Abstract
A label-free optical biosensor based on a nanostructured porous Si is designed for rapid capture and detection of Escherichia coli K12 bacteria, as a model microorganism. The biosensor relies on direct binding of the target bacteria cells onto its surface, while no pretreatment (e.g. by cell lysis) of the studied sample is required. A mesoporous Si thin film is used as the optical transducer element of the biosensor. Under white light illumination, the porous layer displays well-resolved Fabry-Pérot fringe patterns in its reflectivity spectrum. Applying a fast Fourier transform (FFT) to reflectivity data results in a single peak. Changes in the intensity of the FFT peak are monitored. Thus, target bacteria capture onto the biosensor surface, through antibody-antigen interactions, induces measurable changes in the intensity of the FFT peaks, allowing for a 'real time' observation of bacteria attachment.
The mesoporous Si film, fabricated by an electrochemical anodization process, is conjugated with monoclonal antibodies, specific to the target bacteria. The immobilization, immunoactivity and specificity of the antibodies are confirmed by fluorescent labeling experiments. Once the biosensor is exposed to the target bacteria, the cells are directly captured onto the antibody-modified porous Si surface. These specific capturing events result in intensity changes in the thin-film optical interference spectrum of the biosensor. We demonstrate that these biosensors can detect relatively low bacteria concentrations (detection limit of 104 cells/ml) in less than an hour.
Keywords: Bioengineering, Issue 81, analytical chemistry, silicon materials, microbiology, optical materials, Porous Si, optical biosensor, bacteria detection, label-free biosensor, nanostructure, E. coli bacteria
Introduction
Early and accurate identification of pathogenic bacteria is extremely important for food and water safety, environmental monitoring, and point-of-care diagnostics1. As traditional microbiology techniques are time consuming, laborious, and lack the ability to detect microorganisms in "real-time" or outside the laboratory environment, biosensors are evolving to meet these challenges2-5.
In recent years, porous Si (PSi) has emerged as a promising platform for the design of sensors and biosensors6-20. Over the past decade numerous studies regarding PSi-based optical sensors and biosensors were published21,22. The nanostructured PSi layer is typically fabricated by electrochemical anodic etching from a single-crystal Si wafer. The resulting PSi nanomaterials exhibit many advantageous characteristics, such as large surface and free volume, pore sizes that can be controlled and tunable optical properties10,16. The optical properties of the PSi layer, such as photoluminescence8,11 and white light reflectance-based interferometry7,19, are strongly influenced by environmental conditions. Capture of guest molecules/target analytes within the porous layer results in a change in the average refractive index of the film, observed as a modulation in the photoluminescence spectrum or as a wavelength shift in the reflectivity spectrum10.
Although the vast innovation in PSi optical biosensor technology, there are only few reports on PSi-based platforms for bacteria detection6,8,20,23-29. In addition, most of these proof-of-concept studies have demonstrated "indirect" bacteria detection. Thus, generally prior lysis of the cells is required to extract the targeted protein/DNA fragments, characteristic to the studied bacteria29. Our approach is to directly capture the target bacteria cells onto the PSi biosensor. Therefore, monoclonal antibodies, which are specific to target bacteria, are immobilized onto the porous surface. Binding of bacteria cells, via antibody-antigen interactions, onto the surface of the biosensor induce changes in the amplitude (intensity) of the reflectivity spectrum24-26.
In this work, we report on the construction of an optical PSi-based biosensor and demonstrate its application as a label-free biosensing platform for the detection of Escherichia coli (E. coli) K12 bacteria (used as a model microorganism).The monitored optical signal is the light reflected from the PSi nanostructure due to Fabry-Pérot thin film interference (Figure 1A). Changes in the light amplitude/intensity are correlated to specific immobilization of the target bacteria cells onto the biosensor surface, allowing for rapid detection and quantification of the bacteria.
