Abstract
The genus Bradyrhizobium has been considered to be a taxonomically difficult group. In this study, phylogenetics and evolutionary genetics analyses were used to investigate divergence levels among Bradyrhizobium strains nodulating soybeans in China. Eleven genospecies were identified by sequence analysis of three phylogenetic and taxonomic markers (SMc00019, thrA, and truA). This was also supported by analyses of eight genes outside the symbiosis island (“off-island” genes; SMc00019, thrA, truA, fabB, glyA, phyR, exoN, and hsfA). However, seven genes inside the symbiosis island (“island” genes; nifA, nifH, nodC, nodV, fixA, trpD, and rhcC2) showed contrasting lower levels of nucleotide diversity and recombination rates than did off-island genes. Island genes had significantly incongruent gene phylogenies compared to the species tree. Four phylogenetic clusters were observed in island genes, and the epidemic cluster IV (harbored by Bradyrhizobium japonicum, Bradyrhizobium diazoefficiens, Bradyrhizobium huanghuaihaiense, Bradyrhizobium liaoningense, Bradyrhizobium daqingense, Bradyrhizobium sp. I, Bradyrhizobium sp. III, and Bradyrhizobium sp. IV) was not found in Bradyrhizobium yuanmingense, Bradyrhizobium sp. II, or Bradyrhizobium elkanii. The gene flow level of island genes among genospecies is discussed in the context of the divergence level of off-island genes.
INTRODUCTION
Soybeans (Glycine max L.) were first domesticated in China and then introduced into different parts of the planet (1), now with an annual harvest area of 100 million hectares around the world (FAO, 2011). Ninety percent of their production comes from the United States, Brazil, Argentina, China, and India (FAO, 2007 to 2011). One of the key features of soybean is its ability to form symbiotic nitrogen-fixing nodules with diverse rhizobial species (2, 3), implying its important role in sustainable agriculture. It has been recurrently reported that Bradyrhizobium japonicum, Bradyrhizobium elkanii, Bradyrhizobium liaoningense, Bradyrhizobium yuanmingense, and Sinorhizobium fredii could nodulate soybeans (2–5). Recently, Bradyrhizobium huanghuaihaiense, Bradyrhizobium daqingense, Sinorhizobium sojae, and several unnamed species were also found to be effective microsymbionts of soybeans (2, 3, 6–8). Strain USDA110 represents a widely distributed type formerly known as B. japonicum Ia, but it has recently been proposed as a member of the new species Bradyrhizobium diazoefficiens (9).
Recent studies not only suggested differences in the biogeographic distribution of rhizobial species nodulating soybeans but also demonstrated a biased selection of rhizobial species by different genotypes of soybeans (2, 4, 10, 11). Consistent with these findings, comparative genomics of rhizobia revealed that the phyletic distribution of rhizobial functional genes involved in environmental adaptations and symbiotic interactions generally agrees with the phylogeny of rhizobial species (7). Therefore, it is important to distinguish different rhizobial species and even subdivisions of each species. The genus Bradyrhizobium has been considered to be a taxonomically difficult group (12, 13). In contrast to the highly conserved rrs sequences in Bradyrhizobium, sequence analyses of housekeeping genes (atpD, recA, glnII, gyrB, rpoB, dnaK, etc.) have been found to be very useful in this scenario, especially when these genes were concatenated (5, 6, 8, 14–16). Although the phylogeny of the concatenated housekeeping genes was usually considered to represent the species tree, these housekeeping loci showed high rates of intergenic recombination and a limited gap between interspecies and intraspecies sequence similarities (15, 16). Recent developments in rhizobial genomics allowed us to construct a well-resolved, reliable species tree for rhizobia (7, 17). Then, three core genes, SMc00019, truA, and thrA, both alone and in combination, were found to be able to produce phylogenies supporting this predetermined species tree (17). Moreover, these core genes provide a gap of 2% for intraspecies/interspecies average nucleotide identity (ANI) for rhizobia, including Bradyrhizobium (17), implying their potential role as useful markers in taxonomy, phylogeny, and population genetics. In most studies on the phylogenetics and evolutionary genetics of housekeeping genes in Bradyrhizobium, no symbiosis genes were analyzed, or only nifH and/or nodC on the symbiosis island (5, 14–16, 18). On the other hand, studies on the evolution of nodulation and nitrogen fixation genes were mainly focused on phylogenetics of these symbiosis genes (19–21).
In this study, we aimed at providing high-resolution delineations and evolutionary genetics analyses of Bradyrhizobium strains nodulating soybeans in China by studying seven genes (nifA, nifH, nodC, nodV, fixA, trpD, and rhcC2) on the symbiosis island (“island” genes) and eight “off-island” genes (SMc00019, thrA, truA, fabB, glyA, phyR, exoN, and hsfA) (17, 22–32). Strains were assigned to genospecies based on the phylogeny and ANI values of core genes SMc00019, truA, and thrA. Molecular diversity, minimum recombination events, the topology of phylogenetic trees, and the levels of divergence and gene flow were compared between island genes and off-island genes.
MATERIALS AND METHODS
Bacterial strains.
The 272 Bradyrhizobium strains used in this study (see Table S1 and Fig. S1 in the supplemental material) were previously collected from soybean nodules in four ecoregions of China (South China, Huanghuaihai, Northeast China, and Xinjiang) (2, 3, 6, 8, 10, 33). These strains were grown in TY medium at 28°C.
Primers, PCR amplifications, and sequencing.
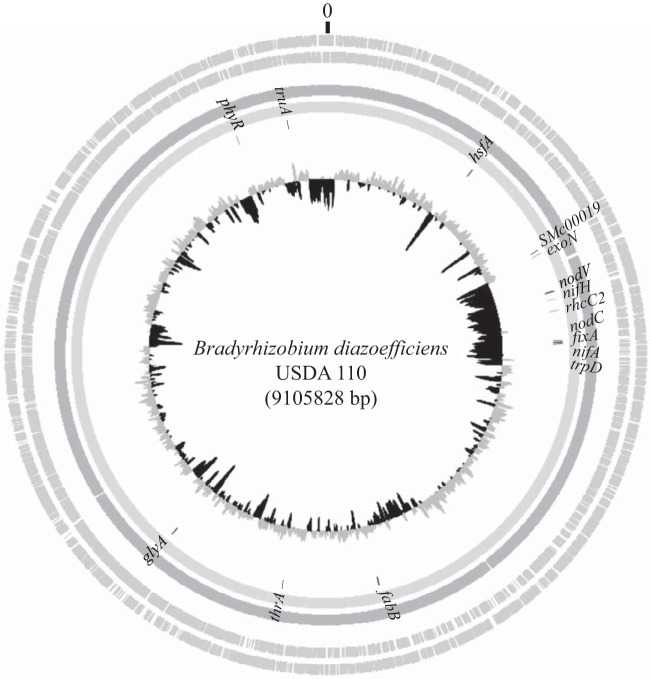
Total template DNA was extracted from each isolate using the GUTC method described by Terefework et al. (34). The PCR amplification of core genes SMc00019, truA, and thrA was performed with the procedure of Zhang et al. (17). The other 12 tested genes have been documented earlier (22–25, 28, 30, 31, 35–37) and were selected by considering their locations in the B. diazoefficiens USDA110 genome (Fig. 1). Briefly, rhcC2, trpD, fixA, nifA, nifH, nodV, and nodC are located within the symbiosis island, and hsfA, exoN, phyR, glyA, and fabB are outside the symbiosis island (Fig. 1). These 12 genes were amplified using primers designed in this study (see Table S2 in the supplemental material). All PCR products were commercially sequenced by BGI, China.
