Abstract
Mit1 is the putative chromatin remodeling subunit of the fission yeast Snf2/histone deacetylase (HDAC) repressor complex (SHREC) and is known to repress transcription at regions of heterochromatin. However, how Mit1 modifies chromatin to silence transcription is largely unknown. Here we report that Mit1 mobilizes histone octamers in vitro and requires ATP hydrolysis and conserved chromatin tethering domains, including a previously unrecognized chromodomain, to remodel nucleosomes and silence transcription. Loss of Mit1 remodeling activity results in nucleosome depletion at specific DNA sequences that display low intrinsic affinity for the histone octamer, but its contribution to antagonizing RNA polymerase II (Pol II) access and transcription is not restricted to these sites. Genetic epistasis analyses demonstrate that SHREC subunits and the transcription-coupled Set2 histone methyltransferase, which is involved in suppression of cryptic transcription at actively transcribed regions, cooperate to silence heterochromatic transcripts. In addition, we have demonstrated that Mit1's remodeling activity contributes to SHREC function independently of Clr3's histone deacetylase activity on histone H3 K14. We propose that Mit1 is a chromatin remodeling factor that cooperates with the Clr3 histone deacetylase of SHREC and other chromatin modifiers to stabilize heterochromatin structure and to prevent access to the transcriptional machinery.
INTRODUCTION
Several primary functions in the cell, including DNA replication, repair, and transcription, are regulated by packaging of the genome with histone and nonhistone proteins into chromatin. The basic unit of chromatin is the nucleosome, which consists of DNA wrapped around an octamer of histone proteins. Covalent modification, positioning, and packaging of nucleosomes then coordinate access to and utilization of genetic information. Chromatin has often been categorized into two classes: euchromatin, characterized as “open,” allowing for higher transcriptional potential, and heterochromatin, compact and thus refractory to transcription. While the properties that define heterochromatin are generally accepted (reviewed in reference 1), the mechanistic details of how heterochromatin-associated factors maintain chromatin structure and silence transcription are largely unknown.
As in many higher organisms, heterochromatin in the fission yeast Schizosaccharomyces pombe can largely be defined by methylation of lysine 9 on the N-terminal tail of histone H3 (H3K9), which is performed by the histone methyltransferase Clr4 (2, 3). H3K9 methylation recruits the HP1-like proteins Swi6 and Chp2, which harbor chromodomains that specifically recognize this modification (4, 5). Swi6 and Chp2 themselves have no known enzymatic activity; however, both are components of protein complexes that are important for heterochromatin maintenance and function (6–8). Chp2 is a constituent of the Snf2/histone deacetylase (HDAC) repressor complex (SHREC), additionally comprised of the Clr1, Clr2, Clr3, and Mit1 proteins, which is required for efficient silencing of heterochromatic transcripts (6, 9). SHREC includes at least two enzymatic functions: Mit1 is an ATPase with homology to Snf2-related chromatin remodeling factors, while Clr3 is a class II histone deacetylase known to target lysine 14 of histone H3 (10). The specific functions of the other two subunits (Clr1 and Clr2) are largely unknown (11–14).
SHREC is similar in composition to Mi-2/nucleosome remodeling and deacetylase (NuRD) complexes found in several higher organisms that have diverse roles within the cell, including regulating gene expression, cell cycle progression, and the response to DNA damage (15). In addition, human NuRD has been shown to associate with and to maintain pericentric heterochromatin (16). The role of NuRD in heterochromatin maintenance seems biologically relevant, since loss of NuRD activity has been proposed to be a contributing factor in natural and diseased aging (17). Homology suggests that SHREC shares two key enzymatic activities with NuRD: ATP-dependent chromatin remodeling activity by chromodomain-helicase-DNA-binding proteins 3 and 4 (CHD3/CHD4, or Mi-2α/Mi-2β) and histone deacetylase activity from histone deacetylase 1 and 2 (HDAC1/HDAC2) subunits (reviewed in reference 18); however, the mechanistic details of how these activities contribute to SHREC function are not fully understood.
Previous studies have implicated a role for Mit1 in positioning nucleosomes (19, 20). In this study, we demonstrated that Mit1 is indeed a nucleosome remodeling factor capable of mobilizing histone octamers in vitro, with characteristics similar to those of Mi-2 remodelers. However, unlike NuRD, Mit1 acts largely as a heterochromatin-specific silencing factor. Interestingly, we found that Mit1 activity is most evident at specific DNA sequences within heterochromatin that are refractory to nucleosome occupancy. Our results suggest that Mit1 is actively mobilizing nucleosomes onto these locations. We documented that Mit1 activity is positively influenced by interactions made through two conserved chromatin association domains of Mit1, the plant homeodomain (PHD) and chromodomain, and showed that Mit1 functions synergistically with the Set2 methyltransferase, previously implicated in stabilizing nucleosome occupancy in transcribed regions (21, 22), to silence heterochromatic transcripts. Finally, we interrogated the interplay between Clr3 histone deacetylase and Mit1 remodeling activities. We demonstrated that Mit1's remodeling activity is critical for SHREC function and that Mit1's role is not limited to facilitating Clr3's deacetylation of Lys14 on histone H3.
MATERIALS AND METHODS
S. pombe strains were grown and manipulated as previously described (23). Strains and plasmids used in this study are listed in Tables S1 and S2 in the supplemental material. Gene deletions and mutagenesis were performed using standard PCR-based procedures. The Mit1 open reading frame (ORF) was cloned into pREP plasmids (24) for episomal expression in yeast. Glutathione S-transferase (GST) fusions were expressed in Escherichia coli and purified after cloning into pGEX-KG (25). Analysis of Mit1 homology was performed using Pfam analysis (26) and the Phyre2 Web server (27).
Transcript analysis by high-resolution tiling array and RT-PCR.
RNA was prepared using a hot phenol extraction method (28). Briefly, 5 × 107 exponentially growing fission yeast cells were resuspended in TES buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 100 mM NaCl, and 0.5% SDS made in diethyl pyrocarbonate [DEPC] H2O) and an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1 [pH 4.7]) and incubated for 1 h at 65°C with occasional vortexing. After centrifugation, the soluble fractions were phenol-chloroform extracted and ethanol precipitated. Trace amounts of DNA were removed using Turbo DNase (Ambion), and RNA was further purified using an RNeasy minikit (Sigma).
Total RNA (7 μg) samples were processed using the GeneChip eukaryotic double-strand whole-transcript protocol (P/N 703064 Rev. 1; Affymetrix). The resulting double-stranded cDNA was fragmented then 3′ end labeled using terminal deoxynucleotidyltransferase with a biotinylated dideoxynucleotide analog. The biotinylated cDNA (4.5 μg) was then hybridized to the Affymetrix S. pombe Tiling 1.0FR array (20-bp resolution). After stringent washing of arrays, bound cDNA targets were visualized by staining with streptavidin-phycoerythrin (PE), followed by scanning using an Affymetrix 3000 7G high-resolution scanner. Raw image (DAT) files were processed using the Affymetrix AGCC software program to generate probe-summarized CEL files for downstream analysis. Probe positions and yeast genes were mapped to the S. pombe genome (2004 version) and visualized using the Integrated Genome Browser (IGB).
Comparison of expression levels in wild-type (WT) and mit1Δ cells was performed using the Affymetrix Tiling Analysis Software v1.1 program. Biological duplicates of each condition were analyzed using the following procedures. Probe intensities from the four arrays were quantile normalized, and the median was scaled to a linear value of 500. Probe-level analysis comparing the two groups was performed as described for the two-sample comparison within the software analysis manual. Parameters used in this analysis included setting the bandwidth to 50 (equivalent to a sliding genome position of 101 bases) and with perfect match/mismatch (PM/MM) summarization. The statistical significance of change was assessed using the two-sided Wilcoxon signed-rank test as described in the software manual. Signal differences and associated P values were exported as binary analysis results (BAR) files and visualized in the Integrated Genome Browser. Interval analysis was also performed using the Tiling Analysis Software to identify contiguous regions of change across the S. pombe genome. BED files were generated that contained genomic regions (minimum size of 150 bases, with a maximum gap of 100 bases) with a greater than 2-fold difference and a P value of <0.05 by the Wilcoxon signed-rank test. Custom Perl scripts (available on request) were used to intersect and merge these BED files with gene annotations obtained from the S. pombe 2004 genome build.
Quantitative PCR (qPCR) analysis of transcripts was performed on reverse-transcribed cDNA using Quantifast SYBR green (Qiagen) and an Eppendorf Mastercycler machine. Primers used for analysis are listed in Table S3 in the supplemental material.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed as previously described (29). Antibodies used were anti-histone H3 C terminus (Abcam ab1791), anti-RNA polymerase II (Pol II; Covance MMS-126R), anti-histone H3 (dimethyl K9) (H3K9me2) (Abcam ab1220), anti-Chp1 (Abcam ab18191), and anti-Swi6 (Thermo Scientific PA1-4977). Primers used for quantitative PCR analysis are listed in Table S3 in the supplemental material.
Biotinylated DNA pulldown.
Pulldowns were performed by incubating 3 μg GST or GST fusion protein with 20 pmol biotin or 601 nucleosome positioning sequence (NPS) generated by PCR with biotinylated or unlabeled primers in 0.5 ml pulldown buffer(20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol [DTT]) for 2.5 h at 4°C with rotation. Streptavidin-Sepharose (GE Healthcare) beads were added, and samples were incubated for an additional hour before washing three times with pulldown buffer. Associated proteins were then analyzed by SDS-PAGE and Western blotting with an antibody against GST (GE Healthcare 27-4577-01).
Calf thymus histone pulldown.
Pulldowns were performed as previously described (30) with some modification. Calf thymus histones (Worthington Biochemical) at 5 μg per reaction were incubated with 2 μg GST or GST fusion protein in CT binding buffer (20 mM Tris-HCl [pH 7.5], 750 mM NaCl, 0.75% NP-40, and 0.5 mM DTT) at 4°C for 4 h. After binding to glutathione-agarose for 1 h, beads were washed 3 times with CT binding buffer with a 5-min rotation for each wash. Bound proteins were then analyzed by SDS-PAGE and Western blotting using antibodies against GST (GE Healthcare 27-4577-01), H3 (Abcam ab1791), H4 (Millipore 05-858), H2A (Active Motif), H2B (Millipore 07-371), H3K4me3 (Active Motif 39160), H3K9me2 (Active Motif 39239), and H3K36me3 (Epigentek A-4042).
Northern analysis of siRNA.
Analysis of small interfering RNA (siRNA) was performed as previously described (31).
EMSA.
Appropriate amounts of GST or GST fusion proteins were incubated with a radiolabeled PCR product (70N0) in electrophoretic mobility shift assay (EMSA) binding buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 50 ng/μl bovine serum albumin [BSA], and 5% glycerol) for 30 min at 4°C. Samples were then loaded onto a 0.5× Tris-borate-EDTA (TBE)–4% polyacrylamide gel and electrophoresed at 200 V for 4 h at 4°C. EMSA with mononucleosomes that lack free DNA ends was performed using mononucleosomes assembled in vitro onto the 0N0 DNA probe.
Coimmunoprecipitation of Mit1-3×V5 and 6×FLAG-Chp2.