Protocol
1. Preparation of Oxidized Porous SiO2
Etch Si wafers (single side polished on the <100> face and heavily doped, p-type, 0.0008 Ω·cm) in a 3:1 (v/v) solution of aqueous HF and absolute ethanol for 30 sEC at a constant current density of 385 mA/cm2. Please note that HF is a highly corrosive liquid, and it should be handled with extreme care.
Rinse the surface of the resulting porous Si (PSi) film with absolute ethanol several times; dry the films under a dry nitrogen gas.
Oxidize the freshly-etched PSi samples in a tube furnace at 800 °C for 1 hr in ambient air (place the sample in the furnace at room temperature, heat the furnace to 800 °C, leave in furnace for 1 hr, turn off the furnace and remove the samples from the furnace only at room temperature).
2. Biofunctionalization of PSiO2 Scaffolds
Incubate an oxidized PSi (PSiO2) sample for 1 hr in mercaptopropyl(trimethoxysilane-3) 95% (MPTS) solution (108 mM in toluene).
Rinse the silane-treated PSiO2 sample with toluene, methanol, and acetone; dry under a dry nitrogen gas.
Incubate the silane-modified PSiO2 sample for 30 min in 1 ml of 100 mM PEO-iodoacetyl biotin solution.
Rinse the biotin-treated PSiO2 sample with 0.1 M phosphate buffered saline (PBS) solution several times.
Incubate the biotin-modified PSiO2 sample for 30 min in 1 ml of 100 μg/ml streptavidin (SA) solution.
Rinse the SA-treated PSiO2 sample with 0.1 M PBS solution several times.
Incubate the resulting SA-modified PSiO2 sample for 30 min in 1 ml of 100 μg/ml biotinylated E. coli monoclonal antibody (Immunoglobulin G, IgG) solution or with 100 μg/ml biotinylated-rabbit IgG (as a model for a monoclonal antibody).
3. Fluorescent Labeling and Fluorescence Microscopy
Incubate the IgG-modified surfaces with 15 μg/ml fluorescein (FITC) tagged anti rabbit IgG for 40 min and with 15 μg/ml fluorescein (FITC) tagged anti mouse IgG as a control.
Rinse the modified samples with 0.1 M PBS solution several times.
Examine the samples under a fluorescence microscope.
4. Bacteria Culture
Cultivate E. coli K12 bacteria in a 10 ml tube with 5 ml of Luria Bertani (LB) medium (medium composition in 1 L of deionized water: 5 g of NaCl, 5 g of yeast extract, and 10 g of tryptone). Incubate the bacteria overnight shaking at 37 °C.
Monitor the bacteria concentration by reading the optical density (OD) at a wavelength of 600 nm. After overnight growth in LB medium, read the OD600 using a spectrophotometer to determine the bacterial concentration. The number of cells is directly proportional to the OD600 measurements (1 OD600 = 108 cells/ml).
5. Bacteria Sensing
Place the IgG-modified PSiO2 and neat PSiO2 (as the control) samples in a custom-made Plexiglas flow cell. Fix the flow cell to ensure that the sample reflectivity is measured at the same spot during all the measurements.
Incubate the samples with E. coli K12 suspension (104 cell/ml) for 30 min at room temperature. Then remove the bacteria suspension by flushing the cell with 0.85% w/v saline solution for 30 min.
Monitor the changes in the reflectivity data throughout the experiment. All optical measurements need to be carried out in an aqueous surrounding. Spectra should be collected using a CCD spectrometer and analyzed by applying fast Fourier transform (FFT), as previously described25,26 .
Confirm the presence of the bacteria on the biosensor surface, by observation of the samples under an upright light microscope immediately after the biosensing experiment.
Representative Results
Oxidized PSi (PSiO2) films are prepared as described in the Protocol Text section. Figure 1B shows a high-resolution scanning electron micrograph of the resulting PSi film after thermal oxidation. The PSiO2 layer is characterized by well-defined cylindrical pores with a diameter in the range of 30-80 nm.