FIG 1.
Locations of the genes used in this study on the genome of Bradyrhizobium diazoefficiens USDA110.
Phylogenetic analyses.
Neighbor-joining trees were reconstructed using MEGA 5 (38). PHYML software (39) in combination with MODELTEST 3.7 (40) was used to build maximum likelihood (ML) trees. Moreover, the Shimodaira-Hasegawa (SH) test (41) was performed to investigate the global phylogenetic congruence of trees inferred from different sets of sequence partitions as implemented in PAUP (42).
Nucleotide polymorphism.
DNASP V5 (43) was used to investigate nucleotide polymorphisms of test genes by calculating statistics as follows: the number of haplotypes (h), defined as sequence types (ST) for each gene and for the concatenated data, and the haplotype diversity (Hd) (44); the nucleotide diversity, π, defined as the average number of nucleotide differences per site between two sequences (44); and πS, the nucleotide diversity for synonymous substitutions (dS), and πN, the nucleotide diversity for nonsynonymous substitutions (dN) (45).
Evolutionary genetics analyses.
The program CLUSTAL W integrated in MEGA 5 was used to align sequences (38). The “No. of differences” method integrated in MEGA 5 was used for calculating the pairwise distance between sequences of a single gene, from which ANI values were obtained using Excel (17). Dxy, the average nucleotide divergence between groups, and Nm, the number of migrants, were estimated by using DNASP (43, 44, 46). Minimal recombination events (Rm) at each locus or for the concatenated sequences were calculated and compared with the values expected under coalescence simulations based on 1,000 genealogy replications with DNASP (43, 47). CLONALFRAME was used to calculate ρ/θ (the relative frequency of the occurrence of recombination compared with point mutation in the history of the lineage) and r/m (the relative impact of recombination compared with point mutation in the genetic diversification of the lineage) as described earlier (48, 49).
Nucleotide sequence accession numbers.
The 1,336 nucleotide sequences obtained in this study were deposited in the GenBank database under accession numbers KF472331 to KF473463, KJ551550 to KJ551556, KF988139 to KF988159, and KF988162 to KF988336 (Table 1).
TABLE 1.
Accession numbers of sequences obtained in this study
| Gene | Accession numbers | Gene function | Reference |
|---|---|---|---|
| SMc00019 | KF473160–KF473235 | Conserved hypothetical protein | 17 |
| truA | KF473388–KF473463 | tRNA pseudouridine synthase A | 17 |
| thrA | KF473236–KF473311, KF988139–KF988159, KF988162–KF988336 | Homoserine dehydrogenase | 17 |
| hsfA | KF472552–KF472627 | Host-specific nitrogen fixation | 22 |
| exoN | KF472331–KF472406 | UTP-glucose-1-phosphate uridylyltransferase | 65 |
| phyR | KF473008–KF473083 | Two-component response regulator | 23 |
| glyA | KF472476–KF472551 | Glycine hydroxymethyltransferase | 24 |
| fabB | KF472856–KF472931 | Beta-ketoacyl acyl carrier protein synthase | 27 |
| rhcC2 | KF473084–KF473159 | Type III secretion system component | 32 |
| trpD | KF473312–KF473387 | Anthranilate phosphoribosyltransferase | 55 |
| fixA | KF472407–KF472475, KJ551550–KJ551556 | Electron transfer flavoprotein FixA | 25 |
| nifA | KF472628–KF472703 | Transcriptional regulator for nitrogen fixation genes | 28 |
| nifH | KF472704–KF472779 | Nitrogenase Fe protein | 25 |
| nodV | KF472932–KF473007 | Two-component regulator | 30 |
| nodC | KF472780–KF472855 | N-Acetylglucosaminyltransferase | 29 |
RESULTS AND DISCUSSION
Eleven genospecies of Bradyrhizobium nodulate soybeans in China.
China is considered to be the domestication center of soybeans and harbors the highest known diversity of rhizobia nodulating soybeans (1–6, 8, 10, 18, 33). However, only a few strains of this rhizobial germplasm were included in an earlier evolutionary genetics study of Bradyrhizobium nodulating soybeans (5). In this study, 272 strains from our earlier studies of soybean rhizobia in four ecoregions of China (see Fig. S1 and Table S1 in the supplemental material) and type strains of related Bradyrhizobium species were subjected to sequence analyses of thrA, a useful phylogenetic and taxonomic marker for rhizobia (17). Based on an intraspecies boundary of 96% ANI (17), and phylogenetic relationships among strains, 9 genospecies were identified: B. japonicum (50 strains), B. diazoefficiens (46 strains), B. elkanii (12 strains), B. yuanmingense (27 strains), B. liaoningense (31 strains), B. daqingense (26 strains), B. huanghuaihaiense (32 strains), Bradyrhizobium sp. I (1 strain), and Bradyrhizobium sp. III (41 strains). For strains with identical thrA sequences, representatives were selected for sequencing the other 14 test genes by considering their geographic origin. Finally, the PCR products of 15 tested loci were successfully obtained for 76 out of 81 representative strains. Therefore, these 76 strains were used in further analyses (see Table S3 in the supplemental material).