Extracts from 1 × 108 exponentially growing fission yeast cells were prepared by addition of coimmunoprecipitation (Co-IP) lysis buffer(10 mM HEPES-NaOH [pH 7.5], 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, 1 mM EGTA, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], and a protease inhibitor cocktail [Roche]) and grinding with a mortar and pestle while frozen using liquid nitrogen. Extracts were subject to ultracentrifugation to remove debris before incubation with anti-V5 affinity gel (Sigma) for 2 h. Beads were washed three times with Co-IP lysis buffer without glycerol, and bound proteins were eluted with 0.4 mg/ml 3×V5 peptide four times for 30 min each. Coimmunoprecipitated proteins were then analyzed by Western blotting with antibodies against V5 (Serotec) and FLAG (Sigma).
Affinity purification of Mit1.
Fission yeast with an endogenous mit1+ deletion and episomally expressing 3×FLAG-Mit1-3×V5 fusion protein under the control of the full-strength nmt1+ promoter were grown to mid-log phase in 8 liters of PMG minimal medium (32) with supplements but lacking uracil to maintain the plasmid. Cells were harvested by centrifugation and subjected to mechanical lysis using a Krups Tipo 203 coffee grinder in the presence of dry ice. Extracts were made by the addition of extraction buffer A (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 8 mM EDTA, 0.2% NP-40, 10% glycerol, 0.1 mM PMSF, and a protease inhibitor cocktail [Roche]) and incubation at 4°C with rotation for 60 min before ultracentrifugation to remove debris. Cleared lysate was adjusted to 150 mM NaCl, 4 mM EDTA, and 0.1% NP-40 before binding to anti-FLAG M2 affinity gel (Sigma). Bound proteins were washed with wash buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% NP-40, and 10% glycerol) and eluted with elution buffer (10 mM Tris-HCl, 150 mM NaCl, and 10% glycerol) containing 0.05% NP-40, 0.1 mM PMSF, and 0.5 mg/ml 3×FLAG peptide. The eluate was incubated with anti-V5 affinity gel (Sigma), washed with wash buffer, and again eluted with elution buffer lacking protease inhibitor and NP-40 but containing 0.5 mg/ml 3×V5 peptide. Mass spectrometry results are presented in Table S4 in the supplemental material.
Nucleosome remodeling assays.
[32P]dCTP-incorporated PCR product was generated using primers that amplify products with distinctly localized nucleosome positioning sequences (70N0, 70N70, and 0N0) from a plasmid containing the 601 nucleosome positioning sequence (p199-1) as a template (33). Mononucleosomes were prepared by first mixing equimolar amounts of histone octamer (human, recombinant; NEB) and the specific labeled PCR product. Assembly was performed by stepwise dilution of the salt concentration from 2 M to 0.25 M at 25°C over the course of 210 min, and material was stored at 4°C.
Nucleosome mobilization was performed by incubating purified remodeler with labeled mononucleosomes in a buffer containing final concentrations of 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 50 ng/μl BSA, and 5 mM ATP for 60 min at 30°C. Reactions were stopped by the addition of plasmid DNA (p199-1) to 300 ng/μl and glycerol to 6%. Samples were incubated on ice for 10 min before electrophoresis on 0.5× TBE–5% native polyacrylamide gels. Gels were dried and exposed to phosphor screens prior to imaging.
ATP hydrolysis assay.
ATP hydrolysis was performed as previously described (34). A total reaction mixture volume of 10 μl, containing normalized amounts of Mit1, mutant, or mock purifications, was incubated with 0.75 μl [α-32P]ATP (3,000 Ci/mmol, 10 mCi/μl; PerkinElmer) in buffer containing final concentrations of 20 mM Tris-HCl (pH 7.5), 70 mM KCl, 15 mM NaCl, 6.5 mM MgCl2, 0.8 mM EDTA, 1 mM DTT, 0.016% NP-40, 1% glycerol, 0.1 mM unlabeled ATP, and 30 ng/μl plasmid DNA (p199-1). After 30 min at 30°C, reactions were stopped by addition of 0.5 μl of 0.5 M EDTA, and reaction mixtures were placed on ice.
Reaction mixtures were then spotted onto polyethyleneimine (PEI)-cellulose thin-layer chromatography plates (Sigma) and resolved in 0.75 M KH2PO4 (pH 3.5). Plates were dried and exposed to a phosphor screen. Screens were analyzed by phosphorimaging (Molecular Dynamics Storm 860). In instances where results were quantified, protein was adjusted to result in approximately 10 to 15% hydrolysis, and densitometry was performed (ImageQuant 3.0; GE Healthcare), subtracting a reaction without remodeler as the background.
In silico prediction of nucleosome occupancy.
Sequences were analyzed by the NuPoP Nucleosome Positioning Prediction Engine (4th order) algorithm (http://nucleosome.stats.northwestern.edu) and the Online Nucleosomes Position Prediction by Genomic Sequence Version 3.0 algorithm (http://genie.weizmann.ac.il/software/nucleo_prediction.html).
In vivo nucleosome scanning assay.
Mononucleosomal DNA was prepared as previously described (35) with some modification. Log-phase cultures of S. pombe (2.5 × 108 cells) were fixed for 20 min in 0.5% paraformaldehyde. Fixation was stopped by the addition of glycine to 125 mM, and pellets were washed extensively with phosphate-buffered saline (PBS). Cell pellets were resuspended in 2 ml preincubation buffer (20 mM citric acid, 20 mM Na2HPO4, 40 mM EDTA, and 30 mM β-mercaptoethanol] for 10 min at 30°C. Cells were permeabilized by the addition of 1 ml sorbitol-Tris buffer (1 M sorbitol, 10 mM β-mercaptoethanol, and 50 mM Tris-HCl [pH 7.5]) with Zymolyase (MP Biomedicals) added to 1 mg/ml and incubation at 30°C with shaking. After washing with sorbitol-Tris buffer lacking Zymolyase, spheroplasts were digested with micrococcal nuclease (Worthington Biochemicals) at 256 U/ml in 0.5 ml NP-40 buffer (1 M sorbitol, 50 mM NaCl, 5 mM MgCl2,1 mM CaCl2, 0.075% NP-40, 0.5 mM spermidine, and 10 mM Tris-HCl [pH 7.5]) for 25 min at 36°C. Reactions were stopped by the addition of 65 μl stop buffer (5% SDS and 100 mM EDTA). RNA was removed from the samples by incubation with RNase A for 90 min at 37°C. Cross-linking was reversed, and proteins were removed by incubation overnight at 65°C in the presence of proteinase K. After phenol-chloroform extraction, mononucleosomal DNA was isolated by gel extraction following agarose gel electrophoresis.
Isolated mononucleosomal and input genomic DNA was subjected to quantitative PCR, analyzed using primers listed in Tables S5 and S6 in the supplemental material, and normalized to amplification by a primer set in the open reading frame of adh1+.
In vitro chromatin reconstitution and nucleosome mapping.
Nucleosomes were assembled on a 2.0-kbp PCR product including the mating-type region REII element by following a salt step-down procedure (see preparation of mononucleosomes) beginning with 10 μg DNA, 7.5 μg histone octamer (NEB), and 2 M NaCl. Reconstitutions (5 μg DNA equivalent) were then incubated in 0.24 ml MNase digestion buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2.5 mM MgCl2, and 2.5 mM CaCl2) with or without 100 U/ml MNase for 10 min at 30°C. Reactions were stopped by the addition of SDS to 0.5% and EDTA to 25 mM. Proteinase K was added and samples were incubated at 37°C overnight. Following phenol-chloroform extraction, mononucleosomal DNA from MNase-digested samples was isolated by agarose gel electrophoresis and analyzed by quantitative PCR relative to undigested input DNA.
Microarray data accession number.
Microarray data obtained in this work can be accessed at the NCBI Gene Expression Omnibus, accession number GSE46649.
RESULTS
Mit1 is required for efficient silencing at regions of heterochromatin.
Mi-2 remodelers play important roles in control of euchromatic gene expression (36, 37). In contrast, Mit1 is known primarily as a heterochromatin silencing factor, but several reports implicate Mit1 in euchromatic control. Like other components of SHREC, Mit1 is localized primarily to regions of heterochromatin, but it is also enriched at some euchromatic loci (9). Mit1 has also been reported to have genome-wide roles in nucleosome positioning, and Mit1 loss was reported to result in expression changes of more than 200 genes (19). Although the genome-wide changes in nucleosome positioning previously linked to Mit1 activity have recently been refuted and attributed to other CHD remodelers (38), it remained possible that Mit1, like Mi-2 remodelers, influences gene expression.
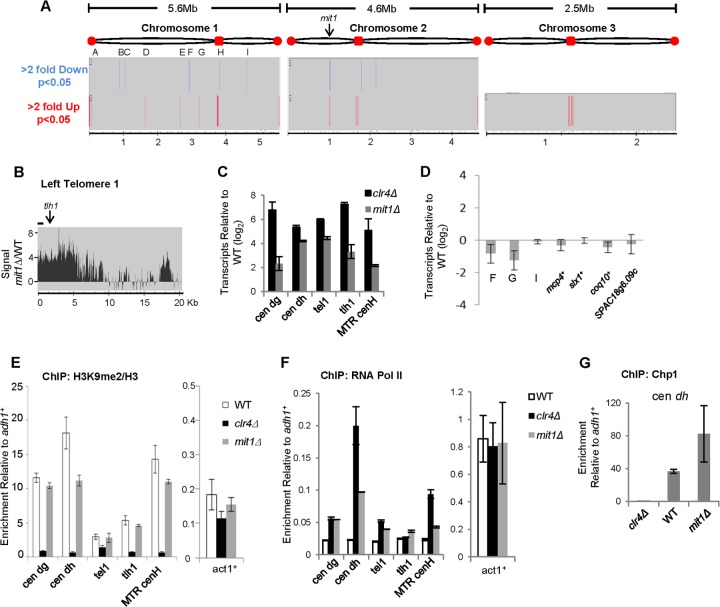
To address whether Mit1 regulates gene expression, we analyzed global transcription changes caused by mit1+ deletion using a genome-wide high-resolution tiling array (Fig. 1A and B). We performed analysis using duplicate biological replicates of cDNA prepared from WT and mit1Δ cells and specified that hits show at least a 2-fold difference in expression between mit1Δ and WT cells, with a significance of P < 0.05 between the replicates. Using these parameters, we observed elevated levels of transcripts in regions normally suppressed by constitutive heterochromatin, including subtelomeres and centromeres in the mit1Δ mutant and relatively few changes in coding regions. Quantitative real-time PCR (qPCR) confirmed an elevation in transcripts from heterochromatic loci for mit1Δ cells (Fig. 1C) (6). Q-PCR analysis was also performed on the nine euchromatic sites on chromosome 1 that our array analysis identified as differentially regulated (sites A to I). Most of these sites are in intergenic rather than coding regions (see Table S7 in the supplemental material), and transcript levels were either very low or undetectable for WT cells, although one transcript (at site G on chromosome 1) showed 2-fold downregulation for mit1Δ cells (Fig. 1D). We conclude from this analysis that many of the euchromatic loci showing differential regulation between mit1Δ and WT samples in our array analysis are regions of low transcriptional activity in WT cells and possibly were detected as differentially regulated because of the relatively permissive P value we employed to maximize detection. Many of the genes and pseudogenes previously identified as being regulated by Mit1 are located in subtelomeric regions. To further confirm our findings and in an effort to resolve differences between our data and a previously published analysis, we also used Q-PCR to reanalyze transcripts from several genes located on chromosome arms previously identified as strongly regulated by Mit1 (19). We found that these genes were not significantly affected by the mit1 deletion (Fig. 1D). We conclude that Mit1 is involved primarily in regulating transcription at regions of heterochromatin and does not have a widespread or significant role in the regulation of steady-state gene expression within euchromatin.