The monoclonal antibody (IgG) molecules are grafted onto the PSiO2 surfaces by using a well-established silanization technology coupled with a biotin-SA system. The detailed synthesis scheme is outlined in Figure 2. First, the thermally oxidized surface is silanized by MPTS, resulting in a thiol-silanized surface (Figure 2c). In the following step, the activated surface is incubated with a SH-reactive biotin linker molecules (Figure 2d), followed by attachment of the SA proteins to the biotin-modified surface via biotin-SA bridges 30 (Figure 2e). The final step in the functionalization of the biosensor surface involves the bioconjugation of biotinylated monoclonal antibodies to the SA (Figure 2f).
The attachment of the antibodies to the PSiO2 surface is confirmed by fluorescent labeling followed by observation of the surface under a fluorescence microscope. In addition, fluorescence studies allow us to characterize the activity and antigenic specificity of the surface-immobilized antibodies (rabbit IgG) by binding of fluorescently tagged anti-rabbit IgG and anti-mouse IgG as a control. Figure 3 summarizes the results of these experiments.
In order to demonstrate bacteria biosensing we have used a specific E. coli IgG instead of the model IgG (rabbit IgG). Again, the approach is to monitor changes in the amplitude (intensity) of the light reflected from the PSiO2 nanostructure. During the sensing experiments, the biosensors are fixed in a flow cell in order to assure that the sample reflectivity is measured at the same spot during all measurements. The entire experiment is carried out in wet environment and the sample reflectivity spectrum is continuously monitored. The biosensors are exposed to E. coli K12 suspension (104 cells/ml) for 30 min, after which a buffer solution is used for washing the biosensor surface (for the removal of unattached bacteria). A FFT spectrum of the biosensor before and after the introduction of the E. coli bacteria (104 cells/ml) is shown in Figure 4a (top). Thus, following exposure to E. coli bacteria (and subsequent rinsing step) an intensity decrease of 7±1% is recorded, while insignificant changes (less than 0.5% decrease in the FFT intensity) are observed for the unmodified PSiO2 surface (Figure 4b, top). Moreover, in order to confirm the presence of captured cells onto the PSiO2 surface, the biosensors are observed under a light microscope immediately after the completion of the biosensing experiment. Figure 4a (bottom) displays immobilized bacteria cells on the biosensor surface, while no cells are observed on the unmodified surfaces (control); see Figure 4b (bottom).
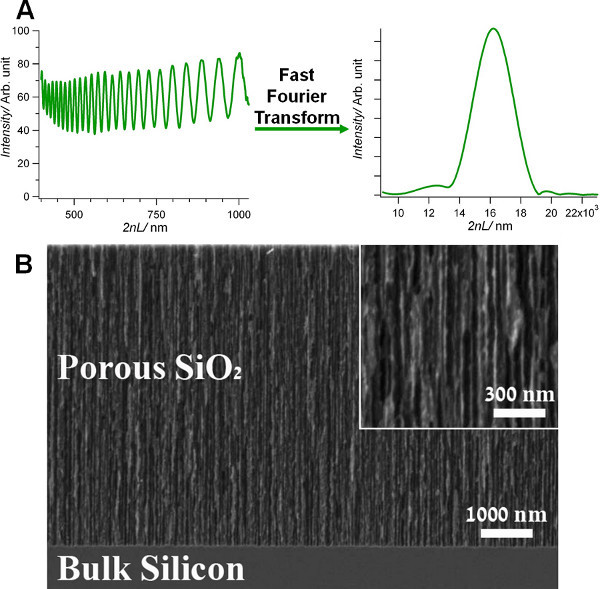 Figure 1. (A) Typical reflectivity spectrum of a typical Fabry-Pérot PSiO2 nanostructure (left) and the corresponding spectrum following a fast Fourier transform (right). The position and magnitude of the resulting single peak are monitored. (B) A cross-sectional high-resolution scanning electron micrograph (secondary electrons) of a PSiO2 layer (etched for 30 sEC at 385 mA/cm2).