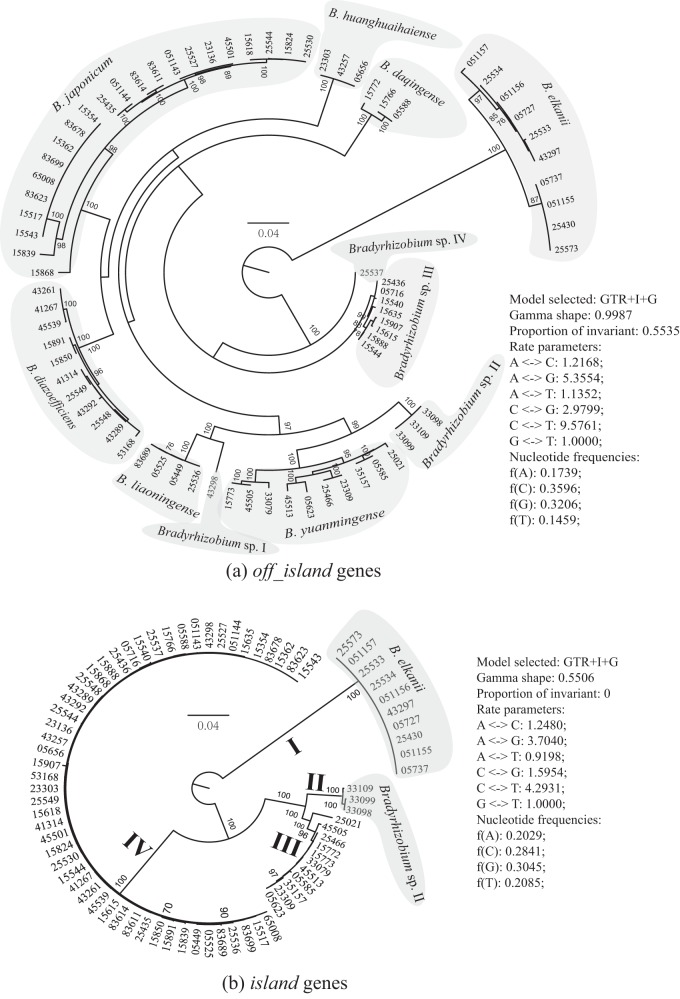
In the ANI analysis of the SMc00019-truA-thrA concatenate, 72/76 test strains were grouped into the same nine species according to the 96% intraspecies boundary (17). They are B. japonicum (containing 22 isolates), B. diazoefficiens (11 isolates), B. daqingense (3 isolates), B. elkanii (10 isolates), B. huanghuaihaiense (3 isolates), B. liaoningense (4 isolates), B. yuanmingense (10 isolates), Bradyrhizobium sp. III (8 isolates), and Bradyrhizobium sp. I (CCBAU 43298). CCBAU 33098, 33099, and 33109 showed a maximum ANI value of 95.6% with the other test genospecies in the SMc00019-truA-thrA concatenate and were consequently considered distinct and named Bradyrhizobium sp. II. CCBAU 25537 had a maximum ANI value of 93.8% with the other genospecies and was named Bradyrhizobium sp. IV. The assignments to the known Bradyrhizobium species were also supported by the well-resolved maximum likelihood tree of the SMc00019-truA-thrA concatenate for the tested strains and related type strains of the corresponding Bradyrhizobium species (see Fig. S2 in the supplemental material), as well as that of all the off-island core genes (Fig. 2a). Out of the 76 strains, 66 were classified into the same species as described in previous studies (2, 3, 10, 33). Eight out of 10 of the strains with a discrepancy in species assignment had previously been classified using only restriction fragment length polymorphism of the 16S-23S rRNA gene intergenic spacer region (IGS-RFLP) or BOX-A1R primer-based repetitive extragenic palindromic PCR (BOX-PCR) clustering. In an earlier study (3), CCBAU 05716 might have been improperly classified into B. japonicum considering its relatively low similarity values with other species in atpD-glnII-recA sequences (<93.4%). CCBAU 33109 was previously identified as B. yuanmingense based on its sequence similarity values of atpD (97.8%), glnII (95.8%), and recA (97.1%) with the type strain CCBAU 10071T of B. yuanmingense (10) but was defined as Bradyrhizobium sp. II in this study by using SMc00019-truA-thrA sequences (ANI <95.64% with other species). So, the sequence-based classification has a great advantage over electrophoresis patterns in terms of data sharing and reinvestigations. Among these Bradyrhizobium species nodulating soybeans, B. japonicum, B. diazoefficiens, and B. elkanii were widely distributed around the world, whereas B. liaoningense, B. daqingense, and B. huanghuaihaiense were so far mainly reported in Asia (2–6, 8–10, 18, 33, 50). Although B. yuanmingense nodulating various legumes has been found in several continents, the biovar nodulating soybeans has so far been found only in Asia (2, 3, 5, 50). The observed high diversity of soybean-nodulating Bradyrhizobium in Asia is consistent with the presence of wild soybeans and the long history of soybean cultivation in this region (51, 52).
FIG 2.
ML tree of off-island genes (a) and island genes (b). Off-island genes: SMc00019, truA, thrA, hsfA, exoN, phyR, glyA, and fabB. Island genes: nifA, nifH, nodC, nodV, fixA, rhcC2, and trpD. In panel b, cluster III includes B. yuanmingense and B. daqingense CCBAU 15772, and cluster IV contains the remaining species except B. elkanii and Bradyrhizobium sp. II. Scale bars indicate 4% substitutions per site.
Molecular diversity of island and off-island genes.
In contrast to some strains such as BTAi1 and ORS278, which use a Nod-independent pathway to form a symbiosis with Aeschynomene species (53), most Bradyrhizobium strains use the Nod-dependent strategy and are characterized by the genomic feature that key symbiotic functions are encoded by genes localized in a symbiosis island region (26, 27). However, our understanding of the diversity of genes in this island is limited to nifH, nifD, and several nod/nol/noe genes, such as nodA, nodC, nodY, nodK, nodZ, nolL, and noeI (14, 18–20, 54). In this study, island genes nifA, nifH, nodC, nodV, fixA, rhcC2, and trpD were sequenced (25, 28–31, 55). As shown in Table 2, the lowest and highest π values are 0.05006 and 0.09329 for nifH and nifA, respectively. This could be partially explained by the large difference between their corresponding πN values (0.0038 for nifH and 0.0617 for nifA) rather than by πS (0.22459 for nifH and 0.22395 for nifA). In line with this observation, regulation mechanisms of nitrogen fixation differ in diverse diazotrophs (56). Moreover, GAF domains of different NifA proteins have a role in regulating NifA activity and seem to have a diverse role in various diazotrophs (56). Therefore, in contrast to the highly conserved nitrogenase component protein NifH, nonsynonymous changes of the regulator gene nifA could be under a different level of selection pressure. Values of πN/πS (dN/dS) for nifA (0.275) and trpD (0.319) are above 0.25, whereas the values for the other test genes are below this boundary, with the value of nifH being the lowest (0.017). This further suggested stronger purifying selection acting on nifH than on nifA. Since the off-island copy of trpD has been shown to be essential for tryptophan biosynthesis (57, 58), the sequenced island copy of trpD in this study might have been subject to relaxed negative selection for new functionalization.
TABLE 2.