FIG 1.
Mit1 is required for efficient silencing at regions of heterochromatin. (A) Genome-wide analysis of transcriptional changes in mit1Δ cells. Genomic regions that have a 2-fold or greater change in expression in mit1Δ cells compared to the wild type were identified. Transcript changes were identified by comparing the hybridization of cDNA from both strain backgrounds (n = 2) to a high-resolution tiling array and filtering for regions that showed contiguous change over greater than 150 bases with a maximum gap size of 100 bases (P < 0.05). No regions were found to be downregulated by Mit1 deletion on chromosome 3. (B) Mit1 promotes silencing at telomere 1L. Representative comparison of hybridization signals at the left telomere of chromosome 1 on a high-resolution microarray of cDNA prepared from mit1Δ and wild-type cells. Elevated levels of transcripts were detected at other chromosome ends, most notably the right telomere of chromosome 2 (data not shown). The line represents the location of tel1 primers on the chromosome end used for transcriptional and ChIP analysis. (C) Confirmation of Mit1's role in heterochromatic silencing by Q-PCR. Q-PCR analysis of elevated transcripts in mit1Δ and clr4Δ cells relative to wild-type levels at regions of heterochromatin normalized to a euchromatic control gene, adh1+. Primer set locations can be found in panel B and Fig. 2C and 5A. (D) Impact of mit1Δ on euchromatic gene expression. Quantitative PCR analysis of transcriptional changes in the mit1Δ strain relative to the wild-type strain for regions of change identified by tiling array for which transcripts could be detected by Q-PCR. Also included are Q-PCR analyses of transcripts for four genes whose expression has previously been identified as being regulated by mit1+ (mcp4+, slx1+, coq10+, and SPAC18g6.09c). (E, F, and G) Loss of Mit1 does not disrupt hallmark heterochromatin proteins but allows access to RNA polymerase II. ChIP experiments used antibodies against H3K9me2 and histone H3 (E), RNA Pol II (F), or Chp1 (G), comparing enrichment of these epitopes at regions of heterochromatin and at act1+ (E and F) relative to a control locus, adh1+. Enrichment for H3K9me2 was normalized to histone H3 in panel E to account for potential histone occupancy changes. Data represent the means of data from duplicate experiments, with error bars reflecting the SEM (n = 2).
It is presently unclear how the SHREC complex acts to silence heterochromatic transcripts. SHREC has been proposed to act downstream of H3K9 methylation in silencing, but SHREC components have also been shown to be important for heterochromatin stability and spreading (7, 9, 11–14, 39). Chromatin immunoprecipitation (ChIP) experiments reveal that histones in heterochromatic regions retain methylation at Lys9 on histone H3 in mit1Δ cells (Fig. 1E) (6, 9, 40). However, consistent with our transcript analyses, RNA Pol II is enriched at sites of heterochromatin in mit1Δ cells (Fig. 1F) (6). Interestingly, at some loci (cen dg, tlh1+), the levels of RNA Pol II recruitment are similar between mit1Δ and clr4Δ cells (which completely lack heterochromatin), even though the accumulation of heterochromatic transcripts in mit1Δ cells is reduced compared with that in clr4Δ cells. These data suggest that silencing pathways, such as cotranscriptional silencing by RNA interference (RNAi), are functional in mit1Δ cells, and act to reduce apparent steady-state levels of heterochromatic transcripts (41–43). Consistent with this, the Chp1 subunit of the RNA-induced transcriptional silencing complex (RITS), an important effector complex in cotranscriptional silencing of heterochromatic transcripts (44), appears to be localized normally in cells lacking mit1+ (Fig. 1G), and centromeric siRNAs are present at elevated levels in cells lacking mit1+ (6). These observations support a model where Mit1/SHREC acts to silence transcription at the level of regulating RNA Pol II accessibility, while other silencing mechanisms that depend on H3K9 methylation remain intact.
Mit1 requires its ATPase activity and conserved chromatin tethering domains to silence transcription.
The behavior of ATP-dependent chromatin remodelers can be influenced greatly by proteins that associate with the remodeler and by the sequences that surround the highly conserved catalytic core (45). In addition to an Snf2-like ATPase domain, Mit1 has a zinc finger-like plant homeodomain (PHD) (9). Through further sequence analysis, we also identified a loosely conserved (Pfam E value of 0.72) chromodomain (CD). Although this domain identification was weak, multiple sequence alignment of this region of Mit1 with known chromodomain-containing proteins revealed a conserved arrangement of aromatic and hydrophobic residues characteristic of the aromatic “caging” and β-bulge of this domain's structure (Fig. 2A) (46). Additionally, secondary structure prediction for this region was aligned to known chromodomains with high confidence, including canonical chromodomain proteins (HP1 family and Suv39h1) and Saccharomyces cerevisiae Chd1 (27) (data not shown).
FIG 2.
Mit1 requires its conserved chromatin tethering domains for silencing. (A) Sequence alignment of Mit1's chromodomain. Sequence alignment of Mit1 with chromodomains of CHD family remodeling enzymes and Suvar39h1. Conserved aromatic (red), acidic (blue), basic (green), and hydrophobic (yellow) residues are indicated. (B) Mit1 has conserved PHD and CD domains. Schematic representation of SpMit1 and Mi-2 conserved motifs including the plant homeodomain (PHD), chromodomain (CD), and SWI/SNF-like ATPase domains. (C) PHD and CD contribute to silencing activity of Mit1. Serial dilution spotting assay on medium lacking leucine (to maintain expression plasmids) with or without 5′-fluoroorotic acid (FOA) of strains containing a centromeric ura4+ reporter gene (otr1::ura4+), indicated in the schematic. Lines underneath the schematic represent the locations of primers used for transcript analyses. Plates were incubated at 32°C. EV, empty vector. (D) Purification of recombinant GST fusions of domains of Mit1. SDS-PAGE and Coomassie staining of purified recombinant GST, GST-PHD(205–273)Mit1, and GST-CD(447–499)Mit1 fusion proteins is shown. (E) Mit1's CD domain can bind DNA in vitro. Biotinylated double-stranded DNA or an equivalent amount of biotin or unlabeled DNA was incubated with recombinant GST or GST fused to the Mit1 chromodomain. The DNA and associated proteins were captured by streptavidin beads and analyzed by Western blotting. (F) Mit1 can bind nucleosomal DNA. EMSA to compare the binding of Mit1 chromodomain to free DNA and nucleosomal DNA. Radiolabeled 147-bp Widom 601 nucleosome positioning sequence (50 ng; lanes 1 to 5) or mononucleosomes without free DNA ends reconstituted by salt dilution on the same sequence (50 ng DNA equivalent; lanes 6 to 10) were incubated with GST (2 μg and 4 μg) or GST-CDMit1 (2 μg and 4 μg) fusion protein. (G) Mit1's PHD domain can bind histone H3 in vitro. Calf thymus histone pulldown experiment comparing the abilities of the GST, GST-PHDMit1, and GST-PHDIng2 proteins to bind to histones. Histone association was monitored by immunoblotting using antibodies specific for different histones or histone modifications. (H) Mit1 PHD and CD are not required to maintain association with Chp2. Immunoprecipitation of wild-type 3×V5-tagged Mit1, Mit1PHDΔ, Mit1CDΔ, and Mit1K587A in strains expressing 6×FLAG-Chp2 and blotting against the V5 and FLAG epitopes. Note that the SDS-PAGE gels used in these experiments do not effectively resolve wild-type Mit1 from Mit1PHDΔ and Mit1CDΔ. The asterisk indicates a nonspecific band. (I) Centromeric association of Chp2 is partially dependent on Mit1. ChIP of Chp2 association with centromeres relative to a euchromatic locus in the indicated strain backgrounds is shown. Error bars reflect the SEM (n = 4).
Both PHD and CD are well recognized as chromatin interaction motifs (47, 48) and are commonly found in chromatin modifying enzymes, including Mi-2 family remodelers (Fig. 2B) (49). We first interrogated whether these domains are required for the silencing function of Mit1, which is required to repress the transcription of a centromeric reporter gene (otr1R::ura4+) (9). We asked whether cells episomally overexpressing Mit1, Mit1 mutated in its ATPase domain, Mit1K587A, or proteins deleted for either the PHD finger (Mit1ΔPHD) or the chromodomain (Mit1ΔCD) could complement the centromeric silencing defect of mit1Δ cells. Expression of wild-type Mit1 efficiently silenced the centromeric ura4+ reporter gene in mit1Δ cells, allowing cells to grow on medium containing 5-fluoroorotic acid (5-FOA), which is toxic to cells expressing ura4+ (Fig. 2C). Mit1K587A-expressing cells were unable to silence this reporter (9). We found that cells expressing Mit1 with deletions of the PHD domain (Δ214–269; Mit1ΔPHD]) or chromodomain (Δ446–502; Mit1ΔCD) were also unable to silence the centromeric reporter, suggesting that these putative chromatin interaction motifs are important for heterochromatic silencing by Mit1. The wild-type and mutant Mit1 proteins were expressed to similar levels (Fig. 2H and data not shown).
The chromodomain of Mit1 binds DNA.
The Mi-2 subfamily of CHD remodelers found in higher organisms are known to utilize their tandem PHD and chromo domains for histone tail and DNA binding, respectively (49, 50). Although Mit1 harbors only a single PHD finger and a loosely conserved chromodomain, we wondered whether these motifs might serve similar roles.
To test this hypothesis, we monitored the ability of Mit1's CD to associate with DNA. First, we tested binding of a recombinant GST fusion protein (Fig. 2D) of Mit1's CD to DNA in solution. GST-CD but not GST alone bound to biotin-labeled double-stranded DNA (dsDNA) and was captured by streptavidin-agarose (Fig. 2E). GST-CD did not bind to biotin alone, nor was the apparent pulldown the result of insoluble aggregation by GST-CD/DNA complexes, since no complex associated with streptavidin beads in the absence of biotin on the DNA.
To determine whether the chromodomain could bind nucleosomal DNA, we used electrophoretic mobility shift assays (Fig. 2F). GST-CD (lanes 4 and 5) specifically caused a slower-migrating shift of a 147-bp-sequence (0N0) radiolabeled dsDNA following native PAGE relative to DNA incubated with GST (lanes 2 and 3) or to the probe alone (lane 1), indicative of DNA binding activity. Nucleosomes were reconstituted onto 0N0 such that they lacked free DNA ends and were tested for association with Mit1 CD. Shifts were seen on incubation with nucleosomal DNA (compare lanes 9 and 10 with lanes 6, 7, and 8), suggesting that the association of Mit1's chromodomain with DNA is not significantly altered by the histone octamer. No DNA binding was observed when the GST-PHD fusion was tested for association with DNA in EMSA (data not shown).
The PHD of Mit1 binds histone H3.