Figure 1. (A) Typical reflectivity spectrum of a typical Fabry-Pérot PSiO2 nanostructure (left) and the corresponding spectrum following a fast Fourier transform (right). The position and magnitude of the resulting single peak are monitored. (B) A cross-sectional high-resolution scanning electron micrograph (secondary electrons) of a PSiO2 layer (etched for 30 sEC at 385 mA/cm2).
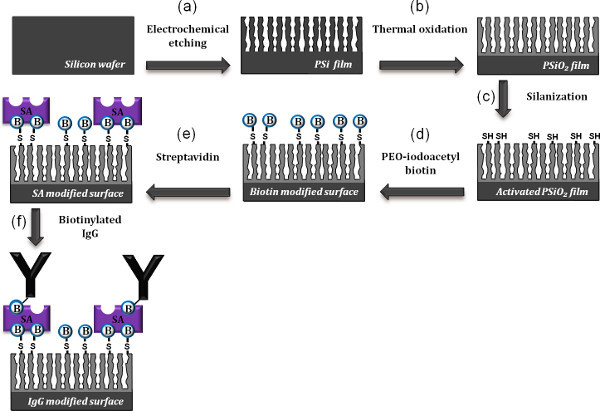 Figure 2. Schematic representation of the synthesis steps followed to biofunctionalize the PSiO2 surface with antibodies. (a) An anodic electrochemical etch is used to prepare a PSi layer from a single-crystal Si wafer. (b) The freshly-etched sample is then thermally oxidized at 800 °C. (c) The PSiO2 surface is initially reacted with MPTS to create free thiol group surface (activated PSiO2 film). (d) Incubation of the activated film with PEO-iodoacetyl biotin to generate biotinylated surface. (e) Incubation of the biotinylated surface with SA. (f) Incubation of the modified surface with biotinylated antibodies. Click here to view larger figure.
Figure 2. Schematic representation of the synthesis steps followed to biofunctionalize the PSiO2 surface with antibodies. (a) An anodic electrochemical etch is used to prepare a PSi layer from a single-crystal Si wafer. (b) The freshly-etched sample is then thermally oxidized at 800 °C. (c) The PSiO2 surface is initially reacted with MPTS to create free thiol group surface (activated PSiO2 film). (d) Incubation of the activated film with PEO-iodoacetyl biotin to generate biotinylated surface. (e) Incubation of the biotinylated surface with SA. (f) Incubation of the modified surface with biotinylated antibodies. Click here to view larger figure.
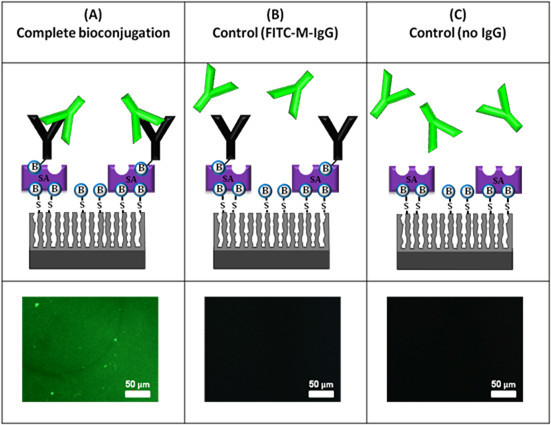 Figure 3. Results of fluorescence labeling experiments to confirm the activity and antigenic specificity of the immobilized IgG. The samples are observed under a fluorescent microscope at a constant exposure time. (A) Complete bioconjugation, PSiO2 + MPTS + biotin + SA + biotinylated-rabbit-IgG and FITC-anti rabbit IgG; (B) control experiment with FITC-anti mouse IgG; (C) control experiment, no IgG, PSiO2 + MPTS + biotin + SA and FITC-anti rabbit IgG.
