Abstract
The role of the negative elongation factor (NELF) in maintaining HIV latency was investigated following small hairpin RNA (shRNA) knockdown of the NELF-E subunit, a condition that induced high levels of proviral transcription in latently infected Jurkat T cells. Chromatin immunoprecipitation (ChIP) assays showed that latent proviruses accumulate RNA polymerase II (RNAP II) on the 5′ long terminal repeat (LTR) but not on the 3′ LTR. NELF colocalizes with RNAP II, and its level increases following proviral induction. RNAP II pause sites on the HIV provirus were mapped to high resolution by ChIP with high-throughput sequencing (ChIP-Seq). Like cellular promoters, RNAP II accumulates at around position +30, but HIV also shows additional pausing at +90, which is immediately downstream of a transactivation response (TAR) element and other distal sites on the HIV LTR. Following NELF-E knockdown or tumor necrosis factor alpha (TNF-α) stimulation, promoter-proximal RNAP II levels increase up to 3-fold, and there is a dramatic increase in RNAP II levels within the HIV genome. These data support a kinetic model for proviral transcription based on continuous replacement of paused RNAP II during both latency and productive transcription. In contrast to most cellular genes, HIV is highly activated by the combined effects of NELF-E depletion and activation of initiation by TNF-α, suggesting that opportunities exist to selectively activate latent HIV proviruses.
INTRODUCTION
HIV persists in the face of highly active antiretroviral therapy (HAART) due to low-level replication in sites that are poorly accessible to drugs and to the development of a latent reservoir in primary memory CD4+ T cells and certain myeloid cells (1, 2). The need to develop novel therapeutic tools to attack the latently infected cells and prevent a rebound of virus after termination of antiviral treatment (3, 4) is a widely recognized goal (5), but a poor understanding of the factors required for the establishment of latency has slowed progress.
Transcription of HIV is regulated by opposing positive and negative cellular elongation factors. In the absence of Tat, short abortive transcripts are induced by the negative elongation factor (NELF) and the 5,6-dichloro-1-beta-d-ribofuranosyl-benzimidazole (DRB) sensitivity-inducing factor (DSIF) (6, 7). The viral transactivator protein, Tat, permits the efficient synthesis of full-length HIV transcripts by recruiting the cellular transcriptional elongation factor P-TEFb (positive transcriptional elongation factor b) (8) to RNA polymerase II (RNAP II) complexes that have transcribed through the HIV transactivation response (TAR) element, an RNA stem-loop structure found at the 5′ end of all viral transcripts. After binding to TAR RNA, the CDK9 subunit of P-TEFb phosphorylates a variety of proteins including the C-terminal domain (CTD) of RNAP II (9), Spt5, a subunit of DSIF that enhances transcriptional elongation (10, 11), and the E (RD) subunit of NELF (12). Tat also recruits the “superelongation complex” containing the AFF4, ENL, AF9, and ELL2 elongation factors to the proviral promoter (13–15). Thus, Tat and P-TEFb are able to stimulate HIV transcription both through the removal of blocks to elongation and by the enhancement of RNAP II processivity. Because Tat functions as part of a positive regulatory circuit, conditions that restrict transcription initiation will, in turn, cause a reduction in Tat levels to below threshold levels and therefore result in dramatically reduced HIV transcription and, eventually, entry into latency (16, 17).
Epigenetic silencing due to recruitment of histone deacetylases (HDACs) and histone methyltransferases (18–21) and to DNA methylation (22, 23) greatly restricts Tat levels during HIV latency. HIV silencing can also arise because of transcriptional interference (24, 25) or because of HIV proviral integration into heterochromatic regions of the genome (26, 27).
Several features of the metabolism of resting CD4 cells additionally ensure that latent proviruses remain transcriptionally inactive for long periods. First, resting cells sequester the initiation factors NF-κB and nuclear factor of activated T cells (NFAT) in the cytoplasm (28–30). Second, CycT1 is expressed at minimal levels, preventing P-TEFb assembly (31–34). After induction of resting T cells, P-TEFb levels are regulated by assembly of the 7SK RNP complex containing the repressor protein HEXIM (34–38). Despite these multiple restrictions, stimulation of memory T cells by cytokines or by T-cell receptor activation provides a powerful signal leading to the resumption of HIV transcription, replication, and spread.
Since previous studies using reporter gene systems demonstrated that NELF enhances the accumulation of short, abortive transcripts (12, 39–42), we decided to measure whether NELF affects promoter-proximal pausing on the HIV long terminal repeat (LTR) during latency. Consistent with an earlier study by Zhang and colleagues (42), small hairpin RNA (shRNA) knockdowns have demonstrated that NELF is a critical factor used to maintain proviral latency. Chromatin immunoprecipitation with high-throughput sequencing (ChIP-Seq) reveals that the HIV LTR shows a novel pattern of RNAP II promoter-proximal pausing, implying a kinetic model for proviral transcription in which there is continuous replacement of paused RNAP II during both latency and productive transcription. In contrast to earlier models, NELF is not required to induce promoter-proximal RNAP II pausing but instead appears to alter the rate of RNAP II escape.
MATERIALS AND METHODS
Western blotting.
Western blot analyses were performed in accordance with standard protocols using the following primary antibodies: NELF subunit E (NELF-E), NELF-A, Spt5 (Santa Cruz Biotechnology), Cobra (NELF-B) (Bethyl Laboratories), THP1 (NELF-C/D) (Acris Antibodies), and tubulin (Santa Cruz). Horseradish peroxidase (HRP)-conjugated secondary antibodies (Dako) were used along with ECL Western blotting detection reagents (GE Healthcare) to detect proteins by chemiluminescence using an ImageQuant LAS 4000. Quantification of the protein bands was accomplished using ImageQuant TL (GE Healthcare).
Flow cytometry.
Cells were analyzed by flow cytometry using either an LSRII or FACSCalibur flow cytometer (Becton, Dickinson), and data were acquired using FacsDIVA and analyzed using the WinList3D or FloJo program.
Construction of shRNA plasmids.
Target sequences intended to suppress the NELF-A, -B, -C/D, and -E subunits, as well as corresponding scrambled controls, were constructed and cloned into the pSuper vector, which provides the RNAP III-transcribed H1 promoter for expression of small hairpin RNA (shRNA) (43). The target sequences and H1 promoter were then subcloned into the pLVTHM lentiviral vector (44), which was modified to express the mCherry fluorescent protein, in place of green fluorescent protein (GFP), to allow for detection of shRNA-expressing cells.
Infection of Jurkat cells.
Vesicular stomatitis virus G protein (VSV-G) pseudotyped shRNA viruses were produced by transient cotransfection of pLVTHM shRNA plasmids with the packaging plasmids pMD.G and pCMVΔR8.9I into 293T cells using Lipofectamine 2000 (Life Technologies) (45). mCherry/d2EGFP (where d2EGFP is a destabilized enhanced GFP) expression was assessed by flow cytometry to confirm shRNA expression. Western blotting was performed to evaluate NELF expression. The mixed populations were then sorted into 100% mCherry-positive populations and subsequently sorted and grown into clonal populations. Clones were subsequently assessed by flow cytometry and Western blot analysis.
Activation and shutdown of Jurkat cells.
Cells were activated with either 10 ng/ml tumor necrosis factor alpha (TNF-α), 5 μg/ml plate-bound anti-CD3, or 5 μg/ml plate-bound anti-CD3 and 1 μg/ml anti-CD28 (BD Biosciences) for 18 h and analyzed for d2EGFP expression by flow cytometry. To initiate shutdown, cells were washed with phosphate-buffered saline (PBS) and resuspended and maintained in RPMI 1640 medium containing 10% fetal bovine serum, penicillin (100 IU/ml), streptomycin (100 μg/ml), and 100 μg/ml Normocin (InvivoGen) at 37°C with 5% CO2. In subsequent days, cells were analyzed by fluorescence-activated cell sorting (FACS) to determine the percentage of d2EGFP-positive cells.
ChIP assays.
Jurkat cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum, penicillin (100 IU/ml), streptomycin (100 μg/ml), and 100 μg/ml Normocin (InvivoGen) at 37°C with 5% CO2. Cells were stimulated with 2 ng/ml to 10 ng/ml TNF-α.
Several closely related ChIP protocols were used during the course of these experiments. The data shown in Fig. 4 and 5 use ChIP protocol 1. After induction of the cells, protein was cross-linked to DNA by adding 0.5% formaldehyde (135 μl of 37% formaldehyde for 10 ml of RPMI medium) for 10 min at room temperature with rotation. The cross-linking reaction was stopped by addition of 125 mM glycine (final concentration) for 5 min, and the cells were chilled on ice; the remainder of the protocol was performed at 4°C. The cells were washed twice with PBS and resuspended at a concentration of 107 cells/ml in CE buffer (10 mM HEPES-KOH [pH 7.9], 60 mM KCl, 1 mM EDTA, 0.5% NP-40 alternative, 1 mM dithiothreitol [DTT]) supplemented with protease inhibitor cocktail [4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), aprotinin, bestatin, E-64, leupeptin, and pepstatin A (Pierce)] for 10 min. The disrupted cells were vortexed for 30 s, and the nuclei were pelleted at 700 × g for 10 min at 4°C. The nuclei were resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) supplemented with protease inhibitor cocktail at a concentration of 5 × 107 cells/ml. Samples could either be stored frozen at −80°C at this stage or processed immediately. Nuclei in 300-μl aliquots were sonicated for 10 pulses of 20 s each (Misonix Sonicator 3000) to generate DNA fragments averaging between 200 and 500 bp, with a maximal length of 1,000 bp. Note that the DNA fragment size recovered following the immunoprecipitation step ranged between 100 and 200 bp as measured by linker-mediated PCR.
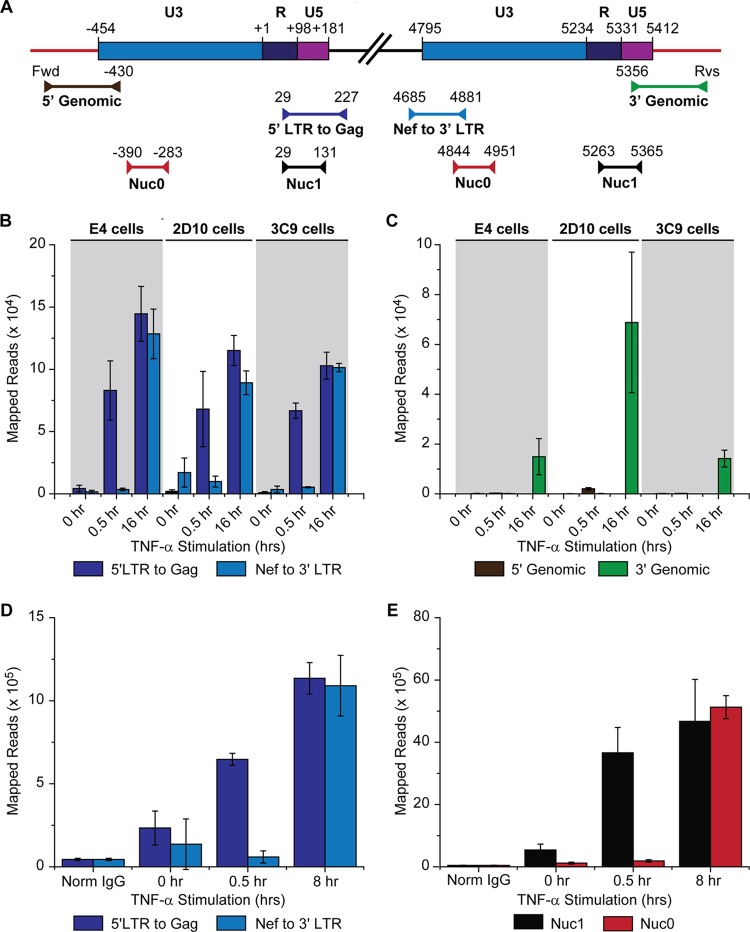
FIG 4.