To test whether Mit1's PHD associates with histones, we assayed the ability of GST-PHD fusion proteins to bind calf thymus histones. As previously demonstrated for the ING2 PHD domain (51), we found that GST-PHD (Mit1) binds histone H3 in preference to other histones (Fig. 2G). Several studies of Mi-2 PHD fingers have shown sensitivity of H3 tail interactions to different methyl marks (52–54). Although we recognize that this is a rather nonphysiological experiment, since binding to histones was tested, rather than binding to nucleosomes, we interrogated the modification status of histone H3 bound by GST-PHD (Mit1) and showed that it was enriched for methyl marks of transcriptional activity, H3K4me3 and H3K36me3, but curiously lacked the heterochromatic hallmark, H3K9me2. This indicates that binding of Mit1 PHD to histone H3 may be inhibited by the H3K9me2 modification, which was a surprising result given that the SHREC complex is recruited to regions of heterochromatin (9). Attempts to further dissect GST-PHD binding specificity were hampered since we were unable to demonstrate specific binding to a modified histone tail peptide array or to peptides in solution (data not shown).
We tested whether loss of Mit1's chromatin association domains specifically impaired Mit1's association with other subunits of the SHREC complex. While it remains a possibility that these deletions impact additional Mit1 interactions, Mit1ΔPHD and Mit1ΔCD maintain association with Chp2 (Fig. 2H), indicating that Chp2's ability to bind Mit1 is retained in these mutants. SHREC can presumably still be targeted to heterochromatin via Chp2's recognition of H3K9 methylation, although we have been unable to directly ChIP Mit1 at these regions. Interestingly, we found that association of Chp2 with centromeric repeats is partially dependent on Mit1 and specifically on its PHD (Fig. 2I). Furthermore, cells expressing Mit1 with the catalytic K587A mutation show no defect in Chp2 association with cen dh, suggesting that for at least the PHD (the role of the CD was not tested), Mit1 domains which interact with elements of chromatin in vitro contribute to the association of Chp2 with chromatin in the cell.
Mit1 is an ATP-dependent nucleosome remodeling factor.
Mit1 has a highly conserved Snf2-related ATPase domain, which suggests Mit1 may modulate the interaction between DNA and histones to reposition nucleosomes on DNA (55). However, to date there has been no demonstration that Mit1 is a bona fide chromatin remodeler. To determine if Mit1 has chromatin remodeling activity, we first purified Mit1 from fission yeast. We found that Mit1 is expressed at low levels and is labile under standard purification conditions. To purify sufficient Mit1, we employed a double epitope tagging strategy incorporating different tags at either end of the protein and overexpressed Mit1 from a full-strength nmt1+ promoter. Sequential affinity purification allowed isolation of full-length Mit1 (Fig. 3A). Mass spectrometry analysis did not identify copurification of other subunits of SHREC under these conditions (see Table S4 in the supplemental material), likely due to the considerable overexpression and dual tagging strategy that was employed to minimize purification of Mit1 breakdown products.
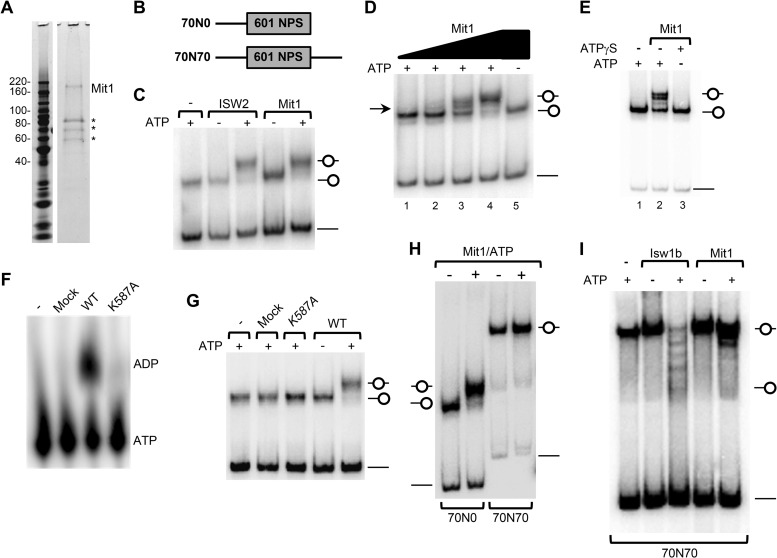
FIG 3.
Mit1 is a directional ATP-dependent nucleosome remodeling factor. (A) Purification of Mit1. An aliquot of the final purification of 3×FLAG-Mit1–3×V5 expressed in mit1Δ cells was analyzed by SDS-PAGE and SYPRO ruby staining. Mit1 was identified as the approximately 170-kDa band with 49% sequence coverage by mass spectrometry. Asterisks represent Mit1 breakdown products and heat shock proteins. See Table S4 in the supplemental material for more details. (B) Schematic representation of short mononucleosomes reconstituted for remodeling experiments. Nucleosomes were positioned by the Widom 601 nucleosome positioning sequence. (C) Mit1 can remodel a mononucleosomal substrate. End-positioned (70N0) mononucleosomes (30 nM) were incubated with ScISW2 (5.0 nM) or S. pombe Mit1 (SpMit1) (2.5 nM) in the presence or absence of ATP and resolved on a native 5% polyacrylamide gel. (D) Titration of Mit1 protein reveals intermediate remodeling events. Nucleosomes were remodeled as for panel C with a titration of Mit1 (0.3 nM, 0.6 nM, 1.25 nM, and 2.5 nM). (E) Mit1 utilizes ATP hydrolysis to remodel a mononucleosomal substrate. Nucleosomes were remodeled as for panel C on addition of ATP but not on addition of the nonhydrolyzable analogue, ATPγS. (F) Mit1 but not Mit1K587A can hydrolyze ATP. ATP hydrolysis assay comparing the activity of wild-type Mit1 to that of Mit1K587A, a mock purification, and a buffer-only control. (G) Mit1K587A cannot mobilize nucleosomes. Nucleosome mobilization assays as performed for panel C, comparing octamer mobilization by wild-type Mit1, Mit1K587A, and mock purifications. (H) Mit1 is a directional remodeler. Nucleosome mobilization assay comparing the remodeling of end-positioned (70N0) and center-positioned (70N70) nucleosomes incubated with or without Mit1 (2.5 nM) and ATP. (I) Comparison of Mit1 and Isw1b remodeling. Center-positioned mononucleosomes (70N70; 30 nM) were incubated with purified ScIsw1b (5 nM) or Mit1 (5 nM) in the presence or absence of ATP.
To determine whether Mit1 has in vitro chromatin remodeling activity, we performed a series of mononucleosome sliding assays which take advantage of the difference in mobility between nucleosomes positioned at the ends and center of short DNA fragments when resolved in native polyacrylamide gels (56). First, we assembled mononucleosomes by salt dialysis onto a 217-bp DNA fragment upon which deposition of the octamer is directed to the DNA end by the 601 nucleosome positioning sequence (57) (Fig. 3B, 70N0). We then incubated the positioned nucleosomes with purified Mit1, or S. cerevisiae ISW2 (ScISW2) as a positive control, in the presence or absence of ATP. ISW2 is known to mobilize histone octamers from the end to the center of short DNA fragments (58). Similar to ScISW2, Mit1 caused an ATP-dependent mobilization of the nucleosome (Fig. 3C). Remodeling by Mit1 was dose dependent (Fig. 3D), since titration of Mit1 generated a species that migrated between bands observed on incubation with no ATP (lane 5) or with saturating Mit1 and ATP (lane 4), which is likely to be a position intermediate (lane 3). In addition, we note that there was no increase in free DNA caused by Mit1 remodeling, suggesting that nucleosome eviction is not a prominent feature of Mit1 remodeling.
We tested whether the chromatin remodeling activity of Mit1 was linked to ATP hydrolysis. Mit1 incubated with a nonhydrolyzable ATP analog, ATPγS, did not change the mobility of an end-positioned nucleosome (Fig. 3E). We also purified a predicted ATPase-dead Mit1K587A protein (mutated in the ATP-binding site of the catalytic core, K587A) and confirmed that it could not hydrolyze ATP (Fig. 3F) (9). This also demonstrated that our purified material was free of contaminating ATPases. Importantly, Mit1K587A showed no sliding activity and could not mobilize an end-positioned nucleosome (Fig. 3G). These results confirm that Mit1 is indeed an ATP-dependent nucleosome remodeling factor.
Next, we addressed whether the remodeling activity of Mit1 was directional, since many other chromatin remodeling enzymes show directional in vitro nucleosome mobilization. Some remodelers, such as ISWI, mobilize histone octamers to the ends of short DNA fragments, while others, including Mi-2 and several CHD family remodelers, preferentially slide nucleosomes toward the center (49, 59). On incubation with saturating amounts of Mit1, essentially all end-positioned nucleosomes were mobilized to the center of the DNA, as evidenced by a discrete slower-migrating species (Fig. 3C, D, and G), suggesting that remodeling by Mit1 is nonrandom and directional. To probe the directionality of remodeling by Mit1, we incubated Mit1 with a centrally positioned mononucleosome (70N70). Consistent with a preference for moving histone octamers away from DNA ends, Mit1 did not move the 70N70 positioned nucleosomes (Fig. 3H and I). This was in contrast to remodeling by S. cerevisiae Isw1b, which mobilizes center-positioned mononucleosomes toward and slightly off the end of DNA fragments (60). Taken together, these data demonstrate that Mit1 is a directional (end-to-center) ATP-dependent chromatin remodeling factor that shares in vitro nucleosome remodeling characteristics with other Mi-2 family enzymes.
Mit1's chromatin tethering domains promote remodeling activity independent of effects on ATPase activity.
We have demonstrated that Mit1's CD can bind to DNA, that the PHD finger binds to histone H3, and that both domains contribute to Mit1's silencing function. We next sought to determine mechanistically how loss of Mit1's chromatin interaction domains impact Mit1 function. First, using purified proteins (Fig. 4A), we tested the ability of the mutant proteins to mobilize end-positioned nucleosomes. We found that Mit1ΔPHD and Mit1ΔCD are compromised for nucleosome mobilization, reflected in the incomplete shift of end-positioned mononucleosomes to a slower-migrating (centered) species in native PAGE (Fig. 4B). Importantly, Mit1ΔPHD and Mit1ΔCD have not lost their directional specificity but have an impaired ability to mobilize histone octamers, since addition of additional enzyme (approximately 8-fold more than the wild-type level) was sufficient to mobilize most of the octamers to the center of the DNA fragment. We did not observe mobilization activity by the catalytic K587A mutant in similar reactions (Fig. 4C).
FIG 4.