Figure 3. Results of fluorescence labeling experiments to confirm the activity and antigenic specificity of the immobilized IgG. The samples are observed under a fluorescent microscope at a constant exposure time. (A) Complete bioconjugation, PSiO2 + MPTS + biotin + SA + biotinylated-rabbit-IgG and FITC-anti rabbit IgG; (B) control experiment with FITC-anti mouse IgG; (C) control experiment, no IgG, PSiO2 + MPTS + biotin + SA and FITC-anti rabbit IgG.
Note: These schematics are for illustration purposes only as conjugation of IgG also occurs inside the pores.
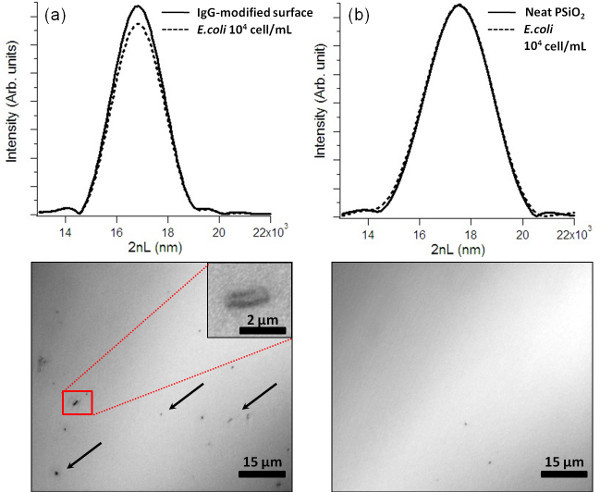 Figure 4. E. coli biosensing experiments and corresponding optical microscope images of the biosensors immediately after the experiments. The biosensors are incubated with 104 cells/ml E. coli suspensions. Key: (a) IgG-modified PSiO2, (b) neat (unmodified) PSiO2.
Figure 4. E. coli biosensing experiments and corresponding optical microscope images of the biosensors immediately after the experiments. The biosensors are incubated with 104 cells/ml E. coli suspensions. Key: (a) IgG-modified PSiO2, (b) neat (unmodified) PSiO2.
Discussion
A label-free optical immunosensor, based on a PSiO2 nanostructure (a Fabry-Pérot thin film) is fabricated, and its potential applicability as a biosensor for bacteria detection is confirmed.
Modifications and troubleshooting
One of the major concerns when designing an immunosensor is the susceptibility of antibodies to undergo undesired conformation changes during deposition and patterning onto the solid substrate, which may lead to a decrease in the biosensor sensitivity 31,32. To minimize this issue, conjugation of the antibodies to the PSiO2 surface is accomplished through biotin-SA bridges. SA and biotin are commonly used in many biotechnological applications due to their high binding affinity (Kd ~10-13 M). The biotin-SA link is one of the most common chemistries for receptor immobilization practiced in biosensor development10,32,33,34. The accessibility of the immobilized antibodies can be regulated by the biotin-SA reaction that provides a flexible linkage between the solid surface and the IgGs31,35. Thus, the PSiO2 is first biofunctionalized with biotin/SA to allow the conjugation of biotinylated antibodies onto the nanostructure.
Limitations of the technique
The main limitations of this biosensing scheme are attributed to the use antibodies as the bacteria capture element. As is with other immunosensors, we are limited to the detection of targets that have available antibodies, the specificity of the used antibodies and their stability36. The type of antibodies to be used depends on the specific application and whenever possible monoclonal antibodies are preferred. Yet, despite the high degree of specificity that monoclonal antibodies provide, the detection of whole bacteria cells below 1,000 cells/ml is still a challenge35. Additionally, the high cost involved in the production of these antibodies should be taken into consideration.
Another limitation is associated with chemical stability of the oxidized PSi transducer in corrosive/aqueous environments. Although, the PSiO2 stability is superior in comparison to PSi thin films, still when conducting long (several hours) flow experiments baseline drifts may be observed14,15,21,24. Moreover, it is favored to work around neutral pH (in the range of 6-8) in order to prevent mechanical instability of the nanostructure.