Molecular diversity for genetic markers
| Gene (length, bp) | No. of seg. sitesa | h/Hdb | πc | πSd | πNe |
|---|---|---|---|---|---|
| Off-island genes | |||||
| SMc00019 (367) | 108 | 25/0.952 | 0.08583 | 0.29780 | 0.03567 |
| truA (512) | 185 | 29/0.962 | 0.10237 | 0.33936 | 0.04514 |
| thrA (479) | 147 | 32/0.966 | 0.08754 | 0.35149 | 0.01980 |
| hsfA (437) | 138 | 28/0.956 | 0.08583 | 0.26083 | 0.04514 |
| exoN (571) | 150 | 34/0.969 | 0.06925 | 0.22485 | 0.03087 |
| phyR (422) | 109 | 29/0.957 | 0.07090 | 0.29060 | 0.01183 |
| glyA (631) | 160 | 32/0.969 | 0.06666 | 0.23850 | 0.02106 |
| fabB (572) | 195 | 31/0.968 | 0.10664 | 0.36170 | 0.04172 |
| Avg | 149 | 30/0.962 | 0.08437 | 0.29564 | 0.03140 |
| Island genes | |||||
| rhcC2 (598) | 159 | 15/0.762 | 0.08314 | 0.26753 | 0.0335 |
| trpD (622) | 204 | 19/0.831 | 0.08242 | 0.18165 | 0.05786 |
| fixA (579) | 147 | 11/0.678 | 0.07184 | 0.20960 | 0.03068 |
| nifA (585) | 186 | 14/0.586 | 0.09329 | 0.22395 | 0.06165 |
| nifH (620) | 115 | 13/0.659 | 0.05006 | 0.22459 | 0.00380 |
| nodV (647) | 145 | 10/0.627 | 0.06514 | 0.20456 | 0.02974 |
| nodC (568) | 132 | 9/0.555 | 0.05746 | 0.18179 | 0.02090 |
| Avg | 155 | 13/0.671 | 0.07190 | 0.21338 | 0.03401 |
seg. sites, segregating sites.
Haplotype number (h) and haplotype diversity (Hd).
π, average number of nucleotide differences per site between two sequences.
πS, nucleotide diversity for synonymous substitutions (dS).
πN, nucleotide diversity for nonsynonymous substitutions (dN).
Compared to off-island genes SMc00019, truA, thrA, hsfA, exoN, phyR, glyA, and fabB (17, 22–24), those island genes showed significantly lower average values of h (t test, P < 0.0001), Hd (P = 0.0002), and πS (P = 0.0027) but no significant differences in π or πN (t test, α = 0.05). To avoid the potential effect of the variations among different species on the estimation of the statistics of molecular diversity, off-island and island genes within each species were also compared. Five species with more than eight strains were analyzed here: B. japonicum (22 strains), B. diazoefficiens (11 strains), B. yuanmingense (10 strains), Bradyrhizobium sp. III (8 strains), and B. elkanii (10 strains). As shown in Table 3, π of off-island genes ranges from 0.00499 to 0.01993. In line with earlier studies on soybean rhizobia from Myanmar, India, Nepal, Vietnam, and eastern North America (5, 18), B. diazoefficiens (π = 0.00669) was found to be among the species with a relatively low level of diversity. Moreover, island genes showed obviously lower π, πS, and πN than off-island genes in all these five species. Intriguingly, the ratio between off-island π and island π varies from 2.46 in B. yuanmingense, through 3.46 in Bradyrhizobium sp. III, 8.92 in B. diazoefficiens, and 24.91 in B. japonicum, to 332.6 in B. elkanii. Similar off-island/island ratios were found for πS and πN. Since off-island nucleotide diversity in B. elkanii (π = 0.01663) is comparable to that of B. japonicum (π = 0.01794) and B. yuanmingense (π = 0.01993), and clearly higher than that of B. diazoefficiens (π = 0.00669) and Bradyrhizobium sp. III (π = 0.00499), the extremely low diversity of island genes in B. elkanii is unlikely to be caused by sampling bias. Instead, these observations suggest a distinct evolutionary history of island genes in B. elkanii. The overall lower diversity of island genes than of off-island genes for soybean Bradyrhizobium might be due to the selection pressure from the legume host. However, different legume genera/species may select island sequence variants, and this would lead to a higher diversity of island genes than of off-island genes, as reported for Bradyrhizobium sampled from 14 legume genera (54).
TABLE 3.
Molecular diversity of Bradyrhizobium nodulating soybean
| Group (no. of strains) | No. of seg. sitesa | h/Hdb | πc | πSd | πNe |
|---|---|---|---|---|---|
| Off-island genes | |||||
| B. japonicum (22) | 247 | 11/0.862 | 0.01794 | 0.06197 | 0.00299 |
| B. diazoefficiens (11) | 79 | 9/0.964 | 0.00669 | 0.02231 | 0.00140 |
| B. yuanmingense (10) | 214 | 8/0.956 | 0.01993 | 0.07080 | 0.00288 |
| Bradyrhizobium sp. III (8) | 44 | 6/0.929 | 0.00499 | 0.01530 | 0.00154 |
| B. elkanii (10) | 136 | 6/0.844 | 0.01663 | 0.05631 | 0.00322 |
| Island genes | |||||
| B. japonicum (22) | 18 | 11/0.883 | 0.00072 | 0.00182 | 0.00038 |
| B. diazoefficiens (11) | 10 | 6/0.873 | 0.00075 | 0.00176 | 0.00041 |
| B. yuanmingense (10) | 144 | 8/0.956 | 0.00810 | 0.02257 | 0.00311 |
| Bradyrhizobium sp. III (8) | 22 | 5/0.786 | 0.00144 | 0.00180 | 0.00132 |
| B. elkanii (10) | 1 | 2/0.200 | 0.00005 | 0.00018 | 0 |
seg. sites, segregating sites calculated with all genes used in this study.
Haplotype number (h) and haplotype diversity (Hd).
π, average number of nucleotide differences per site between two sequences.
πS, nucleotide diversity for synonymous substitutions (dS).
πN, nucleotide diversity for nonsynonymous substitutions (dN).
In addition to point mutation, recombination is also a source of genetic diversity (59). Recent evolutionary genetics studies have shown that recombination could make a contribution comparable to or greater than that of mutation in creating diversity of rhizobia (7, 18, 49). This phenomenon was further supported, in this study, by the r/m values of 1.12 ± 0.03 (average ± standard error of the mean [SEM]) and 1.65 ± 0.13 for off-island and island genes, respectively. However, ρ/θ was 0.096 ± 0.009 (average ± SEM) for island genes, which is less than half of the value for off-island genes (ρ/θ = 0.21 ± 0.005), indicating a lower frequency of recombination in island genes than in off-island genes. This is consistent with the significantly lower average number of recombination events (Rm) per island gene (Rm = 13.7) than the comparable figure for off-island genes (Rm = 30.5; t test, P = 0.00015; see Table S4 in the supplemental material). When we look at the Rm values for B. japonicum, B. diazoefficiens, B. yuanmingense, Bradyrhizobium sp. III, or B. elkanii (Table 4), Rm calculated with island genes is always lower than that obtained with off-island genes for the same species. For the off-island genes, the observed Rm values for B. japonicum, B. yuanmingense, and B. elkanii lie outside the 95% interval (upper limit) of values obtained with coalescence simulations with an intermediate level of recombination, which is consistent with high rates of recombination within each species. B. diazoefficiens and Bradyrhizobium sp. III showed an intermediate level of recombination compared to the simulation data. These observed levels of recombination in off-island genes are comparable to or higher than those reported earlier for B. japonicum, B. diazoefficiens, B. yuanmingense, and B. elkanii nodulating soybeans (5). The discrepancy could be due to either the higher diversity of test strains in this study or different sets of off-island genes used in the two studies. The Rm values calculated from the concatenated sequences include those recombination events between loci. Island loci are much closer together than are off-island loci in this study (Fig. 1), which might lead to biased estimation of recombination events between loci. To exclude this potential bias on Rm estimation, the sum of intragenic Rm values for off-island or island genes was calculated for each species. The resulting values of five species range from 1 to 17 and 0 to 1 for off-island and island genes, respectively. For each species, off-island genes always showed a higher level of recombination than did island genes.