PHD and CD contribute to chromatin remodeling activity in vitro and in vivo but do not impact the ATPase activity of Mit1. (A) Western blot of purified 3×FLAG-Mit1–3×V5 proteins. (B) Mit1 PHD and CD contribute to nucleosome mobilization activity. Nucleosome mobilization assays as performed for Fig. 3C, comparing mobilization by purified wild-type Mit1, Mit1ΔPHD, and Mit1ΔCD proteins (2.5 nM), as well as eluate from a mock purification. (C) Higher concentrations of PHD or CD mutant Mit1 proteins overcome the mobilization defect. End-positioned (70N0) mononucleosomes (30 nM) were remodeled with buffer or increasing concentrations of purified Mit1 (1.5 nM, 3.0 nM, 6.0 nM, and 12.0 nM), Mit1ΔPHD (1.5 nM, 3.0 nM, 6.0 nM, and 12.0 nM), Mit1ΔCD (1.5 nM, 3.0 nM, 6.0 nM, and 12.0 nM), and Mit1K587A (12.0 nM). (D) PHD and CD mutant Mit1 proteins retain ATP hydrolysis activity. ATP hydrolysis assay comparing the activities of purified Mit1 mutants to that of the wild type, as well as buffer-only and mock purifications. A low level of purified proteins was used, so that hydrolysis of ATP to ADP was approximately 10 to 15% for wild-type Mit1. Reactions were quantified by densitometry following thin-layer chromatography (TLC) (see Materials and Methods), and the buffer-only reaction was subtracted as background before making all reactions relative to the hydrolysis observed using purified wild-type Mit1. Error bars represent standard errors of the means (n = 3).
These experiments demonstrate that removal of Mit1's DNA (CD) or H3 (PHD) tethering domain results in a reduced ability to remodel nucleosomes. While their requirement for octamer mobilization is not absolute in vitro, in the context of chromatin these interactions may be essential to maintain efficient interaction with nucleosomes or may modulate how nucleosomes are remodeled, since cells expressing Mit1 lacking these domains have silencing defects similar to those of Mit1K587A or mit1Δ cells (Fig. 2B). Importantly, the remodeling defect did not arise from a reduced ability of the mutant proteins to hydrolyze ATP, since Mit1ΔPHD and Mit1ΔCD hydrolyze ATP at levels similar to those for wild-type Mit1 (Fig. 4D). This suggests that the Mit1 PHD and chromo domains are not required for ATPase activity of the complex but are necessary to promote chromatin association and remodeling by SHREC. It remains possible, however, that in spite of the observed lack of defect in ATP hydrolysis activity of the PHD and CD deletion mutants, the deletions negatively affect the entire structure of Mit1.
Mit1 contributes to the formation of a nucleosome-free region on an intrinsically unfavorable site at REII.
Our data show that Mit1 is required to prevent RNA Pol II access to regions of heterochromatin and that Mit1 is able to mobilize nucleosomes. One mechanism by which heterochromatin can restrict RNA polymerase II accessibility is through altering nucleosomal occupancy to prevent access of transcription factors to their target sequences. Heterochromatin prevents the appearance of nucleosome-free regions (NFRs) at various locations in silenced regions of the genome, and nucleosome occupancy at some of these sites has been shown to depend on Mit1/SHREC (7, 9, 20, 39). Interestingly, the sites that most depend on Mit1 do not map to sites of transcription initiation or to regions particularly enriched for RNA Pol II. Mit1 also did not significantly suppress the formation of any nucleosome-free regions within centromeres (20) despite increased levels of Pol II and elevated transcripts originating from the dg and dh repeats in mit1Δ cells (Fig. 1C and F).
We hypothesized that the DNA sequence may contribute to the location of nucleosome-free-region formation in mit1Δ cells. To test this idea, we analyzed these regions with two in silico nucleosome prediction algorithms that predict nucleosome occupancy based on DNA sequence characteristics (61, 62) We found that some sites that depend on Mit1 for prevention of nucleosome-free regions appear to be particularly refractory to octamer occupancy and lie near positions predicted to have relatively well-positioned nucleosomes, particularly at REII in the mating-type region and telomere 2R (Fig. 5A and B). The predicted low affinity of these sequences is likely due to low G/C content and a high percentage of DNA with 5-mers of exclusively A/T nucleotides (Fig. 5C and D), both of which disfavor nucleosome formation (62, 63). This would suggest that Mit1 may play a critical role in moving nucleosomes onto DNA sequences that are energetically unfavorable for nucleosome positioning.
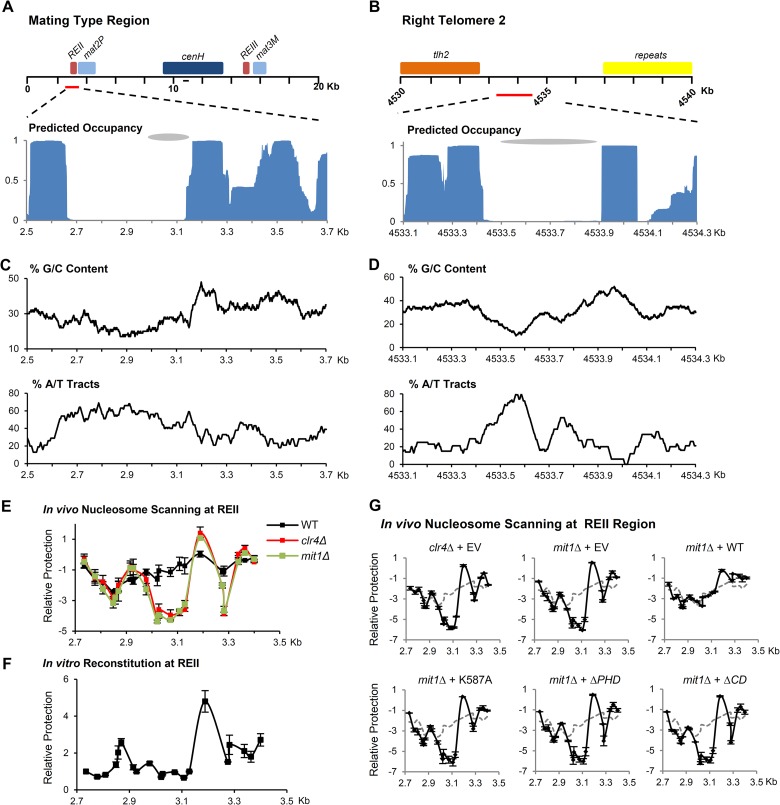
FIG 5.
Mit1 prevents the formation of an NFR on an intrinsically unfavorable site at REII. (A and B) Schematic of the fission yeast mating-type region and right telomere of chromosome 2. Predicted nucleosome occupancy using the Nucleosome Positioning Prediction Engine (NuPoP) for these regions is plotted below. Gray ovals indicate previously identified regions of nucleosome occupancy changes in mit1Δ cells. The line on the schematic represents the location of primers used for analysis in Fig. 1. (C and D) Sequence analysis of nucleosome-depleted regions. Sequence was analyzed for G/C content and the presence of A/T tracts and reported as the percentage of nucleotides in 100-bp windows that are either G or C or within A/T tracts defined as five or more consecutive A or T nucleotides. (E) In vivo nucleosome scanning of REII at the mating-type locus. A nucleosome scanning experiment compared the relative protection of the region surrounding the REII silencing element from digestion by micrococcal nuclease in wild-type, mit1Δ, and clr4Δ backgrounds. Mononucleosomal DNA was analyzed by Q-PCR, normalized to amplification within adh1+, and compared to that of the wild type. Data are plotted on a log2 scale; error bars represent SEM (n = 2). (F) In vitro nucleosome scanning at REII using reconstituted chromatin. A nucleosome scanning experiment used mononucleosomal DNA isolated from in vitro reconstitution of histones onto REII region synthetic DNA. Error bars represent SEM (n = 3). (G) Mutant Mit1 proteins cannot suppress NFR formation at REII in mit1Δ cells. A nucleosome scanning experiment was performed as for panel E, comparing wild-type cells transformed with empty vector (dashed line) to clr4Δ or mit1Δ cells transformed with empty vector and mit1Δ cells transformed with vectors expressing Mit1, Mit1K587A, Mit1ΔPHD, and Mit1ΔCD. Data are plotted on a log2 scale; error bars represent SEM (n = 2).
We used an in vivo PCR-based nucleosome scanning method to confirm the existence of an NFR at REII that forms in clr4Δ and mit1Δ cells. In short, mononucleosomal DNA was prepared following micrococcal nuclease digestion of chromatin prepared from wild-type and mutant cells and was subjected to Q-PCR with primers that amplify 18 overlapping fragments spanning the region. Using this assay, regions occupied by nucleosomes will be well represented in the DNA sample, whereas regions with low nucleosome occupancy or poorly positioned nucleosomes will amplify less. In wild-type cells, evenly dispersed and poorly positioned nucleosomes span this region (Fig. 5E). In contrast, in clr4Δ and mit1Δ cells, an NFR clearly forms and is evident as a strong trough in amplification over the REII sequence. In contrast to the “fuzzy” nucleosome occupancy found in wild-type cells at this locus, a strongly positioned nucleosome is observed in the absence of silencing factors at the downstream sequence adjacent to the nucleosome-free region at 3.2 kb. The position of this nucleosome relative to the NFR corresponds to a sharp transition in the G/C content of the underlying DNA sequence.
To investigate the influence of sequence on nucleosome occupancy, we assembled chromatin in vitro by salt dilution on synthetic DNA, including the REII region. Nucleosome occupancy in this cell-free reconstitution system greatly resembled both the predicted occupancy and interestingly the occupancy observed in clr4Δ and mit1Δ cells (Fig. 5F). These results suggest that silencing factors act to override the contribution of DNA sequence to nucleosome positioning at this site.
Next, we assayed whether Mit1 or Mit1 chromatin interaction mutants could correct NFR formation at REII in mit1Δ cells. In vivo nucleosome scanning assays were performed with mit1Δ cells transformed with vectors expressing wild-type or mutant Mit1 proteins (Fig. 5G). These experiments revealed that while episomally expressed wild-type Mit1 could complement for nucleosome occupancy at REII in mit1Δ cells, all three mutant Mit1 proteins (Mit1K587A, Mit1ΔPHD, and Mit1ΔCD) were unable to eliminate the NFR. These data demonstrate that the PHD and CD motifs of Mit1 that are required for efficient octamer mobilization in vitro are also essential for the nucleosome positioning function of Mit1 in vivo.
Mit1 functions synergistically with Set2 for maintenance of transcriptional silencing.
Given that the PHD domain of Mit1 associates preferentially with K4 and K36 but not K9 methyl-marked histone H3, we sought to determine whether we could place Mit1 in a genetic pathway linking it to methyltransferase activity. In fission yeast, H3K4 methylation is mediated by Set1 and is not required for heterochromatin silencing (64). Set2, which mediates H3K36 methylation, is believed to make minor contributions to heterochromatin silencing and methylates histones within centromeric repeats when heterochromatin is disrupted during S phase (65, 66). To test whether Mit1 functions in the same pathway as Set2 to repress aberrant transcription, we assessed genetic epistasis. We found that Set2 strongly synergizes with Mit1 to block accumulation of centromeric and subtelomeric transcripts relative to two control genes, act1+ and adh1+ (Fig. 6A), with transcript levels approaching those found in clr4Δ cells that completely lack heterochromatin. In contrast combining deletions of mit1 and set1 resulted in no additive or synergistic effects, which may indicate that Mit1 and Set1 function within the same pathway or that there is no synthetic genetic interaction. We also found that there was strong accumulation of centromeric transcripts in cells expressing the Mit1K587A, Mit1ΔPHD, and Mit1ΔCD Mit1 mutants in the set2Δ background and that set2Δ showed synthetic interactions with deletions of other SHREC components (data not shown). Together these data suggest that Set2 functions in conjunction with Mit1 and SHREC to suppress transcript accumulation from regions of heterochromatin.