Of course that when dealing with real food or water samples, which are contaminated with bacteria, an appropriate pretreatment process has to be considered prior to the sensing step. The pretreatment should be designed in accordance to the specific characteristics of studied sample. For example, preparation procedures such as dispersion, filtration, pH balance, etc., could be considered. However, these pretreatment techniques are beyond the scope of this work.
Significance with respect to existing methods
To date, detection and identification of bacterial contaminations rely mainly on classical microbiology culturing techniques or on rapid techniques in microbiology, such as biochemical kits, ELISA (enzyme-linked immunosorbent assay), and PCR (polymerase chain reaction) assays2,3. These methods are laborious, time-consuming and require several days to obtain results, thus ''real-time'' assessment of water and food safety remain unfeasible2,4,5. In the present work, we demonstrate the capability of PSi-based biosensors to overcome the drawbacks of these techniques. The present biosensor allows for a simple and relatively sensitive detection of bacteria via a ''direct cell capture'' approach, by monitoring changes in its optical interference spectrum. Our preliminary results display a low detection limit of 104 cell/ml E. coli and a rapid response time of several minutes. For comparison, the detection limit of current state-of-the-art surface Plasmon resonance (SPR) biosensors is in the range of 102-106 cell/ml37-40. In terms of response time, our biosensors response to bacteria exposure is comparable to that of SPR techniques. Thus, the major strengths of this biosensing scheme are the simplicity of the assay and rapid detection allowing for "real-time" identification of the target bacteria.
Future applications
Several approaches are currently being explored to improve the detection limit and sensitivity of the biosensor, including optimization of the antibody concentration and orientation, use antibody fragments instead of whole antibodies. Additionally, we are studying the potential of aptamers as alternative capture probes. In conclusion, the work presented here provides a generic biosensing platform that can be translated to many biosensing applications of a variety of microorganisms.
Critical steps within the protocol
In order to achieve optimal performance of the biosensor, it is highly important to confirm the biofunctionalization of the PSiO2 scaffolds (see section 3 in the protocol text, Fluorescent Labeling and Fluorescence Microscopy). The fluorescence studies allow us to confirm the attachment of the antibodies to the transducer (PSiO2) surface and to characterize the activity and antigenic specificity of the attached antibodies, by binding of fluorescently tagged anti-rabbit IgG and anti-mouse IgG as a control.
Moreover, in order to eliminate false results in the optical readout, it is essential to properly rinse the surface of the biosensor following exposure to the target bacteria (for the removal of bacteria cells that are nonspecifically bound or reside on the surface) and to conduct control experiments with neat PSiO2. The importance of the control experiment is for the elimination of unspecific bacteria adsorption on the biosensor surface (see section 5 in the protocol text, Bacteria Sensing).
Disclosures
The authors declare no competing financial interests.
Acknowledgments
This work was supported by the Israel Science Foundation (grant No. 1118/08 and grant No. 1146/12) and the Minna Kroll Memorial Research Fund. E.S gratefully acknowledges the financial support of the Russell Berrie Nanotechnology Institute.