TABLE 4.
Recombination within species
| Group (no. of strains) | Rm | Coalescence simulationa |
||
|---|---|---|---|---|
| Rm avg | 95% confidence interval | P ≤ observed Rm | ||
| Off-island genes | ||||
| B. japonicum (22) | 20 | 3.44 | 1, 7 | 1.00 |
| B. diazoefficiens (11) | 6 | 4.35 | 1, 8 | 0.86 |
| B. yuanmingense (10) | 22 | 9.42 | 3, 16 | 1.00 |
| Bradyrhizobium sp. III (8) | 5 | 3.27 | 0, 7 | 0.89 |
| B. elkanii (10) | 5 | 0.85 | 0, 3 | 1.00 |
| Island genes | ||||
| B. japonicum (22) | 0 | 1.07 | 0, 3 | 0.29 |
| B. diazoefficiens (11) | 0 | 0.058 | 0, 2 | 0.58 |
| B. yuanmingense (10) | 4 | 0.001 | 0, 0 | 1.0 |
| Bradyrhizobium sp. III (8) | 1 | 0.0 | 0, 0 | 1.0 |
| B. elkanii (10) | 0 | 0.008 | 0, 0 | 0.992 |
Neutral coalescence simulations given the number of segregating sites with an intermediate level of recombination.
Phylogenetic analysis of off-island and island genes.
Molecular diversity analyses imply a different evolutionary history for island genes than for the off-island genes. This view was further proved by the results of tree topology comparisons in the maximum likelihood framework (Table 5). All the gene trees of island genes (nifA, nifH, nodC, nodV, fixA, rhcC2, and trpD) were significantly different (P < 0.001) from the species tree based on the SMc00019-truA-thrA concatenate, whereas no significantly incongruent signals could be detected for the gene tree of each off-island gene compared to the species tree (P > 0.05). This result is similar to earlier findings on nifH, nodA, nodC, nodY, nodK, nodZ, nolL, and noeI (14, 18–20), where Bradyrhizobium formed a monophyletic clade in the phylogeny of these symbiosis genes but showed a topology that was incongruent with the reference species tree. In the well-resolved ML tree of the concatenated off-island genes (Fig. 2a), 11 genospecies were clearly identified. In contrast, the tested strains formed only four clusters in the ML tree of island genes (Fig. 2b), i.e., cluster I (B. elkanii), cluster II (Bradyrhizobium sp. II), cluster III (including B. yuanmingense and B. daqingense CCBAU 15772), and cluster IV, containing the remaining eight genospecies. CCBAU 15772, 15766, and 15773 were isolated from the same sampling site (see Table S1 in the supplemental material), but 15766 and 15772 belong to B. daqingense whereas strain 15773 belongs to B. yuanmingense. Strain 15772 might have obtained the typical island genes of B. yuanmingense from strain 15773, or vice versa. It was hypothesized that dissemination of nodulation and nitrogen fixation genes within the Bradyrhizobium lineage mainly occurred through vertical transmission, with a limited role for lateral gene transfer (19, 20). In this study, the grouping of strains belonging to cluster I, II, or III in the island phylogeny (Fig. 2b) closely followed the off-island phylogeny (Fig. 2a). Therefore, in addition to nodulation and nitrogen fixation genes, other genes (such as fixA, rhcC2, and trpD) in the symbiosis island may also be mainly disseminated through vertical transmission in B. elkanii, B. yuanmingense, and Bradyrhizobium sp. II. However, the epidemic cluster IV of island genes in eight genospecies might be viewed as a bona fide example of lateral gene transfer. Moreover, nifA, nifH, nodC, nodV, fixA, rhcC2, and trpD could be disseminated together, as supported by the similar phylogeny among these island genes (data not shown). Congruent phylogeny among nifH and nodulation genes was also reported in earlier studies of Bradyrhizobium (19, 20). The transfer of island genes into certain chromosomal backgrounds has also been observed in Bradyrhizobium strains isolated from other legume genera (54). A similar situation has been found with symbiosis plasmids in Rhizobium and Sinorhizobium species (60–62).
TABLE 5.
SH test of gene phylogeny with the reference phylogeny based on SMc00019, truA, and thrA
| Gene | −ln La | Difference in −ln Lb | Pc |
|---|---|---|---|
| Off-island genes | |||
| SMc00019 | 7,374.87 | 331.96 | 0.281 |
| truA | 7,333.84 | 290.93 | 0.314 |
| thrA | 7,266.93 | 224.02 | 0.373 |
| hsfA | 7,600.97 | 558.07 | 0.11 |
| exoN | 7,477.30 | 434.40 | 0.198 |
| phyR | 7,699.41 | 656.50 | 0.071 |
| glyA | 7,459.06 | 416.15 | 0.213 |
| fabB | 7,658.13 | 615.22 | 0.087 |
| All off-island genes | 7,111.34 | 68.48 | 0.404 |
| Island genes | |||
| rhcC2 | 15,655.20 | 8,612.30 | 0.000* |
| trpD | 13,532.42 | 6,489.51 | 0.000* |
| fixA | 14,849.94 | 7,807.03 | 0.000* |
| nifA | 15,863.37 | 8,820.46 | 0.000* |
| nifH | 14,704.16 | 7,661.25 | 0.000* |
| nodV | 14,933.56 | 7,890.65 | 0.000* |
| nodC | 15,866.32 | 8,823.41 | 0.000* |
| All island genes | 12,205.51 | 5,162.61 | 0.000* |
−ln L, negative log-likelihood values correspond to those for the constrained topology.
Score differences between unconstrained and constrained trees.
Significance of difference in −ln L scores achieved by constrained and unconstrained trees, as assessed by SH test. *, P < 0.05.
Genetic divergence and gene flow.