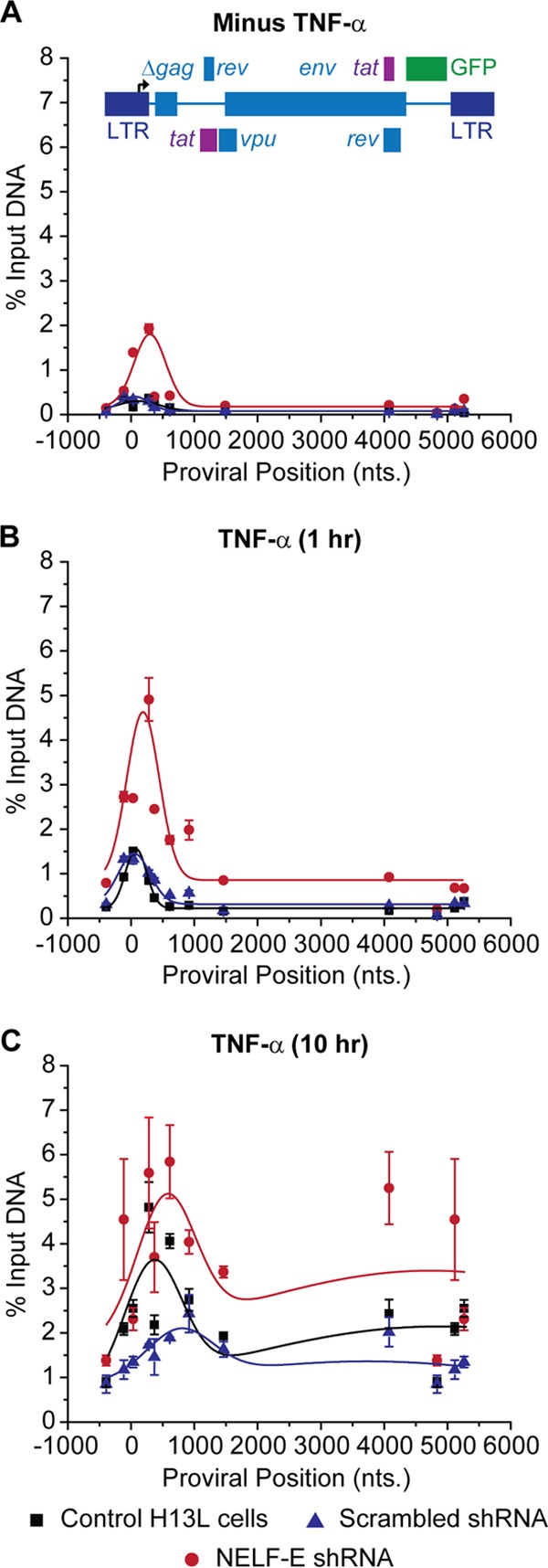
Distribution of RNAP II on the HIV genome following knockdown of NELF-E. ChIP analyses were performed using NELF-E shRNA (clone N2G10) and its scrambled control (clone N2F2) in unstimulated cells (minus TNF-α) (A), cells stimulated with 2 ng/ml TNF-α for 1 h (B), and cells stimulated with 2 ng/ml TNF-α for 10 h (C). Error bars represent the standard deviations of triplicate real-time PCR determinations for each primer set.
FIG 5.
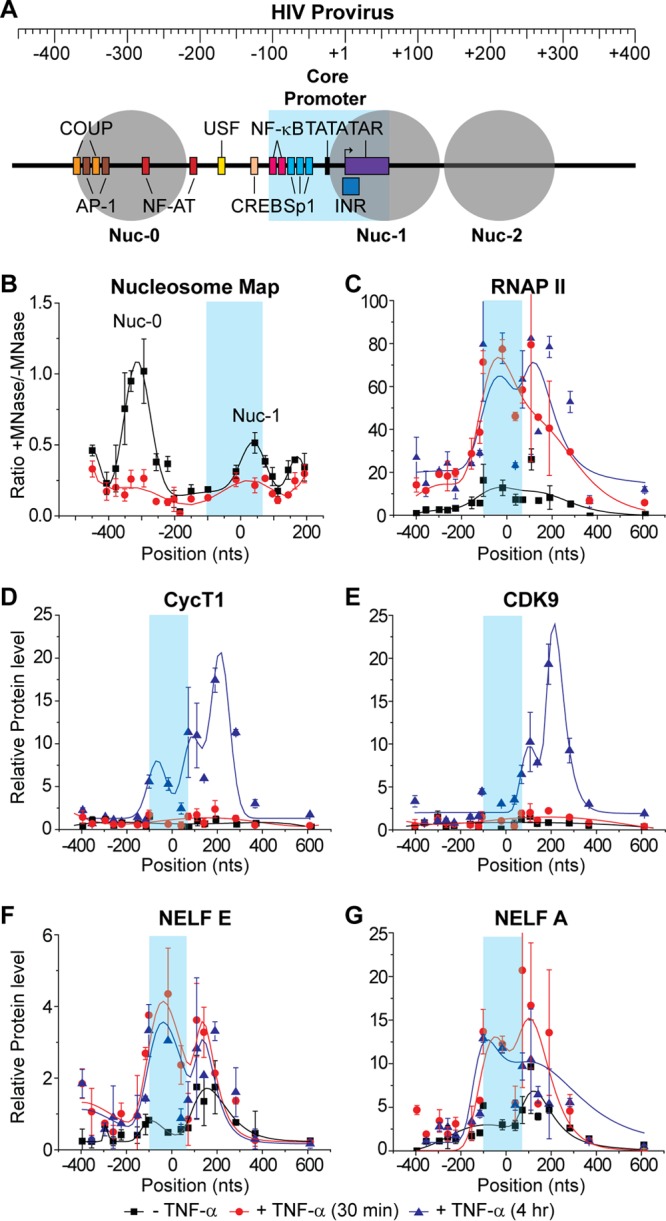
Distribution of RNAP II and associated transcription factors on the HIV genome following TNF-α activation. ChIP assays were used to measure the density of RNA polymerase along the proviral genome from the core promoter region in the LTR (shaded in blue) to 600 nt downstream of the transcription start site. (A) Schematic map of the HIV LTR showing nucleosomes and transcription factor binding sites. (B) Micrococcal nuclease protection assay. (C to G) ChIP analyses were performed using antibodies against the indicated proteins: RNAP II, CycT1, CDK9, NELF-E, and NELF-A. Error bars represent standard deviations of triplicate real-time PCR determinations for each primer set.
For chromatin immunoprecipitations, 100 μl of the sonicated sample was diluted with 900 μl of ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 150 mM NaCl) supplemented with protease inhibitor cocktail. Specific primary antibody or control antiserum was added (typically at a 1/500 dilution), and the samples were incubated for a minimum of 2 h to a maximum of 16 h at 4°C, with rotation. Following incubation with the primary antibody, complexes were purified using protein A-Sepharose beads. Immediately prior to use, the protein A-Sepharose beads were preincubated in 0.5 mg/ml bovine serum albumin (BSA) and 0.125 mg/ml calf thymus DNA, washed in PBS, and resuspended in ChIP dilution buffer. Fifty microliters of beads was added to each sample and incubated with rotation for 2 h at 4°C. The protein A-Sepharose beads were recovered by centrifugation (1,280 × g for 4 min) and washed with 1 ml of each of the following wash buffers in the following order: (i) once with low-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl); (ii) once with high-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl); (iii) once with LiCl buffer (0.25 M LiCl, 1% NP-40 alternative, 1% Na-deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1); and (iv) twice with TE buffer (10 mM Tris-HCl, pH 8.1, 0.1 mM EDTA).
The washed pellets were resuspended in 250 μl of freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3). After pellets were vortexed briefly to mix them, they were incubated at room temperature for 1 h with rotation. The agarose beads were recovered by centrifugation at 1,280 × g for 4 min, and the supernatant fraction was transferred to another tube. A second elution for 15 min was performed, 20 μl of 5 M NaCl was added to the combined eluates (total volume, 500 μl), and the cross-links were reversed by incubation at 65°C for 12 h. Ten microliters of 0.5 M EDTA, 10 μl of 2 M Tris-HCl (pH 6.5), and 2 μl of 10 mg/ml of proteinase K were added to the combined eluates, and the samples were incubated for 2 h at 50°C. The DNA was recovered by phenol-chloroform extraction and ethanol precipitation using either glycogen or 20 μg of tRNA as a carrier. The pellets were resuspended in 250 μl of water, and 5 μl was used for real-time PCR.
For input control DNA, a separate 50-μl aliquot of the sonicated nuclei was diluted with 450 μl of SDS lysis buffer, and 20 μl of 5 M NaCl was added. The cross-links were reversed at 65°C for 12 h.
DNA levels in the immunoprecipitated and input DNA control samples were quantified by real-time PCR as previously described (19, 21, 46, 47).
Antibodies used for immunoprecipitations were anti-RNAP II (sc-899), anti-NELF-E (sc-32912), anti-NELF-A (sc-32911), anti-CDK9 (sc-484), and anti-cyclin T1 (sc-10275) from Santa Cruz Biotechnology.
High-resolution MNase nucleosomal mapping.
Nucleosome positions were monitored using a modification of the method of Petesch and Lis (48). Cells were cross-linked according to ChIP protocol 1 described above, washed twice in PBS, and then suspended in hypertonic buffer A (300 mM sucrose, 3 mM MgCl2, 10 mM Tris, pH 7.4) to 2.5 × 106 cells/ml. Cells were incubated on ice for 5 min, another 2 ml of buffer A plus 0.2% NP-40 was added, and cells were disrupted by vigorous pipetting. The nuclei were collected by centrifugation at 4°C for 5 min at 720 × g and resuspended in 1 ml of buffer A plus 10 mM CaCl2. The equivalent of 5.0 × 106 cells was used per individual micrococcal nuclease (MNase) reaction. MNase (NEB) was added so that 0, 2, 5, 12.5, 50, or 200 total units was used per 100 μl of reaction mixture and digested for 30 min at 37°C. Reactions were stopped with the addition of EGTA and NaCl to final concentrations of 20 mM and 200 mM, respectively. After overnight incubation at 65°C, each reaction mixture was treated with RNase A and purified using a QiaQuick PCR kit (Qiagen). Eluted product was run on a 1% agarose gel to confirm MNase digestion. Samples that were treated with 2 (undigested) and 50 (digested) units of MNase were then processed similar to ChIP samples for real-time qPCR analysis.
ChIP assays using Ion Torrent sequencing.
Jurkat T cells were stimulated with 10 ng/ml of TNF-α for 0.5, 8, or 16 h. The samples used for the experiments shown in Fig. 3 were processed using a streamlined and partially automated protocol (ChIP protocol 2). Cells were harvested and cross-linked with 1% formaldehyde for 15 min as described in ChIP protocol 1. Nuclei from 2 × 106 cells were collected and washed twice with PBS, lysed, resuspended in 100 μl, SDS lysis buffer, and sonicated in a Bioruptor ultrasonicator (Diagenode) for two 20-min runs using a cycle of 30-s on and 30-s off. The sonicated lysates were diluted 10-fold with low-SDS ChIP dilution buffer, and 100 μl-ChIP reaction mixtures were assembled in 96-well plates using an EpMotion pipetting robot and 7 μl of RNA polymerase II antibody (sc-899; Santa Cruz Biotechnology).