References
- Velusamy V, et al. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010;28(2):232–23. doi: 10.1016/j.biotechadv.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Doyle MP, Beuchat LR, Montville TJ, editors. Food Microbiol.: Fundamentals and Front. Vol. 2. Washington: ASM Press; 2001. [Google Scholar]
- Radke SM, Alocilja EC. A microfabricated biosensor for detecting foodborne bioterrorism agents. IEEE Sens. J. 2005;5(4):744. [Google Scholar]
- Glynn B, et al. Current and emerging molecular diagnostic technologies applicable to bacterial food safety. Int. J. of Dairy Technol. 2006;59(2):126. [Google Scholar]
- Leonard P, et al. Advances in biosensors for detection of pathogens in food and water. Enzyme Microb. Technol. 2003;32(1):3. [Google Scholar]
- Alvarez SD, et al. Using a porous silicon photonic crystal for bacterial cell-based biosensing. Physica Status Solidi a-Applications and Materials Science. 2007;204(5):1439. [Google Scholar]
- Archer M, et al. Electrical porous silicon microarray for DNA hybridization detection. Micro- and Nanosystems. 2004;782:385. [Google Scholar]
- Chan S, Horner SR, Fauchet PM, Miller BL. Identification of Gram Negative Bacteria Using Nanoscale Silicon Microcavities. J. Am. Chem. Soc. 2001;123:11797. doi: 10.1021/ja016555r. [DOI] [PubMed] [Google Scholar]
- Dancil K-PS, Greiner DP, Sailor MJ. In: Canham LT, Sailor MJ, Tanaka K, Tsai CC, editors. Development of a Porous Silicon Based Biosensor; Proceedings of the Mat. Res. Soc. Symp; Boston, MA. 1999. pp. 557–562. [Google Scholar]
- D'Auria S, et al. Nanostructured silicon-based biosensors for the selective identification of analytes of social interest. J Phys - Condens Matter. 2006;18(33):S2019. [Google Scholar]
- de Leon SB, et al. Neurons culturing and biophotonic sensing using porous silicon. Appl Phys Lett. 2004;84(22):4361. [Google Scholar]
- Janshoff A, et al. Macroporous p-type silicon Fabry-Perot layers. Fabrication, characterization, and applications in biosensing. J. Am. Chem. Soc. 1998;120(46):12108. [Google Scholar]
- Orosco MM, Pacholski C, Miskelly GM, Sailor MJ. Protein-coated porous silicon photonic crystals for amplified optical detection of protease activity. Adv. Mater. 2006;18:1393. [Google Scholar]
- Pacholski C, et al. Biosensing using porous silicon double-layer interferometers: reflective interferometric Fourier transform spectroscopy. J. Am. Chem. Soc. 2005;127(33):11636. doi: 10.1021/ja0511671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholski C, et al. Reflective Interferometric Fourier Transform Spectroscopy: A Self-Compensating Label-Free Immunosensor Using Double-layers of Porous SiO2. J. Am. Chem. Soc. 2006;128:4250. doi: 10.1021/ja056702b. [DOI] [PubMed] [Google Scholar]
- Sailor MJ, Link JR. Smart Dust: nanostructured devices in a grain of sand. Chem. Comm. 2005. p. 1375. [DOI] [PubMed]
- Schwartz MP, Alvarez SD, Sailor MJ. Porous SiO2 interferometric biosensor for quantitative determination of protein interactions: Binding of protein a to immunoglobulins derived from different species. Anal. Chem. 2007;79(1):327. doi: 10.1021/ac061476p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MichaelP, et al. The smart petri dish: A nanostructured photonic crystal for real-time monitoring of living cells. Langmuir. 2006;22:7084. doi: 10.1021/la060420n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MP, Buriak JM. Chemical and biological applications of porous silicon technology. Adv. Mater. 2000;12(12):859. [Google Scholar]
- Zhang D, Alocilja EC. Characterization of nanoporous silicon-based DNA biosensor for the detection of Salmonella enteritidis. IEEE Sens J. 2008;8(5-6):775. [Google Scholar]