As island genes are considered to be typical accessory genes, the acquisition of island genes does not require homology to integrate into the recipient genome (63, 64). Therefore, the evolutionary fate of such genes may be only loosely coupled with that of species where they are found (64). However, in the phylogenetic analyses of this study, the epidemic cluster IV of island genes was not found in B. elkanii, B. yuanmingense, and Bradyrhizobium sp. II (Fig. 2b). Could the transmission patterns of island genes be related to the genetic divergence level of off-island genes? As shown in Table 6 and Table 3, Dxy values (the average nucleotide divergence between genospecies) are all higher than the within-genospecies divergence (π) for off-island genes in representative species, including B. elkanii, B. yuanmingense, B. japonicum, B. diazoefficiens, and Bradyrhizobium sp. III, reflecting genetic differentiation among these genospecies. However, Dxy values calculated for island genes among B. japonicum, B. diazoefficiens, and Bradyrhizobium sp. III were similar to or lower than π for each genospecies. Notably, the Dxy values of off-island genes between B. japonicum, B. diazoefficiens, and Bradyrhizobium sp. III were among the lowest values in this study. Moreover, those island Dxy values higher than corresponding π values showed a positive linear relationship with off-island Dxy values (Pearson coefficient = 0.9898, P < 0.0001). When the off-island Dxy values were lower than 0.075, island Dxy values dropped dramatically (Table 6). Similarly, a clear gap between intraspecies and interspecies ANI values could be observed for those calculated with off-island genes, but no intraspecies/interspecies ANI gaps of island genes were found among B. japonicum, B. diazoefficiens, and Bradyrhizobium sp. III (Table 6). The relationships between island and off-island ANI values were similar to those between island and off-island Dxy. The high level of Nm calculated with island genes among three less divergent species in off-island genes (B. japonicum, B. diazoefficiens, and Bradyrhizobium sp. III) further supported a potential relationship between the gene flow of island genes and the divergence of off-island genes (Table 6). Taken together, the different levels of gene flow in island genes among different genospecies defined by off-island genes might imply that island genes require the correct off-island background to function. This view was also supported by earlier phylogenetic analyses of nodulation and nitrogen fixation genes in Bradyrhizobium and the comparative genomics of soybean rhizobia (7, 19, 20). On the other hand, the cooccurrence of these Bradyrhizobium species in sympatry (see Table S3 in the supplemental material) precludes the potential influence of geographic isolation on the observed phenomenon.
TABLE 6.
Genetic divergence and gene flow between populations
| Variable and population | Value for populatione: |
||||
|---|---|---|---|---|---|
| B. elkanii | B. yuanmingense | B. japonicum | B. diazoefficiens | Bradyrhizobium sp. III | |
| Dxya | |||||
| B. elkanii | 0.16470 | 0.17608 | 0.17612 | 0.17626 | |
| B. yuanmingense | 0.14851 | 0.11565 | 0.11572 | 0.11548 | |
| B. japonicum | 0.15427 | 0.08286 | 0.00083 | 0.00123 | |
| B. diazoefficiens | 0.14773 | 0.08248 | 0.07296 | 0.00121 | |
| Bradyrhizobium sp. III | 0.14737 | 0.07691 | 0.07361 | 0.07496 | |
| ANIb | |||||
| B. elkanii | 0.9708/0.9997c | 0.8361 | 0.8245 | 0.8242 | 0.8242 |
| B. yuanmingense | 0.8551 | 0.9678/0.9695 | 0.8859 | 0.8854 | 0.8857 |
| B. japonicum | 0.8485 | 0.9202 | 0.9653/0.9976 | 1 | 1 |
| B. diazoefficiens | 0.8546 | 0.9197 | 0.9431 | 0.9866/0.9980 | 0.9997 |
| Bradyrhizobium sp. III | 0.8546 | 0.9260 | 0.9280 | 0.9270 | 0.9906/0.995 |
| Nmd | |||||
| B. elkanii | 0.01 | 0.00 | 0.00 | 0.00 | |
| B. yuanmingense | 0.07 | 0.02 | 0.02 | 0.02 | |
| B. japonicum | 0.06 | 0.15 | 4.28 | 4.99 | |
| B. diazoefficiens | 0.04 | 0.10 | 0.10 | 6.07 | |
| Bradyrhizobium sp. III | 0.04 | 0.10 | 0.09 | 0.04 | |
Dxy is the average nucleotide divergence between groups.
The maximum interspecies ANI values are shown in the upper and lower triangles.
Minimum intraspecies ANI values (off-island/island) calculated with off-island and island genes, respectively.
Nm, number of migrants.
The underlined values were calculated with island genes, and the other values were calculated with off-island genes.
Conclusions.
Based on three useful phylogenetic and taxonomic markers (SMc00019, thrA, and truA), 76 representative Bradyrhizobium strains nodulating soybean in China were grouped into 11 genospecies. This was confirmed by analyses of eight off-island genes (SMc00019, thrA, truA, fabB, glyA, phyR, exoN, and hsfA). However, island genes (nifA, nifH, nodC, nodV, fixA, trpD, and rhcC2) showed characteristics contrasting with those of off-island genes in terms of nucleotide diversity and the rate of recombination. Variations in related statistics were also observed between different island genes (such as nifA and nifH) or between different genospecies. Although phylogenetic analyses suggested a different evolutionary history of island genes in contrast to off-island genes, variations in the gene flow level of island genes among different genospecies might imply that island genes require the correct off-island background to function.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Peter W. Young for language revision and comments.
This study was funded by the National Natural Science Foundation of China (31200002).
Footnotes
Published ahead of print 14 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00044-14.
REFERENCES
- 1.Qiu LJ, Chen PY, Liu ZX, Li YH, Guan RX, Wang LH, Chang RZ. 2011. The worldwide utilization of the Chinese soybean germplasm collection. Plant Genet. Resour. 9:109–122. 10.1017/S1479262110000493 [DOI] [Google Scholar]
- 2.Zhang YM, Li Y, Jr, Chen WF, Wang ET, Tian CF, Li QQ, Zhang YZ, Sui XH, Chen WX. 2011. Biodiversity and biogeography of rhizobia associated with soybean plants grown in the North China Plain. Appl. Environ. Microbiol. 77:6331–6342. 10.1128/AEM.00542-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li QQ, Wang ET, Zhang YZ, Zhang YM, Tian CF, Sui XH, Chen WF, Chen WX. 2011. Diversity and biogeography of rhizobia isolated from root nodules of Glycine max grown in Hebei province, China. Microb. Ecol. 61:917–931. 10.1007/s00248-011-9820-0 [DOI] [PubMed] [Google Scholar]