FIG 3.
RNAP II accumulates on the 5′ LTR but not on the 3′ LTR in latently infected cells. (A) Location of primers used for ChIP assays (Table 1 gives primer sequences). (B) ChIP analysis of RNAP II levels on the 5′ and 3′ proviral LTR using LTR-specific primers. Analysis was performed on E4, 2D10, and 3C9 cells induced for 0, 0.5, or 16 h with 2 ng/ml TNF-α. (C) Analysis of the same samples as used for the experiment shown in panel B using genomic primers on the 5′ and 3′ LTR. (D) RNAP II levels on the 5′ and 3′ LTR of E4 cells induced for 0, 0.5, or 8 h using LTR-specific primers. (E) Analysis of the same samples as described for panel D using nonspecific primers for the Nuc-0 and Nuc-1 regions of the LTR. Readout for this assay was from Ion Torrent sequencing of PCR products. Error bars represent the standard deviations of triplicate ChIP assays (21, 46). Fwd: forward genomic primer; Rvs, reverse genomic primer.
The antibody and ChIP reaction mixtures were mixed slowly for 5 min at room temperature. The entire reaction mixture was pipetted into a preblocked protein A- and G-coated 96-well plate (Pi-15138; Thermo Scientific Pierce) using an EpMotion pipetting robot and incubated at room temperature for 30 min with mixing at 500 rpm. After samples were allowed to bind to the plates, residual buffer was removed, and samples were washed with radioimmunoprecipitation assay (RIPA) buffer, followed by two washes with TE buffer. Elution was performed using 0.1 M NaHCO3–1% SDS buffer containing EDTA and proteinase K, and samples were incubated at 65°C with shaking at 800 rpm for 30 min. DNA was purified using Axygen PCR cleanup magnetic beads (MAG-PCR-CL-50; Corning) according to the manufacturer's protocol.
Each experimental sample was amplified using eight multiplexed and bar-coded primers in one PCR. The multiplex primer sets (Table 1) (see Fig. 3 diagram) used in each amplification reaction had unique bar codes associated with them to identify them during the sequencing analysis. A total of 12 available bar codes permitted pooling of up to 12 multiplexed reaction mixtures containing eight primers each for sequencing on a single 314 sequencing chip.
TABLE 1.
LTR-specific PCR primers
| Target (cell line) and primer namea | Primer sequence | Genomic locationb |
Size (bp) | ||
|---|---|---|---|---|---|
| Chr | 5′ coordinate | 3′ coordinate | |||
| CD2AP gene (E4) | |||||
| Fwd U3 LTR | CTTTGGGAGTGAATTAGCCCTTCC | ||||
| Rvs CD2AP | GGGCAGACAACACACTGAGAC | 6 | 47561609 | 47561628 | 189 |
| Fwd CD2AP | AATTGACAACAGACTCTCGTG | 6 | 47561548 | 47561568 | 146 |
| Rvs U5 LTR | TGTGTGCCCGTCTGTTGTGTG | ||||
| SNX4 gene (3C9) | |||||
| Fwd SNX4 | CACACGGGCAGACAACACACT | 3 | 125198548 | 125198567 | 152 |
| Rvs U3 LTR | TCAGGGAAGTAGCCTTGTGTGT | ||||
| Fwd U5 LTR | TGTGTGCCCGTCTGTTGTGTG | 171 | |||
| Rvs SNX4 | CCATGACAAAAGGGACTAGAAA | 3 | 125198654 | 125198675 | |
| MSRB1 gene (2D10) | |||||
| Fwd LTR | TCAGGGAAGTAGCCTTGTGTGT | 210 | |||
| Rvs MSRB1 | GGAAAGGCGGGAGCTGATGA | 16 | 1991455 | 1991455 | |
| Fwd MSRB1 | TGTGCATACTTCGAGCGGCT | 16 | 1991346 | 1991365 | 169 |
| Rvs LTR | TGTGTGCCCGTCTGTTGTGTG | ||||
| 3′ LTR specific | |||||
| Nef Fwd | ACAAGAGGAGGAAGAGGTGGGT | 196 | |||
| Nuc-0 Rvs | GCCCTGGTGTGTAGTTCTGCCA | ||||
| 5′ LTR specific | |||||
| Fwd Nuc-1 | CTGGGAGCTCTCTGGCTAACTA | 198 | |||
| Rvs Gag | GCTCCTCTGGTTTCCCTTTCGC | ||||
The PCRs were monitored using real-time quantitative PCR (qPCR) and stopped during the log phase of amplification, typically after 30 to 35 cycles of amplification. The multiplexed samples were pooled, column purified, and gel size selected to remove any remaining primer. A total of 300 pg of the amplified PCR products was then sequenced using an Ion Torrent PGM sequencer (Life Technologies) according to the manufacturer's protocol.
The sequencing information was analyzed as follows. First, primer sequences were trimmed off. The remaining sequences were then deconvoluted based on their bar code sequence and each bar-coded set was mapped to a synthetic DNA sequence comprising the relevant proviral and control DNA sequences. Data were analyzed using Genomics Workbench, version 6.0, software. Control genes used for quality control purposes included glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TNF-α, IκBα, and EGR2 (46).
ChIP-Seq sample preparation.
Preparation of DNA for the ChIP-Seq experiments shown in Fig. 6 to 8 (ChIP protocol 3) followed a modified version of the protocol of Schmidt et al. (49) and was essentially a scaled-up version of ChIP protocol 1 using 25 × 106 cells for each sample. The DNA was sheared by sonication using a Bioruptor Ultrasonicator (Diagenode) for at least two 20-min runs using a 30-s on and 30-s off cycle. Tubes were rotated between runs, and sonication was monitored in real time by running aliquots of DNA on an agarose gel. Sonication was stopped when fragments were sheared to approximately 200 bp, with a maximum of 500 bp. The final DNA pellet was dissolved in TE buffer, and sequencing libraries were made from the DNA using a TruSeq DNA sample preparation kit from Illumina. Libraries were sequenced on an Illumina Genome Analyzer IIx. The final DNA size of the immunoprecipitated samples ranged between 100 and 200 bp, as confirmed by agarose gel electrophoresis.
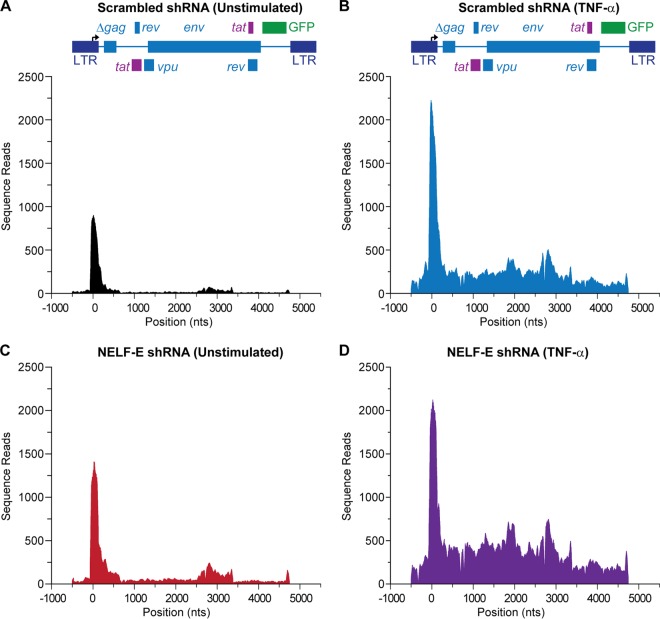
FIG 6.
Knockdown of NELF-E expression selectively enhances elongation from the HIV LTR. The distribution of RNA polymerase on HIV was measured by ChIP-Seq assays performed using the parental line E4 and clones carrying the NELF-E shRNA (N1C6) and its scrambled control (N1D9) in unstimulated control (N1D9) cells (A), control cells stimulated by 10 ng/ml TNF-α for 60 min (B), unstimulated cells expressing NELF-E shRNA (N1C6) (C), and cells expressing NELF-E shRNA and activated by 10 ng/ml TNF-α for 60 min (D). Note that the downstream distribution of RNAP II uniformly increases when NELF-E is knocked down, but there is only minimal impact of NELF-E depletion on promoter-proximal pausing.
FIG 8.
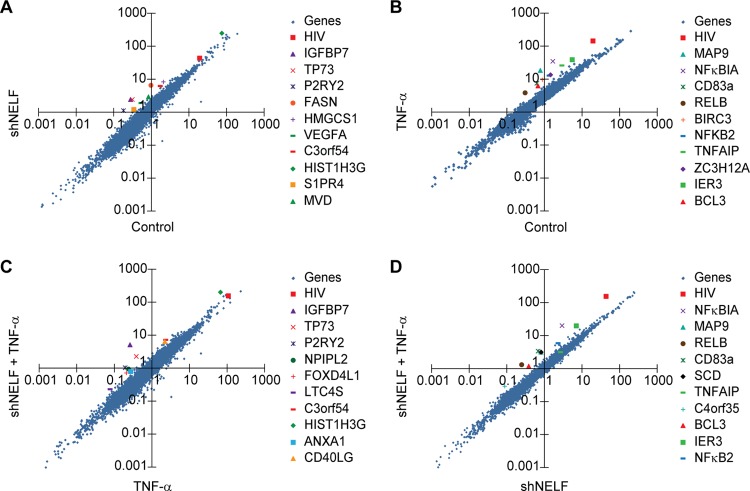
Relative RNAP II accumulation on cellular genes and the HIV provirus. RNAP II reads from the ChIP-Seq data sets were mapped to individual gene coordinates and then totaled within each gene boundary. Two experimental conditions were compared in each analysis and plotted using log-log plots. (A to D) RNAP II distribution per gene region in the presence of NELF-E shRNA (N1C6) and its scrambled control (N1D9) under unstimulated conditions (A), in the presence and absence of TNF-α in scrambled control cells (B), in the presence of NELF-E shRNA and its scrambled control after stimulation by TNF-α (C),and in the presence of NELF-E shRNA in the presence and absence of TNF-α (D). For each condition the HIV provirus (red square) and the top 10 highest-scoring genes are indicated. The HIV “gene” has highly upregulated RNAP II binding patterns compared to those of cellular genes when the provirus is stimulated with TNF-α in the presence and absence of NELF.
Analysis of sequencing results and RNAP II distribution.
Sequences obtained from the Illumina sequencer were first subjected to quality control procedures, and the sequence bases were called and verified by Illumina's proprietary consensus assessment of sequence and variation (CASAVA) software. The resulting sequence reads were filtered to remove primer sequences, trimmed to a uniform read length of 36 bp, and outputted as a FASTQ file.
As shown in Table 2, four data sets were obtained in parallel from two separate cell lines: control E4 cells superinfected by a lentiviral vector expressing a scrambled shRNA (clone N1D9) and E4 cells superinfected by a lentiviral vector expressing an shRNA targeting NELF-E (clone N1C6). These cells were analyzed before and after NF-κB induction by TNF-α, resulting in four separate experimental conditions. Each separate sample run yielded between 30 million and 40 million 36-bp reads, and two experimental replicates were combined to yield a total of 60 million to 80 million reads per sample. The combined reads in the FASTQ format were then mapped/aligned to the hg19/GRCh37 build of the human genome and to the HIV-1 reporter vector sequence as shown in Fig. 6. The results were output in a bowtie file format (50).
TABLE 2.