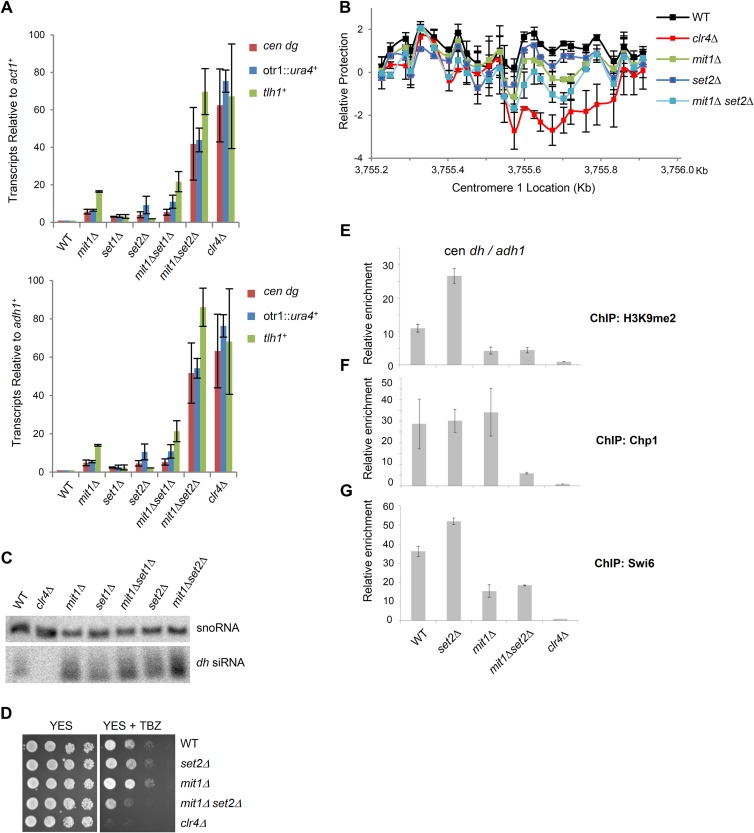
FIG 6.
Mit1 and Set2 synergize to silence heterochromatin transcripts. (A) Mit1 and Set2 cooperate to silence heterochromatin. Quantitative real-time PCR analysis of transcripts from a centromeric transgene (otr1::ura4+), centromeric repeats (cen dh), and subtelomeres (tlh1+) in cDNA prepared from the indicated strain backgrounds. Transcript levels were normalized to act1+ transcripts (top) or adh1+ (bottom) and to transcripts in WT cells. Error bars represent SEM (n = 2). (B) In vivo nucleosome scanning of a centromeric dh sequence. Nucleosome scanning experiment comparing the relative protection of part of the dh centromeric repeat from digestion by micrococcal nuclease in wild-type, mit1Δ, set2Δ, mit1Δset2Δ, and clr4Δ backgrounds. Mononucleosomal DNA was analyzed by Q-PCR, normalized to amplification within adh1+, and compared to that of the wild type. Data are plotted on a log2 scale. Error bars represent SEM (n = 2). (C) Monitoring of RNAi. Northern blot of purified small RNA using probes corresponding to cen dh sequences and snoR69 for a loading control. (D) Thiabendazole sensitivity plating assay. Serial dilution assay of wild-type and mutant strains plated on YES medium or YES medium containing 15 μg/ml TBZ. Plates were incubated at 25°C. (E to G) ChIP analysis for strains lacking Mit1 and Set2. Immunoprecipitated and input chromatin was analyzed by Q-PCR for relative enrichment of centromeric dh sequence relative to adh1+ in immunoprecipitated chromatin with anti-H3K9me2 (E), anti-Chp1 (F), or anti-Swi6 (G). ChIPs are normalized to a clr4Δ strain. Error bars represent SEM (n = 2).
Mit1 is not thought to significantly contribute to removal of nucleosome-free regions within centromere 1 (20). Given the strong synthetic interaction between Set2 and Mit1 for silencing of centromeric transcripts, we tested whether the combined loss of both factors results in altered nucleosome occupancy within centromeric sequences. We focused on analyzing a region of centromeric dh sequence that includes a Clr4-dependent NFR (20). In vivo PCR-based nucleosome scanning was performed on a 0.7-kb region, using 26 sets of real-time PCR primers. In agreement with previous microarray-based studies, cells lacking clr4 show a clear NFR centered at 3,755.6 kb on chromosome 1 (Fig. 6B) (20). We found that mit1Δ cells show reduced nucleosome occupancy at this site, and cells lacking set2 also have a somewhat decreased signal. Importantly, the mit1Δ set2Δ double mutant shows a greater defect in nucleosome occupancy than does the mit1Δ mutant. Thus, our analysis has uncovered a role for Mit1 in helping to prevent NFR formation at a centromeric locus, and this phenotype is exacerbated in the mit1Δ set2Δ compound mutant.
The large increase in centromeric transcript accumulation in mit1Δ set2Δ cells prompted us to investigate whether posttranscriptional silencing mechanisms are intact in this genetic background. RNAi appears fully functional in cells lacking either set2+ or mit1+, since these strains accumulate high levels of dh siRNAs (Fig. 6C). Surprisingly, levels of siRNA production were maintained in the mit1Δ set2Δ double mutant. Given this lack of effect on the RNAi pathway but the strong accumulation of centromeric transcripts in mit1Δ set2Δ mutant cells, we asked whether centromere function was affected. Many mutants that are deficient in chromosome segregation are sensitive to the microtubule-destabilizing drug thiabendazole (67). Plating assays with cells on medium containing thiabendazole demonstrated that while cells lacking clr4+ display sensitivity (Fig. 6D), the mit1Δ set2Δ compound mutant showed only intermediate thiabendazole sensitivity, indicative of some disruption of heterochromatin function compared with that with the mit1Δ mutation alone.
ChIP assays also revealed that H3K9me2 levels at centromeres and subtelomeres were similar in mit1Δ mutant and mit1Δ set2Δ compound mutant cells and were only slightly decreased compared with those in wild-type cells (Fig. 6E). Interestingly however, we found association of Chp1 with sites of heterochromatin was more severely impacted in cells lacking both Set2 and Mit1 than in either single mutant (Fig. 6F), whereas Swi6 association with centromeres was similar between mit1Δ cells and mit1Δ set2Δ cells (Fig. 6G). These results indicate that heterochromatin silencing by SHREC is partially redundant with the Set2-mediated repression of transcription that functions more generally at Pol II transcribed regions, and together these factors prevent transcription and disruption of heterochromatin.
SHREC's role in NFR prevention is not limited to deacetylation of H3K14.
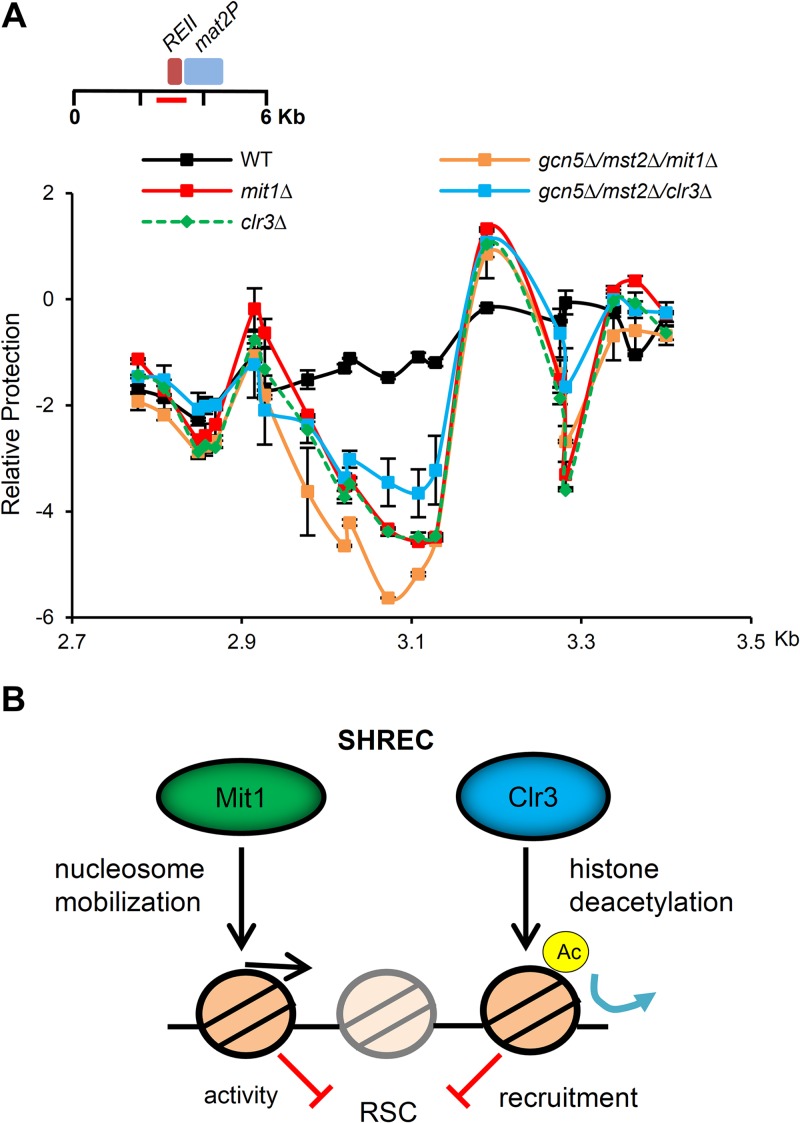
In addition to the chromatin remodeling activity of Mit1, it has been proposed that the SHREC subunit Clr3 is important for eliminating nucleosome-free regions within heterochromatin. Lysine 14 of histone H3 is thought to be the primary target of Clr3 deacetylation (10), and it has been hypothesized that this modification may destabilize heterochromatin by recruiting the Remodels Structure of Chromatin complex (RSC) to the region (20). Recruitment of RSC chromatin remodeling activity to promoters is frequently associated with nucleosome eviction and transcriptional activation (68, 69). Although we have demonstrated that Mit1 is a chromatin remodeling factor, it remained possible that rather than modifying chromatin structure and nucleosome positioning directly, the true nature of Mit1's contribution to heterochromatin silencing is facilitation of Clr3 activity. We hypothesized that if deacetylation of H3K14 was the sole function of SHREC, preventing H3K14 acetylation may suppress the formation of nucleosome-free regions observed in SHREC mutants. The fission yeast genome contains two genes encoding histone acetyltransferases responsible for H3K14 acetylation, gcn5+ and mst2+ (70). Importantly, while H3K14 acetylation is essentially undetectable in gcn5Δ mst2Δ strains, they remain competent for centromeric silencing, suggesting that heterochromatin is intact (71). We combined deletion of gcn5 and mst2 with deletion of the catalytic subunits of SHREC and analyzed nucleosome occupancy at the mating-type REII locus. We found that combined deletion of gcn5+ and mst2+ only partially rescued the NFR formation seen in the clr3Δ strain and did not suppress the mit1Δ phenotype at all (Fig. 7A). This experiment demonstrates that deacetylation of Lys14 on histone H3 is not the sole contribution of SHREC to nucleosome positioning.
FIG 7.