- 4.Shiro S, Matsuura S, Saiki R, Sigua GC, Yamamoto A, Umehara Y, Hayashi M, Saeki Y. 2013. Genetic diversity and geographical distribution of indigenous soybean-nodulating bradyrhizobia in the United States. Appl. Environ. Microbiol. 79:3610–3618. 10.1128/AEM.00236-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinuesa P, Rojas-Jimenez K, Contreras-Moreira B, Mahna SK, Prasad BN, Moe H, Selvaraju SB, Thierfelder H, Werner D. 2008. Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic continent. Appl. Environ. Microbiol. 74:6987–6996. 10.1128/AEM.00875-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang YM, Li Y, Jr, Chen WF, Wang ET, Sui XH, Li QQ, Zhang YZ, Zhou YG, Chen WX. 2012. Bradyrhizobium huanghuaihaiense sp. nov., an effective symbiotic bacterium isolated from soybean (Glycine max L.) nodules. Int. J. Syst. Evol. Microbiol. 62:1951–1957. 10.1099/ijs.0.034546-0 [DOI] [PubMed] [Google Scholar]
- 7.Tian CF, Zhou YJ, Zhang YM, Li QQ, Zhang YZ, Li DF, Wang S, Wang J, Gilbert LB, Li YR, Chen WX. 2012. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc. Natl. Acad. Sci. U. S. A. 109:8629–8634. 10.1073/pnas.1120436109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JY, Wang R, Zhang YM, Liu HC, Chen WF, Wang ET, Sui XH, Chen WX. 2013. Bradyrhizobium daqingense sp. nov., isolated from soybean nodules. Int. J. Syst. Evol. Microbiol. 63:616–624. 10.1099/ijs.0.034280-0 [DOI] [PubMed] [Google Scholar]
- 9.Delamuta JRM, Ribeiro RA, Ormeño-Orrillo E, Melo IS, Martínez-Romero E, Hungria M. 2013. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 63:3342–3351. 10.1099/ijs.0.049130-0 [DOI] [PubMed] [Google Scholar]
- 10.Man CX, Wang H, Chen WF, Sui XH, Wang ET, Chen WX. 2008. Diverse rhizobia associated with soybean grown in the subtropical and tropical regions of China. Plant Soil 310:77–87. 10.1007/s11104-008-9631-3 [DOI] [Google Scholar]
- 11.Yang S, Tang F, Gao M, Krishnan HB, Zhu H. 2010. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U. S. A. 107:18735–18740. 10.1073/pnas.1011957107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willems A, Coopman R, Gillis M. 2001. Comparison of sequence analysis of 16S-23S rDNA spacer regions, AFLP analysis and DNA-DNA hybridizations in Bradyrhizobium. Int. J. Syst. Evol. Microbiol. 51:623–632 [DOI] [PubMed] [Google Scholar]
- 13.Willems A, Coopman R, Gillis M. 2001. Phylogenetic and DNA-DNA hybridization analyses of Bradyrhizobium species. Int. J. Syst. Evol. Microbiol. 51:111–117 [DOI] [PubMed] [Google Scholar]
- 14.Vinuesa P, Silva C, Werner D, Martinez-Romero E. 2005. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 34:29–54. 10.1016/j.ympev.2004.08.020 [DOI] [PubMed] [Google Scholar]
- 15.Menna P, Barcellos FG, Hungria M. 2009. Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. Int. J. Syst. Evol. Microbiol. 59:2934–2950. 10.1099/ijs.0.009779-0 [DOI] [PubMed] [Google Scholar]
- 16.Rivas R, Martens M, de Lajudie P, Willems A. 2009. Multilocus sequence analysis of the genus Bradyrhizobium. Syst. Appl. Microbiol. 32:101–110. 10.1016/j.syapm.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 17.Zhang YM, Tian CF, Sui XH, Chen WF, Chen WX. 2012. Robust markers reflecting phylogeny and taxonomy of rhizobia. PLoS One 7:e44936. 10.1371/journal.pone.0044936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang J, Bromfield ESP, Rodrigue N, Cloutier S, Tambong JT. 2012. Microevolution of symbiotic Bradyrhizobium populations associated with soybeans in east North America. Ecol. Evol. 2:2943–2961. 10.1002/ece3.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moulin L, Béna G, Boivin-Masson C, Stepkowski T. 2004. Phylogenetic analyses of symbiotic nodulation genes support vertical and lateral gene co-transfer within the Bradyrhizobium genus. Mol. Phylogenet. Evol. 30:720–732. 10.1016/S1055-7903(03)00255-0 [DOI] [PubMed] [Google Scholar]
- 20.Menna P, Hungria M. 2011. Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidences for the theory of monophyletic origin and spread and maintenance by both horizontal and vertical transfer. Int. J. Syst. Evol. Microbiol. 61:3052–3067. 10.1099/ijs.0.028803-0 [DOI] [PubMed] [Google Scholar]
- 21.Aserse AA, Räsänen LA, Aseffa F, Hailemariam A, Lindström K. 2012. Phylogenetically diverse groups of Bradyrhizobium isolated from nodules of Crotalaria spp., Indigofera spp., Erythrina brucei and Glycine max growing in Ethiopia. Mol. Phylogenet. Evol. 65:595–609. 10.1016/j.ympev.2012.07.008 [DOI] [PubMed] [Google Scholar]
- 22.Oh HS, Son O, Chun JY, Stacey G, Lee MS, Min KH, Song ES, Cheon CI. 2001. The Bradyrhizobium japonicum hsfA gene exhibits a unique developmental expression pattern in cowpea nodules. Mol. Plant Microbe Interact. 14:1286–1292. 10.1094/MPMI.2001.14.11.1286 [DOI] [PubMed] [Google Scholar]
- 23.Gourion B, Sulser S, Frunzke J, Francez-Charlot A, Stiefel P, Pessi G, Vorholt JA, Fischer HM. 2009. The PhyR-sigma(EcfG) signalling cascade is involved in stress response and symbiotic efficiency in Bradyrhizobium japonicum. Mol. Microbiol. 73:291–305. 10.1111/j.1365-2958.2009.06769.x [DOI] [PubMed] [Google Scholar]
- 24.Rossbach S, Hennecke H. 1991. Identification of glyA as a symbiotically essential gene in Bradyrhizobium japonicum. Mol. Microbiol. 5:39–47. 10.1111/j.1365-2958.1991.tb01824.x [DOI] [PubMed] [Google Scholar]
- 25.Mao C, Qiu J, Wang C, Charles TC, Sobral BW. 2005. NodMutDB: a database for genes and mutants involved in symbiosis. Bioinformatics 21:2927–2929. 10.1093/bioinformatics/bti427 [DOI] [PubMed] [Google Scholar]
- 26.Kaneko T, Maita H, Hirakawa H, Uchiike N, Minamisawa K, Watanabe A, Sato S. 2011. Complete genome sequence of the soybean symbiont Bradyrhizobium japonicum strain USDA6T. Genes 2:763–787. 10.3390/genes2040763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:225–256. 10.1093/dnares/9.6.225 [DOI] [PubMed] [Google Scholar]
- 28.Fischer HM, Alvarez-Morales A, Hennecke H. 1986. The pleiotropic nature of symbiotic regulatory mutants: Bradyrhizobium japonicum nifA gene is involved in control of nif gene expression and formation of determinate symbiosis. EMBO J. 5:1165–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Göttfert M, Lamb JW, Gasser R, Semenza J, Hennecke H. 1989. Mutational analysis of the Bradyrhizobium japonicum common nod genes and further nod box-linked genomic DNA regions. Mol. Gen. Genet. 215:407–415. 10.1007/BF00427037 [DOI] [PubMed] [Google Scholar]
- 30.Göttfert M, Grob P, Hennecke H. 1990. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U. S. A. 87:2680–2684. 10.1073/pnas.87.7.2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krause A, Doerfel A, Göttfert M. 2002. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 15:1228–1235. 10.1094/MPMI.2002.15.12.1228 [DOI] [PubMed] [Google Scholar]