ChIP-Seq data sets
| ChIP sample (cell type and infection condition)a | Clone | Alignment to genome reference sequence hg19b |
||
|---|---|---|---|---|
| Total no. of reads | No. of mapped reads | % mapped | ||
| E4 + shNELF | N1C6 | 73,960,658 | 54,109,686 | 73.16 |
| E4 + shNELF + TNF-α | N1C6 | 74,824,356 | 55,239,884 | 73.83 |
| E4 + Scr shRNA | N1D9 | 67,786,174 | 45,009,143 | 66.40 |
| E4 + Scr shRNA + TNF-α | N1D9 | 77,884,760 | 56,105,949 | 72.04 |
E4 cells were infected with a lentiviral vector expressing either a scrambled (Scr) shRNA (controls) or an shRNA targeting NELF (shNELF) and were either untreated or treated with TNF-α for 60 min.
Data were deposited in the GEO database under accession number GSE47481. hg19, human genome build 19.
Model-based analysis for ChIP-Seq (MACS) algorithms (51) were used to identify regions of RNAP II enrichment, and these regions were extracted using Unix and Pearl scripts. RNAP II distribution plots were generated based on the mapped reads.
Following the convention of Brannan et al. (52), we defined the escape index (EI), which provides an estimate of the flux of polymerase II (Pol II) from the promoter region into the body of the provirus, as the ratio of the body density/promoter density, where the body density is the number of reads per base in a range between bases −454 and +300 from the transcription start site (TSS) and the promoter density is the Pol II density at base +301 from the TSS and poly(A) site.
For the genome-wide analyses, gene coordinates as defined by Ensembl and/or the University of California, Santa Cruz (UCSC), bioinformatics group were cataloged into a file. Sequence reads in the bowtie files were counted and separated into the coordinates as defined in the cataloged file. Reads were totaled within each gene boundary. Two experimental conditions were compared in each analysis: polymerase distribution per gene region in the presence and absence of NELF and polymerase distribution per gene region with and without TNF-α stimulation. Log10-log10 plots were generated from this analysis.
Curve fitting.
Origin Pro (version 9.5) was used to generate curve fits for each of the data sets shown in Fig. 7 and in Fig. S9 and S10 in the supplemental material. The mapped reads fitted to a series of overlapping Gaussian distributions. The derivative of the original data set was used to objectively identify the numbers and approximate positions of the peaks. To perform the fits, the baseline, y0, was fixed, and sequential iterations (until minimal χ2 values were obtained) were used to find optimal parameters for the peak center, xcn, amplitude, A, and peak width, w, for each peak.
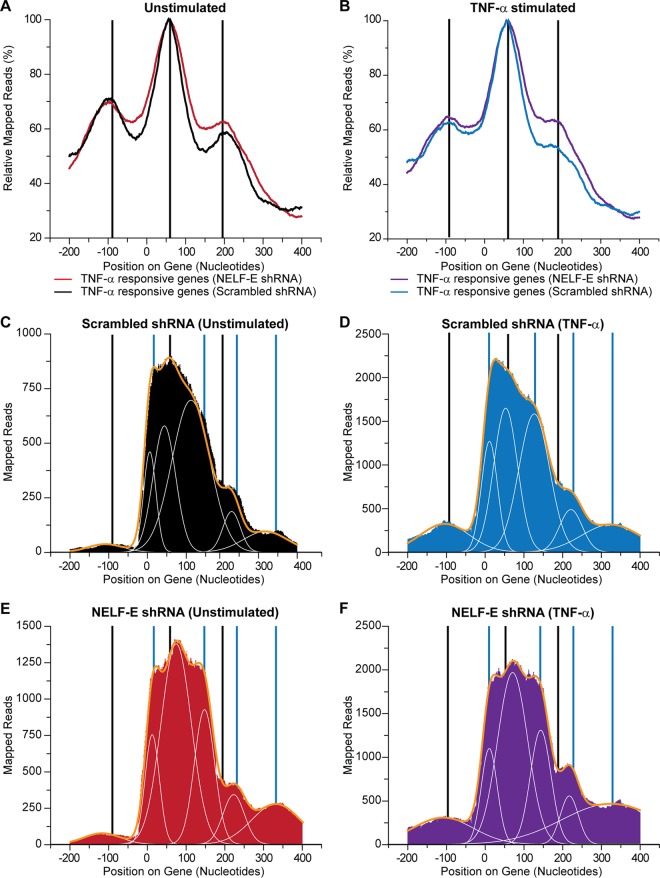
FIG 7.
Promoter-proximal pause sites on the HIV LTR. (A) The average RNAP II distribution relative to the transcription start site for the 500 genes that are most highly activated by TNF-α in unstimulated control (N1D9) cells (A) and control cells stimulated by TNF-α (B). (C to F) RNAP II accumulation in HIV promoter region in unstimulated control (N1D9) cells (C), control (N1D9) cells stimulated by TNF-α (D), unstimulated cells expressing NELF-E shRNA (N1C6) (E), and cells expressing NELF-E shRNA and stimulated by TNF-α (F). Black vertical lines indicate the sites of RNAP II accumulation on averaged cellular genes. Blue vertical lines indicate additional sites of RNAP II accumulation on the HIV LTR. The major peaks were fitted to a series of Gaussian curves (white lines) and summed (yellow lines).
Nucleotide sequence accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (53) and are accessible through GEO series accession number GSE47481.
RESULTS
NELF restricts HIV transcription in latently infected cells.
To investigate the contribution made by NELF to the maintenance of HIV latency, shRNA sequences targeting NELF subunits (and corresponding scrambled controls) were introduced into latently infected Jurkat T-cell lines (E4 and 2D10) (21). The shRNAs were carried by a lentiviral self-inactivating (SIN) vector modified to express mCherry as a marker for infection (pLVTHM) (44). The SIN vector deleted essentially the U3 region and prevented its function as a promoter and therefore does not contribute to the ChIP signals in the experiments described below. Both latent cell lines carry single integrated proviruses with a d2EGFP reporter in place of the Nef gene. The provirus in E4 cells carries the wild-type HIVNL4-3 Tat sequence, whereas the provirus in 2D10 cells contains the H13L variant which has a reduced affinity for CycT1 and promotes entry into latency (21, 54, 55).
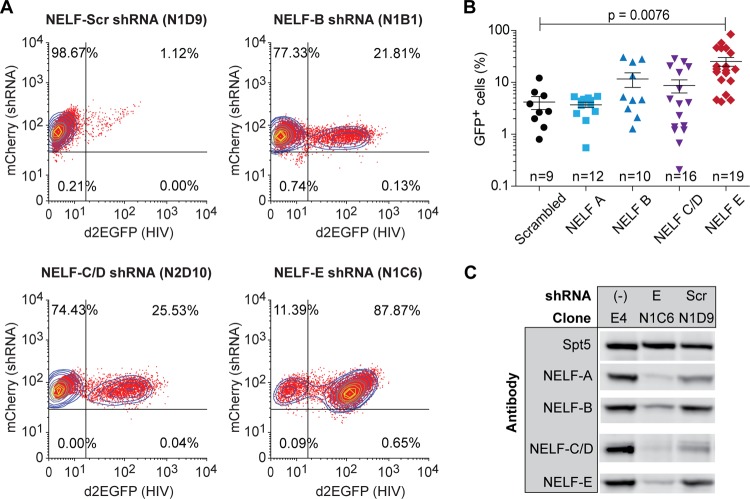
In the experiment shown in Fig. 1 mCherry-positive clones were analyzed for HIV gene expression levels. Flow cytometry of representative clones carrying shRNAs to the NELF-B (COBRA1), NELF-C/D (TH-1L), and NELF-E (RD) subunits showed that each shRNA can induce reactivation of the latent proviruses in 20% to 80% of the cells. However, there was wide variation in proviral reactivation efficiency in multiple clones carrying the same shRNAs (Fig. 1B), and statistically significant induction of proviruses compared to the scrambled shRNA control was seen only for the NELF-B and NELF-E knockdowns. Consistent with our results, Zhang and colleagues (42) have previously shown that a small interfering RNA (siRNA) to NELF-B can induce transcription from the integrated provirus in the U1 cell line.
FIG 1.
Silencing of the NELF subunits activates latent proviral transcription. (A) Flow cytometry. Clones of E4 cells superinfected with lentiviral vectors expressing shRNA targeting individual NELF subunits (A, B, C/D, and E) were analyzed by two-color flow cytometry. mCherry, shRNA marker; d2EGFP, HIV expression marker. (B) HIV reactivation in clones infected with lentiviral vectors expressing shRNAs targeting NELF subunits. (C) Western blot analysis of NELF subunit expression in the N1C6 clone expressing shRNA to NELF-E. Scr, scrambled shRNA.
Western blot analyses (Fig. 1C) showed that NELF-E shRNA induced the concomitant degradation of each of the NELF subunits, consistent with the observation of Narita et al. (56) that NELF-E is required for stable assembly of the NELF complex. The NELF-B, NELF-C/D, and NELF-A shRNAs were less potent and led to only partial knockdown of the targeted subunits and were therefore not analyzed further (see Fig. S1 in the supplemental material). It seems likely that these potency differences were due to differences in their knockdown efficiencies rather than to biological differences between the NELF subunits.
Silencing of NELF-E affects the rate and extent of reversion to latency.
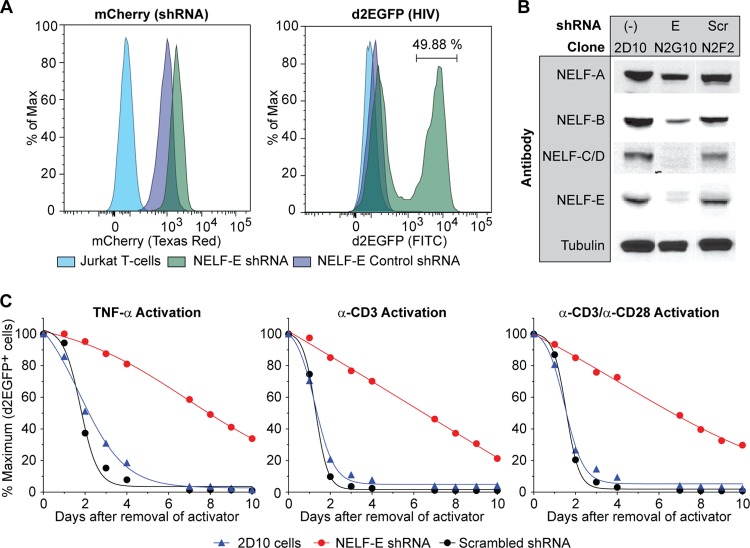
In cells that carry inducible proviruses, activation signals need to be continuously present in order to maintain high levels of HIV transcription (21, 46, 57, 58). To measure the contribution of NELF to the establishment of HIV latency, the rate at which the activated cells carrying NELF-E shRNA reenter latency was compared to control cells carrying scrambled shRNA or the parental 2D10 cell line (21). As shown in Fig. 2A an NELF-E shRNA clone (N2G10) was constitutively reactivated, whereas the scrambled control cells (N2F2) remained latent. Western blotting (Fig. 2B) confirmed the knockdown of NELF-E and other NELF subunits in the N2G10 cells.
FIG 2.