- Bonanno LM, Segal E. Nanostructured porous silicon-polymer-based hybrids: from biosensing to drug delivery. Nanomedicine. 2011;6(10):1755. doi: 10.2217/nnm.11.153. [DOI] [PubMed] [Google Scholar]
- Jane A, Dronov R, Hodges A, Voelcker NH. Porous silicon biosensors on the advance. Trends Biotechnol. 2009;27(4):230. doi: 10.1016/j.tibtech.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Li S, Huang J, Cai L. A porous silicon optical microcavity for sensitive bacteria detection. Nanotechnology. 2011;22(42):425502. doi: 10.1088/0957-4484/22/42/425502. [DOI] [PubMed] [Google Scholar]
- Massad-Ivanir N, Shtenberg G, Segal E. In: Nano Bio-Technology for Biomedical and Diagnostics Research. Zahavy E, Ordentlich A, Yitzhaki S, Shafferman A, editors. Vol. 733. Netherlands: Springer; 2012. [Google Scholar]
- Massad-Ivanir N, et al. Engineering Nanostructured Porous SiO2 Surfaces for Bacteria Detection via "Direct Cell Capture". Anal. Chem. 2011;83(9):3282–32. doi: 10.1021/ac200407w. [DOI] [PubMed] [Google Scholar]
- Massad-Ivanir N, Shtenberg G, Zeidman T, Segal E. Construction and characterization of porous SiO2/hydrogel hybrids as optical biosensors for rapid detection of bacteria. Adv Funct Mater. 2010;20(14):2269–22. [Google Scholar]
- Mathew FP, Alocilja EC. Porous silicon-based biosensor for pathogen detection. Biosens. Bioelectron. 2005;20(8):1656. doi: 10.1016/j.bios.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ouyang H, Archer M, Fauchet PM. Frontiers in Surface Nanophotonics. 2007;133:49. [Google Scholar]
- Ouyang H, DeLouise LA, Miller BL, Fauchet PM. Label-free quantitative detection of protein using macroporous silicon photonic bandgap biosensors. Anal. Chem. 2007;79(4):1502–15. doi: 10.1021/ac0608366. [DOI] [PubMed] [Google Scholar]
- Hermanson GT. Bioconjugate Techniques. 1st. Academic Press; 1996. [Google Scholar]
- Piervincenzi RT, Reichert WM, Hellinga HW. Genetic engineering of a single-chain antibody fragment for surface immobilization in an optical biosensor. Biosensors and Bioelectronics. 1998;13(3-4):305. doi: 10.1016/s0956-5663(97)00130-9. [DOI] [PubMed] [Google Scholar]
- Saerens D, Huang L, Bonroy K, Muyldermans S. Antibody Fragments as Probe in Biosensor Development. Sensors. 2008;8(8):4669. doi: 10.3390/s8084669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtenberg G, et al. Picking up the Pieces: A Generic Porous Si Biosensor for Probing the Proteolytic Products of Enzymes. Anal. Chem. 2013;85(3):1951. doi: 10.1021/ac303597w. [DOI] [PubMed] [Google Scholar]
- Bonanno LM, DeLouise LA. Steric Crowding Effects on Target Detection in an Affinity Biosensor. Langmuir. 2007;23(10):5817. doi: 10.1021/la063659c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banada PP, Bhunia AK. In: Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems. Mohammed E, Zourob S, Turner APF, editors. Springer; 2008. p. 567. [Google Scholar]
- Poma A, Whitcombe M, Piletsky S. In: Designing receptores for the next generation of biosensors. Whitcombe MJ, Piletsky SA, editors. Springer; 2013. p. 105. [Google Scholar]
- Dudak FC, Boyaci IH. Development of an immunosensor based on surface plasmon resonance for enumeration of Escherichia coli in water samples. Food Res. Int. 2007;40(7):803. [Google Scholar]
- Dudak FC, Boyaci IH. Rapid and label-free bacteria detection by surface plasmon resonance (SPR) biosensors. Biotechn J. 2009;4(7):1003. doi: 10.1002/biot.200800316. [DOI] [PubMed] [Google Scholar]
- Skottrup PD, Nicolaisen M, Justesen AF. Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron. 2008;24(3):339. doi: 10.1016/j.bios.2008.06.045. [DOI] [PubMed] [Google Scholar]
- Taylor AD, Ladd J, Homola J, Jiang S. Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems. Springer; 2008. p. 83. [Google Scholar]