- 32.Okazaki S, Zehner S, Hempel J, Lang K, Gottfert M. 2009. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 295:88–95. 10.1111/j.1574-6968.2009.01593.x [DOI] [PubMed] [Google Scholar]
- 33.Han LL, Wang ET, Han TX, Liu J, Sui XH, Chen WF, Chen WX. 2009. Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil 324:291–305. 10.1007/s11104-009-9956-6 [DOI] [Google Scholar]
- 34.Terefework Z, Kaijalainen S, Lindstrom K. 2001. AFLP fingerprinting as a tool to study the genetic diversity of Rhizobium galegae isolated from Galega orientalis and Galega officinalis. J. Biotechnol. 91:169–180. 10.1016/S0168-1656(01)00338-8 [DOI] [PubMed] [Google Scholar]
- 35.Preisig O, Anthamatten D, Hennecke H. 1993. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc. Natl. Acad. Sci. U. S. A. 90:3309–3313. 10.1073/pnas.90.8.3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quelas JI, López-García SL, Casabuono A, Althabegoiti MJ, Mongiardini EJ, Pérez-Giménez J, Couto A, Lodeiro AR. 2006. Effects of N-starvation and C-source on Bradyrhizobium japonicum exopolysaccharide production and composition, and bacterial infectivity to soybean roots. Arch. Microbiol. 186:119–128. 10.1007/s00203-006-0127-3 [DOI] [PubMed] [Google Scholar]
- 37.Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, Fischer HM, Hennecke H. 2007. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 20:1353–1363. 10.1094/MPMI-20-11-1353 [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 40.Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- 41.Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114–1116. 10.1093/oxfordjournals.molbev.a026201 [DOI] [Google Scholar]
- 42.Swofford DL. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 43.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 44.Nei M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, NY [Google Scholar]
- 45.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 46.Hudson RR, Slatkin M, Maddison WP. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudson RR, Kaplan NL. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. 10.1534/genetics.106.063305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian CF, Young JPW, Wang ET, Tamimi SM, Chen WX. 2010. Population mixing of Rhizobium leguminosarum bv. viciae nodulating Vicia faba: the role of recombination and lateral gene transfer. FEMS Microbiol. Ecol. 73:563–576. 10.1111/j.1574-6941.2010.00909.x [DOI] [PubMed] [Google Scholar]
- 50.Appunu C, Sasirekha N, Prabavathy VR, Nair S. 2009. A significant proportion of indigenous rhizobia from India associated with soybean (Glycine max L.) distinctly belong to Bradyrhizobium and Ensifer genera. Biol. Fertil. Soils 46:57–63. 10.1007/s00374-009-0405-8 [DOI] [Google Scholar]
- 51.Li Y, Guan R, Liu Z, Ma Y, Wang L, Li L, Lin F, Luan W, Chen P, Yan Z, Guan Y, Zhu L, Ning X, Smulders MJ, Li W, Piao R, Cui Y, Yu Z, Guan M, Chang R, Hou A, Shi A, Zhang B, Zhu S, Qiu L. 2008. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor. Appl. Genet. 117:857–871. 10.1007/s00122-008-0825-0 [DOI] [PubMed] [Google Scholar]
- 52.Wen Z, Ding Y, Zhao T, Gai J. 2009. Genetic diversity and peculiarity of annual wild soybean (G. soja Sieb. et Zucc.) from various eco-regions in China. Theor. Appl. Genet. 119:371–381. 10.1007/s00122-009-1045-y [DOI] [PubMed] [Google Scholar]
- 53.Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, Bena G, Hannibal L, Fardoux J, Kojadinovic M, Vuillet L, Lajus A, Cruveiller S, Rouy Z, Mangenot S, Segurens B, Dossat C, Franck WL, Chang WS, Saunders E, Bruce D, Richardson P, Normand P, Dreyfus B, Pignol D, Stacey G, Emerich D, Vermeglio A, Medigue C, Sadowsky M. 2007. Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316:1307–1312. 10.1126/science.1139548 [DOI] [PubMed] [Google Scholar]
- 54.Parker MA. 2012. Legumes select symbiosis island sequence variants in Bradyrhizobium. Mol. Ecol. 21:1769–1778. 10.1111/j.1365-294X.2012.05497.x [DOI] [PubMed] [Google Scholar]
- 55.Göttfert M, Rothlisberger S, Kundig C, Beck C, Marty R, Hennecke H. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405–1412. 10.1128/JB.183.4.1405-1412.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621–631. 10.1038/nrmicro954 [DOI] [PubMed] [Google Scholar]
- 57.Kuykendall LD, Hunter WJ. 1995. Symbiotic ineffectiveness of trpCD deletion mutants of Bradyrhizobium japonicum. Soil Biol. Biochem. 27:1035–1039. 10.1016/0038-0717(95)00013-5 [DOI] [Google Scholar]
- 58.Kuykendall LD, Hunter WJ. 1997. The sequence of a symbiotically essential Bradyrhizobium japonicum operon consisting of trpD, trpC and a moaC-like gene. Biochim. Biophys. Acta Gene Struct. Expr. 1350:277–281. 10.1016/S0167-4781(96)00237-0 [DOI] [PubMed] [Google Scholar]
- 59.Guttman DS, Dykhuizen DE. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380–1383. 10.1126/science.7973728 [DOI] [PubMed] [Google Scholar]
- 60.Guo HJ, Wang ET, Zhang XX, Li QQ, Zhang YM, Tian CF, Chen WX. 2014. Replicon-dependent differentiation of symbiosis-related genes in Sinorhizobium nodulating Glycine max. Appl. Environ. Microbiol. 80:1245–1255. 10.1128/AEM.03037-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ormeño-Orrillo E, Menna P, Almeida LG, Ollero FJ, Nicolás MF, Pains Rodrigues E, Shigueyoshi Nakatani A, Silva Batista JS, Oliveira Chueire LM, Souza RC, Ribeiro Vasconcelos AT, Megías M, Hungria M, Martínez-Romero E. 2012. Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.). BMC Genomics 13:735. 10.1186/1471-2164-13-735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.González V, Acosta JL, Santamaría RI, Bustos P, Fernández JL, Hernández González IL, Díaz R, Flores M, Palacios R, Mora J, Dávila G. 2010. Conserved symbiotic plasmid DNA sequences in the multireplicon pangenomic structure of Rhizobium etli. Appl. Environ. Microbiol. 76:1604–1614. 10.1128/AEM.02039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiedenbeck J, Cohan FM. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35:957–976. 10.1111/j.1574-6976.2011.00292.x [DOI] [PubMed] [Google Scholar]
- 64.Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323:741–746. 10.1126/science.1159388 [DOI] [PubMed] [Google Scholar]
- 65.Becker BU, Kosch K, Parniske M, Müller P. 1998. Exopolysaccharide (EPS) synthesis in Bradyrhizobium japonicum: sequence, operon structure and mutational analysis of an exo gene cluster. Mol. Gen. Genet. 259:161–171. 10.1007/s004380050801 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.