Knockdown of NELF-E by shRNA inhibits resilencing of HIV proviruses. (A) Flow cytometry using 2D10 cell-derived clones expressing NELF-E shRNA (N2G10) and a scrambled control (N2F2). mCherry expression was used as an shRNA marker, and d2EGFP expression was used as a marker of HIV transcription. Max, maximum. (B) Western blot analysis of NELF subunit expression in cells transfected with shRNA to NELF-E (N2G10) or a scrambled RNA control (N2F2). (C) Silencing of activated cells. 2D10 cells and clones carrying NELF-E shRNA (N2G10) and its scrambled control (N2F2) were stimulated overnight with 10 ng/ml TNF-α, 5 μg/ml anti-CD3, or 5 μg/ml anti-CD3 and 1 μg/ml anti-CD28. d2EGFP expression was monitored by flow cytometry between 0 and 10 days after removal of the activators. The percentage of d2EGFP-expressing cells immediately after activation was normalized. α, anti.
Cells were activated overnight with either TNF-α, anti-CD3 antibody, or anti-CD3 plus anti-CD28 antibody. After removal of the stimuli, HIV gene expression showed a biphasic decay pattern (Fig. 2C). The scrambled control N2F2 and 2D10 cells showed a half-life (t1/2) between 1.5 and 1.9 days and became fully silenced within 5 to 6 days. Depletion of NELF dramatically slowed the silencing of the activated proviruses, with the t1/2 varying between 4.0 and 6.9 days for the different activation conditions.
None of the NELF-E depleted clones ever became completely silenced but instead reverted to the starting condition of the population, with between 20 and 50% spontaneously activated cells. In clonal populations of latently infected cells, a small fraction of the cells became spontaneously activated due to stochastic events such as fluctuations in the cellular levels of NF-κB, Tat, and other transcription factors (17, 21, 59, 60). The NELF-depleted cell populations have a characteristically elevated background because of their reduced level of suppression.
RNAP II accumulates at the 5′ LTR of latent proviruses.
We next used ChIP assays to measure the distribution of RNAP II along the proviral genome (46, 57). The presence of duplicated LTRs is often ignored, but it creates a challenge for any ChIP experiment performed on HIV proviruses (46, 57, 58, 61) since there is no a priori way to assign sequence reads to individual LTRs.
To distinguish between the two LTRs, we developed primer sets that carried one primer in unique 5′ or 3′ chromosomal flanking sequences and a second primer within the LTR (Fig. 3A). Proviral integration sites were sequenced following the ligation-mediated PCR method of Wu et al. (62). The genome positions for the integration sites are given in Table 1 and shown diagrammatically in Fig. S2 in the supplemental material. We also developed primer sets that carried one primer in unique viral sequences of Gag or Nef that are adjacent to the 5′ or 3′ LTR and a second primer within the LTR (Fig. 3A and Table 1).
The data shown in Fig. 3 was obtained using an improved ChIP method using Ion Torrent sequencing to quantitate the PCR-amplified DNA fragments (see Fig. S3 and S4 in the supplemental material). The results correlate well with qPCR measurements. However, there are numerous advantages to the sequencing method, including greater sensitivity, reduced background since only correctly amplified products are scored, and an ability to extensively multiplex samples, increasing throughput.
Three latently infected cell clones which have unique proviral insertion sites (E4, 2D10, and 3CG) were examined using LTR-specific primers (Fig. 3B). Essentially all of the RNAP II is associated with the 5′ LTR during latency or after a 30-min induction by TNF-α. However, after a 16-h induction by TNF-α, there is significant accumulation of RNAP II at the downstream LTR, consistent with our previous observation that there is a 4-h lag prior to the induction of Tat from latent proviruses (46). The complementary analysis using the chromosome-specific primers (Fig. 3C) on the same samples also showed that significant levels of RNAP II accumulation on the 3′ LTR are seen only after 16 h of induction. A similar set of experiments is shown in Fig. 3D and E using E4 cells induced for 0.5 h or 8 h with TNF-α. High levels of RNAP II are seen on the 3′ LTR only after 8 h of induction. Similarly, when the nonspecific primers to the nucleosomes Nuc-1 and Nuc-0 are used, enhanced RNAP II levels at Nuc-0 are seen only after 8 h of stimulation, which can be attributed to accumulation of RNAP II on the 3′ LTR due to transcription initiating on the 5′ LTR.
Primer sets with similar amplification efficiencies were selected and optimized for this analysis, and the data are presented as mapped reads. As shown in Fig. S5 and S6 in the supplemental material, it is possible to convert the sequencing reads into percent input DNA concentrations, as typically done for ChIP experiments using qPCR, or to mathematically correct for the minor differences in primer efficiency. Thus, as long as early time points are evaluated, one can confidently assign RNAP II signals to the 5′ LTR. Our conclusion that RNAP II accumulates almost exclusively at the 5′ LTR in latently infected cells is consistent with the findings of Gallastegui et al. (63), who showed that there are high levels of RNA transcripts initiating from the 5′ LTR in latently infected cell lines.
Silencing of NELF-E specifically enhances the elongation capacity of RNAP II along the HIV proviral genome.
In the experiments shown in Fig. 4, the effect of NELF-E knockdown on the distribution of RNAP II was analyzed using clones derived from 2D10 cells. The unstimulated cells do not express Tat and are restricted for initiation because nuclear NF-κB levels are extremely low (21, 46, 58). Nonetheless, RNAP II accumulates downstream of the promoter (Fig. 4A). Depletion of NELF-E results in a 4-fold increase in the amount of RNAP II near the promoter. The majority of these transcription complexes accumulate around the TAR element and immediately downstream of the TAR region (+50 to + 300).
Stimulation of the cells with TNF-α for 60 min resulted in the influx of NF-κB into the nucleus and led to a 2-fold increase in RNAP II in the promoter-proximal region of the HIV LTR (Fig. 4B). Under these conditions, HIV elongation remains restricted by low levels of Tat (21, 46, 58), and relatively little RNAP II is found in the distal regions of the provirus. The NELF-E-depleted cells showed a similar 2- to 4-fold increase in RNAP II in the promoter-proximal region of HIV provirus compared to the levels in unstimulated cells, and significantly higher levels of RNAP II accumulated downstream.
After stimulation of the cells with TNF-α for 10 h (Fig. 4C), newly synthesized Tat (21, 46, 58) induced high levels of RNAP II downstream of the HIV promoter. Once again, depletion of NELF-E resulted in a 4-fold increase in the amount of RNAP II in the TAR region and a 2-fold increase in the amount of RNAP II in the downstream regions. After activation by TNF-α, there was also a strong increase in the amounts of RNAP II associated with the promoter, suggesting that when there is a higher turnover of polymerase at the promoter-proximal pause sites, there is also increased occupancy of the promoter by preinitiation complexes.
NELF is recruited to the HIV LTR in latently infected and induced cells.
High-density chromatin immunoprecipitation assays (ChIP) and micrococcal nuclease (MNase) protection assays were used to characterize changes in key transcription factors and chromatin structure before and after HIV induction by TNF-α in E4 cells. The assays were performed using a series of 18 primer pairs spanning the −395 to +770 region of the provirus that amplified fragments of approximately 100 bp.
HIV proviruses have a well-defined nucleosomal structure near the start site of transcription (64) (Fig. 5A). The latent E4 cells showed a high degree of nucleosome structure at the expected sites as measured by MNase protection assays (Fig. 5B). After induction by TNF-α for 30 min, there was a significant rearrangement of the nucleosomal structure and a loss of protection from both the Nuc-1 and Nuc-0 regions. These results are consistent with nucleosomal mapping studies by Rafati et al. (65) and illustrate that stable, presumably heterochromatic, structures are formed on latent proviral genomes.
As shown in Fig. 5C, the latent proviruses carried low levels of RNAP II in the region of the transcription start site (TSS) and downstream of the promoter between +0 and +400. Induction of the cells for 30 min by TNF-α led to the recruitment of RNAP II to the promoter and promoter-proximal region. At these early times Tat levels remain restricted in the latently infected cells (21, 46, 58). After 4 h of exposure to TNF-α, newly synthesized Tat stimulates transcriptional elongation (46), and enhanced levels of RNAP II can be found in both the promoter-proximal region and throughout the proviral genome.
In parallel, we measured the distribution of the P-TEFb subunits, CycT1 (Fig. 5D) and CDK9 (Fig. 5E). There was no significant recruitment of P-TEFb to the latent provirus until Tat was induced at 4 h. Both the CycT1 and CDK9 distributions are coincident and indicate association with RNAP II downstream of the TAR element between +200 and +400. A smaller peak of the P-TEFb subunits is also seen in the TAR region between +50 and +150.
In the latently infected cells, both the NELF-E (Fig. 5F) and NELF-A (Fig. 5G) subunits were found associated with RNAP II in the promoter-proximal region and near the TSS. Following induction by TNF-α, both NELF subunits accumulated in parallel with RNAP II at the TSS and in the promoter-proximal region.
NELF-E restricts HIV transcription at a variety of pause sites.
ChIP-Seq experiments were performed to map the distribution of RNAP II throughout the HIV proviral genome at the highest possible resolution. In the analyses shown in Fig. 6, we compared RNAP II distributions on the HIV provirus from control E4 cells and the corresponding NELF-E knockdown cells (clone N1C6) in the presence and absence of TNF-α induction for 60 min. As shown in Table 2, between 45 million and 56 million reads per sample were aligned to the proviral sequence and human genome.
It is striking that, even in the latently infected cells, high levels of RNAP II were found in the promoter-proximal region (Fig. 6A). Approximately 2-fold more RNAP II accumulated in the promoter-proximal region following TNF-α activation of the HIV LTR (Fig. 6B), and there was a concomitant increase in RNAP II throughout the body of the provirus. Knockdown of NELF-E also increased the RNAP II levels near the promoter and along the entire HIV genome (Fig. 6C). TNF-α activation of the NELF-E shRNA-treated cells led to maximal RNAP II levels in the body of the provirus but did not increase the RNAP II levels near the promoter, presumably because these were already at maximal levels following TNF-α induction (Fig. 6D).
The increase of RNAP II throughout the proviral genome following NELF-E knockdown, while considerably smaller than that achieved after TNF-α induction, is indicative of ongoing active transcription and is likely to account for the fraction of activated cells detected by the flow cytometry methods described earlier. However, it is difficult to make comparisons between the results of the two assays since reporter assays measure the fraction of cells that are activated, whereas the ChIP assay measures the steady state of RNAP II on the gene. If there is high turnover of RNAP II on the gene, even low levels of RNAP II detected by ChIP assays can give rise to high levels of gene expression.
The impact of induction by TNF-α or knockdown of NELF on the flux of Pol II from the promoter region into the body of a gene can be estimated using the escape index (EI) defined by Brannan et al. (52), which is the ratio of RNAP II in the body of the provirus (+301 to +5248) to the RNAP II at the 5′ LTR (−454 to + 300). In the latently infected cells the EI of 0.427 shows more than 2-fold accumulation of RNAP II at the promoter relative to the gene body. After NELF knockdown the EI increased to 1.002, consistent with dramatically enhanced RNAP II escape. TNF-α induction resulted in an increase of the EI to 1.872, which was further increased to an EI of 2.976 after NELF knockdown.
Figure 6A and C also illustrate that in latently infected cells, there was significant accumulation of RNAP II in the region around +2275, which corresponds to the Rev-response element (RRE), the largest RNA secondary structure element in the HIV genome (66). RNAP II accumulation in this region increased 2- to 3-fold following NELF-E knockdown (Fig. 6C).
In contrast to the HIV provirus, mapping of reads to the unique regions of the pLVTHM vector for delivery of shRNA showed relatively constant levels of RNAP II accumulation under all four experimental conditions (see Fig. S8 in the supplemental material).
Data from the cellular genes collected in this experiment provided an important additional measure of the consistency of the four data sets (see Fig. S9 to S13 in the supplemental material). The selected genes represent examples of constitutively expressed, highly NELF-restricted or highly TNF-α-responsive genes. HIV is unusual in being both NELF-restricted and TNF-α responsive (see below).
