydrofuranoxydans sp. nov. Int. J. Syst. Evol. Microbiol. 56:1535–1538. 10.1099/ijs.0.64199-0 [DOI] [PubMed] [Google Scholar]
- 48.Monod J. 1941. On the new phenomenon of complex growth in bacterial cultures. C. R. Hebd. Acad. Sci. 212:934–936 [Google Scholar]
- 49.Wong TY, Pei H, Bancroft K, Childers GW. 1995. Diauxic growth of Azotobacter vinelandii on galactose and glucose: regulation of glucose transport by another hexose. Appl. Environ. Microbiol. 61:430–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao YL, Lv ZM, Min H, Lv ZH, Jiao HP. 2009. Isolation, identification and characterization of a novel Rhodococcus sp. strain in biodegradation of tetrahydrofuran and its medium optimization using sequential statistics-based experimental designs. Bioresour. Technol. 100:2762–2769. 10.1016/j.biortech.2009.01.006 [DOI] [PubMed] [Google Scholar]
- 51.Raj CBC, Ramkumar N, Siraj AHJ, Chidambaram S. 1997. Biodegradation of acetic, benzoic, isophthalic, toluic and terephthalic acids using a mixed culture: effluents of PTA production. Process Saf. Environ. 75:245–256. 10.1205/095758297529129 [DOI] [Google Scholar]
- 52.Guyot JP, Macarie H, Noyola A. 1990. Anaerobic-digestion of a petrochemical waste-water using the UASB process. Appl. Biochem. Biotechnol. 24/25:579–589 [Google Scholar]
- 53.Furuya T, Hirose S, Osanai H, Semba H, Kino K. 2011. Identification of the monooxygenase gene clusters responsible for the regioselective oxidation of phenol to hydroquinone in mycobacteria. Appl. Environ. Microbiol. 77:1214–1220. 10.1128/AEM.02316-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lan RS, Smith CA, Hyman MR. 2013. Oxidation of cyclic ethers by alkane-grown Mycobacterium vaccae JOB5. Remediation 23:23–42 [Google Scholar]
- 55.Sales CM, Grostern A, Parales JV, Parales RE, Alvarez-Cohen L. 2013. Oxidation of the cyclic ethers 1,4-dioxane and tetrahydrofuran by a monooxygenase in two Pseudonocardia species. Appl. Environ. Microbiol. 79:7702–7708. 10.1128/AEM.02418-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brow CN, O'Brien Johnson R, Johnson RL, Simon HM. 2013. assessment of anaerobic toluene biodegradation activity by bssA transcript/gene ratios. Appl. Environ. Microbiol. 79:5338–5344. 10.1128/AEM.01031-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prior SD, Dalton H. 1985. Acetylene as a suicide substrate and active-site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 29:105–109. 10.1111/j.1574-6968.1985.tb00843.x [DOI] [Google Scholar]
- 58.Hyman MR, Wood PM. 1985. Suicidal inactivation and labeling of ammonia mono-oxygenase by acetylene. Biochem. J. 227:719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sei K, Kakinoki T, Inoue D, Soda S, Fujita M, Ike M. 2010. Evaluation of the biodegradation potential of 1,4-dioxane in river, soil and activated sludge samples. Biodegradation 21:585–591. 10.1007/s10532-010-9326-3 [DOI] [PubMed] [Google Scholar]
- 60.Li M, Mathieu J, Yang Y, Fiorenza S, Deng Y, He Z, Zhou J, Alvarez PJ. 2013. Widespread distribution of soluble di-iron monooxygenase (SDIMO) genes in Arctic groundwater impacted by 1,4-dioxane. Environ. Sci. Technol. 47:9950–9958. 10.1021/es402228x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