Prevention of H3K14 acetylation is not sufficient to rescue nucleosome occupancy changes observed in SHREC-deficient cells. (A) Histone acetyltransferase mutant strains cannot suppress NFR formation at REII in SHRECΔ cells. The nucleosome scanning experiment was performed as for Fig. 5E, comparing wild-type cells to clr3Δ and mit1Δ cells as well as the gcn5Δ mst2Δ clr3Δ and gcn5Δ mst2Δ mit1Δ triple deletion mutants. Data are plotted on a log2 scale, error bars represent SEM (n = 2). We note that the occupancy profiles for mit1Δ and clr3Δ are largely overlapping. (B) Model for SHREC contribution to nucleosome occupancy at regions of heterochromatin. We propose a model where SHREC, as well as other chromatin modifiers, such as Set2/Clr6, Spt6, and Asf1/HIRA, prevent localized transient and steady-state nucleosome depletion in heterochromatic domains. SHREC performs this function using distinct but likely related activities in nucleosome mobilization and histone deacetylation in part to oppose the activity of the RSC complex (20). Elimination of NFRs is then important for efficient transcriptional silencing by restricting access to Pol II in regions of heterochromatin.
DISCUSSION
Here we have demonstrated for the first time that Mit1 is a bona fide chromatin remodeling factor that shows directional specificity for moving nucleosomes. The usage of conserved chromatin tethering domains to mobilize histone octamers on DNA and basic remodeling characteristics of purified Mit1 are analogous to the documented activity of Drosophila and human Mi-2 (72–74), suggesting that in addition to related composition between the Mit1/SHREC and Mi-2/NuRD complexes, Mit1 and Mi-2 also modify chromatin similarly. It is important to note that our in vitro studies were restricted to analysis of Mit1 in isolation rather than in the context of the SHREC complex. The other subunits of SHREC may impact some aspects of Mit1's remodeling activity in vivo and perhaps in vitro as well, although remodeling by recombinant Mi-2 is similar to that of the purified NuRD complex (75).
Unlike the case with NuRD, we find that Mit1 does not seem to be required for regulation of gene expression. Nucleosome positioning in euchromatin in fission yeast is regulated by fellow CHD family chromatin remodeling enzymes, Hrp1 and Hrp3 (38, 76). Our data suggest that Mit1 mobilizes nucleosomes within regions of heterochromatin and that DNA sequence may contribute at sites where the loss of Mit1 is most notable. Interestingly, fission yeast apparently lack ISWI family chromatin remodelers, which have been found to direct nucleosomes onto intrinsically refractory sequences in S. cerevisiae (77). We propose that Mit1 is a heterochromatin-specific factor that performs a similar function to silence transcription in S. pombe.
Previous work mapping nucleosome occupancy within fission yeast heterochromatin has demonstrated a role for silencing factors in preventing localized nucleosome depletion. The significance of preventing nucleosome-free-region formation is not always clear, especially at those sites that require Mit1 activity within the mating-type region and subtelomeres, since these sites do not correlate with known transcription start sites and do not appear to be “hot spots” for RNA Pol II recruitment. Furthermore, despite its clear role in limiting access to RNA Pol II and preventing the accumulation of centromeric transcripts, Mit1 plays a minor role in the retention of steady-state nucleosome levels at centromeric repeats (Fig. 6B) (20).
Our bioinformatic analysis of previously identified nucleosome-free regions that form in the yeast mating-type locus and at telomere 2R revealed that these sequences are refractory to nucleosome occupancy relative to neighboring sequences. We speculate that while Mit1 acts extensively throughout heterochromatin, nucleosome-free regions are more readily observable at particular locations because nucleosomes are particularly difficult to position at these sites. Furthermore, the AT-rich sequence at these locations may be influencing local nucleosome positioning by acting as an intrinsic barrier to nucleosome occupancy (78) and may contribute to nucleosome-free-region formation and organization of neighboring nucleosomes in the absence of silencing factors. It is presently unclear if these sequences are directly causative of nucleosome depletion, since not all AT-rich sequences become nucleosome depleted in the absence of Mit1 (Fig. 5A, 2.9 kb; also data not shown). However, the observation that sequences that are most dependent on Mit1 to prevent nucleosome depletion are intrinsically refractory to the histone octamer suggests that this characteristic contributes at least in part to their formation.
Our experiments demonstrate that the PHD of Mit1 is required for full remodeling activity of Mit1 and that it contributes to chromatin association of SHREC. Since the in vitro experiments were performed with unmodified nucleosomes and loss of the PHD reduced mobilization, it is possible that the PHD finger contributes to the stabilization or activity of SHREC at heterochromatic regions through general interactions with histone H3. Our attempts to further dissect how the PHD associates with histones were thwarted by our inability to detect binding of the recombinant PHD to modified histone peptide arrays. We speculate that Mit1's PHD in isolation may have low affinity for histone H3 or require a more extensive interaction interface with histone H3 than peptides provide. The influence of modifications to histone H3 binding might be revealed only in a more physiological context, such as full-length Mit1 binding to chromatin, rather than free histones or peptides in solution.
Intriguingly, we have demonstrated that SHREC and a Set2-mediated pathway are partially redundant for suppressing transcription from heterochromatic domains and that Mit1 and Set2 synergize to prevent NFR formation within centromeres. Set2 is thought to prevent aberrant transcription in euchromatin by mediating repression of intragenic cryptic transcription through recruitment of HDAC activity (22, 79–81) and through suppression of nucleosome turnover in transcribed coding regions (82, 83) without causing detectable changes in nucleosome positioning (84). An interesting possibility is that both enzymes contribute to transient remodeling of chromatin (for example, at centromeres) that is not effectively captured by many of the techniques currently used to assess nucleosome positioning. An alternate possibility is that Mit1 remodeling activity is impacting transcriptional elongation or termination, especially since sites where Mit1 influences remodeling do not map to known promoters. Such a role for Mit1 may explain the synergy with Set2, since Set2 pathway mutants exhibit changes in transcriptional elongation (80). In addition to SHREC and Set2, many related chromatin modifiers, such as Asf1/HIRA, Sir2, and Spt6, contribute to transcriptional silencing in ways that are presumably overlapping but not entirely defined mechanistically (7, 85–87). We suggest that partial redundancy between chromatin modifiers in the context of constitutive heterochromatin, such as we describe for Mit1 and Set2, minimizes the effects of loss of Mit1 except at sites that are particularly prone to nucleosome loss.
The SHREC histone deacetylase subunit Clr3 has recently been shown to be important for suppressing histone turnover within regions of heterochromatin (88). While these subunits of SHREC may physically interact, the contributions of Clr3 to steady-state nucleosome positioning extend beyond that of Mit1, suggesting that their activities may not entirely overlap. Our experiments suggest that H3K14 deacetylation by SHREC is likely not the sole contribution of SHREC to nucleosome occupancy, since deletion of the histone acetyltransferases required for this mark does not rescue the phenotype of mit1Δ. Clr3-mediated deacetylation of H3K14 is thought to act in part by limiting recruitment of the RSC chromatin remodeling complex to heterochromatin (20). Interestingly, the in vitro remodeling properties of RSC are opposite to those we describe for Mit1, including center-to-end octamer mobilization and eviction under some conditions (89, 90). We propose that while Clr3 acts to prevent recruitment of the RSC complex, Mit1 may antagonize its activity within heterochromatin (Fig. 7B). Counteracting RSC has been suggested as a possible mechanism for the suppression of aberrant transcription by the Isw2 chromatin remodeling complex at some budding yeast promoters (91, 92). It seems likely that Mit1 may perform an analogous function at fission yeast heterochromatin. Future studies aimed at understanding the cooperation between nucleosome mobilization by Mit1 and histone deacetylation by Clr3 will be important for developing a comprehensive model for how SHREC acts to silence transcription.
This study demonstrates that Mit1 is a heterochromatin-specific silencing factor that shares many in vitro activity and binding characteristics with Mi-2 family remodelers of higher organisms (73, 93). Given the known association of Mi-2/NuRD with centromeres and implications in heterochromatin function (16, 17, 94), it will be interesting to determine whether NuRD repositions nucleosomes similarly in regions of mammalian centromeric heterochromatin, especially considering the repetitive AT-rich nature of these satellite sequences (95).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Bedford, K. Gould, S. Forsburg, G. Thon, J. Nakayama, S. Jia, and S. Francesconi for plasmids and strains and Hartwell center staff (St. Jude Children's Research Hospital) for peptide synthesis, mass spectrometry, and tiling array services. We thank Marc Buhler for sharing updated genomic mapping data for the array analyses and Jon Obenauer for implementing the revisions. We thank Thomas Schalch, Paul Brindle, Nick Laribee, and Ben Alper for useful discussions.
This work was funded by NIA fellowship NRSA F31-AG038153 (to K.M.C.), NIH R01 GM084045 (to J.F.P.), NIH R01 GM070864 (to B.B.), Cancer Center support grant CCSG 2 P30 CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
Footnotes
Published ahead of print 24 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01609-13.