Promoter-proximal pausing.
The high number of reads mapping to the HIV LTR obtained in ChIP data sets allowed us to make a detailed map of the promoter-proximal pause sites on the HIV LTR. Since the reads from the majority of cellular promoters are typically 10% or less than the HIV reads (see Fig. 8), it is often difficult to make direct comparisons. We therefore used an averaged set of TNF-α-responsive genes for comparison to the HIV provirus (Fig. 7A and B). We also had sufficient reads to produce detailed maps using the TNF-α-responsive JUN gene and the NELF-restricted SNGH3 gene (see Fig. S14 and S15 in the supplemental material).
In the latently infected E4 cells, RNAP II accumulated in a broad distribution, with several obvious shoulders at a series of sites immediately downstream of the transcription start site (Fig. 7C and D). We used curve fitting to accurately define the location of the potential pause sites. The latent proviruses showed five major sites of accumulation centered near the transcription start site (nucleotide [nt] +11), an early pause site (nt +50), and a late pause site (nt +119) downstream of the TAR element and two minor pause sites at nt +227 and nt +318, which match those seen in the cellular genes and are consistent with RNAP II pausing at the borders of nucleosomes.
Following induction of transcription by TNF-α there was a >2-fold increase in the levels of RNAP II associated with the HIV LTR (Fig. 7D). TNF-α induction did not significantly shift the location of any of the downstream RNAP II accumulation sites. However, the proportion of RNAP II accumulating near the transcription start site (nt +11) increased relative to that of the latent provirus, suggesting that there was enhanced initiation. It is notable that TNF-α induced a peak at −100, which corresponds precisely to the negative-strand transcription site seen on the cellular genes and is a feature not previously documented for the HIV promoter. The amount of negative-strand transcription from the HIV LTR is 3-fold to 10-fold lower than that of the cellular genes, suggesting that the HIV LTR is unusually polarized toward positive-strand transcription.
Knockdown of NELF-E by shRNA (Fig. 7E) also induced RNAP II accumulation near the promoter, but in contrast to TNF-α, there was enhanced accumulation at the first promoter-proximal pause site and a shift of the peak downstream from +50 to +70. There is no significant shift in the other promoter-proximal pause sites. Combining TNF-α induction and NELF-E knockdown (Fig. 7F) resulted in a combined pattern, with TNF-α inducing enhanced RNAP II at the −100 and +11 sites and NELF knockdown inducing a shift of the early promoter-proximal pause site from +50 to +70.
As shown in Fig. 7A and B, the major peaks from the averaged cellular promoters were at a promoter-proximal pause site (nt +57) and a downstream arrest site at nt +200. There was also evidence for negative-strand transcription, with peaks at nt −100. This pattern of promoter-proximal pausing is consistent with previous reports of paused RNAP II in the range of nt +40 to +60 obtained from ChIP-Seq and global run-on sequencing (GRO-Seq) experiments (52, 67–73).
The JUN and SNGH3 genes (see Fig. S14 and S15 in the supplemental material) showed slightly different positions for promoter-proximal paused RNAP II; however, in contrast to the HIV LTR, the cellular promoters did not show significant shifts in the position of the RNAP II following TNF-α induction or NELF-depletion and had relatively greater RNAP II accumulation at the first promoter-proximal site and much higher levels of RNAP II in the upstream negative-strand region. Thus, the accumulation of RNAP II at the TSS and the additional pausing after TAR RNA are features of HIV transcription that are not commonly observed on cellular promoters.
HIV is more highly responsive to TNF-α and NELF-E depletion than the majority of cellular genes.
To evaluate the relative response of HIV compared to that of cellular genes, the total number of normalized reads per gene were compared (Fig. 8). Figure 8A shows the pairwise comparison of the NELF-E knockdown response without TNF-α stimulation (scrambled control versus cells expressing shNELF-E). Among the 20,000 genes surveyed, the HIV provirus ranked 90th for activation by NELF-E knockdown in the absence of TNF-α. The genes that exhibited the highest upregulation following TNF-α induction were known NF-κB-responsive genes. The HIV provirus ranked 4th in the RNAP II occupancy ratio for TNF-α-activated genes in the presence of NELF (Fig. 8B). Similarly, RNA Pol II occupancy along the HIV provirus ranked 792nd for NELF-E knockdown in the presence of TNF-α (Fig. 8C) and a remarkable 4th for TNF-α-activated genes in the absence of NELF-E (Fig. 8D).
Thus, the HIV LTR is a rare promoter that is disproportionately sensitive to both TNF-α induction and NELF restriction. Transcription activation due to the removal of NELF by P-TEFb and TNF-α stimulation can therefore be anticipated to work synergistically to increase transcription rates and allow for viral emergence from latency under conditions where most cellular genes are relatively unaffected.
DISCUSSION
NELF is required to maintain HIV proviral latency.
Previous studies have shown that latent HIV proviruses produce only minimal levels of mRNA due to a combination of chromatin restrictions (21, 64, 74), the absence of NF-κB (58, 75), restricted levels of Tat (76), and a failure to recruit TFIIH (57). Nonetheless, early elongation complexes can be detected paused near the promoter of latent proviruses by ChIP assays (19, 21, 46, 47, 58, 61, 76) and DNA footprinting (42). Furthermore, latently infected cells accumulate short abortive transcripts due to the premature dissociation of transcription complexes from the proviral template (39, 77).
In this paper we have shown that a major silencing mechanism used to preserve HIV latency, despite the accumulation of early elongation complexes, is the recruitment of the negative elongation factor NELF. NELF contains four subunits: NELF-A (WHSC2), NELF-B (COBRA1), NELF-C or its alternatively spliced homologue NELF-D (TH-1), and NELF-E (RD) (34, 62, 78). Knockdown of NELF-E resulted in efficient depletion of each of the other NELF subunits in the NELF complex and strongly activated latent proviruses, without any additional extracellular stimulation (Fig. 1). Less efficient knockdown and activation profiles were obtained using shRNAs to NELF-B and NELF-C/D. Our results are consistent with an earlier report from the Zhang et al. showing that knockdown of NELF-B is able to induce transcription from an integrated HIV provirus (42).
Latent HIV proviruses accumulate RNAP II at the 5′ LTR.
ChIP assays performed using primers that distinguish between the 5′ and 3′ viral LTRs demonstrated that latent proviruses exclusively accumulate RNAP II at the 5′ LTR immediately downstream of the transcription start site. This seems to be a universal feature of the latent proviruses since similar results were obtained using three independent clones with proviruses in unique chromosomal sites and different orientations with respect to cellular promoters.
The presence of RNAP II at the 5′ LTR is inconsistent with models for HIV latency that postulate that RNAP II initiating at the host promoter disrupts the formation of initiation complexes and transcription factors on the 5′ LTR of the provirus, causing transcriptional interference (25). However, it is consistent with models for latency, suggesting that heterochromatic structures on the LTR help limit efficient RNAP II escape and productive transcription during latency (19, 21, 63). The mechanisms leading to inactivation of the 3′ LTR are unknown, but we postulate that this is associated with the formation of a gene loop structure between the 5′ LTR promoter and 3′ LTR poly(A) signal (79).
NELF is present on both latent and actively transcribing HIV proviruses.
Detailed maps of the distribution of RNAP II, P-TEFb, and NELF on the HIV provirus before and after proviral induction by TNF-α show that RNAP II transcription complexes accumulate near the TSS of latent HIV proviruses and at multiple sites further downstream, reaching as far as +400. NELF is present in the promoter-proximal region of the latent proviruses as well as on induced proviruses following a distribution pattern that closely matches that of the RNAP II (Fig. 3). Our data extend the results of Zhang et al. (42) who demonstrated that the NELF-B subunit is recruited to the HIV provirus in latently infected U1 cells. Using ChIP assays, NELF-B was mapped to a fragment between −155 and +186 of the transcription start site (42).
It is interesting that the ratio of NELF to RNAP II is much higher on the latent proviruses than on the induced proviruses, suggesting that NELF is part of stable complex accumulating on the latent provirus. This is consistent with observations that NELF and DSIF cooperatively bind to RNAP II in the promoter-proximal region of cellular genes (7, 80, 81). Recent data suggest that DSIF binds the elongation complex via association with the nascent transcript and subsequently recruits NELF (82, 83).
Efficient P-TEFb recruitment to the HIV LTR is dependent upon Tat.
D'Orso et al. (61, 84) have recently developed the provocative hypothesis that P-TEFb is recruited to an upstream region in the HIV promoter in a catalytically inactive state bound to the inhibitory 7SK small nuclear ribonucleoprotein (snRNP). In their model Tat is recruited to the DNA template before the TAR element is synthesized, and as the RNAP II transcribes through the TAR region, there is an exchange that leads to displacement of the 7SK RNA and the activation of the P-TEFb kinase. Key experiments supporting this idea come from ChIP and ChIP-Seq data showing HEXIM in upstream sites on both HIV and cellular promoters (61, 84, 85).
Our ChIP data obtained using latently infected cells are somewhat inconsistent with these results. We do not see any recruitment of the P-TEFb subunits CycT1 and CDK9 over background levels to the latent proviruses. Similarly, there is no detectable P-TEFb recruitment following a 30-min induction by TNF-α, which leads to chromatin remodeling and RNAP II recruitment but is at a time prior to the increased production of Tat (46). It is only after 4 h, when Tat levels have significantly increased, that there is substantial recruitment of the P-TEFb subunits to the LTR.
One possible explanation for these discrepancies is that D'Orso and Frankel (61) performed their experiments in HeLa cells, which typically display a much higher level of basal HIV transcription and less restriction by heterochromatic structures than the latently infected Jurkat T-cell lines that we have used in our studies. It therefore possible that the P-TEFb recruitment they observed in the absence of Tat is due to residual transcription in the HeLa cell system, perhaps mediated by the high levels of Brd4 that they have found associated with the HIV provirus. It is also possible that the 7SK RNP complex is only loosely associated with promoters and can be detected only after extensive cross-linking (85).
Unique patterns of RNAP II pausing on the HIV LTR.
Promoter-proximal pausing is a general feature of eukaryotic transcription (86–91) and is generally regarded as a mechanism that allows genes to remain in a poised state for rapid gene expression in response to activating signals. The position of these sites is remarkably constant across genomes and across species. Typically, the major RNAP II pause sites are found in the +20 to +60 region for positive-strand transcription and in the −90 to −110 region for negative-strand transcription.
Since the HIV LTR, even in latently infected cells, accumulates significantly more RNAP II than the average cellular gene, we took advantage of the large number of reads mapped to the LTR to develop detailed maps of promoter-proximal pause sites. The latent HIV provirus shows a more extensive and complex pattern of RNAP II accumulation in the promoter-proximal region than is typically seen on cellular genes. First, there is accumulation of RNAP II near the TSS (nt +11) that probably corresponds to occluded preinitiation complexes. This is consistent with our earlier observation that latent proviruses are relatively deficient in TFIIH and accumulate hypophosphorylated RNAP near the transcription start site (57). RNAP II also accumulates at nt +50, which corresponds to the major pause site seen in the cellular genes (52, 68, 69). Zhang et al. (42) also reported that RNAP II transcription complexes pause on latent proviruses prior to TAR since they are able to identify paused transcription complexes at approximately +41 to +45 using elegant permanganate footprinting experiments.
When transcription is induced by depletion of NELF, the +50 pause site is displaced approximately 20 nt downstream. This downstream shift may reflect rearrangements in nucleosomal structure following HIV transcription induction (Fig. 2) and suggests that NELF depletion may enhance repositioning of nucleosome 1 by the chromatin remodeling factor, PBAF (polybromo-associated BAF), as reported by Rafati et al. (65). Similarly, Natarajan et al. (34) have recently suggested that NELF has direct effects on promoter-proximal pausing and chromatin structure by NELF recruiting Pcf11, a transcription termination factor, and NCoR1-GPS2-HDAC3, a corepressor complex that is associated with creation of heterochromatic structures.
In addition to the pause site at +50, there is a late pause site (+120) which is immediately downstream of the TAR region and which appears to be a site for accumulation of P-TEFb, NELF, and other transcription factors. Finally, there are two minor pause sites at nt +220 and +330 that match pause sites seen in the cellular genes and that are consistent with RNAP II pausing at the borders of nucleosomes (69).
Depletion of NELF-E or stimulation of initiation by TNF-α enhances the accumulation of transcription complexes throughout the length of the genome, but rather than reducing promoter-proximal pausing, as traditionally postulated, NELF depletion actually increases RNAP II density near the promoter. The HIV provirus shows a remarkable 6.9-fold variation in the escape index (EI), depending on the induction conditions. Depletion of NELF alone produces a 2.5-fold increase in the escape index, demonstrating that NELF provides an efficient surveillance mechanism for limiting RNAP II escape from the promoter region. In the downstream region, RNAP II pause sites can be seen throughout the genome with an underlying periodicity which is suggestive of pausing primarily at nucleosomal boundaries. RNAP II also accumulates at regions of high RNA secondary structure, such as the RRE.
A limitation of the ChIP-Seq assay is that it does not formally identify actively transcribing RNAP II or the transcription orientation of polymerases. The complementary nuclear run-on assay (GRO-Seq) both measures RNAP II that is competent to elongate and can determine the direction of transcription (86, 92). Detailed comparisons of the results of the two methods have demonstrated that over 80% of the polymerase found by ChIP-Seq can be accounted for by the signal from the GRO-Seq data set. From this we infer that the majority of the RNAP II that we see on the HIV LTR is transcriptionally competent.
In summary, the accumulation of RNAP II at the TSS and the additional pausing after TAR RNA are features of HIV transcription that are unique to HIV and reflect both the efficiency of the LTR as a promoter and the extensive attenuation of transcription seen during proviral latency.
Mechanism of action of NELF in HIV transcription.
A striking feature of elongation control of the HIV promoter is that the restriction of elongation in the absence of Tat is stronger than typically seen in cellular promoters, whereas the efficiency of elongation in the presence of Tat is much stronger (46). For example, we reported previously that in the presence of Tat, elongation efficiencies on HIV proviruses range from 60 to 80%, whereas cellular genes typically remain in the 20 to 30% range (46).
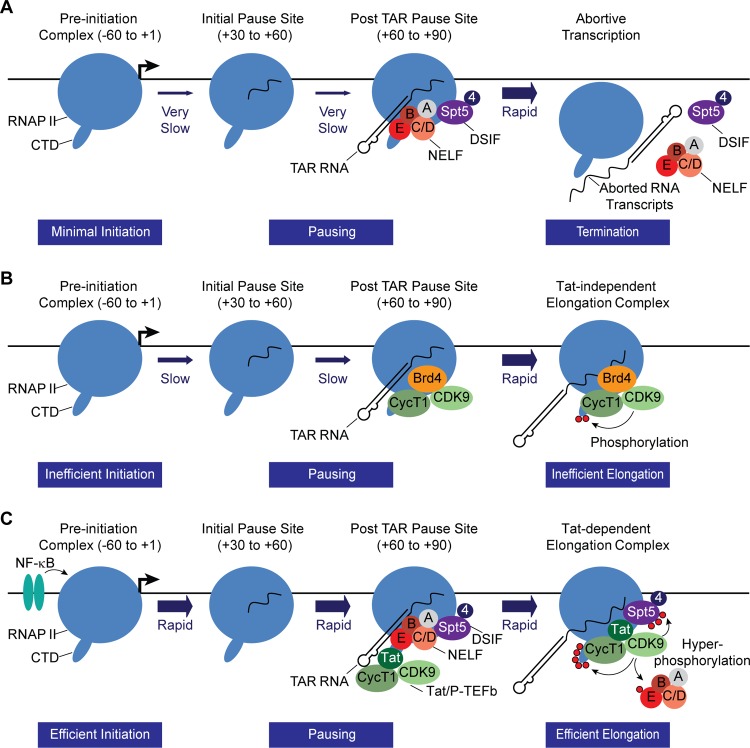
The data presented in this paper, as summarized diagrammatically in Fig. 9, are consistent with a kinetic model for the control of HIV latency involving multiple promoter-proximal pause sites to strongly attenuate HIV transcription during latency. NELF, which is believed to force premature termination over a range of several hundred nucleotides (93), is likely to limit the escape of transcription complexes from the promoter-proximal pause site. The additional pause sites that we have detected after the TAR element may further serve to attenuate transcription from the HIV LTR.
FIG 9.
Kinetic model for the restriction of HIV transcription from latent proviruses by NELF. (A) Latent HIV provirus. In latent proviruses transcription elongation is very inefficient due to absence of the transcription elongation factor NF-κB as well as chromatin restrictions (not shown for simplicity). However, a significant number of proviruses carry RNAP II paused in the promoter-proximal region. The small number of transcription complexes that are able initiate and elongate through TAR are subject to additional elongation restrictions by NELF which forces premature termination. (B) Partially activated HIV provirus in the absence of the NELF. In the absence of NELF, promoter-proximal pausing remains relatively constant, but there is enhanced escape of paused complexes. (C) NF-κB and Tat-activated transcription. Initiation is strongly induced by NF-κB, which removes chromatin restrictions near the promoter through recruitment of histone acetyltransferases. Under these circumstances promoter clearance is also much more efficient, and there is an enhanced accumulation of elongation complexes in the promoter-proximal region. After the transcription through the TAR element, both NELF and the Tat/P-TEFb complex (the superelongation complex factors are not shown for simplicity) are recruited to the elongation complex via binding interactions with TAR RNA. This activates the CDK9 kinase and leads to hyperphosphorylation of the CTD of RNA polymerase II, Spt5, and NELF-E. The phosphorylation of NELF-E leads to its release. Although the promoter is transcribing more rapidly than in the latent condition, there is relatively little change in the amount of RNAP II that accumulates in the promoter-proximal region due to its rapid replacement by newly initiated transcription complexes.
Processing of nascent RNA transcripts by nucleases leads to the accumulation of short transcripts corresponding to the TAR RNA stem-loop structure (39). Recently, Wagschal et al. (94) have proposed that TAR RNA processing is controlled by the microprocessor, the termination factors Setx and Xrn2, and the 3′ to 5′ exoribonuclease, Rrp6. After an initial cleavage by Drosha that releases TAR RNA, Rrp6 generates a small RNA that can enhance chromatin-mediated restriction of the HIV promoter (94). It is interesting to speculate that depletion of NELF, which results in enhanced escape of RNAP II from the promoter-proximal region, might have an indirect effect on chromatin remodeling due to a reduction in the levels of processed short RNA transcripts as well as its interactions with the NCoR1 repressive complex (34).
NELF restricts elongation only in the presence of the DRB (5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole) sensitivity-inducing factor (DSIF), which contains the Spt4 and Spt5 subunits. A genome-wide location analysis based on ChIP analyses showed that NELF (NELF-A) and DSIF (Spt5) colocalize on the genome on most genes (95). NELF and DSIF are known to cooperatively bind to RNAP II in the promoter-proximal region of cellular genes (7, 80, 81). Recent data suggest that DSIF binds the elongation complex via association with the nascent transcript and subsequently recruits NELF (82, 83). In the case of the HIV provirus, the recruitment of NELF may be further enhanced by the ability of NELF-E to bind directly to TAR RNA (12, 78, 96). Our ChIP experiments show that the majority of NELF-E and -A subunits (and presumably any other NELF subunits required to form a functional complex) preferentially associate with paused RNA transcription complexes downstream of the TAR region (Fig. 5), consistent with the recently discovered high NELF-E binding element (NBE) present within the loop region of the transactivation response element (TAR) of HIV-1 RNA (96). However, NELF can also be detected in the pre-TAR region, consistent with its less efficient recruitment via RNAP II binding.
The switch from promoter-proximal pausing to productive elongation is mediated by the positive transcription elongation factor (P-TEFb). Specifically, phosphorylation of NELF-E by P-TEFb forces dissociation of NELF from TAR and is likely to lead to a release of paused transcription elongation complexes (12). In the absence of NELF (Fig. 9B), this “fail-safe” surveillance mechanism is disrupted, and this leads to enhanced stochastic firing of the HIV promoter. This, in turn, will lead to Tat production and eventual escape from latency. Assuming that recruitment of new RNAP II to the preinitiation complexes is at least partially occluded by RNAP II restricted to the promoter-proximal region, the more rapid clearance of the promoter in the absence of NELF could then lead to the enhanced accumulation of NELF at the promoter, as we have observed. Since productive elongation still requires P-TEFb, we hypothesize, as outlined in Fig. 9B, that P-TEFb recruitment via Brd4 permits a few transcription complexes to complete transcription of the HIV genome prior to new synthesis of Tat and the eventual escape from latency.
NF-κB, which induces rapid chromatin rearrangements on the HIV LTR (Fig. 5) and the removal of heterochromatin blocks (19, 65), also enhances recruitment of the RNAP II to the promoter and increases the level of RNAP II associated with the promoter and a corresponding increase in RNAP II at the proximal pause sites (Fig. 9C). The system becomes fully activated only once newly synthesized Tat is able to direct P-TEFb and the associated superelongation complex (13, 15, 97, 98) to the elongating RNA polymerase and stimulate exceptionally efficient transcriptional elongation and escape from the promoter-proximal pause sites. The activation of the enzymatic activity of P-TEFb by Tat not only leads to hyperphosphorylation of Spt5 (9) and the RNAP II CTD (9) but also ensures efficient removal of NELF (12), turning the HIV LTR into one of the most powerful transcription machines that is known to exist.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sheldon Bai for help in the ChIP-Seq analysis, Mudit Tyagi for help in preparing ChIP-Seq samples, David Alvarez and Michael Greenberg for help in the development of the sequence-based ChIP assay, and our colleagues Nick Funderburg and Doug Bazdar for assistance in the multicolor flow cytometry. We also thank present and former Karn laboratory members Hongxia Mao, Richard Pearson, Joseph Hokello, Julia Friedman, and Kara Lassen for their help and useful discussions.
This work was supported by Public Health Service grants R01-AI067093 and DP1-DA028869 to J.K. and the Martin Delaney CARE Collaboratory (U19-AI096113). We also thank the Case Western Reserve University/University Hospitals Center for AIDS Research (NIH grant P30-AI036219) for provision of flow cytometry services.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 17 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01013-13.