REFERENCES
- 1.Elgin SC, Grewal SI. 2003. Heterochromatin: silence is golden. Curr. Biol. 13:R895–R898. 10.1016/j.cub.2003.11.006 [DOI] [PubMed] [Google Scholar]
- 2.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113. 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- 3.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- 4.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124. 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- 5.Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J. 2008. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol. Cell. Biol. 28:6973–6988. 10.1128/MCB.00791-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. 2008. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 32:778–790. 10.1016/j.molcel.2008.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI. 2011. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol. Cell 41:56–66. 10.1016/j.molcel.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI. 2009. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 106:8998–9003. 10.1073/pnas.0813063106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128:491–504. 10.1016/j.cell.2006.12.035 [DOI] [PubMed] [Google Scholar]
- 10.Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K. 2002. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 22:2170–2181. 10.1128/MCB.22.7.2170-2181.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjerling P, Ekwall K, Egel R, Thon G. 2004. A novel type of silencing factor, Clr2, is necessary for transcriptional silencing at various chromosomal locations in the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res. 32:4421–4428. 10.1093/nar/gkh780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thon G, Klar AJ. 1992. The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131:287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thon G, Cohen A, Klar AJ. 1994. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138:29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekwall K, Ruusala T. 1994. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics 136:53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazzio TG, Rando OJ. 2012. NURDs are required for diversity. EMBO J. 31:3036–3037. 10.1038/emboj.2012.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sims JK, Wade PA. 2011. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol. Biol. Cell 22:3094–3102. 10.1091/mbc.E11-03-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. 2009. Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol. 11:1261–1267. 10.1038/ncb1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denslow SA, Wade PA. 2007. The human Mi-2/NuRD complex and gene regulation. Oncogene 26:5433–5438. 10.1038/sj.onc.1210611 [DOI] [PubMed] [Google Scholar]
- 19.Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. 2010. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 17:251–257. 10.1038/nsmb.1741 [DOI] [PubMed] [Google Scholar]
- 20.Garcia JF, Dumesic PA, Hartley PD, El-Samad H, Madhani HD. 2010. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 24:1758–1771. 10.1101/gad.1946410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. 2013. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 41:2881–2893. 10.1093/nar/gks1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592. 10.1016/j.cell.2005.10.023 [DOI] [PubMed] [Google Scholar]
- 23.Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF. 2005. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol. Cell. Biol. 25:2331–2346. 10.1128/MCB.25.6.2331-2346.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsburg SL. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955–2956. 10.1093/nar/21.12.2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan KL, Dixon JE. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262–267. 10.1016/0003-2697(91)90534-Z [DOI] [PubMed] [Google Scholar]
- 26.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 28.Leeds P, Peltz SW, Jacobson A, Culbertson MR. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303–2314. 10.1101/gad.5.12a.2303 [DOI] [PubMed] [Google Scholar]
- 29.Hansen KR, Hazan I, Shanker S, Watt S, Verhein-Hansen J, Bahler J, Martienssen RA, Partridge JF, Cohen A, Thon G. 2011. H3K9me-independent gene silencing in fission yeast heterochromatin by Clr5 and histone deacetylases. PLoS Genet. 7:e1001268. 10.1371/journal.pgen.1001268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang PY, Hom RA, Musselman CA, Zhu L, Kuo A, Gozani O, Kutateladze TG, Cleary ML. 2010. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J. Mol. Biol. 400:137–144. 10.1016/j.jmb.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partridge JF, Debeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. 2007. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol. Cell 26:593–602. 10.1016/j.molcel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- 32.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823. 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- 33.Sengupta SM, VanKanegan M, Persinger J, Logie C, Cairns BR, Peterson CL, Bartholomew B. 2001. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem. 276:12636–12644 [DOI] [PubMed] [Google Scholar]
- 34.Shen X, Xiao H, Ranallo R, Wu WH, Wu C. 2003. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299:112–114. 10.1126/science.1078068 [DOI] [PubMed] [Google Scholar]
- 35.Lantermann A, Stralfors A, Fagerstrom-Billai F, Korber P, Ekwall K. 2009. Genome-wide mapping of nucleosome positions in Schizosaccharomyces pombe. Methods 48:218–225. 10.1016/j.ymeth.2009.02.004 [DOI] [PubMed] [Google Scholar]
- 36.Lai AY, Wade PA. 2011. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11:588–596. 10.1038/nrc3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu G, Wade PA. 2012. NuRD and pluripotency: a complex balancing act. Cell Stem Cell 10:497–503. 10.1016/j.stem.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pointner J, Persson J, Prasad P, Norman-Axelsson U, Stralfors A, Khorosjutina O, Krietenstein N, Svensson JP, Ekwall K, Korber P. 2012. CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe. EMBO J. 31:4388–4403. 10.1038/emboj.2012.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. 2005. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell 20:173–185. 10.1016/j.molcel.2005.10.002 [DOI] [PubMed] [Google Scholar]
- 40.Marina DB, Shankar S, Natarajan P, Finn KJ, Madhani HD. 2013. A conserved ncRNA-binding protein recruits silencing factors to heterochromatin through an RNAi-independent mechanism. Genes Dev. 27:1851–1856. 10.1101/gad.226019.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buhler M, Haas W, Gygi SP, Moazed D. 2007. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129:707–721. 10.1016/j.cell.2007.03.038 [DOI] [PubMed] [Google Scholar]
- 42.Schalch T, Job G, Shanker S, Partridge JF, Joshua-Tor L. 2011. The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS. Nat. Struct. Mol. Biol. 18:1351–1357. 10.1038/nsmb.2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837. 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 44.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–676. 10.1126/science.1093686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller-Planitz F, Klinker H, Ludwigsen J, Becker PB. 2013. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat. Struct. Mol. Biol. 20:82–89 [DOI] [PubMed] [Google Scholar]
- 46.Okuda M, Horikoshi M, Nishimura Y. 2007. Structural polymorphism of chromodomains in Chd1. J. Mol. Biol. 365:1047–1062. 10.1016/j.jmb.2006.10.039 [DOI] [PubMed] [Google Scholar]
- 47.Bienz M. 2006. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 31:35–40. 10.1016/j.tibs.2005.11.001 [DOI] [PubMed] [Google Scholar]
- 48.Eissenberg JC. 2012. Structural biology of the chromodomain: form and function. Gene 496:69–78. 10.1016/j.gene.2012.01.003 [DOI] [PubMed] [Google Scholar]
- 49.Marfella CG, Imbalzano AN. 2007. The Chd family of chromatin remodelers. Mutat. Res. 618:30–40. 10.1016/j.mrfmmm.2006.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall JA, Georgel PT. 2007. CHD proteins: a diverse family with strong ties. Biochem. Cell Biol. 85:463–476. 10.1139/O07-063 [DOI] [PubMed] [Google Scholar]
- 51.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442:96–99. 10.1038/nature04835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP. 2011. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J. Biol. Chem. 286:11779–11791. 10.1074/jbc.M110.208207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musselman CA, Mansfield RE, Garske AL, Davrazou F, Kwan AH, Oliver SS, O'Leary H, Denu JM, Mackay JP, Kutateladze TG. 2009. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem. J. 423:179–187. 10.1042/BJ20090870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Musselman CA, Ramirez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, Mackay JP, Wade PA, Hagman J, Kutateladze TG. 2012. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc. Natl. Acad. Sci. U. S. A. 109:787–792. 10.1073/pnas.1113655109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hota SK, Bartholomew B. 2011. Diversity of operation in ATP-dependent chromatin remodelers. Biochim. Biophys. Acta 1809:476–487. 10.1016/j.bbagrm.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meersseman G, Pennings S, Bradbury EM. 1992. Mobile nucleosomes—a general behavior. EMBO J. 11:2951–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lowary PT, Widom J. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19–42. 10.1006/jmbi.1997.1494 [DOI] [PubMed] [Google Scholar]
- 58.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686–697. 10.1101/gad.13.6.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson CL. 2000. ATP-dependent chromatin remodeling: going mobile. FEBS Lett. 476:68–72. 10.1016/S0014-5793(00)01673-2 [DOI] [PubMed] [Google Scholar]
- 60.Gangaraju VK, Bartholomew B. 2007. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol. Cell. Biol. 27:3217–3225. 10.1128/MCB.01731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366. 10.1038/nature07667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xi L, Fondufe-Mittendorf Y, Xia L, Flatow J, Widom J, Wang JP. 2010. Predicting nucleosome positioning using a duration Hidden Markov Model. BMC Bioinformatics 11:346. 10.1186/1471-2105-11-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626–630. 10.1126/science.1112178 [DOI] [PubMed] [Google Scholar]
- 64.Noma K, Grewal SI. 2002. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 99(Suppl. 4):16438–16445. 10.1073/pnas.182436399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. 2008. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451:734–737. 10.1038/nature06561 [DOI] [PubMed] [Google Scholar]
- 66.Morris SA, Shibata Y, Noma K, Tsukamoto Y, Warren E, Temple B, Grewal SI, Strahl BD. 2005. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot. Cell 4:1446–1454. 10.1128/EC.4.8.1446-1454.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109(Part 11):2637–2648 [DOI] [PubMed] [Google Scholar]
- 68.Kuryan BG, Kim J, Tran NN, Lombardo SR, Venkatesh S, Workman JL, Carey M. 2012. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc. Natl. Acad. Sci. U. S. A. 109:1931–1936. 10.1073/pnas.1109994109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lorch Y, Griesenbeck J, Boeger H, Maier-Davis B, Kornberg RD. 2011. Selective removal of promoter nucleosomes by the RSC chromatin-remodeling complex. Nat. Struct. Mol. Biol. 18:881–885. 10.1038/nsmb.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, Kallgren SP, Reddy BD, Kuntz K, Lopez-Maury L, Thompson J, Watt S, Ma C, Hou H, Shi Y, Yates JR, III, Bahler J, O'Connell MJ, Jia S. 2012. Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. J. Biol. Chem. 287:4386–4393. 10.1074/jbc.M111.329417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reddy BD, Wang Y, Niu L, Higuchi EC, Marguerat SB, Bahler J, Smith GR, Jia S. 2011. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 25:214–219. 10.1101/gad.1993611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, Brehm A. 2002. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 21:2430–2440. 10.1093/emboj/21.10.2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Muller J, Becker PB. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19:4332–4341. 10.1093/emboj/19.16.4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED. 2012. The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. J. Mol. Biol. 422:3–17. 10.1016/j.jmb.2012.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang HB, Zhang Y. 2001. Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res. 29:2517–2521. 10.1093/nar/29.12.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shim YS, Choi Y, Kang K, Cho K, Oh S, Lee J, Grewal SI, Lee D. 2012. Hrp3 controls nucleosome positioning to suppress non-coding transcription in eu- and heterochromatin. EMBO J. 31:4375–4387. 10.1038/emboj.2012.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Whitehouse I, Tsukiyama T. 2006. Antagonistic forces that position nucleosomes in vivo. Nat. Struct. Mol. Biol. 13:633–640. 10.1038/nsmb1111 [DOI] [PubMed] [Google Scholar]
- 78.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18:1073–1083. 10.1101/gr.078261.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joshi AA, Struhl K. 2005. Eaf3 chromodomain interaction with methylated H3–K36 links histone deacetylation to Pol II elongation. Mol. Cell 20:971–978. 10.1016/j.molcel.2005.11.021 [DOI] [PubMed] [Google Scholar]
- 80.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593–605. 10.1016/j.cell.2005.10.025 [DOI] [PubMed] [Google Scholar]
- 81.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298–1306. 10.1128/MCB.22.5.1298-1306.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. 2012. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 19:884–892. 10.1038/nsmb.2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. 2012. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489:452–455. 10.1038/nature11326 [DOI] [PubMed] [Google Scholar]
- 84.Hennig BP, Bendrin K, Zhou Y, Fischer T. 2012. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 13:997–1003. 10.1038/embor.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kiely CM, Marguerat S, Garcia JF, Madhani HD, Bahler J, Winston F. 2011. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 31:4193–4204. 10.1128/MCB.05568-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alper BJ, Job G, Yadav RK, Shanker S, Lowe BR, Partridge JF. 2013. Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast. EMBO J. 32:2321–2335. 10.1038/emboj.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kato H, Okazaki K, Iida T, Nakayama J, Murakami Y, Urano T. 2013. Spt6 prevents transcription-coupled loss of posttranslationally modified histone H3. Sci. Rep. 3:2186. 10.1038/srep02186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aygun O, Mehta S, Grewal SI. 2013. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol. 20:547–554. 10.1038/nsmb.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Vugt JJ, de Jager M, Murawska M, Brehm A, van Noort J, Logie C. 2009. Multiple aspects of ATP-dependent nucleosome translocation by RSC and Mi-2 are directed by the underlying DNA sequence. PLoS One 4:e6345. 10.1371/journal.pone.0006345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lorch Y, Maier-Davis B, Kornberg RD. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. U. S. A. 103:3090–3093. 10.1073/pnas.0511050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450:1031–1035. 10.1038/nature06391 [DOI] [PubMed] [Google Scholar]
- 92.Hartley PD, Madhani HD. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137:445–458. 10.1016/j.cell.2009.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guschin D, Wade PA, Kikyo N, Wolffe AP. 2000. ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry 39:5238–5245. 10.1021/bi000421t [DOI] [PubMed] [Google Scholar]
- 94.Helbling Chadwick L, Chadwick BP, Jaye DL, Wade PA. 2009. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma 118:445–457. 10.1007/s00412-009-0207-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hayden KE. 2012. Human centromere genomics: now it's personal. Chromosome Res. 20:621–633. 10.1007/s10577-012-9295-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.