REFERENCES
- 1.Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10:525–531. 10.1016/j.molmed.2004.09.006 [DOI] [PubMed] [Google Scholar]
- 2.Shen L, Siliciano RF. 2008. Viral reservoirs, residual viremia, and the potential of highly active antiretroviral therapy to eradicate HIV infection. J. Allergy Clin. Immunol. 122:22–28. 10.1016/j.jaci.2008.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 94:13193–13197. 10.1073/pnas.94.24.13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. 10.1126/science.278.5341.1295 [DOI] [PubMed] [Google Scholar]
- 5.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. 10.1126/science.1165706 [DOI] [PubMed] [Google Scholar]
- 6.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, Endoh M, Yamada T, Handa H. 2003. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 23:1863–1873. 10.1128/MCB.23.6.1863-1873.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41–51. 10.1016/S0092-8674(00)80713-8 [DOI] [PubMed] [Google Scholar]
- 8.Wei P, Garber ME, Fang S-M, Fischer WH, Jones KA. 1998. A novel cdk9-associated c-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop specific binding to TAR RNA. Cell 92:451–462. 10.1016/S0092-8674(00)80939-3 [DOI] [PubMed] [Google Scholar]
- 9.Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. 2002. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol. Cell. Biol. 22:4622–4637. 10.1128/MCB.22.13.4622-4637.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. 2002. Spt5 cooperates with Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22:1079–1093. 10.1128/MCB.22.4.1079-1093.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov D, Kwak YT, Guo J, Gaynor RB. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970–2983. 10.1128/MCB.20.9.2970-2983.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24:787–795. 10.1128/MCB.24.2.787-795.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 38:428–438. 10.1016/j.molcel.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37:429–437. 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell 38:439–451. 10.1016/j.molcel.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A, Weinberger LS. 2009. Stochastic gene expression as a molecular switch for viral latency. Curr. Opin. Microbiol. 12:460–466. 10.1016/j.mib.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182. 10.1016/j.cell.2005.06.006 [DOI] [PubMed] [Google Scholar]
- 18.du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, Bertrand E, Marcello A, Emiliani S, Kiernan R, Benkirane M. 2007. Suv39H1 and HP1γ are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26:424–435. 10.1038/sj.emboj.7601517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, Karn J. 2011. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of zeste 2 (EZH2). J. Virol. 85:9078–9089. 10.1128/JVI.00836-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. 2009. A limited group of class I histone deacetylases act to repress human immunodeficiency virus type-1 expression. J. Virol. 83:4749–4756. 10.1128/JVI.02585-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. 2008. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 82:12291–12303. 10.1128/JVI.01383-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554. 10.1371/journal.ppat.1000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495. 10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Y, Lin YB, An W, Xu J, Yang HC, O'Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF. 2008. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4:134–146. 10.1016/j.chom.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenasi T, Contreras X, Peterlin BM. 2008. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4:123–133. 10.1016/j.chom.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 79:6610–6619. 10.1128/JVI.79.11.6610-6619.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherrill-Mix S, Lewinski MK, Famiglietti M, Bosque A, Malani N, Ocwieja KE, Berry CC, Looney D, Shan L, Agosto LM, Pace MJ, Siliciano RF, O'Doherty U, Guatelli J, Planelles V, Bushman FD. 2013. HIV latency and integration site placement in five cell-based models. Retrovirology 10:90. 10.1186/1742-4690-10-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosque A, Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65. 10.1182/blood-2008-07-168393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. 1997. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6:235–244. 10.1016/S1074-7613(00)80326-X [DOI] [PubMed] [Google Scholar]
- 30.Nabel G, Baltimore DA. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713. 10.1038/326711a0 [DOI] [PubMed] [Google Scholar]
- 31.Garriga J, Peng J, Parreno M, Price DH, Henderson EE, Grana X. 1998. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene 17:3093–3102. 10.1038/sj.onc.1202548 [DOI] [PubMed] [Google Scholar]
- 32.Ghose R, Liou LY, Herrmann CH, Rice AP. 2001. Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4+ T lymphocytes by combination of cytokines. J. Virol. 75:11336–11343. 10.1128/JVI.75.23.11336-11343.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liou LY, Herrmann CH, Rice AP. 2002. Transient induction of cyclin T1 during human macrophage differentiation regulates human immunodeficiency virus type 1 Tat transactivation function. J. Virol. 76:10579–10587. 10.1128/JVI.76.21.10579-10587.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natarajan M, Schiralli-Lester GM, Lee C, Missra A, Wasserman GA, Steffen M, Gilmour DS, Henderson AJ. 2013. NELF coordinates RNA polymerase II pausing, premature termination and chromatin remodeling to regulate HIV transcription. J. Biol. Chem. 288:25995–26003. 10.1074/jbc.M113.496489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija Peterlin B, Zhou Q. 2007. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35:2003–2012. 10.1093/nar/gkm063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. 2004. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 23:2608–2619. 10.1038/sj.emboj.7600275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. 2007. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 35:4347–4358. 10.1093/nar/gkm443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971–982. 10.1016/S1097-2765(03)00388-5 [DOI] [PubMed] [Google Scholar]
- 39.Adams M, Sharmeen L, Kimpton J, Romeo JM, Garcia JV, Peterlin BM, Groudine M, Emerman M. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. U. S. A. 91:3862–3866. 10.1073/pnas.91.9.3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ping Y-H, Rana TM. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951–12958. 10.1074/jbc.M006130200 [DOI] [PubMed] [Google Scholar]
- 41.Ratnasabapathy R, Sheldon M, Johal L, Hernandez N. 1990. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 4:2061–2074. 10.1101/gad.4.12a.2061 [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Klatt A, Gilmour DS, Henderson AJ. 2007. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem. 282:16981–16988. 10.1074/jbc.M610688200 [DOI] [PubMed] [Google Scholar]
- 43.Brummelkamp TR, Bernards R, Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553. 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- 44.Wiznerowicz M, Trono D. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957–8961. 10.1128/JVI.77.16.8957-8951.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. 10.1126/science.272.5259.263 [DOI] [PubMed] [Google Scholar]
- 46.Kim YK, Mbonye U, Hokello J, Karn J. 2011. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 410:896–916. 10.1016/j.jmb.2011.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyagi M, Pearson RJ, Karn J. 2010. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84:6425–6437. 10.1128/JVI.01519-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petesch SJ, Lis JT. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134:74–84. 10.1016/j.cell.2008.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. 2009. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48:240–248. 10.1016/j.ymeth.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, Bentley DL. 2012. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol. Cell 46:311–324. 10.1016/j.molcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar R, Domrachev M, Lash AE. 2002. Gene Expr. Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emiliani S, Fischle W, Ott M, van Lint C, Amella CA, Verdin E. 1998. Mutations in the Tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yukl S, Pillai S, Li P, Chang K, Pasutti W, Ahlgren C, Havlir D, Strain M, Gunthard H, Richman D, Rice AP, Daar E, Little S, Wong JK. 2009. Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology 387:98–108. 10.1016/j.virol.2009.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell 26:349–365. 10.1016/j.molcel.2007.04.011 [DOI] [PubMed] [Google Scholar]
- 57.Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J. 2006. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 25:3596–3604. 10.1038/sj.emboj.7601248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams SA, Kwon H, Chen LF, Greene WC. 2007. Sustained induction of NF-κB is required for efficient expression of latent human immunodeficiency virus type 1. J. Virol. 81:6043–6056. 10.1128/JVI.02074-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burnett JC, Miller-Jensen K, Shah PS, Arkin AP, Schaffer DV. 2009. Control of stochastic gene expression by host factors at the HIV promoter. PLoS Pathog. 5:e1000260. 10.1371/journal.ppat.1000260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinberger LS, Shenk T. 2007. An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer. PLoS Biol. 5:e9. 10.1371/journal.pbio.0050009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D'Orso I, Frankel AD. 2010. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 17:815–821. 10.1038/nsmb.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu X, Li Y, Crise B, Burgess SM. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749–1751. 10.1126/science.1083413 [DOI] [PubMed] [Google Scholar]
- 63.Gallastegui E, Millan-Zambrano G, Terme JM, Chavez S, Jordan A. 2011. Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J. Virol. 85:3187–3202. 10.1128/JVI.01920-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Lint C, Emiliani S, Ott M, Verdin E. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112–1120 [PMC free article] [PubMed] [Google Scholar]
- 65.Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. 2011. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 9:e1001206. 10.1371/journal.pbio.1001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716. 10.1038/nature08237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. 2009. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. U. S. A. 106:18207–18212. 10.1073/pnas.0910177106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13:720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, Young RA, Price DH. 2012. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell 45:38–50. 10.1016/j.molcel.2011.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. 2012. Defining the status of RNA polymerase at promoters. Cell Rep. 2:1025–1035. 10.1016/j.celrep.2012.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143:540–551. 10.1016/j.cell.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilchrist DA, Fromm G, Gdos Santos Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. 2012. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 26:933–944. 10.1101/gad.187781.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22:1921–1933. 10.1101/gad.1643208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868–1877. 10.1093/emboj/cdg188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen BK, Feinberg MB, Baltimore D. 1997. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J. Virol. 71:5495–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mbonye U, Karn J. 2011. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Curr. HIV Res. 9:554–567. 10.2174/157016211798998736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sheldon M, Ratnasabapathy R, Hernandez N. 1993. Characterization of the inducer of short transcripts, a human immunodeficiency virus type 1 transcriptional element that activates the synthesis of short RNAs. Mol. Cell. Biol. 13:1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22:2918–2927. 10.1128/MCB.22.9.2918-2927.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. 2008. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol. Cell 29:56–68. 10.1016/j.molcel.2007.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343–356. 10.1101/gad.12.3.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. 1999. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274:8085–8092. 10.1074/jbc.274.12.8085 [DOI] [PubMed] [Google Scholar]
- 82.Cheng B, Price DH. 2008. Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic Acids Res. 36:e135. 10.1093/nar/gkn630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Missra A, Gilmour DS. 2010. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc. Natl. Acad. Sci. U. S. A. 107:11301–11306. 10.1073/pnas.1000681107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.D'Orso I, Jang GM, Pastuszak AW, Faust TB, Quezada E, Booth DS, Frankel AD. 2012. Transition step during assembly of HIV Tat:P-TEFb transcription complexes and transfer to TAR RNA. Mol. Cell. Biol. 32:4780–4793. 10.1128/MCB.00206-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. 2013. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153:855–868. 10.1016/j.cell.2013.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848. 10.1126/science.1162228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130:77–88. 10.1016/j.cell.2007.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. 2007. RNA polymerase is poised for activation across the genome. Nat. Genet. 39:1507–1511. 10.1038/ng.2007.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nechaev S, Adelman K. 2008. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 7:1539–1544. 10.4161/cc.7.11.6006 [DOI] [PubMed] [Google Scholar]
- 90.Saunders KO, Freel SA, Overman RG, Cunningham CK, Tomaras GD. 2010. Epigenetic regulation of CD8+ T-lymphocyte mediated suppression of HIV-1 replication. Virology 405:234–242. 10.1016/j.virol.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39:1512–1516. 10.1038/ng.2007.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Core LJ, Lis JT. 2008. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319:1791–1792. 10.1126/science.1150843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601–42609. 10.1074/jbc.M104967200 [DOI] [PubMed] [Google Scholar]
- 94.Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, Boyer F, Parrinello H, Berkhout B, Terzian C, Benkirane M, Kiernan R. 2012. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell 150:1147–1157. 10.1016/j.cell.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445. 10.1016/j.cell.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pagano JM, Kwak H, Waters CT, Sprouse RO, White BS, Ozer A, Szeto K, Shalloway D, Craighead HG, Lis JT. 2014. Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet. 10:e1004090. 10.1371/journal.pgen.1004090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chou S, Upton H, Bao K, Schulze-Gahmen U, Samelson AJ, He N, Nowak A, Lu H, Krogan NJ, Zhou Q, Alber T. 2013. HIV-1 Tat recruits transcription elongation factors dispersed along a flexible AFF4 scaffold. Proc. Natl. Acad. Sci. U. S. A. 110:E123–131. 10.1073/pnas.1216971110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schulze-Gahmen U, Upton H, Birnberg A, Bao AK, Chou S, Krogan NJ, Zhou Q, Alber T. 2013. The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat. eLife 2:e00327. 10.7554/eLife.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.